Abstract
Both genetic variants and brain region abnormalities are recognized as important factors for complex diseases (e.g., schizophrenia). In this paper, we investigated the correspondence between single nucleotide polymorphism (SNP) and brain activity measured by functional magnetic resonance imaging (fMRI) to understand how genetic variation influences the brain activity. A group sparse canonical correlation analysis method (group sparse CCA) was developed to explore the correlation between these two datasets which are high dimensional-the number of SNPs/voxels is far greater than the number of samples. Different from the existing sparse CCA methods (sCCA), our approach can exploit structural information in the correlation analysis by introducing group constraints. A simulation study demonstrates that it outperforms the existing sCCA. We applied this method to the real data analysis and identified two pairs of significant canonical variates with average correlations of 0.4527 and 0.4292 respectively, which were used to identify genes and voxels associated with schizophrenia. The selected genes are mostly from 5 schizophrenia (SZ)-related signalling pathways. The brain mappings of the selected voxles also indicate the abnormal brain regions susceptible to schizophrenia. A gene and brain region of interest (ROI) correlation analysis was further performed to confirm the significant correlations between genes and ROIs.
Keywords: Group sparse CCA, SNP, fMRI, Feature selection, Imaging genetics
1. Introduction
Schizophrenia is a complex disease and considered to be caused by the interplay of a number of genetic factors (e.g., change of gene regulation, and alteration of mRNA and SNP) and environmental effects. Genetic factors play an important role in causing schizophrenia disease. People born from a family with a history of schizophrenia have higher risks of schizophrenia than those without a family schizophrenia history. In recent years, many studies have focused on exploring critical genes associated with the schizophrenia. Many potential genetic variants have been reported as possible risk factors such as the G72/G30 gene locus on chromosome 13q (Badner and Gershon, 2002; Abecasis et al., 2004) Gene DISC1 variation (Callicott et al., 2005; Porteous et al., 2006) and copy number variations on gene GRIK3, EFNA5, AKAP5 and CACNG2 (Wilson et al., 2006; Sutrala et al., 2007). In addition to genetic studies, fMRI has also been widely used for the study of schizophrenia because of its capability to identify functional abnormalities within brain regions of schizophrenic patients (Jansma et al., 2004; Li et al., 2007; Meda et al., 2008; Szycik et al., 2009).
Genetic variants and brain region abnormalities are both important markers for the study of schizophrenia. Combining both data can not only contribute to a better understanding of biological mechanisms on brain structure and function but also have the potential to improve the diagnosis and treatments of complex diseases. However, current imaging genetics studies either take brain imaging measurements as endophenotypes to study the associated genetic variants or investigate the effects of a small set of candidate genetic variants on the whole brain measurements (Hamid et al., 2009; Le Cao et al., 2009; Wiley, 2011). It is still challenging to explore the relationship between a large amount of genetic variants and a large number of brain imaging measurements. Therefore, correlative analysis approaches for large-scale multi-modal data analysis are highly demanded.
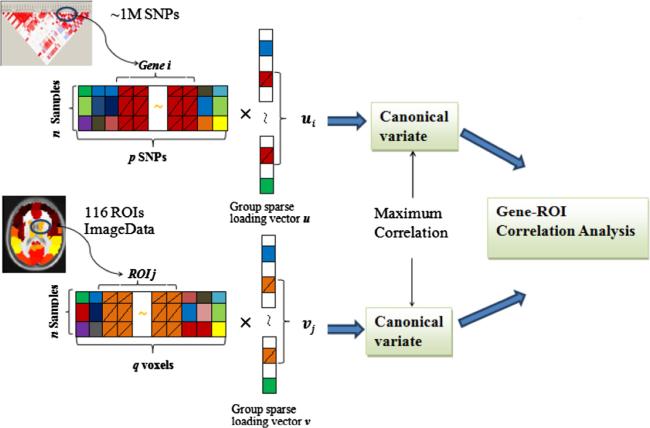
In this work, we aim to study the effects of multiple SNPs or genes on functional brain activity in schizophrenia. An effective multivariate statistical method is needed. Canonical Correlation Analysis (CCA (Hotelling, 1936)) or Partial Least Squares regression (PLSR (Le Cao et al., 2008)) have been proposed to analyze multi-modal datasets. The CCA aims to maximize the correlation between the linear combinations of variables from two data sets, e.g., a linear combination of SNPs and a linear combination of voxels. However, the method will have the over-fitting issues in analyzing high dimensional data such as SNP and brain imaging data as shown in Fig. 1. Thousands of SNPs with linkage disequilibrium (LD) are detected to reflect the genetic variant at different locus. The number of voxels included in the whole brain fMRI image is also very large (e.g., 53 × 63 × 46). Traditional CCA will perform poorly in such a case due to the multi-collinearity (linear dependence) problem, and thus having computational difficulty (Park-homenko et al., 2009). To address above issue, sparse CCA (sCCA) methods, mostly using the l – 1 norm (CCA-l1) or the combination of l – 1 and l – 2 norm (CCA-elastic net) penalties, have been developed by introducing the sparse penalties into the traditional CCA model (Waaijenborg et al., 2008; Le Cao et al., 2009; Parkhomenko et al., 2009; Witten et al., 2009; Witten and Tibshirani, 2009; Boutte and Liu, 2010). Despite the success, they didn't account for group structures within the data in the analysis (e.g., multiple SNPs within the same gene, a group of voxels within the same region, a group of voxels within the same ROI, etc.), which often exist or are implied by the biological mechanism. For example, SNPs within the same gene have similar functions and act together at the gene or pathway level to affect the brain activity. These SNP effects can be added up to a larger difference (Tyekucheva et al., 2011). Several previous works have shown the benefit of accounting for the group effect of features in the sCCA models (Chen and Liu, 2012; Chen et al., 2013; Lin et al., 2013). However, to our knowledge, little work has been reported to incorporate the group effect into the sCCA model for fMRI and SNP data integration. Motivated by this fact, in this paper, we developed a group sparse CCA model based integration method by imposing the sparse group lasso penalty on the CCA model for the integrative analysis of SNP and fMRI data; please refer to Fig. 1 for illustration. This method has the following advantages: (1) A group of features (voxels/SNPs) will be inspected during the correlation analysis, which can study the joint effects of multiple SNPs on the regions of voxels; (2) feature selection will be performed at both group and single feature level. Irrelevant groups of features as well as single feature within each group can be removed. Our group sparse CCA method can both exploit group information in the correlation analysis while filter out noisy features within the group simultaneously.
Fig. 1.
A schematic illustration of combining both fMRI image and SNP data by the group sparse CCA model to identify correlated genes and ROIs.
The group sparse CCA can estimate the correlation between canonical variates, corresponding to a set of significant SNPs or brain imaging voxels. Based on the estimates, we provided a gene-ROI correlation analysis to further confirm the significance of the correlations between genes and brain functions in ROIs.
The rest of the paper is organized as follows. The proposed group sparse CCA model and algorithm are introduced in the section of theory. The group sparse CCA based integration method for SNP and fMRI data is described in the section of method. The validation and comparison of our model with other sCCA models on both simulated and real data analysis are presented in the section of results. The pathway analysis results and limitations of the proposed method are discussed finally.
2. Theory
In this section, we first introduces CCA model, based on which the group sparse model is presented. Then we propose a numerical algorithm based on block coordinate descent to solve the model. Finally, we show that the general model we propose can include several existing sCCA models and hence the numerical algorithm can also be applied for their efficient solutions.
2.1. Group sparse CCA
We consider two sets of data X and Y with n samples, where X has p variables and Y has q variables. Assuming and the columns of X and Y have been standardized such that the mean of each column is zero and the 2-norm is one. The variance matrices and covariance matrices of X and Y are denoted by , , (or ) respectively. The CCA model aims to find two loading vectors or projections α, β, to maximize the correlation between the linear combinations of variables in X and Y, αtX and βtY as shown in the following equation:
| (1) |
There are over-fitting issues in Eq. (1) due to the small samples but high dimensional variables (i.e., ), which also might result in the ill-condition of the matrices and . So, sparse penalties such as the l – 1 norm, elastic net, have been imposed on the loading vectors in Eq. (2) in CCA analysis.
| (2) |
where Ψ(·) and Φ(·) denote the penalized function on α and β respectively, which are often taken to be l – 1 norm, Ψ(α) = ∥α∥1, Φ(β) = ∥β∥1. These sparse penalties can result in a large number of features/variables to zero in α and β. In addition, the constrained conditions in Eq. (2) are relaxed compared to Eq. (1) so that we can first find the alternative solution in a closed region and then obtain the solution of the optimization problem satisfying , (see optimization algorithms in Section 2.2). Similar to conventional CCA, Sparse CCA can also perform multiple pairs of canonical variates extraction iteratively as the following procedure: maximizing the correlation between two data sets by extracting the first pair of sparse loading vectors as well as pair of canonical variates; then removing the effects of the first pair of canonical variates and finding the second sparse loading vectors that maximizes the correlation but is irrelevant to the first pair. This process will not stop until the rth projection pair is gained (r = rank (XtY)).
Group information of variables is expected to be incorporated in the Eq. (2) by changing the penalized function since in many applications, a set of variables often form a group (e.g., SNPs spanning a gene, genes in a pathway). Therefore, we propose a group sparse CCA model by introducing the sparse group lasso penalty into Eq. (2). For simplicity, we will only consider non-overlapping groups in this paper and assume that variables in X and Y are partitioned into L and H disjoint groups respectively. The following model, namely group sparse CCA (or CCA-sparse group), is proposed to consider group structures existed in the data:
| (3) |
where , are the group penalties to account for joint effects of features within the same group. This model is more realistic in many cases, e.g., multiple SNPs rather than individual SNPs from the same gene work together as a group to be associated with a disease. The group penalty uses the nondifferentiability of ∥αl∥2 (or ∥βh∥2) at αl = 0 (βh = 0) to set the coefficients of the group to be 0; then the entire group of features will be removed to achieve the group sparsity. While we consider group effects, we can still keep the selection of individual variable/feature. So the l – 1 norm penalties on the individual features (i.e., ∥α∥1 and ∥β∥1) are imposed. λ1 and λ2 are the tuning parameters to control the group sparsity while τ1 and τ2 are used to control individual feature sparsity. ωl and μh are the weights to adjust for the group size differences. We set them to be , where si is the ith group size.
In a particular case, since sparse group lasso penalty is a combination of l – 1 norm and group lasso penalty, Eq. (3) can be reduced to the CCA-group model with only group lasso penalty (τ1 = τ2 = 0) and CCA-l1 model without group lasso penalty (λ1 = λ2 = 0). As discussed above, CCA-group model can tend to select features group by group and keep all features within a selected group while CCA-l1 will ignore group effect among features.
2.2. Iterative optimization algorithm
To solve Eq. (3), we rewrite Eq. (3) based on the singular vector decomposition of matrix K and impose sparse penalties on vectors u, v
| (4) |
where , and di is the positive square root of the ith eigenvalue of KtK. ui vi are the ith eigenvectors of KtK corresponding to di. , should be satisfied when the solution of the optimization problem is obtained. The loading vectors can then be derived by
| (5) |
The matrices and in Eq. (5) might be ill-conditioned because of the high dimensionality of data. We adopt Witten and Tibshirani's (2009) method by replacing the covariance matrices with identity matrices I and hence penalizing the vectors u, v instead of the loading vectors α, β.
Since the problem in Eq. (4) is still not a convex optimization with respect to u and v, for simplicity, we decouple the problem into two simple biconvex optimizations by fixing u and v alternatively. An iterative algorithm is then derived to solve the problem, as shown in Table 1.
Table 1.
The iterative algorithm of group sparse CCA.
| 1. Initialize u0 and v0 by traditional CCA decomposition, ∥u0∥2 = 1, ∥v0∥2 = 1. |
| 2. Solve Uj, Vj using the following iterations until it convergence: |
| (a) |
| (b) |
| (c) dj ← tr (Kvj (uj)t) or tr (Kuj (vj)t) |
| 3. Update the remaining matrix K ← K – tr(Kvut)uvt; go to Step (1) to obtain the next pair of loading vectors (u, v). |
Taking the solution of (a) for example, one can find the solution with the following Lagrange form formulation:
| (6) |
where Δ is the parameter to make . This is converted into a simple optimization problem with separable objection function and sparse group lasso penalty. A block coordinate decent algorithm has been developed to solve this problem efficiently as shown in Table 2.
Table 2.
Block coordinate decent for group sparse CCA.
| Input: iteration step j, uj, vj, ∥uj∥2 = 1, ∥vj∥2 = 1, λ1 , τ1. |
| Output: u j +1 |
| Solve uj using block coordinate decent until it convergence: |
| 1. For each group k = 0 to L |
| 2. Softk(Kv) = S((Kv)k, τ1), where S(·) is the soft-thresholding function. |
| 3. . |
| 4. Else |
| 5. . |
| 6. . |
| 7. End if |
| 8. End for |
| 9. . |
| 10. Repeat (1-9), until ∥uj+1 – uj∥2 ≤ ε, else uj = uj+1. |
The coordinate decent algorithm has been shown to be effective in solving generalized linear regression models (Wu et al., 2009; Friedman et al., 2010a, 2010b), especially for underdetermined system. It estimates parameter one by one by fixing the other parameters unchanged. Similarly, instead of estimating parameter individually, block coordinate decent algorithm will estimate a block of parameters each time while fixing the other blocks of parameters. In data set X, each group k = 1,2,. . .,L, will be inspected one by one. If a group is selected, we will then select each feature in the group by the coordinate decent with the soft-thresholding. Since the optimization is convex, the optimal solution of Eq. (6) is determined by a sub-gradient equation and will converge to a global minimum.
The CCA-group model is a special case of group sparse CCA when τ1 = 0; the soft-threshold operator in Step (2) (Table 2) need to be changed to the simpler form:
| (7) |
The general sparse CCA formula Eq. (3) that we propose can include a variety of sparse CCA models used before (Le Cao et al., 2009; Witten and Tibshirani, 2009), e.g., CCA-l1 (Eq. (8)) and CCA-elastic net (Eq. (9)) models. The coordinate decent algorithm can also be applied to solve both models.
| (8) |
| (9) |
where δ1, δ2 are the parameters, controlling the trade-off between l – 2 norm and l – 1 norm penalties.
3. Method
We applied group sparse CCA to investigate the association of functional brain regions with genetic variations as shown in Fig. 1. Components extracted from fMRI represent brain regions expressing the functional difference in different subjects. Components from SNP data are linear combinations of SNPs from different genes that may have associations with the disease. After preprocessing, the collected SNPs and ROI-based voxels are both still high dimensional with a large number of features compared to the number of samples. We then used the group sparse CCA to estimate two group loading vectors u and v, from which a pair of canonical variates is obtained. The loading vectors for each component reflect the effect size of the features on the correlation. Then u and v were also used to perform gene-ROI correlation analysis to identify the significantly correlated genes and ROIs.
3.1. Group sparse CCA for fMRI and SNP data analysis
We represented fMRI data collected from participants as a set of spatial voxels. These voxels were divided into 116 ROIs based on the aal (automated anatomical labelling) template (Tzourio-Mazoyer et al., 2002). These ROIs were assumed to be spatially independent but the voxels within each ROI may correlate with each other. These voxels were grouped by ROIs so that we can perform the whole brain analysis (Ng and Abugharbieh, 2011). For SNP data, we extracted those SNPs from preselected 74 reported SZ-risk genes. These SNPs were grouped at gene level (Liu et al., 2013). Hence the X and Y matrices can be constructed as follows:
where L = 74 indicates the number of genes in SNP data; H = 116 is the number of ROIs used in fMRI data, and n is the number of samples. lk and ht are the number of SNPs and voxels contained in the kth gene and tth ROI respectively. Under this data structure, two sparse loading vectors u and v (or α and β) were also grouped in the same way. This way the original data were projected into a low dimensional space.
Four tuning parameters (λ1; τ1; λ2; τ2) were used in the model to control the group sparsity (λ1; λ2) and individual feature sparsity (τ1, τ2). A cross-validation can be used to select the optimal parameters but is time-consuming. Therefore, we divided the cross-validation into two steps: (1) using the CCA-group method to select optimal λ1; λ2 by considering only the group penalty; (2) selecting the optimal τ1, τ2 based on the selected λ1; λ2: In summary, the procedure of performing group sparse CCA on the data analysis is as follows:
Decompose matrix K in Eq. (5) using SVD to initialize the loading vectors and normalize the vectors with the l – 2 norm. Use the two-step cross validation to obtain the optimal tuning parameters.
Perform the sub-optimization analysis with respect to each loading vector (u or v) in each modality alternatively by the block coordinate decent algorithm. A stopping criterion is assessed for each modality.
Two loading vectors are updated and the stopping criteria for both loading vectors are assessed. If they are satisfied, the solution is obtained, go to Step 4. Otherwise, go back to Step 2.
Test the significance of the predict correlation (corrj) by permutation. If the correlation is significant, go to Step 5. Otherwise, there is no significant correlation between the current two data sets.
Perform gene-ROI analysis to explore the pair-wise correlation between each gene and ROI.
Calculate the remaining matrix and repeat step (1–4) to extract the next pair of loading vectors until r pairs of loading vectors are obtained or the extracted predict correlation (corrj) is not significant.
3.2. Tuning parameters selection
The k-fold cross validation was recommended by Waaijenborg and Zwinderman (2009) and Parkhomenko et al. (2009) for parameter selection. Parkhomenko et al. (2009) adopted a criterion that maximizes the mean absolute canonical correlation value of the testing set as shown in the following equation:
| (10) |
Waaijenborg et al. (2008) considered the mean difference between the canonical correlations of the training and testing subsets as in the following equation:
| (11) |
This criterion determines the number of variables which tend to have the same correlations in both training and testing subsets. It is sensitive to the correlation sign change that if the correlation of testing subset changes sign it would be penalized more than when the sign would not change.
Witten and Tibshirani (2009) proposed another permutation based method for optimizing parameters. The whole data set (including both training and testing data sets) will be used to compute the non-permuted correlation d0 and then X is permuted with T times to calculate the permuted correlation di=1,. . .,T. The parameters having the largest p-value as in Eq. (12) will be selected.
| (12) |
Here, û–i and v̂–i are estimated loading vectors from training data set. X–i and Yi are training data set in which subset j is deleted. Xi and Yi are the testing data set. Based on these three criteria, we performed a simulation study using our proposed group sparse CCA model and other sCCA models to choose the optimal criterion (the details are described in Section 4.1).
3.3. Gene-ROI correlation analysis
After the parameters were determined, we obtained the estimates of vectors u, v as well as the corresponding canonical variates. We used these estimates to measure the strength and significance of the correlation between genes and ROIs. The null hypothesis of no correlation between a gene and ROI can be written as H0 : ρ11 = ρ12 = · · · ρck = ρsisj = 0 versus the alternative hypothesis HA : ∃c; k > 0; ρck ≠ 0, where si and sj are the number of SNPs and voxels in Genei and ROIj respectively; ρck is the correlation between the cth SNP and the kth voxel. To test the hypothesis, we first calculated the pair-wise correlation between the SNPs from Genei and voxels from ROIj. SNPs and voxels were indicated by non-zero coefficients in u and v respectively Then, to avoid the potential bias due to varying gene or ROI size, we averaged the correlation of each SNP-voxel pair as the test statistics as follows.
| (13) |
By comparing our observed statistic ρij with the null statistics with T times permutations of the samples, we can evaluate the significance of the correlation by
| (14) |
This gene-ROI correlation analysis can also be applied to the gene–gene correlation and ROI-ROI correlation studies.
3.4. Statistical evaluations
Although our CCA-group sparse method aims to obtain a higher correlation with fewer features selected, in our real data both number of voxels and SNPs are much larger than that of subjects. In order to test the significance of the correlation obtained by the CCA-sparse group method, we took the permutation-based testing. We first obtained u and v based on the optimal parameters, and then permutated the order of subjects in SNP data randomly while keeping the subject order in fMRI data unchanged. The correlation for each permutation can be calculated by the same u and v, which is expected to estimate the null distribution of the correlation. The p-value can be estimated by large number of permutations. The lower the p-value, the more significant the detected correlation is.
4. Results
4.1. Simulation
To assess the performance of the proposed group sparse CCA method, we first simulated two correlated data sets and then we compared group sparse CCA with the other penalized CCA methods such as the CCA-group and CCA-l1 on these simulated data.
Two data sets of SNP data X with p SNPs and fMRI data Y with q voxels were simulated. To correlate the SNPs with the voxels, a latent model similar to (Parkhomenko et al., 2009) was used. We first set a latent variable Υ = {γi|i = 1, . . ., n} with normal distribution to have the similar effect on the associated SNPs and voxels in the two data sets. Then X and Y data set were simulated from a multivariate normal distribution by XtX and respectively, where the vectors xi ∊ Rp, yi ∊ Rq are the observations of the ith sample in X and Y. α = [α1, . . ., αj, . . ., αp], β = [β1, . . ., βk, . . ., βq], αj ≠ 0, βk ≠ 0, if xj, yk are the correlated variables; otherwise, the variables would be considered as noise with zero means (αj = 0, βk = 0). γi is the ith observation of Υ. , , are the variances of Υ and noise variable, which were used to control the correlation between SNPs and voxels. and were the variance–covariance matrices of each data set. They were used to simulate the group effect within each data set. For each data set, we set the correlation between the correlated variables within the same group as from the uniform distribution U(0.1,0.3).
The samples of two data sets were simulated from the multivariate normal distribution. Then SNP data X will be converted into categorical variables with three levels (e.g., –1, 0, 1) by specifying the minor allele frequency (MAF) p from the uniform distribution Unif(0.2,0.4). The probability of an observation being from a particular level was set to be p2, p(1 – p) and (1 – p)2 respectively. For each variable, the expected number of observations in each level was n1 = p2n, n2 = p(1 – p)n, n3 = (1 – p)2n. We sorted the observations in each variable and assign value 1 to those n1 observations with the highest value, –1 to those n3 observations with lowest value and 0 in between (Simon and Tibshirani, 2012). To simulate the fMRI data Y with a brain structure similar to that of real data, we referred to the standard aal template with 116 brain ROIs containing 54277 voxels. The group structure of voxels is based on the ROIs. Voxels correlated with SNPs were generated by latent variable model. Irrelevant voxels within each ROI were simulated from where and are mean and covariance matrix estimated from the tth ROI in real data.
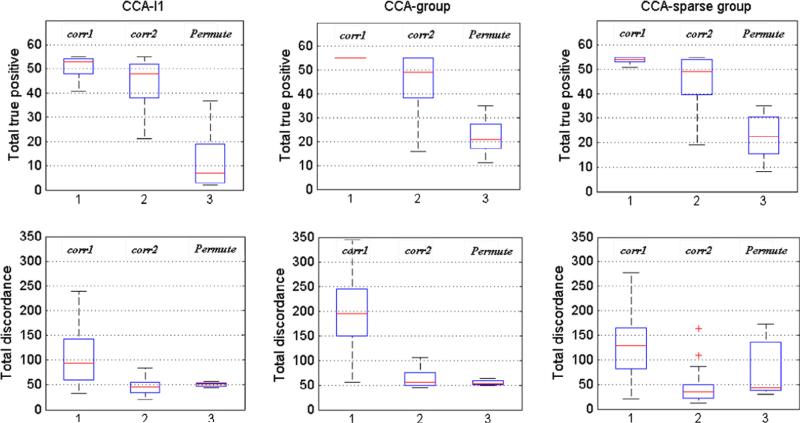
First, we compared the three parameter optimization criteria by applying them to the simulation study. Two data sets X with 200 SNPs and Y with 200 voxels were simulated. Both data sets were divided into 20 groups with group size 10. 20 SNPs from 4 groups in set X and 30 voxels from 5 groups in set Y were set to be correlated. Standard deviation σγ = 1 and σe = 0.3. The sample size was 100 and 50 simulations were replicated. The total true positive (TTP) and total discordance (TD: the total false positive plus the total false negative) were used for the comparison. Fig. 2 shows the box-plot of TTP and TD by using different criteria. It can be seen that, by all the methods, Δcorr1 tends to give higher TTP but also introduces higher TD; the use of ppermute can cause the decrease of TD rapidly at the cost of low TTP; and Δcorr2 is a trade-off between these two criteria by keeping a high TTP and decreasing TD to a low level. Hence the last one is chosen as our parameter selection criterion.
Fig. 2.
The total true positive number (TPP) and total discordance (TD) of estimating correlations using CCA-l1, CCA-group and CCA-sparse group methods using three different parameter selection criteria: corr1 (Eq. (10)) proposed by Parkhomenko et al. (2009), corr2 (Eq. (11)) proposed by Waaijenborg et al. (2008) and permutation based criterion (Eq. (12)) proposed by Witten and Tibshirani (2009).
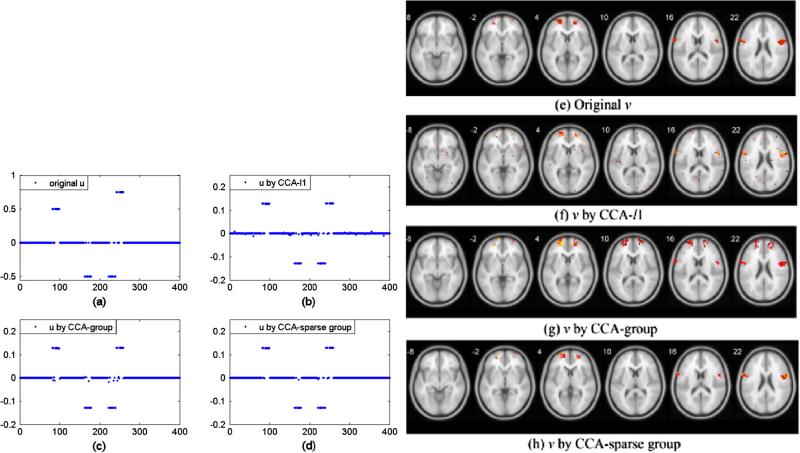
Second, we compared the performance of three methods in the simulation. SNP data X with 400 SNPs are divided into 20 groups evenly. 4 groups were randomly selected and 15 SNPs randomly selected from these 4 groups were generated to be correlated with voxels, as shown in Fig. 3(a). For the sake of simplicity, 5% voxels randomly from the first 4 regions with totally 214 voxels were associated with SNPs (Fig. 3(e)). Fig. 3(b–d, f–h) shows the results of recovered loading vectors u and v by CCA-l1, CCA-group and CCA-sparse group methods. A 5-fold cross-validation was used to select the optimal parameters. From Table 3, it can be seen that the CCA-sparse group method can better estimate true u and v than other two methods. For the SNP data, all three methods can identify true SNPs (60) while CCA-l1 has more false positives (36, shown in Fig. 3(b)) than those of CCA-group (20) and CCA-sparse group (4). For the fMRI data, CCA-l1 also misses out more true voxels (29 out of 214) in Fig. 3(f) when selecting v. The CCA-group, however, can better recover all the groups with true features. Nevertheless, it selects all features from the group, which results in very high false positive (4097, shown in Fig. 3(g)). CCA-sparse group can not only identify the group structure to find more true variables than CCA-l1 but also remove those noisy features in the group, leading to the least false positive (113 false voxels versus 912 by CCA-l1 and 4097 by CCA-group).
Fig. 3.
A comparison of the performance of group sparse CCA and three sCCA methods. (a) True u; (b–d) u recovered by CCA-l1, CCA-group and CCA-sparse group respectively. (e) True v; (f–h) v recovered by three sCCA methods.
Table 3.
The count of true positive (TP), false positive (FP) and discordance by three methods.
| Model | TP (voxels/SNPs) | FP (voxels/SNPs) | Discordance (voxels/SNP) |
|---|---|---|---|
| CCA-/1 | 185/60 | 912/36 | 941/36 |
| CCA-group | 214/60 | 4097/20 | 4097/20 |
| CCA-sparse group | 200/60 | 113/4 | 127/4 |
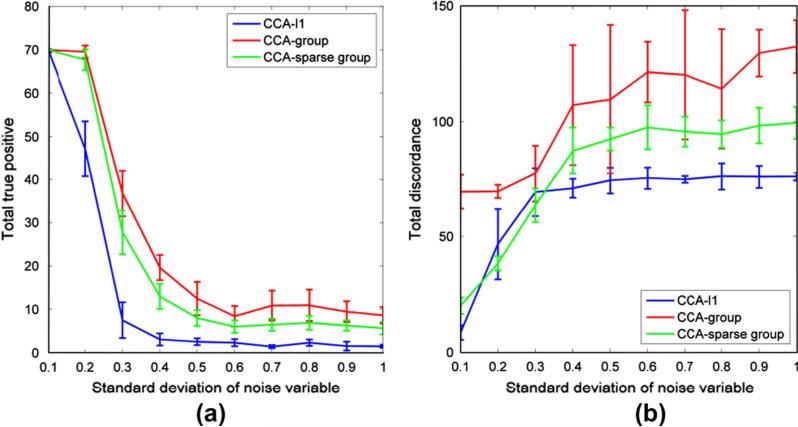
Finally, we evaluated the performance of three models with respect to different noise levels, which affected the values of correlation between two data sets. 200 SNPs from 20 genes were simulated in data set X. The group size was 10. 50 voxels from 5 ROIs were randomly selected to be correlated with 20 SNPs in X. The sample size was 150. According to the correlation estimate in (Parkhomenko et al., 2009), we simulated different level of correlations between two data sets by changing the standard deviation of the noise variable σe from 0.1 to 1 with interval 0.1. From the results in Fig. 4, we can see that when the true correlation increases with the decrease of noise variance, more true variables can be recovered with less total discordance by all three methods. The CCA-group model can recover the most correlated variables but has the highest total discordance. When σe is larger than 0.6 (the estimated highest correlation is 0.39), the number of TTP and TD is relatively unchanged. When σe decreases, more true variables can be identified by all methods and the TD of CCA-l1 and CCA-sparse group decreases rapidly.
Fig. 4.
A comparison of three methods for different correlation level influenced by noise. (a) The value of total true positive obtained by three methods when the standard deviation of noise increases from 0.1 to 1, showing that the highest correlation of true variables between two data sets is within the range from 0.958 to 0.18. (b) The total discordance by three methods when the standard deviation of noise changes from 0.1 to 1.
4.2. Experimental results
In this study, subject recruitment and data collection were conducted by The Mind Clinical Imaging Consortium (MCIC). Two types of data (SNP and fMRI) were collected from 208 subjects including 92 schizophrenia patients (age: 34 ± 11, 22 females) and 116 healthy controls (age: 32 ± 11, 44 females). All of them were provided written informed consents. Healthy participants were free of any medical, neurological or psychiatric illnesses and had no history of substance abuse. By the clinical interview of patients for DSM IV-TR Disorders (22) or the Comprehensive Assessment of Symptoms and History, patients met criteria for DSM-IV-TR schizophrenia (23). Antipsychotic history was collected as part of the psychiatric assessment.
4.2.1. fMRI data collection and preprocessing
fMRI data were collected during a sensor motor task, a block-design motor response to auditory stimulation. The images were acquired on a Siemens3T Trio Scanner and 1.5 T Sonata with echo-planar imaging (EPI) sequences taking parameters (TR = 2000 ms, TE = 30 ms (3.0 T)/40 ms (1.5 T), field of view = 22 cm, slice thickness = 4 mm, 1 mm skip, 27 slices, acquisition matrix = 64 × 64, flip angle = 90°). Data were pre-processed with SPM5 (http://www.fil.ion.ucl.ac.uk/spm) and were realigned, spatially normalized and resliced to 3 × 3 × 3 mm, smoothed with a 10 × 10 × 10 mm3 Gaussian kernel, and analyzed by multiple regression considering the stimulus and their temporal derivatives plus an intercept term as repressors. Finally the stimulus-on versus stimulus-off contrast images were extracted with 53 × 63 × 46 voxels and all the voxels with missing measurements were excluded. 116 ROIs were extracted based on the aal brain atlas, which resulted in 41236 voxels left for analysis.
4.2.2. Genotyping and preprocessing
A blood sample was obtained for each participant and DNA was extracted. Genotyping for all participants was performed at the Mind Research Network using the Illumina Infinium HumanOmni1-Quad assay covering 1,140,419 SNP loci. Bead Studio was used to make the final genotype calls. PLINK software package (http://pngu.mgh.harvard.edu/~purcell/plink) was used to perform a series of standard quality control procedures, resulting in the final dataset spanning 777,635 SNP loci. Each SNP was categorized into three clusters based on their genotype and was represented with discrete numbers: 0 for ‘BB’ (no minor allele), 1 for ‘AB’ (one minor allele) and 2 for ‘AA’ (two minor alleles)’. SNP with >20% missing data were deleted and missing data were further imputed. SNPs with minor allele frequency <1% were removed. To reflect the influence of genetic variation on brain behavior, SNPs included in top 75 schizophrenia genes listed on the SZGene database (http://www.szgene.org/) were selected for the analysis. This procedure yielded 3082 SNPs, which were annotated with 74 genes. There was no SNP found in the remaining one gene.
4.2.3. Correlation analysis
The fMRI voxels were grouped based on ROIs while SNPs were grouped by genes. Then we applied our CCA-sparse group method to the analysis of the correlation between two data sets. 208 subjects were randomly divided into two subsets: 150 subjects for training and the remaining 58 ones for testing. In training data set, we fit three models: (i) CCA-group lasso, i.e., using group lasso penalty on genes and ROIs, (ii) CCA-l1, i.e., only imposing l – 1 norm on the effects of all SNPs and voxels, and (iii) CCA-sparse group model, i.e., using both group-level and single feature level regularizations. The optimal parameters were obtained from training data by 5-fold cross validation. The models were estimated as well as the features were selected from the training data using the optimal parameters. Then, these estimated models were applied to test data to predict the correlation between two data sets.
To have a stable feature selection, we performed random sampling from 208 subjects repeatedly for B times, selected the same proportion of subjects for training and test data sets, and fitted three models on each sub-sample. We assumed that those SNPs and voxels selected more frequently are more valuable for exploring the correlation between two data sets. For each sub-sample, ûb, v̂b were estimated, b = 1,2,. . .,B. A measure of feature importance can be computed by frequency of their appearance defined as the selection probability in (15). A set of features with high value will be selected by a cut-off threshold.
| (15) |
where is taken to be (or ), i.e., the loading coefficient of sCCA model corresponding to the jth SNP (or voxel), and I(·) is indicator function.
By the algorithm in Section 2.2, we can derive several pairs of canonical variates by iteratively implementing the CCA decomposition. Here, we only show the first two pairs of canonical variates. B = 50 replicates were performed. To compare the performance, the test correlations cor(Xtestû, Y testv̂) based on the estimated loading vectors (û, v̂) in training data by three methods were calculated. The numbers of selected features were also used for comparison. Fig. 5(a) shows that CCA-group method chose the largest number of SNPs and voxels compared to the other two methods while CCA-sparse group method selected the least features due to the double sparsity constraints on loading vectors. As shown in Table 4, CCA-sparse group generally performs better than the other two methods by obtaining higher predict correlations with comparable low variance. The low variance of these correlations demonstrates the robustness of the estimation. The correlation variance by CCA-sparse group method is also comparable with that of the other two methods. In addition, the small training and test correlation difference (Δcorr = 0.146, 0.1331, 0.1471 for three methods respectively) indicates the advantage of using criterion Eq. (11) for tuning parameters selection, i.e., the stability of the criterion regardless of the method used.
Fig. 5.
The comparison of the number of features in loading vectors u and v selected by three methods. (a) The number of SNPs and voxels selected by three methods with 50 sub-samplings. (b–c) Show the top ranked SNPs (b) and voxels (c) in the first pair of canonical variates with cut-off threshold = 0.3 selection probability.
Table 4.
Correlation coefficients based on the variants estimated by three models in the test data.
| CCA-group lasso | CCA-l1 | CCA-sparse group | |
|---|---|---|---|
| Test sample correlation (mean ±SD) | |||
| 1st pair | 0.4497±(0.043) | 0.4235±(0.057) | 0.4527±(0.048) |
| 2nd pair | 0.4137±(0.034) | 0.3959±(0.045) | 0.4292±(0.033) |
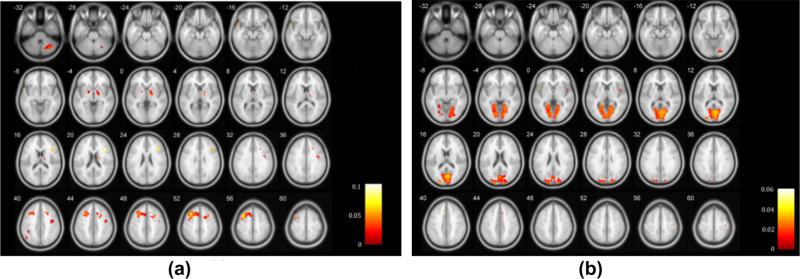
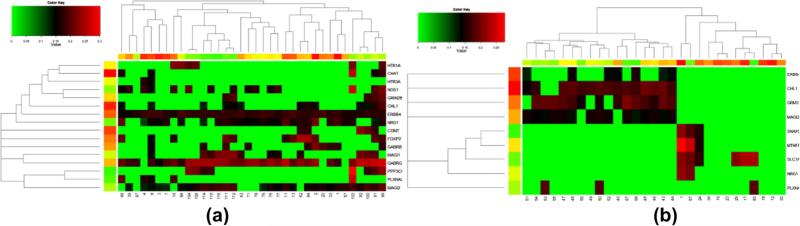
We further discuss the selected features. Choosing SNPs and voxels by ranking the selection probabilities (pj in Eq. (14)) with a cut-off threshold 0.3, we summarized those top ranked SNPs and voxels in two pairs of variates by three methods in Fig. 5(b and c). There are a high proportion of overlapped SNPs and voxels selected by these methods. Table 5 lists top ranked SNPs and corresponding genes in two pairs of variates by CCA-sparse group method. The first pair contained 51 SNPs from 16 genes correlated with 756 voxels from 36 ROIs with the average correlation 0.4527, p < 0.001. 30 SNPs from genes ERBB4 and MAGI2 and 212 voxels from ROIs 7 and 8 were selected by all methods. The other pair had19 SNPs from 9 genes correlated with 1558 voxels from 29 ROIs, and the average correlation was 0.4292, p < 0.001. 3 SNPs from MAGI2 and 251 voxels from ROIs 47, 51 were overlapped by all three methods. Table 6 listed the top ranked brain ROIs in the first and second pair of variates respectively. Fig. 6 shows the brain mapping of genomic correlated ROIs. Several brain regions have been reported to be susceptible to schizophrenia from other neuroimaging studies (Shenton et al., 2001; Kumari et al., 2002; Onitsuka et al., 2004; Torrey, 2007; Bellani et al., 2010; Pinault, 2011). They include superior, middle, and medial frontal gyrus, inferior parietal lobule, superior temporal gyrus, thalamus, parahippocampal gyrus, cingulate gyrus, and lingual gyrus, which provided additional evidences that these disease relevant brain regions may be affected by the correlated genomic variations.
Table 5.
The list of genes selected in the first and second pair of canonical variates.
| Canonical variate | Gene ID | SNPs number |
|---|---|---|
| 1st pair | CHAT CHL1 COMT ERBB4 FOXP2 GABRB2 GABRG2 GRIN2B HTR1A HTR3A MAGI1 MAGI2 NOS1 NRG1 PLXNA2 PPP3CC | 51 |
| 2nd pair | CHL1 ERBB4 GRM3 MAGI2 MTHFR NRG1 PLXNA2 SLC1A2 SNAP29 | 19 |
Table 6.
The correlated brain ROIs selected in two pairs of canonical variates.
| 1st pair |
2nd pair |
||||
|---|---|---|---|---|---|
| Brain region | Brodmann area | L/R volume (cm3) | Brain region | Brodmann area | L/R volume (cm3) |
| Superior Frontal Gyrus | 6, 8 | 3.1/0.1 | Lingual Gyrus | 17, 18, 19 | 5.4/6.5 |
| Inferior Parietal Lobule | 40 | 0.6/* | Posterior Cingulate | 18, 23, 30, 31 | 2.3/2.9 |
| Medial Frontal Gyrus | 6, 8, 32 | 0.7/0.3 | Parahippocampa Gyrus | 18, 19, 30 | 1.2/0.4 |
| Precentral Gyrus | 6 | 0.3/1.2 | Precuneus | 19, 31 | 0.7/0.8 |
| Middle Frontal Gyrus | 6, 8 | 4.5/0.6 | Superior Temporal Gyrus | 22 | */0.1 |
| Thalamus | * | 0.1/0.4 | Cingulate Gyrus | 32 | 0.5/0.2 |
| Cingulate Gyrus | 6 | 0.3/1.2 | Cuneus | 17, 18, 19, 23, 30 | 8.1/9.3 |
| Culmen | * | 0.1/0.5 | Middle Occipital Gyrus | 18 | 0.8/0.1 |
| Declive | * | 0.2/0.6 | Insula | 13 | 0.1/0.2 |
Fig. 6.
Maps showing regions correlated with genetic factors in the first (a) and second (b) canonical variates.
4.2.4. Gene-ROI correlation analysis
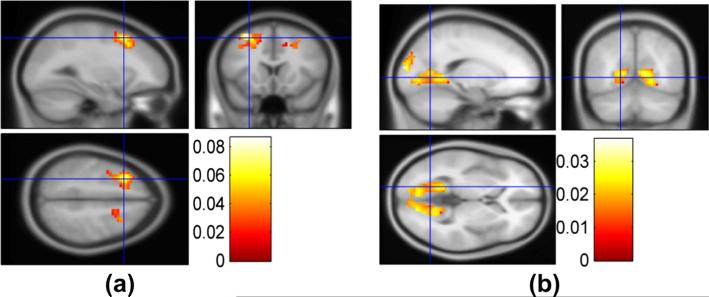
Based on the two pairs of canonical variates, we further explored and verified the correlation between genes and ROIs. We found that some regions of brain might be correlated with a set of genes. For each gene-ROI, gene-gene and ROI-ROI correlation, 10000 permutations (as mentioned in Method section) were performed to test the significance. In Fig. 7(a), the absolute values of significant gene-ROI correlation are 0.1862 ± 0.0317 (mean ± SD, p < 0.005). As shown in Fig. 8(a), Gene ERBB4, NRG1, MAGI2 and GABRG2 show correlations with some ROIs such as 3, 4, 7, 8 (the index of ROI is defined by the aal template (Tzourio-Mazoyer et al., 2002)) with the correlation (ρ = 0.1822, 0.1483, 0.1335,0.1782, p < 0.004) respectively. These ROIs mostly consist of superior, middle, medial front gyrus and pre-central gyrus located at frontal lobe which contains primary motor cortex and has been suspected to have abnormal changes in schizophrenia patients (Honey et al., 2005; Kiehl et al., 2005). ROIs 75, 76, 77, 11 and 13(not shown) are also found to be correlated with gene ERBB4, NRG1, and MAGI2 with the correlation value of 0.1822, 0.1649 and 0.1541 respectively. These ROIs are mainly located at thalamus (right) which plays a critical role in coordinating the pass of information between brain regions. Many studies show the association between dysfunction of thalamus with schizophrenia (Kiehl and Liddle, 2001; Clinton and Meador-Woodruff, 2004; Sui et al., 2011). These three genes are also correlated with each other, which may have the similar effects on the ROIs. In addition, ROIs 91, 92, 99, 100, 103, 111, 112 are correlated with many genes (e.g. GRIN2B, CHL1, ERBB4, NRG1, FOXP2, MAGI1, GABRG2, MAGI2). These ROIs are located at declive of cerebellum and culmen. Their relationship with schizophrenia is not clear yet but several previous publications have reported significant difference of these regions between normal control and schizophrenia patients (Kim et al., 2009) and there are models of schizophrenia which discuss importance of the cerebellum (Andreasen and Pierson, 2008).
Fig. 7.
Heatmap of the gene-ROI correlations derived from the first (a) and second (b) pairs of canonical variates.
Fig. 8.
(a) ROIs 3, 4, 7, 8 which are identified to be mainly correlated with genes ERBB4, NRG1, MAGI1 and GABRG2. (b) ROIs 40, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 67, 68 which are found to correlate with Gene ERBB4, CHL1, GRM3 and MAGI2. Note: the index of ROI is given by the aal template (Tzourio-Mazoyer et al., 2002).
In the second pair of canonical variates (Fig. 7(b)), the absolute value of significant gene-ROI correlation is 0.1713 ± 0.0308 (mean ± SD, p < 0.005). There are two patterns apparent in the figure. The first pattern corresponds to ROIs 40, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 67, 68, which are located at occipital lobe including lingual gyrus, posterior cingulate, precuneus, parahippocampagyrus and superior, middle and inferior occipital gyrus as shown in Fig. 8(b). Four genes ERBB4, CHL1, GRM3 and MAGI2 were significantly correlated with these ROIs at 0.1776, 0.1356, 0.1762 and 0.1429, p < 0.005 respectively. These critical regions have been widely studied and are shown to have potential relationship with schizophrenia (Kiehl et al., 2005; Fransson and Marrelec, 2008; Kim et al., 2009). The second pattern are ROIs 1, 24, 57 from precentral gyrus at parietal lobe corresponding to genes SNPA29, MTHFR, SLC1A and NRG1 with the correlation of 0.2251, 0.1899, 0.1816 and 0.21 respectively.
5. Discussion and conclusion
In this paper, we proposed a novel method to explore the relationship between genomic data and fMRI brain imaging data by considering the group effects of the variables in the data. We introduced the group sparse CCA method and the numerical implementation based on the regularized SVD and block coordinate decent algorithm. The performance of group sparse CCA model was compared with other sCCA models in the simulation study, showing that our group sparse CCA method could better recover the true correlations with lower false positive and total discordance. Then we applied the method to correlation analysis between the SNP data and fMRI imaging data. Two pairs of canonical variates with significant correlations were identified. There are 5 pathways implied by these identified genes, which may involve with the biological processes related to schizophrenia. The SZ-risk genes correlated with brain regions have also been reported to be susceptible to schizophrenia, which further validates the results of our method.
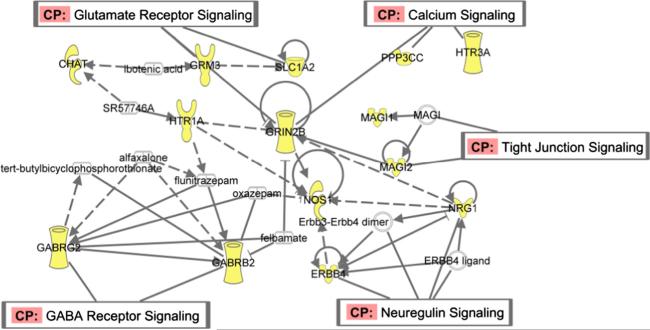
We identified two pairs of canonical variates with significant correlations and further verified those linked components between gene-ROI with significant pair-wise correlation. Those identified genes are from the preselected 74 genes from α database which were reported to be susceptible to SZ. Some of these genes may not directly show group difference in our fMRI data. Therefore, in this section, we focus on those group-discriminating genetic factors associated with motor response task-related brain function disruption in schizophrenia. We use the difference of minor allele frequency (MAF) between case and control group to evaluate the effects of the SNPs in the gene correlated with ROI component. Significantly higher MAF in case group indicates the positive discriminating effect while lower MAF indicates the negative discriminating effect. Pathway analysis of these important genes is performed through Ingenuity Pathway Analysis (IPA: Ingenuity Systems, http://www.ingenuity.com). We found those selected genes involving in 5 pathways as shown in Fig. 9.
Neuregulin signalling pathway: NRG1 and ERBB4. These two genes have significant effects in both pairs of canonical variates. It has been reported that the NRG1-ERBB4 modulates some plausible neurobiological mechanisms, i.e., neuronal plasticity in human brain which may be altered in SZ (Buonanno, 2010). In our results, SNPs ‘rs10090544’, ‘rs16878394’, ‘rs7843384’(NRG1) and’rs16847732’, ‘rs16847769’, ‘rs2008506’ in ERBB4 exhibit higher MAFs in patients;rs2466063(NRG1) and the MAFs in’rs11903508’, ‘rs16846111’, ‘rs16846352’in ERBB4 are lower in patients indicating negative effect.
Glutamate receptor signalling: GRIN2B, GRM3 and SLC1A2. These genes are involved in encoding ionotropic glutamate receptors and histamine receptors which may regulate the neurotransmitter transmission associated with SZ (Bishop et al., 2005). Several researches have supported the possibility of GRIN2B conferring the susceptibility to schizophrenia (Ohtsuki et al., 2001; Qin et al., 2005). GRM3 may be involved in the pathophysiology of schizophrenia, and its associated cognitive impairment, especially at prefrontal and hippocampal regions (Egan et al., 2004). In our results, ‘rs2299218’, ‘rs802432’ in GRM3 and ‘rs2284425’ in GRIN2B have lower MAFs in patients showing negative effects while ‘rs3026164’(GRIN2B), ‘rs802425’ (GRM3) and ‘rs11033095’ (SLC1A2) have positive effects on the disease.
GABA receptor signalling: GABRB2 and GABRG2. GABA neuronal dysfunction has been found to associate with cognitive impairment of schizophrenia. We identified a SNP ‘rs153303’ (GABRB2) with lower MAF in schizophrenia, showing negative effect while ‘rs211037’ (GABRG2) has higher MAF in patients with a possible opposite influence.
Calcium signalling pathway: HTR3A, ERBB4 and PPP3CC. Recentbiochemical research supports that schizophrenia may be produced by alterations in various intracellular molecules as long as these alterations lead to abnormal functioning of some central intracellular regulatory pathways and one of unifying elements of molecular changes in schizophrenia is their association with potential altered Ca2+signalling (Lidow, 2003).‘rs1150219’ (HTR3A) and ‘rs7010861’ (PPP3CC) both show higher MAFs in patients in the results.
Tight junction: MAGI1, MAGI2. These genes attend the coding for MAGI proteins which may relate to SZ by influencing the development and communication between nerve cells (Karlsson et al., 2012). In our results, ‘rs9311944’, ’rs6809559’ (MAGI1), and ‘rs10230275’, ’rs10229284’, ’rs1330490’ (MAGI2) present higher MAFs in patients while ‘rs9821646’, ’rs2061937’(MAGI1), ’rs10230752’, ’rs10953782’(MAGI2) exhibit lower MAFs in patients.
Fig. 9.
Functional network built based on the genes identified within the two pairs of canonical variates.
The tuning parameters selection criteria used in this work are also important. The performance of Waaijenborg et al.'s criterion is better than those of other two criteria in terms of feature selection accuracy, which is consistent with the conclusion from (Waaijenborg and Zwinderman, 2009). However, these parameter selection criteria are not directly used for the purpose of feature selection; a better criterion can be developed. In addition, the selection of tuning parameters is via cross-validation, relying on large training data, which is often not available. Alternatively, Bayesian information criterion and Akaike information criterion (Posada and Buckley, 2004) can be used for the optimal parameter selection.
Acknowledgement
This work is partially supported by both NSF and NIH. It is also supported by Shanghai Eastern Scholarship Program.
References
- Abecasis GR, Burt RA, et al. Genomewide scan in families with schizophrenia from the founder population of Afrikaners reveals evidence for linkage and uniparental disomy on chromosome 1. Am. J. Hum. Genet. 2004;74(3):403–417. doi: 10.1086/381713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen NC, Pierson R. The role of the cerebellum in schizophrenia. Biol. Psychiatr. 2008;64(2):81–88. doi: 10.1016/j.biopsych.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol. Psychiatr. 2002;7(4):405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- Bellani M, Ferro A, et al. The potential role of the parietal lobe in schizophrenia. Epidemiol. Psichiatr. Soc. 2010;19(2):118–119. [PubMed] [Google Scholar]
- Bishop JR, Ellingrod VL, et al. Association between the polymorphic GRM3 gene and negative symptom improvement during olanzapine treatment. Schizophr. Res. 2005;77(2–3):253–260. doi: 10.1016/j.schres.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Boutte D, Liu J. Sparse canonical correlation analysis applied to fMRI and genetic data fusion.; IEEE International Conference on Bioinformatics and Biomedicine; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonanno A. The neuregulin signaling pathway and schizophrenia: from genes to synapses and neural circuits. Brain Res. Bull. 2010;83(3–4):122–131. doi: 10.1016/j.brainresbull.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callicott JH, Straub RE, et al. Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc. Natl. Acad. Sci. USA. 2005;102(24):8627–8632. doi: 10.1073/pnas.0500515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liu H. An efficient optimization algorithm for structured sparse CCA, with applications to eQTL Mapping. Stat. Biosci. 2012;4:3–26. [Google Scholar]
- Chen J, Bushman FD, et al. Structure-constrained sparse canonical correlation analysis with an application to microbiome data analysis. Biostatistics. 2013;14(2):244–258. doi: 10.1093/biostatistics/kxs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinton SM, Meador-Woodruff JH. Thalamic dysfunction in schizophrenia: neurochemical, neuropathological, and in vivo imaging abnormalities. Schizophr. Res. 2004;69(2–3):237–253. doi: 10.1016/j.schres.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Egan MF, Straub RE, et al. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc. Natl. Acad. Sci. USA. 2004;101(34):12604–12609. doi: 10.1073/pnas.0405077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransson P, Marrelec G. The precuneus/posterior cingulate cortex plays a pivotal role in the default mode network: evidence from a partial correlation network analysis. Neuroimage. 2008;42(3):1178–1184. doi: 10.1016/j.neuroimage.2008.05.059. [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, et al. A Note on the Group Lasso and a Sparse Group Lasso. 2010 < http://www.arxiv.org/pdf/1001.0736>.
- Friedman J, Hastie T, et al. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010b;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- Hamid JS, Hu P, et al. Data integration in genetics and genomics: methods and challenges. Hum. Genom., Proteom. 2009 doi: 10.4061/2009/869093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey GD, Pomarol-Clotet E, et al. Functional dysconnectivity in schizophrenia associated with attentional modulation of motor function. Brain. 2005;128(Pt 11):2597–2611. doi: 10.1093/brain/awh632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotelling H. Relations between two sets of variates. Biometrika. 1936;28:321–377. [Google Scholar]
- Jansma JM, Ramsey NF, et al. Working memory capacity in schizophrenia: a parametric fMRI study. Schizophr. Res. 2004;68(2–3):159–171. doi: 10.1016/S0920-9964(03)00127-0. [DOI] [PubMed] [Google Scholar]
- Karlsson R, Graae L, et al. MAGI1 copy number variation in bipolar affective disorder and schizophrenia. Biol. Psychiatr. 2012;71(10):922–930. doi: 10.1016/j.biopsych.2012.01.020. [DOI] [PubMed] [Google Scholar]
- Kiehl KA, Liddle PF. An event-related functional magnetic resonance imaging study of an auditory oddball task in schizophrenia. Schizophr. Res. 2001;48(2–3):159–171. doi: 10.1016/s0920-9964(00)00117-1. [DOI] [PubMed] [Google Scholar]
- Kiehl KA, Stevens MC, et al. Abnormal hemodynamics in schizophrenia during an auditory oddball task. Biol. Psychiatr. 2005;57(9):1029–1040. doi: 10.1016/j.biopsych.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Mathalon DH, et al. Auditory oddball deficits in schizophrenia: an independent component analysis of the fMRI multisite function BIRN study. Schizophr. Bull. 2009;35(1):67–81. doi: 10.1093/schbul/sbn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari V, Gray JA, et al. Procedural learning in schizophrenia: a functional magnetic resonance imaging investigation. Schizophr. Res. 2002;57(1):97–107. doi: 10.1016/s0920-9964(01)00270-5. [DOI] [PubMed] [Google Scholar]
- Le Cao KA, Rossouw D, et al. A sparse PLS for variable selection when integrating omics data. Stat. Appl. Genet. Mol. Biol. 2008;7 doi: 10.2202/1544-6115.1390. Article 35. [DOI] [PubMed] [Google Scholar]
- Le Cao KA, Martin PGP, et al. Sparse canonical methods for biological data integration: application to a cross-platform study. BMC Bioinform. 2009;10:34. doi: 10.1186/1471-2105-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Branch CA, et al. FMRI study of language activation in schizophrenia, schizoaffective disorder and in individuals genetically at high risk. Schizophr. Res. 2007;96(1–3):14–24. doi: 10.1016/j.schres.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidow MS. Calcium signaling dysfunction in schizophrenia: a unifying approach. Brain Res. Brain Res. Rev. 2003;43(1):70–84. doi: 10.1016/s0165-0173(03)00203-0. [DOI] [PubMed] [Google Scholar]
- Lin D, Zhang J, et al. Group sparse canonical correlation analysis for genomic data integration. BMC Bioinform. 2013;14:245. doi: 10.1186/1471-2105-14-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Huang J, et al. Incorporating group correlations in genome-wide association studies using smoothed group Lasso. Biostatistics. 2013;14(2):205–219. doi: 10.1093/biostatistics/kxs034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meda SA, Bhattarai M, et al. An fMRI study of working memory in first-degree unaffected relatives of schizophrenia patients. Schizophr. Res. 2008;104(1–3):85–95. doi: 10.1016/j.schres.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng B, Abugharbieh R. Generalized sparse regularization with application to fMRI brain decoding. Inform. Process. Med. Imag. 2011;22:612–623. doi: 10.1007/978-3-642-22092-0_50. [DOI] [PubMed] [Google Scholar]
- Ohtsuki T, Sakurai K, et al. Mutation analysis of the NMDAR2B (GRIN2B) gene in schizophrenia. Mol. Psychiatr. 2001;6(2):211–216. doi: 10.1038/sj.mp.4000808. [DOI] [PubMed] [Google Scholar]
- Onitsuka T, Shenton ME, et al. Middle and inferior temporal gyrus gray matter volume abnormalities in chronic schizophrenia: an MRI study. Am. J. Psychiatr. 2004;161(9):1603–1611. doi: 10.1176/appi.ajp.161.9.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhomenko E, Tritchler D, et al. Sparse canonical correlation analysis with application to genomic data integration. Stat. Appl. Genet. Mol. Biol. 2009;8(1) doi: 10.2202/1544-6115.1406. [DOI] [PubMed] [Google Scholar]
- Pinault D. Dysfunctional thalamus-related networks in schizophrenia. Schizophr. Bull. 2011;37(2):238–243. doi: 10.1093/schbul/sbq165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteous DJ, Thomson P, et al. The genetics and biology of DISC1 – an emerging role in psychosis and cognition. Biol. Psychiatr. 2006;60(2):123–131. doi: 10.1016/j.biopsych.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Posada D, Buckley TR. Model selection and model averaging in phylogenetics: Advantages of akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst. Biol. 2004;53(5):793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Qin S, Zhao X, et al. An association study of the N-methyl-D-aspartate receptor NR1 subunit gene (GRIN1) and NR2B subunit gene (GRIN2B) in schizophrenia with universal DNA microarray. Eur. J. Hum. Genet. 2005;13(7):807–814. doi: 10.1038/sj.ejhg.5201418. [DOI] [PubMed] [Google Scholar]
- Shenton ME, Dickey CC, et al. A review of MRI findings in schizophrenia. Schizophr. Res. 2001;49(1–2):1–52. doi: 10.1016/s0920-9964(01)00163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon N, Tibshirani R. Standarization and the group lasso penalty. Stat. Sin. 2012;22:983–1001. doi: 10.5705/ss.2011.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Pearlson G, et al. Discriminating schizophrenia and bipolar disorder by fusing fMRI and DTI in a multimodal CCA+ joint ICA model. Neuroimage. 2011;57(3):839–855. doi: 10.1016/j.neuroimage.2011.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutrala SR, Goossens D, et al. Gene copy number variation in schizophrenia. Schizophr. Res. 2007;96(1–3):93–99. doi: 10.1016/j.schres.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Szycik GR, Munte TF, et al. Audiovisual integration of speech is disturbed in schizophrenia: an fMRI study. Schizophr. Res. 2009;110(1–3):111–118. doi: 10.1016/j.schres.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Torrey EF. Schizophrenia and the inferior parietal lobule. Schizophr. Res. 2007;97(1–3):215–225. doi: 10.1016/j.schres.2007.08.023. [DOI] [PubMed] [Google Scholar]
- Tyekucheva S, Marchionni L, et al. Integrating diverse genomic data using gene sets. Genome Biol. 2011;12(10):R105. doi: 10.1186/gb-2011-12-10-r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N, Landeau B, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Waaijenborg S, Zwinderman AH. Correlating multiple SNPs and multiple disease phenotypes: penalized non-linear canonical correlation analysis. Bioinformatics. 2009;25(21):2764–2771. doi: 10.1093/bioinformatics/btp491. [DOI] [PubMed] [Google Scholar]
- Waaijenborg S, Hamer PCVDW, et al. Quantifying the association between gene expressions and DNA-Markers by penalized canonical correlation analysis. Stat. Appl. Genet. Mol. Biol. 2008;7(1) doi: 10.2202/1544-6115.1329. [DOI] [PubMed] [Google Scholar]
- Wiley HS. Integrating multiple types of data for signaling research: challenges and opportunities. Sci. Signal. 2011;4(160):pe9. doi: 10.1126/scisignal.2001826. [DOI] [PubMed] [Google Scholar]
- Wilson GM, Flibotte S, et al. DNA copy-number analysis in bipolar disorder and schizophrenia reveals aberrations in genes involved in glutamate signaling. Hum. Mol. Genet. 2006;15(5):743–749. doi: 10.1093/hmg/ddi489. [DOI] [PubMed] [Google Scholar]
- Witten DM, Tibshirani RJ. Extensions of sparse canonical correlation analysis with applications to genomic data. Stat. Appl. Genet. Mol. Biol. 2009;8(1) doi: 10.2202/1544-6115.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten DM, Tibshirani R, et al. A penalized matrix decomposition, with applications to sparse principal components and canonical correlation analysis. Biostatistics. 2009;10(3):515–534. doi: 10.1093/biostatistics/kxp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Chen YF, et al. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics. 2009;25(6):714–721. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]