Abstract
Cancer-testis (CT) antigens are potential targets for cancer immunotherapy because of their restricted expression in immune-privileged germ cells and various malignancies. Current application of CT-based immunotherapy has been focused on CT expression-rich tumors such as melanoma and lung cancers. In this study, we surveyed CT expression using the Cancer Genome Atlas (TCGA) datasets for ten common cancer types. We show that, CT expression is specific and enriched within certain cancer molecular subtypes. For example, HORMAD1, CXorf61, ACTL8 and PRAME are highly enriched in the basal subtype of breast cancer; MAGE and CSAG are most frequently activated in the magnoid subtype of lung adenocarcinoma; and PRAME is highly upregulated in the ccB subtype of clear cell renal cell carcinoma. Analysis of CT gene expression and DNA methylation indicates that some CTs are regulated epigenetically while others are controlled primarily by tissue- and subtype-specific transcription factors. Our results suggest that although for some CTs expression is associated with patient outcome, not many are independent prognostic markers. Thus, CTs with shared expression pattern are heterogeneous molecules with distinct activation modes and functional properties in different cancers and cancer subtypes. These data suggest a cancer subtype-orientated application of CT antigen as biomarkers and immunotherapeutic targets.
Keywords: cancer-testis antigen, tumor subtype, immunotherapy, prognosis, biomarker
Introduction
The cancer-testis (CT) antigens are characterized by their spontaneous immunogenicity and distinct expression patterns normally restricted to germ cells of the testis and placenta but frequently are activated in tumor cells (1, 2). T cells and antibodies against CT proteins are detectable in cancer patients (3–7), suggesting that the abnormal expression of CT antigens in tumors could induce adaptive immune response. More than one hundred CT antigen genes have been identified (8). Among these, CT-X genes form clusters on the X chromosome (e.g., MAGE, SSX, and SPANX gene families) and encode the most immunogenic CT proteins. Other CT genes are single-copy genes located on various autosomes. The expression frequency of CT genes varies greatly in cancers. Some cancers, such as colon, renal carcinoma, and glioblastoma, are CT-poor, with detectable CT expression in fewer than 20% of tumors. CT-rich cancer types, such as lung carcinoma and melanoma, can have CT expression frequencies greater than 50%. Within a cancer type, CT expression is heterogeneous in tumor cells and varies among different tumor grades. For example, higher frequency CT gene expression has been reported in more advanced stages of non-small cell lung cancer (9, 10). In bladder cancer, the expression of the MAGE gene family is most frequently found in the invasive forms (11), and the expression of NY-ESO-1 is correlated with higher nuclear grade (12).
Whether the reactivation of the CT genes in cancers represents a causal or correlative event is not clear and is under active investigation. Clinicopathological analyses have linked CT expression frequently with worse prognosis and less frequently with improved outcome in different cancer types (13–17); however, the molecular and cellular function of CT antigens is not well understood. For example, MAGEA11 is proposed to act as an oncogene by inhibiting prolyl hydroxylase 2 (PHD2) which down regulates the tumor-promoting hypoxia-inducible factor alpha (HIF1-α) in prostate cancer cells (18–20), MAGEA4 in the same gene family is suggested to be a tumor repressor by inducing apoptosis in non-small cell lung cancer (21–23).
Due to their limited expression in normal tissues and their wide distribution in tumors, CT antigens are promising targets for cancer immunotherapy. However, clinical trials based on strategies targeting two well-characterized CT antigens (MAGEA3 and NY-ESO-1) have shown limited success in cancer patients (24). Recently two parallel phase II studies, using heterologous prime-boost vaccination with rV-NY-ESO-1 and rF-NY-ESO-1, have reported promising clinical benefit for patients with melanoma and ovarian cancer that are at high risk for relapse (25). Moreover, adoptively transferred autologous T cells transduced with a T-cell receptor (TCR) directed against NY-ESO-1 have mediated tumor regression in patients with synovial cell sarcoma (26).
In this study, we performed a comprehensive survey of the expression of CT antigens in multiple human cancers from the Cancer Genome Atlas (TCGA) RNAseq datasets and identified multiple tumor subtype-specific CT antigens that can be further studied as potential biomarkers and targets for immunotherapy.
Materials and Methods
Molecular profiling datasets and data preprocessing
Level 3 RNAseq data (RNAseq RPKM or RNAseqV2 RSEM), level 3 Agilent microarray data, level 2 DNA methylation data (Infinium Human Methylation 450), and clinical data for multiple cancers were downloaded from the TCGA data portal (ref. 27; https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm). For DNA methylation data, M values were calculated as the log2 ratio of methylated intensity over unmethylated intensity (28). For RNAseq data, RSEM (RNA-Seq by Expectation Maximization) values were used for most analyses since they produced similar results as those from RPKM (reads per kilobase per million mapped reads) but providing more coverage on tumor samples. RPKM values were used only in the expression heatmaps (as in Figure 2) for comparing the expression levels between genes. The breast cancer dataset NKI-295 (29) was downloaded from http:/microarray-pubs.stanford.edu//wound_NKI/explore.htm. Due to ambiguous reads mapping to sets of genes with nearly 100% identity, only one gene of each set was used as a mapping target in RNAseq expression calculation. They include: CTAG1B for CTAG1A and CTAG1B; MAGEA2 for MAGEA2 and MAGEA2B; MAGEA9B for MAGEA9 and MAGEA9B; XAGE1D for XAGE1A, XAGE1B, XAGE1C, XAGE1D; GAGE12F for GAGE12F, GAGE12G and GAGE12I; GAGE12D for GAGE12C, GAGE12D and GAGE12E; SPANXB2 for SPANXB1 and SPANXB2.
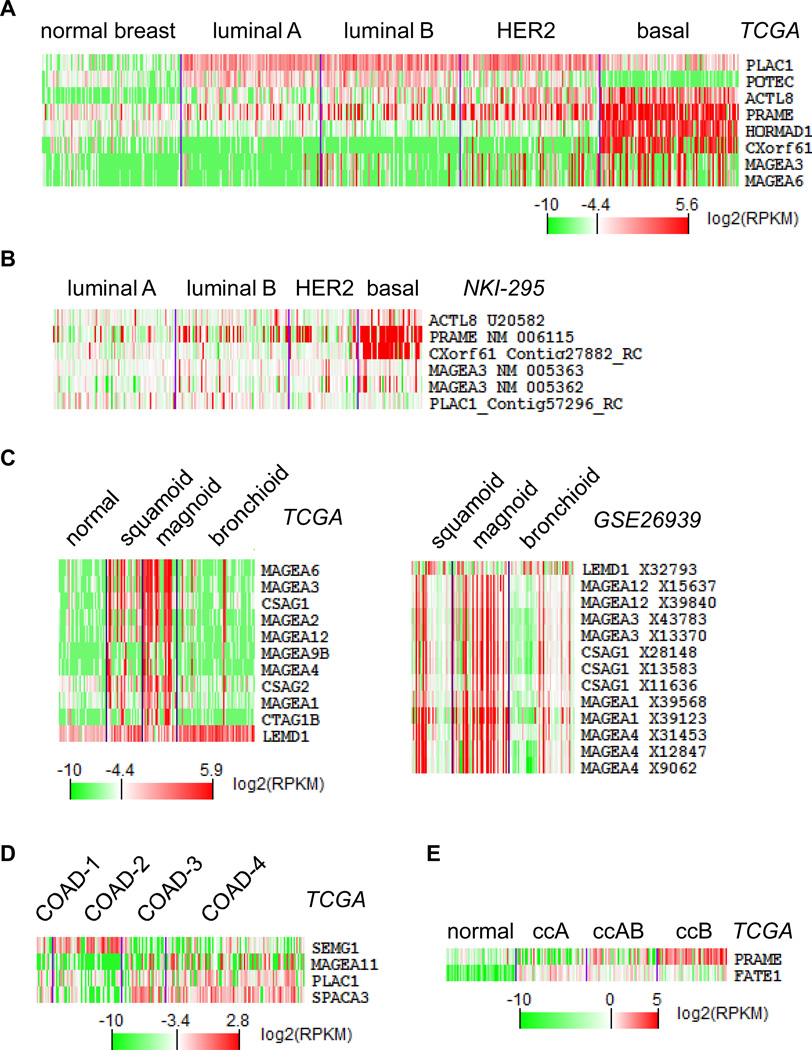
Figure 2.
Cancer subtype-specific CT gene expression. Gene expression heatmaps of top cancer subtype-specific CT genes (see Methods) were generated using TCGA RNAseq log2(RPKM) values for (A) breast cancer, (C) lung adenocarcinomas, (D) colorectal cancer, and (E) renal clear cell carcinomas. (B), Gene expression heatmap from a microarray dataset (NKI-295) generated by cluster and TreeView software using medium removal of normalized data to validate basal-specific CT genes found in TCGA (A). Similarly, right panel of (C) is validation result from GSE26939 for TCGA lung adenocarcinoma results.
Compilation of CT gene list
A list of 240 CT genes was queried from the current CT database (8). A search of the expression database generated from twelve normal somatic tissues by the Illumina BodyMap project (GEO accession: GSE30611) further narrowed this list down to 129 genes that have restricted expression in germ cells except for the minor expression in the brain.
Identification of CT gene overexpression in cancers and cancer subtypes
We examined CT gene expression in 318 normal tissue samples collected from autologous sites of six types of tumors (see supplemental Table S2) using RNAseqV2 RSEM values to set a baseline for each CT gene as the median normal expression level plus 3 times standard deviation. Any tumor with RSEM values above the baseline for a particular gene was considered positive for expression of that gene. To identify CT genes that are significantly expressed in cancer subtypes, we estimated CT gene expression percentage in each cancer subtype as described above. Only genes with >30% frequency of expression within a cancer subtype were used for the study. We used the ANOVA test to identify candidate CT genes with a cancer subtype-specific expression pattern. Based on different samples sizes, the P value cutoffs were set to 1e-12, 1e-8, 1e-5, and 0.02 for the BRCA, KIRC, LUAD, and COAD dataset, respectively.
Determination of tumor molecular subtypes using consensus clustering
Cancer molecular subtypes were determined by consensus k-means clustering of gene expression data using either Agilent microarray data or RNAseq RSEM values. Results using data from these two platforms from the same tumor correspond well with each other. The top 5,000 variably expressed genes with the highest Median Absolute Deviation (MAD) values were used to perform consensus clustering using the GenePattern website at http://genepattern.broadinstitute.org. The conditions used were k-means max 5 clusters with 500 rounds of resampling iterations. Validation of clustering results for known breast cancer and glioblastoma subtypes were performed using clustering analysis of genes from PAM50 (30) and the 840-gene list (31), respectively (see supplemental Figure S1).
Cluster analysis and statistical analysis
Clustering analyses were performed using the Cluster and TreeView software (http://rana.lbl.gov/EisenSoftware.htm). ANOVA test was conducted in R (http://www.r-project.org). For survival analysis, patients were stratified using the “k-means” function from the R software to two expression groups (high-expresser and low-expresser). Kaplan-Meier plots were drawn using the “survival” package from the R software. Multivariate analysis was carried out in SPSS Statistics 22.
Identification of DNA methylation sites regulating gene expression
Infinium Human Methylation450 probes covering a gene and its 3kb promoter region were collected. Pearson correlation was calculated between M values for each methylation probe and logged RNAseq RSEM values. This was done for each gene within each tumor type tested, and for each methylation probe a median Pearson coefficient was generated across all tumor types. For each gene, the probe with the least median Pearson coefficient was chosen as the best probe showing inversed correlation between methylation and gene expression.
Results
CT gene expression in human cancers
We compiled a set of 129 CT genes, which includes 82 CT-X and 47 non-X CT genes (see Methods and supplemental Table S1). RNAseq data on nearly 3,500 tumor samples along with 318 normal tissues were obtained from the TCGA data repository, comprising ten cancer types including breast invasive carcinoma, lung adenocarcinoma, lung squamous carcinoma, colon adenocarcinoma, ovarian serous adenocarcinoma, clear cell renal cell carcinoma (ccRCC), head and neck squamous carcinoma, endometrial carcinoma, skin cutaneous melanoma, and glioblastoma (see supplemental Table S2). For RNAseq data, RSEM values were utilized for most analyses since these generated similar results as those from RPKM but with greater data coverage for tumor samples (32). RPKM values were used only in expression heatmaps for comparing expression levels between genes.
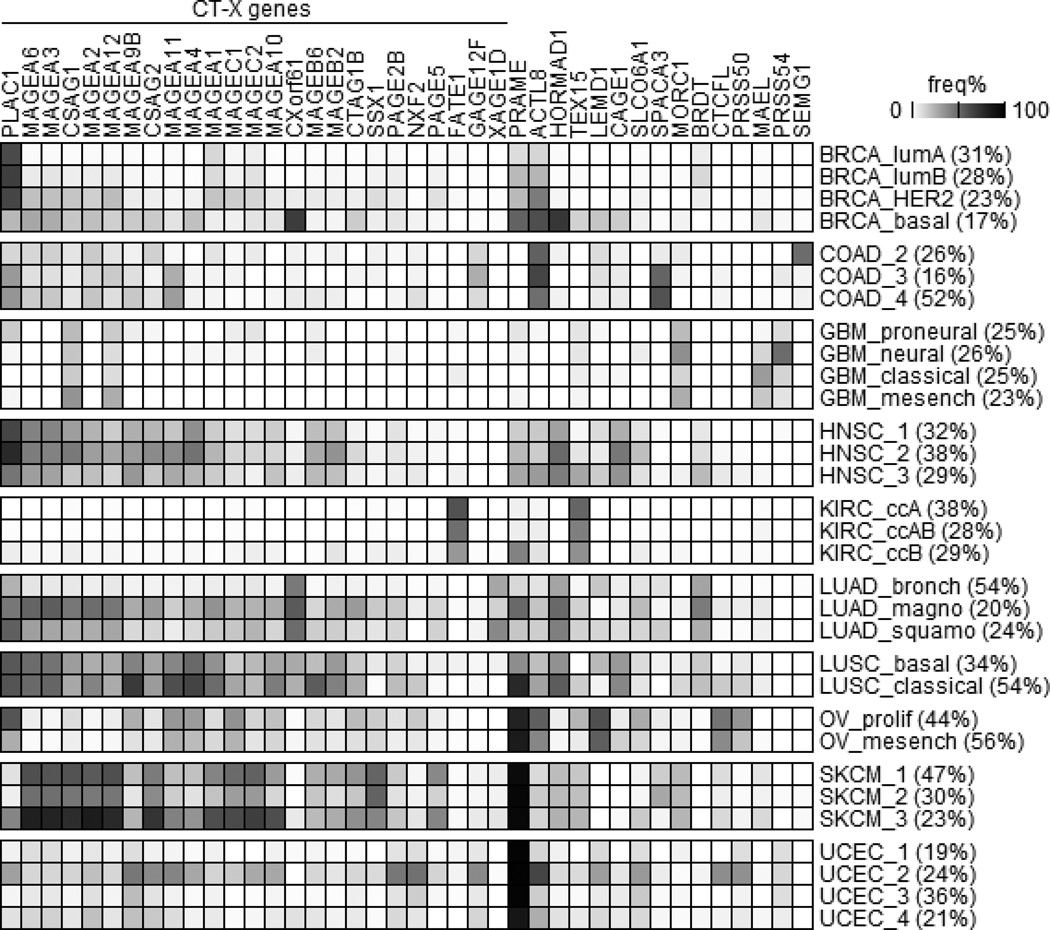
To identify CT genes with restricted expression in tumors we first examined CT gene expression in all normal tissue samples collected from autologous sites of six types of tumors (see supplemental Table S2) and set a baseline for each CT gene as the median normal expression level plus 3 times standard deviation (see Methods). Tumors with expression above the baseline for a particular CT gene were used for further analysis. Figure 1 summarizes the frequency of CT gene overexpression in a panel of human cancers, covering genes having a >15% expression frequency in at least one tumor type. This analysis identified 35 CT-X genes and 19 non-X CT genes for further study. PLAC1 is the most frequently expressed CT-X gene and PRAME is the most frequently expressed non-X CT gene in all cancers. These results also confirmed that glioblastoma and ccRCC are CT-poor tumors, while melanomas and lung carcinomas have frequent CT gene expression. A summary of CT gene overexpression frequencies is shown in supplemental Table S3.
Figure 1.
CT gene expression frequency in human cancers. CT gene expression frequencies were calculated (see Methods) and genes having a >15% expression frequency in at least one tumor were plotted. Codes for cancer types: BRCA, breast; COAD, colon; GBM, glioblastoma; HNSC, head and neck; KIRC, kidney/renal clear cell carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian; SKCM, skin cutaneous melanoma; and UCEC, endometrial carcinoma.
Enriched expression of CT genes in molecular subtypes of cancers
Since CT genes are often not prevalently overexpressed in cancers, and cancers are known to be intrinsically heterogeneous, we examined if CT genes are activated within particular cancer subtypes. To this end, we defined molecular subtypes of all ten cancers included in this study by consensus clustering of gene expression using the RNAseq RSEM values (see Methods). As expected, our analysis identified known molecular subtypes for breast, lung, and brain tumors (supplemental Figure S1). For example, our analysis separated breast cancers into luminal A/B, HER2-enriched, and basal subtypes (33), glioblastomas into proneural, neural, classical, and mesenchymal subtypes (31), and lung adenocarcinomas into bronchioid, magnoid, and squamoid subtypes (34). For ccRCC, besides the previously known subtypes ccA and ccB, we identified a new subtype named ccAB which has a gene expression pattern between those of the ccA and ccB classes (35,36). Many of these molecular subtypes are known to have prognostic value, which we confirmed in lung, kidney, and endometrial cancers (supplemental Figure S1), but not in breast cancer where basal subtype was a well-known poor prognosis group (data not shown).
We next used the ANOVA test to identify CT gene expression enriched within cancer subtypes (see Methods). In breast cancer, we confirmed that CXorf61 and HORMAD1 were specific for the basal subtype and PLAC1 for the non-basal subtype (37,38). We also identified additional subtype-specific genes such as ACTL8 and PRAME for the basal subtype and POTEC for the non-basal subtype (Figure 2A, supplemental Table S4). In lung adenocarcinomas, a set of MAGE genes are most frequently overexpressed in the magnoid subtype and least expressed in the bronchioid subtype (Figure 2C left panel). Similarly, SEMG1 overexpression is enriched in the colon cancer COAD-1 subtype, SPACA3 is enriched in the COAD-2, -3 subtypes, and PRAME is enriched in the ccB subtype of ccRCC (Figure 2D–E). We validated our findings using external microarray gene expression datasets available for breast cancer and lung adenocarcinomas 34,39). This confirmed that basal breast cancers have higher expression of CXorf61, PRAME, ACTL8, and MAGEA3 (Figure 2B) and the expression of MAGEA1, 3, 4, 12 and CSAG1 is higher in magnoid lung adenocarcinomas (Figure 2C right panel).
Figure 3 highlights this updated landscape of CT gene expression in various cancers including subtype information, covering 40 genes with >30% expression frequency in at least one subtype of one cancer. These results indicate that genes such as CXorf61, HORMAD1, and SEMG1 exhibit cancer subtype-specific overexpression. In addition, particular cancer subtypes such as magnoid of lung adenocarcinoma and UCEC-2 of endometrial cancer exhibit overall increased CT gene expression compared to other subtypes of the same cancer. Therefore, CT gene expression may be restricted to particular cancer subtypes even when its expression frequency in a particular cancer is low. These genes represent potential immunotherapy targets for those specific cancer subtypes.
Figure 3.
CT gene expression frequency in cancer subtypes. Cancer molecular subtypes were determined using consensus clustering (see Methods) and genes having a >30% overexpression frequency in at least one cancer subtype were plotted. For each cancer subtype, its percentage within the corresponding cancer was labeled in parentheses.
Regulation of CT gene expression by DNA methylation
Previous studies have shown that epigenetic modifications including promoter hypomethylation and histone deacetylation have important roles in CT gene activation (40–44). We analyzed TCGA methylation microarrays data (Infinium human Met450) compare the DNA methylation status with CT gene expression. This analysis focused on eight tumor types for which sufficient data were available (with more than 50 samples having both RNAseq and methylation data) on Met450 probes covering the entire gene structure and the 3 kb upstream promoter region for each of the 35 CT genes listed in Figure 3.
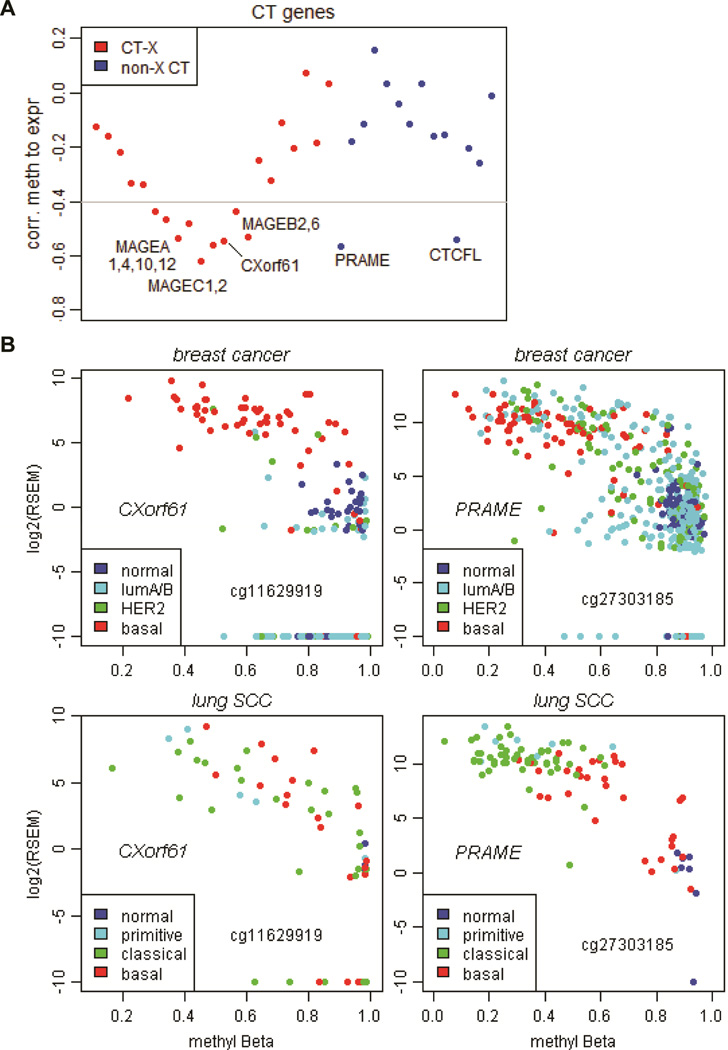
The negative correlations between CT gene methylation and expression across cancer types are estimated and shown in Figure 4A, reporting the least median Pearson correlation coefficient from all Met450 probes for one CT gene in eight tumors (see Methods and supplemental Table S5). This analysis confirmed that the transcription of the CT-X genes (e.g. MAGEs, CXorf61) is regulated primarily by promoter DNA methylation (45). On the other hand, non-X CT gene expression correlated less well with DNA methylation except for PRAME and CTCFL. A detailed analysis of the CXorf61 gene expression and DNA methylation in breast cancers confirmed that it is more hypomethylated and highly expressed in basal tumors (Fig.4B left). However, there are non-basal breast tumors that were equally methylated yet with less expression, indicating that there are other subtype-specific mechanisms inhibiting CXorf61 expression in non-basal tumors. There is a correlation between CXorf61 gene expression and DNA methylation in lung squamous carcinomas, even though it is not expressed in a subtype-specific manner.
Figure 4.
Correlation of CT gene expression to DNA methylation. (A) Plot of the least median Pearson correlation coefficients between gene expression (using logged RNAseq RSEM values) and DNA methylation (using Met450 M values) for CT-X genes (red) and non-X CT genes (blue) (see Methods). (B) Scatter plots of CXorf61 and PRAME gene expression (logged RSEM values) versus DNA methylation (beta values) in breast cancers and lung squamous cell carcinomas. Samples were labeled in colors to mark different cancer subtypes as shown in the legend. Infinium Met450 probes under study were marked on the figure, which can be used to locate exact chromosomal coordinates of the DNA methylation sites.
Similarly, PRAME expression and DNA methylation are correlated in both breast cancer and lung squamous carcinomas, with better subtype-specificity in lung squamous carcinomas (Fig.4B right). Thus, activation of CT genes which are enriched in certain cancer subtypes is likely to be controlled by both DNA methylation and other subtype-specific mechanisms.
Prognostic value of CT genes in ccRCC
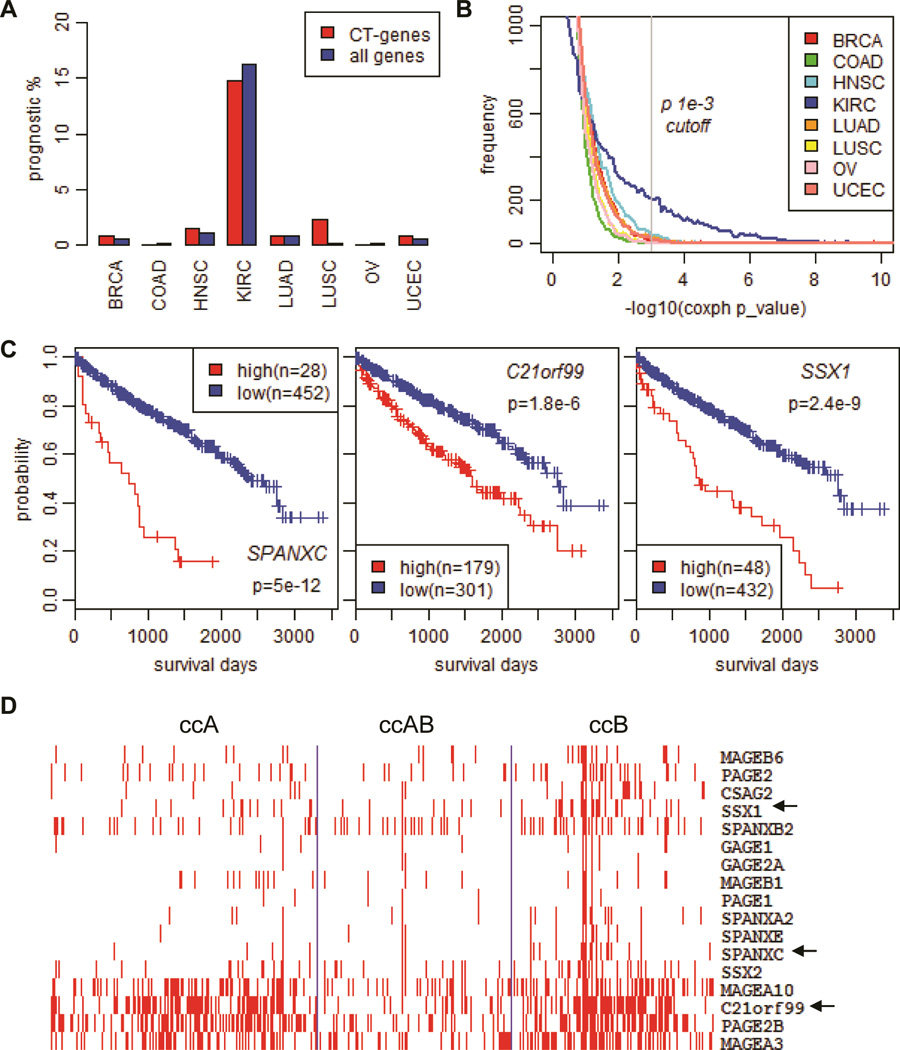
Correlations between CT expression and cancer prognosis have been reported in various studies (46,47). In our analysis, we examined all 129 CT genes for their prognostic values with TCGA data by univariate Cox proportional hazard regression test in eight cancers using the same sample size of 200 randomly selected samples. This analysis revealed that approximately 15% of CT genes are potentially prognostic in ccRCC, a percentage much higher than those found in other cancer types (Figure 5A and supplemental Table S6). Results from Kaplan-Meier survival analysis on the top three candidate prognostic genes are shown in Figure 5C, confirming that overexpression of SPANXC, C21orf99, and SSX1 are indicators of poor prognosis for ccRCC. When the same test was performed on all known genes, we found that more prognostic genes in ccRCC than in other cancers (Figure 5A and 5B). Indeed, cluster analysis of ~3,000 prognostic genes found in ccRCC clearly identified two major types of prognostic genes, those highly expressed in the ccB subtype (poor prognostic genes) and those down-regulated in the ccB subtype (supplemental Figure S2), which has significantly shortened survival compared with the ccA and ccAB subtypes (supplemental Figure S1A). A closer examination of CT gene expression associated with poor prognosis within the ccA, ccAB, and ccB subtypes also identified many CTs including the SPANXC and SSX1/2 genes with enriched expression in the ccB subtype (Figure 5D). Therefore, a higher percentage of prognostic CTs seen in ccRCC are not unexpected. A previous study on conventional RCC identified a 259-gene prognostic gene expression signature, of which 45% of the genes overlap with our findings (data not shown) (48).
Figure 5.
Prognostic value of CT genes in clear cell renal cell carcinomas (ccRCC). (A) Percentages of CT genes and all genes found to be prognostic in human cancers. (B) Histogram of gene density along survival analysis Coxph p values (in −log10) for eight tumors all using 200 randomly picked samples. (C) Kaplan-Meier analyses of patient outcome stratified by expression of SPANXC, C21orf99, and SSX1 genes in ccRCC (see Methods). (D) Expression of top poor prognostic CT genes in ccRCC subtypes. Heatmap was generated using cluster and TreeView software after medium removal using RNAseq RSEM values.
ANOVA tests showed that the expression of the poor prognosis CTs (SPANXC, SSXs, C21orf99 and PRAME) is significantly associated with higher tumor grade/stage while the expression of the better prognosis CT (FATE1) is significantly associated with lower tumor grade/stage (supplemental Figure S2B). This is in agreement with a previous report that the ccB subtype comprised higher grade tumors (36). In the multivariate analysis with histological grade, pathological stage and metastatic status added as confounding variables, CT association with prognosis is either diminished or weakened even as the significance remains for some CTs such as SPANXC and SSX1. Similar results of prognostic feature were observed in other cancer types (supplemental Table S7). Therefore, a significant proportion of CT genes show prognostic values in ccRCC that can be attributed partly to their coincidental overexpression in the poor prognostic ccB subtype. As these CT genes can be treated as potential prognostic markers, they are unlikely to be the causal factor of poor prognosis in ccRCC.
Discussion
Although CT genes are increasingly promising as targets for cancer immunotherapy approaches, a limitation is the selection of adequate tumor-specific targets, especially for CT-poor cancers. Previous studies of CT expression analyses generated by RT-PCR, immunohistochemistry (IHC) or microarray platforms have particular limitations. For example, RT-PCR and IHC could not provide a genome-wide unbiased context and microarray probes did not cover all CTs and could not discriminate close members within a CT-X gene cluster. The large TCGA datasets generated by the next generation sequencing technology has enabled better characterization of tumor-specific CT antigens as biomarkers and proficient identification of promising immunotherapy targets within cancers and cancer subtypes. In this study, we analyzed the large datasets of expression, methylation and clinicopathological features available from the TCGA database to create a landscape of CT gene re-activation in cancers.
In particular, we refined the previous notion of CT-poor or CT-rich cancers based on the enrichment of CTs in specific subtypes. The first suggestion of subtype-specific CT gene expression in a CT-poor tumor type was described in the estrogen receptor (ER)-negative and basal breast cancer molecular subtypes (38). Although in that study the analysis was limited to CT-X genes that were present on the arrays, it showed the possibility of identifying subsets of patients that could potentially benefit from CT-based immunotherapy approaches. This raises the possibility of using subtype-specific CT antigen information in clinical trial design within the context of several other factors such as the antigenicity of each CT, the specificity of gene expression, and the heterogeneity of CT expression within a tumor. A caveat to this approach is that in tumors heterogeneous for CT expression, antigen-negative tumor cells might escape from immune intervention. Thus, finding additional options for CT-based immunotherapy would expand the reservoir for polyvalent vaccines in order to enhance immune response and reduce the chance of tumor cell escape from immunotherapy.
The control of CT expression was also examined in this study with the integration of the TCGA methylation data. As expected, CT-X genes cluster on the chromosome and usually are co-activated; for example, the MAGEA (2, 3, 6, 12) and CSAG (1, 2, 3) genes are within 70 kb (151, 869, 311 – 151,938, 240) at Xq28 and they are frequently co-expressed. A similar pattern was observed for MAGEA4 and MAGEA10, which are ~200 kb apart from 151,092,137 to 151,302,983. Co-expression of CT genes within a physical proximity in all cancer types (Figure 1 and Figure 3) can be partially explained by shared local epigenetic regulation driven by demethylation and histone modification. However, even in regions with high correlation, discordance is observed at the gene level. For example, some tumors express MAGEAs but not CSAGs and vice versa. It is not surprising that FATE1, a CT gene 100 kb upstream of MAGEA4, is rarely co-expressed with MAGE4, given its R value of −0.2 for expression and methylation.
Understanding the mechanisms of CT upregulation is of clinical importance as correlations between CT expression and epigenetic changes could lead to combinatorial therapies. For example, a combined regimen of demethylation agents and HDAC inhibitors might increase the expression of a number of CT genes thus leading to improved immune responses.
Expression of many CTs shows association with prognosis in this study. However, our results do not address if survival effects might relate to any biological driver effect of CT genes. Instead, our data suggest that this may relate to subtype-specific or grade-associated expression. Among the ten cancers studied, we identified the most significant subtype effects in ccRCC. For this cancer, patients with subtype ccB have a significantly worse survival rate than those with ccA or ccAB. The largest number of prognostic CTs is identified in ccRCC and most of them exhibit a subtype-specific expression pattern. Multivariate cox regression analysis with other clinical parameters shows that the prognostic value of CT expression could be attributed to its association with higher tumor grade/stage. Among the breast cancer molecular subtypes, the HER2-overexpressing and basal-like breast cancers have been shown to link to poorer outcomes (49,50). In the TCGA dataset, however, this link was not significant for the HER2 or basal subgroups. The median follow-up for the dataset is approximately 430 days (14 months), which is much shorter than that in other existing datasets. Thus, it is not surprising that our data did not reveal prognostic CTs in breast cancer, even though a large number of subtype-specific CTs are identified in the TCGA breast cancer dataset.
In this study, we have utilized the extensive TCGA RNAseq datasets as the indicator of gene expression. Future serological experiments will be needed to investigate the correlation between messenger RNA, protein expression and antibody response in serum. Our data provide support for additional studies toward the development of enhanced approaches for cancer immunotherapy based on subtype-specific expression of the CT genes as either potential diagnostic markers or immunotherapeutic targets.
Supplementary Material
Acknowledgement
We would like to acknowledge the TCGA research network for providing the cancer genomics data used in this study.
Financial support: This study is supported in part by the TCGA grant U24CA143883 from NCI/NIH, and funds from Ludwig Institute for Cancer Research.
W.K.A.Y has received a research grant from Daiichi Sankyo, honoraria from Actelion, and serves as a consultant to Novartis and Merck.
Footnotes
The other authors declare no conflict of interest.
References
- 1.Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 2.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 3.Jager D, Jager E, Knuth A. Immune responses to tumour antigens: implications for antigen specific immunotherapy of cancer. J Clin Pathol. 2001;54:669–674. doi: 10.1136/jcp.54.9.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li G, Miles A, Line A, Rees RC. Identification of tumour antigens by serological analysis of cDNA expression cloning. Cancer Immunol Immunother. 2004;53:139–143. doi: 10.1007/s00262-003-0471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mischo A, Kubuschok B, Ertan K, Preuss KD, Romeike B, Regitz E, et al. Prospective study on the expression of cancer testis genes and antibody responses in 100 consecutive patients with primary breast cancer. Int J Cancer. 2006;118:696–703. doi: 10.1002/ijc.21352. [DOI] [PubMed] [Google Scholar]
- 6.Groeper C, Gambazzi F, Zajac P, Bubendorf L, Adamina M, Rosenthal R, et al. Cancer/testis antigen expression and specific cytotoxic T lymphocyte responses in non small cell lung cancer. Int J Cancer. 2007;120:337–343. doi: 10.1002/ijc.22309. [DOI] [PubMed] [Google Scholar]
- 7.Bricard G, Bouzourene H, Martinet O, Rimoldi D, Halkic N, Gillet M, et al. Naturally acquired MAGE-A10- and SSX-2-specific CD8+ T cell responses in patients with hepatocellular carcinoma. J Immunol. 2005;174:1709–1716. doi: 10.4049/jimmunol.174.3.1709. [DOI] [PubMed] [Google Scholar]
- 8.CT database. http://www.cta.lncc.br/index.php.
- 9.Gure AO, Chua R, Williamson B, Gonen M, Ferrera CA, Gnjatic S, et al. Cancer-testis genes are coordinately expressed and are markers of poor outcome in non-small cell lung cancer. Clin Cancer Res. 2005;11:8055–8062. doi: 10.1158/1078-0432.CCR-05-1203. [DOI] [PubMed] [Google Scholar]
- 10.Konishi J, Toyooka S, Aoe M, Omura Y, Washio K, Tsukuda K, et al. The relationship between NY-ESO-1 mRNA expression and clinicopathological features in non-small cell lung cancer. Oncol Rep. 2004;11:1063–1067. doi: 10.3892/or.11.5.1063. [DOI] [PubMed] [Google Scholar]
- 11.Nishiyama T, Tachibana M, Horiguchi Y, Nakamura K, Ikeda Y, Takesako K, et al. Immunotherapy of bladder cancer using autologous dendritic cells pulsed with human lymphocyte antigen-A24-specific MAGE-3 peptide. Clin Cancer Res. 2001;7:23–31. [PubMed] [Google Scholar]
- 12.Kurashige T, Noguchi Y, Saika T, Ono T, Nagata Y, Jungbluth A, et al. Ny-ESO-1 expression and immunogenicity associated with transitional cell carcinoma: correlation with tumor grade. Cancer Res. 2001;61:4671–4674. [PubMed] [Google Scholar]
- 13.Freitas M, Malheiros S, Stavale JN, Biassi TP, Zamuner FT, de Souza Begnami M, et al. Expression of cancer/testis antigens is correlated with improved survival in glioblastoma. Oncotarget. 2013;4:636–646. doi: 10.18632/oncotarget.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou X, Yang F, Zhang T, Zhuang R, Sun Y, Fang L, et al. Heterogeneous expression of CT10, CT45 and GAGE7 antigens and their prognostic significance in human breast carcinoma. Jpn J Clin Oncol. 2013;43:243–250. doi: 10.1093/jjco/hys236. [DOI] [PubMed] [Google Scholar]
- 15.von Boehmer L, Keller L, Mortezavi A, Provenzano M, Sais G, Hermanns T, et al. MAGE-C2/CT10 protein expression is an independent predictor of recurrence in prostate cancer. PLoS One. 2011;6:e21366. doi: 10.1371/journal.pone.0021366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Duin M, Broyl A, de Knegt Y, Goldschmidt H, Richardson PG, Hop WC, et al. Cancer testis antigens in newly diagnosed and relapse multiple myeloma: prognostic markers and potential targets for immunotherapy. Haematologica. 2011;96:1662–1669. doi: 10.3324/haematol.2010.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svobodova S, Browning J, MacGregor D, Pollara G, Scolyer RA, Murali R, et al. Cancer-testis antigen expression in primary cutaneous melanoma has independent prognostic value comparable to that of Breslow thickness, ulceration and mitotic rate. Eur J Cancer. 2011;47:460–469. doi: 10.1016/j.ejca.2010.09.042. [DOI] [PubMed] [Google Scholar]
- 18.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 19.Kimmins S, Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 2005;434:583–589. doi: 10.1038/nature03368. [DOI] [PubMed] [Google Scholar]
- 20.Aprelikova O, Pandolfi S, Tackett S, Ferreira M, Salnikow K, Ward Y, et al. Melanoma antigen-11 inhibits the hypoxia-inducible factor prolyl hydroxylase 2 and activates hypoxic response. Cancer Res. 2009;69:616–624. doi: 10.1158/0008-5472.CAN-08-0811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peikert T, Specks U, Farver C, Erzurum SC, Comhair SA. Melanoma antigen A4 is expressed in non-small cell lung cancers and promotes apoptosis. Cancer Res. 2006;66:4693–4700. doi: 10.1158/0008-5472.CAN-05-3327. [DOI] [PubMed] [Google Scholar]
- 22.Sakurai T, Itoh K, Higashitsuji H, Nagao T, Nonoguchi K, Chiba T, et al. A cleaved form of MAGE-A4 binds to Miz-1 and induces apoptosis in human cells. J Biol Chem. 2004;279:15505–15514. doi: 10.1074/jbc.M310437200. [DOI] [PubMed] [Google Scholar]
- 23.Nagao T, Higashitsuji H, Nonoguchi K, Sakurai T, Dawson S, Mayer RJ, et al. MAGE-A4 interacts with the liver oncoprotein gankyrin and suppresses its tumorigenic activity. J Biol Chem. 2003;278:10668–10674. doi: 10.1074/jbc.M206104200. [DOI] [PubMed] [Google Scholar]
- 24.Vansteenkiste J, Zielinski M, Linder A, Dahabreh J, Gonzalez EE, Malinowski W, et al. Adjuvant MAGE-A3 Immunotherapy in Resected Non-Small-Cell Lung Cancer: Phase II Randomized Study Results. J Clin Oncol. 2013;31:2396–2403. doi: 10.1200/JCO.2012.43.7103. [DOI] [PubMed] [Google Scholar]
- 25.Odunsi K, Matsuzaki J, Karbach J, Neumann A, Mhawech-Fauceglia P, Miller A, et al. Efficacy of vaccination with recombinant vaccinia and fowlpox vectors expressing NY-ESO-1 antigen in ovarian cancer and melanoma patients. Proc Natl Acad Sci U S A. 2012;109:5797–5802. doi: 10.1073/pnas.1117208109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbins PF, Morgan RA, Feldman SA, Yang JC, Sherry RM, Dudley ME, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol. 2011;29:917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.TCGA data portal. ( https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm)
- 28.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The breast cancer dataset NKI-295 was downloaded. from http:/microarray-pubs.stanford.edu//wound_NKI/explore.htm. [Google Scholar]
- 30.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Network CGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkerson MD, Yin X, Walter V, Zhao N, Cabanski CR, Hayward MC, et al. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLoS One. 2012;7:e36530. doi: 10.1371/journal.pone.0036530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Creighton CJ, Morgan M, Gunaratne PH, Wheeler DA, Gibbs RA, Gordon Robertson A, et al. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;4:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brannon AR, Reddy A, Seiler M, Arreola A, Moore DT, Pruthi RS, et al. Molecular Stratification of Clear Cell Renal Cell Carcinoma by Consensus Clustering Reveals Distinct Subtypes and Survival Patterns. Genes Cancer. 2010;1:152–163. doi: 10.1177/1947601909359929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adelaide J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565–11575. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- 38.Grigoriadis A, Caballero OL, Hoek KS, da Silva L, Chen YT, Shin SJ, et al. CT-X antigen expression in human breast cancer. Proc Natl Acad Sci U S A. 2009;106:13493–13498. doi: 10.1073/pnas.0906840106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 40.De Smet C, Lurquin C, Lethe B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19:7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oi S, Natsume A, Ito M, Kondo Y, Shimato S, Maeda Y, et al. Synergistic induction of NY-ESO-1 antigen expression by a novel histone deacetylase inhibitor, valproic acid, with 5-aza-2'-deoxycytidine in glioma cells. J Neurooncol. 2009;92:15–22. doi: 10.1007/s11060-008-9732-0. [DOI] [PubMed] [Google Scholar]
- 42.Sigalotti L, Fratta E, Coral S, Tanzarella S, Danielli R, Colizzi F, et al. Intratumor heterogeneity of cancer/testis antigens expression in human cutaneous melanoma is methylation-regulated and functionally reverted by 5-aza-2'-deoxycytidine. Cancer Res. 2004;64:9167–9171. doi: 10.1158/0008-5472.CAN-04-1442. [DOI] [PubMed] [Google Scholar]
- 43.Goelz SE, Vogelstein B, Hamilton SR, Feinberg AP. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science. 1985;228:187–190. doi: 10.1126/science.2579435. [DOI] [PubMed] [Google Scholar]
- 44.Kim R, Kulkarni P, Hannenhalli S. Derepression of Cancer/testis antigens in cancer is associated with distinct patterns of DNA hypomethylation. BMC Cancer. 2013;13:144. doi: 10.1186/1471-2407-13-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 46.Shigematsu Y, Hanagiri T, Shiota H, Kuroda K, Baba T, Mizukami M, et al. Clinical significance of cancer/testis antigens expression in patients with non-small cell lung cancer. Lung Cancer. 2009;68:105–110. doi: 10.1016/j.lungcan.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 47.Caballero OL, Chen YT. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 2009;100:2014–2021. doi: 10.1111/j.1349-7006.2009.01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao H, Ljungberg B, Grankvist K, Rasmuson T, Tibshirani R, Brooks JD. Gene expression profiling predicts survival in conventional renal cell carcinoma. PLoS Med. 2006;3:e13. doi: 10.1371/journal.pmed.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.