Background: Gib2 interacts with Gpa1, Gpg1, Gpg2, Cin1, and Pkc1. Gib2 overexpression promotes cAMP levels in cells lacking Gpa1.
Results: Gib2 regulates cAMP through a novel mechanism involving functions of Ras1 and Cac1.
Conclusion: Gib2 functions as a scaffolding adaptor protein.
Significance: This study reveals a novel mechanism of cAMP signaling and identifies Gib2 as a key element in fungal regulatory networks.
Keywords: Adenylate Cyclase (Adenylyl Cyclase), Cyclic AMP (cAMP), G Proteins, Ras, Signal Transduction, Gβ-like/RACK1, Fungal Growth and Virulence, Protein Interactive Network
Abstract
Gβ-like/RACK1 functions as a key mediator of various pathways and contributes to numerous cellular functions in eukaryotic organisms. In the pathogenic fungus Cryptococcus neoformans, noncanonical Gβ Gib2 promotes cAMP signaling in cells lacking normal Gpa1 function while displaying versatility in interactions with Gα Gpa1, protein kinase Pkc1, and endocytic intersectin Cin1. To elucidate the Gib2 functional mechanism(s), we demonstrate that Gib2 is required for normal growth and virulence. We show that Gib2 directly binds to Gpa1 and Gγ Gpg1/Gpg2 and that it interacts with phosphodiesterase Pde2 and monomeric GTPase Ras1. Pde2 remains functionally dispensable, but Ras1 is found to associate with adenylyl cyclase Cac1 through the conserved Ras association domain. In addition, the ras1 mutant exhibits normal capsule formation, whereas the ras1 gpa1 mutant displays enhanced capsule formation, and the ras1 gpa1 cac1 mutant is acapsular. Collectively, these findings suggest that Gib2 promotes cAMP levels by relieving an inhibitory function of Ras1 on Cac1 in the absence of Gpa1. In addition, using GST affinity purification combined with mass spectrometry, we identified 47 additional proteins that interact with Gib2. These proteins have putative functions ranging from signal transduction, energy generation, metabolism, and stress response to ribosomal function. After establishing and validating a protein-protein interactive network, we believe Gib2 to be a key adaptor/scaffolding protein that drives the formation of various protein complexes required for growth and virulence. Our study reveals Gib2 as an essential component in deciphering the complexity of regulatory networks that control growth and virulence in C. neoformans.
Introduction
Cryptococcus neoformans is a major cause of fungal meningoencephalitis in immunodeficient individuals. This disease currently afflicts approximately one million people, reflecting major occurrences in areas such as sub-Saharan Africa and other developing countries and nearly 600,000 fatalities worldwide (1). C. neoformans is a basidiomycetous organism with a defined life cycle and a bipolar mating system consisting of α and a mating types. The ability of the fungus to grow at mammalian body temperature, the prevalence of the α mating type, and the production of melanin pigment and a polysaccharide capsule are key factors contributing to virulence (2–4).
Canonical heterotrimeric G proteins, consisting of Gα, Gβ, and Gγ, regulate myriad cellular functions including growth, morphogenesis, production of secondary metabolites, and/or virulence in the lower eukaryotic organisms such as fungi (4–6). Upon activation by binding of the ligands to the specific G protein-coupled receptor, Gα undergoes structural conformational changes due to exchange of GDP for GTP, and Gα dissociates from the Gβγ heterodimeric complex (7). The activated Gα will then resume the inactive GDP-bound state upon GTP hydrolysis, blocking further signaling by reassociation with Gβγ. In C. neoformans, mating and virulence are linked to and regulated by two distinct and parallel G protein signal transduction pathways (8, 9). Gβ Gpb1 governs pheromone-responsive mating through association with Gαs Gpa2 and Gpa3 and Gγs Gpg1 and Gpg2 as heterotrimeric complexes, whereas Gα Gpa1 controls a conserved cyclic adenosine monophosphate (cAMP)-dependent signaling pathway that regulates the production of virulence factors such as melanin, capsule, and virulence (8, 10, 11). This pathway consists of several conserved components including the Gα subunit (Gpa1), adenylyl cyclase (Cac1), protein kinase A catalytic and regulatory subunits (Pka1 and Pkr1, respectively), and phosphodiesterases (Pde1 and Pde2) (12–15). In comparison with Saccharomyces cerevisiae Ras1 and Ras2 proteins that play a prominent role in promoting cAMP levels through the activation of adenylyl cyclase Cyr1 and Schizosaccharomyces pombe Ras1 that regulates the mating process (for a review, see Ref. 6), Ras1 was shown to regulate primarily thermal responses and cell polarization during budding in C. neoformans (16). Ras2 shares certain functions with Ras1, but its regulatory role appears to be minor (16). Despite Cac1 containing a conserved Ras association (RA)4 domain, the role of Ras1 or Ras2 in Cac1-cAMP regulation has not been established in C. neoformans.
We previously identified in a yeast two-hybrid (Y2H) assay that Gib2 (Gpa1-interacting β) binds to Gpa1 and found that overexpression of Gib2 positively regulates cAMP in strains lacking normal Gpa1 function (17). In addition, Gib2 exhibits the ability to associate with other proteins including Smg1, a multicopy suppressor of melanin defects due to gpa1 mutation; the Pkc1 kinase; and the human intersectin ITSN1 homolog Cin1 (17). Structural modeling of Gib2 showed that it contains a seven-WD domain (WD-40) motif similar to the mammalian Gβ-like/receptor for activated protein C kinase 1 (RACK1) protein and budding and fission yeast S. cerevisiae Asc1/S. pombe Cpc2 (17).
Mammalian Gβ-like/RACK1 proteins (RACK1 hereafter) are scaffold proteins that integrate many different cellular processes through interactions with various proteins including the activated protein kinase C (PKC), Src tyrosine kinase, and integrin (18–23). RACK1 interacts with a subset of heterotrimeric Gtαβγ (Gtα type) and/or heterodimeric Gβγ proteins (24–26). Studies also showed that RACK1 interacts with Pde4D5, an isoform of Pde4D, to suppress cAMP degradation (27). Moreover, RACK1 was shown to be the 40 S ribosomal core component that interacts with many proteins involved in protein translation and post-translational modifications (28, 29).
The budding yeast S. cerevisiae RACK1 homolog Asc1 functions as a guanine nucleotide dissociation inhibitor to inhibit the functions of Gpa2 and Cyr1 (30). Asc1 was further characterized as a ribosomal core protein that associates with the 40 S ribosome and regulates stress resistance and adhesion-dependent growth (31, 32). In fission yeast S. pombe, the cpc2 (Cpc2 is a RACK1 homolog) mutant displayed delayed cell cycle progression (33). In basidiomycetous Ustilago maydis, disruption of RAK1 (Rak1 is a RACK1/Asc1/Cpc2 homolog) affects cellular growth, cell wall integrity, cell fusion, and virulence (34). Collectively, these findings indicate that RACK1/Asc1/Cpc2 proteins share the ability to modulate multiple pathways and cellular responses in the lower eukaryotic fungi.
We previously identified Gib2 using a Y2H screening and found that Gib2 not only binds to Gpa1 and Gpg1/Gpg2 but also modulates cAMP levels when overexpressed (17). Gib2 also associates with proteins such as Pkc1 and Smg1, suggesting that it could modulate multiple pathways similarly to RACK1/Asc1/Cpc2. To further characterize Gib2 and identify the mechanism involved in Gib2-regulated cAMP signaling, we show here that Gib2 directly binds to Gpa1 and Gpg1/Gpg2, and the binding is not mediated by Ste4 and Ste18 of the yeast host. Gib2 is required for growth at body temperature and full virulence of C. neoformans. In addition, Gib2 promotes cAMP signaling by interacting with Ras1, which interacts with Cac1 through the conserved RA domain to suppress its activity. Moreover, through glutathione S-transferase (GST) affinity purification combined with mass spectrometry, we identified 47 additional proteins that interact with Gib2. Finally, we established and validated a Gib2-protein interactive network to illustrate that Gib2 is a multifunctional adaptor protein important in growth, differentiation, and virulence of C. neoformans.
EXPERIMENTAL PROCEDURES
Strains, Media, and Plasmids
C. neoformans var. neoformans (serotype D) JEC21 (MATα) and JEC20 (MATa) and var. grubii (serotype A) H99 (MATα) strains were used as parental strains (35, 36). Key strains and oligonucleotide primers for PCR amplification are listed in Tables 1 and 2, respectively.
TABLE 1.
Description and genotypes of major C. neoformans strains used in this study
| Strains | Genotype | Source/Ref. |
|---|---|---|
| Serotype D | ||
| JEC21 | MATα | Kwon-Chung et al. (35) |
| JEC20 | MATa | Kwon-Chung et al. (35) |
| BAC21 | MATα gpa1 | A gift from J. A. Alspaugh |
| BAC20 | MATa gpa1 | A gift from J. A. Alspaugh |
| GYS1 | MATa pde1::NAT | This study |
| GYS5 | MATa pde2::NAT | This study |
| GYS44 | MATa pde1::URA pde2::NAT | This study |
| GYS9 | MATα gpa1 pde1 | This study |
| GYS14 | MATα gpa1 pde2 | This study |
| GYS41 | MATα gpa1 pde1::URA pde2::NAT | This study |
| GYS85 | MATa gpa1 pde1::NEO GAL7-GIB2 | This study |
| GYS581 | MATa gpa1 pde1::URA pde2::NAT GAL7-GIB2 | This study |
| GYS186 | MATα ras1::NEO | This study |
| GYS174 | MATα gpa1 ras1::NEO | This study |
| GYS195 | MATa gpa1 ras1::NAT | This study |
| GYS247 | MATa gpa1 ras2::NEO | This study |
| GYS53 | MATα cac1::NAT | This study |
| GYS58 | MATa cac1::NAT | This study |
| GYS39 | MATa gpa1 cac1::URA | This study |
| GYS226 | MATa gpa1 cac1::URA ras1::NEO | This study |
| GYS72 | MATa gpa1 cac1 GAL7-GIB2 | This study |
| GYS155 | MATa gpa1 ras1 GAL7-GIB2 | This study |
| Serotype A | ||
| H99 | MATα | Perfect et al. (36) |
| PWC555 | MATα, 2 | This study |
| PWC556 | MATα, 3 | This study |
TABLE 2.
Key primers for PCR amplification used in this study
| PW590 | 5′-ATAGATATCTCTGCCACAGGGAAG-3′ |
| PW593 | 5′-GATCGCCTTTTACTGGGCGAATG-3′ |
| PW596 | 5′-TTTCATATGATGCCTTTATCCTTAACTGC-3′ |
| PW599 | 5′-TTTGAATTCTCATAGATGGTTACTACTGG-3′ |
| PW612 | 5′-CCAGCTCACATCCTCGCAGCCCAATGTTCCTGTAGTTCCG-3′ |
| PW613 | 5′-CTACATCTCTTCTATAAGCTTACGCCATAGCCTACAGACGAAC-3′ |
| PW614 | 5′-CCAGCTCACATCCTCGCAGCTGACCCACAAGCTCATCAG-3′ |
| PW615 | 5′-CTACATCTCTTCTATAAGCTTTTTGCAGAGCCGACCAATTG-3′ |
| PW638 | 5′-TGCCACACGCATAGCTGGAAG-3′ |
| PW639 | 5′-GATTCGTCGTCCGATTCGTCG-3′ |
| PW704 | 5′-AGCCATATTGCAGCGCTTCAC-3′ |
| PW705 | 5′-CGTCGATGATGACGATTCGTC-3′ |
| PW718 | 5′-CATCCTACCCATTATGGATCC-3′ |
| PW719 | 5′-AAAGGCGCGCCAACATTGGCACCCAACTGAGC-3′ |
| PW720 | 5′-AAAGCGGCCGCCAGAGTGCGAAAAAGATCC-3′ |
| PW721 | 5′-CATGCACAATGAGCTTTTGGTC-3′ |
| PW812 | 5′-AGGTAGCCGGATCAAGCGTATG-3′ |
| PW1013 | 5′-ACGACAAGCTTATATTCACGCAG-3′ |
| PW1014 | 5′-GAGGAATTCGGAGTGCGTAGAGCTATAGAG-3′ |
| PW1015 | 5′-TTGAGCAGAATGAGGAACTTGGTC-3′ |
| PW1027 | 5′-TCTGCAGAGCGAACGATATCTGC-3′ |
| PW1050 | 5′-ATCACCTATTCTGACACTTTG-3′ |
| PW1051 | 5′-AAGCTTGGACTGTTATGGCAGTC-3′ |
| PW1052 | 5′-GTGTTTGGTCAAGAAGATCAAG-3′ |
| PW1053 | 5′-ACTCTGTCGAGCCTGATGTAG-3′ |
| PW1756 | 5′-AGAATTCATGTCTGACACTAAGGGTGCGGCC-3′ |
| PW1757 | 5′-AAAGCTTTTAGATGAGGTCGGCAACGTTAAGA-3′ |
| PW1762 | 5′-ACTGCAGTTAGATGAGGTCGGCAACGTTAAGA-3′ |
| PW1767 | 5′-TCTCTGCATTGGATAAGGATTTAC-3′ |
| PW1768 | 5′-TTAGTCGTTCTTTCCTAACTCGTCGAG-3′ |
Yeast extract-peptone-dextrose, synthetic medium, 10% V8 agar (pH 5.0) for mating, and Niger seed agar for melanin production were prepared following the standard protocols or as described previously (9, 37).
Rapid amplification of cDNA ends for var. neoformans PDE2 was performed as described previously (37). Partial cDNA for var. neoformans PDE2 and full-length cDNA for RAS1, RAS2, and CAC1 were synthesized by RT-PCR and inserted into the pGBKT7 plasmid (BD Biosciences) according to the standard protocols (38). cDNA for the Cac1-RA domain was amplified using primers PW1767 and PW1868, and eIF4A cDNA was amplified with primers PW1756/PW1757 or PW1762. C. neoformans var. neoformans GIB2 cDNA was previously inserted into pGADT7 (17).
Construction of Mutant Strains
The gib2 mutant allele was generated by incorporating a SmaI restriction site into the GIB2 allele with primers PW225 and PW226, and the NAT and NEO gene cassettes were inserted into the SmaI site, generating the gib2::NAT and gib2::NEO mutant alleles, respectively. Mutant strains were obtained by transformation of var. grubii H99 with the gib2::NAT or gib2::NEO allele and of var. neoformans JEC21 with the gib2::NAT allele. The gib2::NAT mutant of H99 was complemented by reintroduction of a 2.0-kb wild-type GIB2 linked to the NEO marker (pGS200) (39).
The following mutant alleles are var. neoformans (serotype D)-specific. To disrupt the PDE1 gene, the upstream and downstream fragments overlapping with the 5′ and 3′ termini of the NAT gene were first amplified with primers PW590 and PW612 and primers PW613 and PW593, respectively. The upstream and downstream split markers were synthesized by overlap PCR with primers PW590 and PW639 and primers PW638 and PW593, respectively. The same approach was used to generate the pde1::NEO mutant strain. The pde2::NAT allele was generated using primer sets PW596 and PW614, PW615 and PW599, PW596 and PW639, and PW638 and PW599, respectively. The pde1 pde2 mutant was obtained by transformation of the pde1::NEO mutant with the pde2::NAT allele.
For RAS1 disruption, two ligation reactions were performed. One involved linking the HindIII fragment (upstream; amplified with PW1027 and PW1013) to the NEO fragment (KpnI/HindIII), and the other involved linking the EcoRI fragment (downstream; amplified with PW1014 and PW1015) to the NEO marker (EcoRI). The ras1::NEO mutant allele was amplified with primers PW1027 and PW638 and primers PW812 and PW1015. For RAS2 disruption, the upstream HindIII fragment (amplified with PW1050 and PW1051) and the downstream EcoRI fragment (amplified with PW1052 and PW1053) were each linked to NEO through ligation reactions. The ras2::NEO mutant allele was amplified with primers PW1050 and PW638 and primers PW812 and PW1053 using the two fragments as the templates. To disrupt the CAC1 gene, two ligations were also performed. One linked the upstream AscI fragment amplified with PW718 and PW719 to the URA5 fragment (AscI/NotI; pGS007), and the other linked the downstream NotI fragment (amplified with PW720 and PW721) with the URA5 fragment. The cac1::URA5 mutant allele was amplified with primers PW718 and PW705 and primers PW704 and PW721.
The URA5, NAT, and NEO gene markers were amplified from the plasmids pGS007 (a derivative of pCnTel1), pGMC200, and pGS063, respectively (39–41). PW1756, PW1757, and PW1762 were used to synthesize eIF4A cDNA, and PW1767 and PW1768 were used to synthesize RA domain (Cac1) cDNA.
Phenotypic Characterization
Growth was assessed by spotting serially diluted cell cultures on medium plates. Effects on cAMP signaling were judged on the formation of the melanin pigment and the polysaccharide capsule, two well established virulence factors. Melanin formation was observed in cells growing on Niger seed agar, and capsules were induced by growing cells in Dulbecco's modified Eagle's medium (DMEM) at 30 °C for 72 h as described previously (37, 42). Cells were stained with India ink for capsule visualization using a Zeiss microscope (Axio Imager A2) equipped with a digital camera (42).
Virulence tests were carried out in female A/JCr (The Jackson Laboratory) mice using a murine inhalation model, and mouse survival was analyzed by the Kaplan-Meier method using Prism 4.0 software (GraphPad Software, Inc.) as described previously (37). Where applicable, the brains of euthanized animals were collected, homogenized in glass homogenizers, stained with India ink, and visualized under a microscope. Animal testing was carried out under protocol number 171, which was approved by the Institutional Animal Care and Use Committee of the Research Institute for Children following the guidelines from the American Association for Laboratory Animal Science. Transient cAMP levels after starvation and glucose triggering were detected using the Amersham Biosciences cAMP Biotrak system (RPN225, GE Healthcare) following the same method we described previously (17).
Yeast Two-hybrid Assay, Co-immunoprecipitation (Co-IP), GST-Gib2 Fusion Construct, and GST Affinity Purification
The two-hybrid assay was carried out in both S. cerevisiae PJ69-4A and AH109 host strains as described previously (17, 43). The Δste4 mutant versions of PJ69-4A and AH109 were used for Gib2-Gpa1 and Gib2-Gpg2 interactions. pET-41a(+) plasmids (Novagen) with the GST fusion genes (GST-GIB2 and GST-RA) were respectively transformed into Escherichia coli RosettaTM 2(DE3), and protein expression was induced for 4 h at room temperature following the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 0.2 mm (17). To extract GST and GST-Gib2 fusion proteins, cells were suspended in PBS buffer, pH 7.3 containing 1 mm EDTA, 0.5% Nonidet P-40, and protease inhibitors (Roche Applied Science) and incubated for 30 min at 4 °C with gentle shaking. Samples were centrifuged at 13,000 rpm for 20 min at 4 °C, and supernatants were recovered. For protein purification, supernatants were mixed with glutathione-Sepharose resin (Amersham Biosciences) for 1.5 h and packed into a column. The fusion protein was verified by SDS-PAGE analysis and Western blotting with the anti-GST antibody (Santa Cruz Biotechnology, SC-138) and anti-Gib2 antibody (17). Ras1 and eIF4A fusion proteins were expressed using the pRSET-B vector (Invitrogen), and co-IPs were performed following the methods described previously (17).
For GST affinity purification, 400 μl of the GST-Gib2 protein extract was added to glutathione-Sepharose resin (Amersham Biosciences) and washed with buffer C (50 mm Tris, pH 7.5, 150 mm NaCl, 10 mm MgCl2, 1 mm EDTA, 1 mm PMSF, and proteinase inhibitors (Roche Applied Science)). The protein extracts (H99 and JEC20 gpa1) were respectively preconditioned in buffer A (50 mm NaHepes, pH 7.5, 150 mm NaCl, 10 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 25 mm β-glycophosphate, 100 μm Na3VO4, 10 μm GDP, and protease inhibitor mixture) for 30 min at room temperature before addition to the resin containing GST-Gib2. The mixture was incubated overnight with gentle rotation at 4 °C, precipitated, and washed twice with PBS buffer containing 0.2% Nonidet P-40 and 1 mm PMSF. 50 μl of SDS-containing gel loading buffer was added to the resin and briefly boiled, and 20 μl was analyzed by SDS-PAGE and Western blotting using the anti-GST antibody. A second SDS-PAGE was performed, gels were stained with Coomassie Blue stain, and protein bands were sliced and collected. The gel slices were cut into small pieces, digested with trypsin, and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Separate affinity purification for controls including GST-embedded resin and extraction buffer were run simultaneously (GST-Gib2 + extracts, GST-Gib2 + extraction buffer, GST + extracts, and GST + extraction buffer), and each sample was done in duplicate.
LC-MS/MS Analysis and Database Search
Protein samples were reduced by dithiothreitol (DTT), alkylated with iodoacetamide, and trypsin-digested overnight at 37 °C. The digestion was stopped by adding 5% formic acid, and the solvents were then evaporated in a speed vacuum (44). The dried samples were suspended in 2% acetonitrile (containing 0.1% formic acid) and subjected to LC-electrospray ionization-MS/MS analysis on a Finnigan LTQ ion trap mass spectrometer (ThermoFinnigan, San Jose, CA) as described previously (44). Briefly, the resuspended sample was loaded into a C18 trap column for desalting before being eluted into a reverse-phase C18 analytical column for LC separation and MS detection. The acquired raw data were processed using BioWorks (version 3.3) (Thermo Electron). Parameters for the SEQUEST database search against the UniProtKB/TrEMBL database were set as follows: differential mass increase of +57.02 Da on cysteinyl residue and +15.99 Da on methionine residue. The number of missed cleavage sites was set to 2. The search results were filtered with cross-correlation (XCorr) scores set as 2.0, 3.0, and 3.5 for singly, doubly, and triply charged peptides, respectively.
Establishing and Validating a Gib2 Interactome
To generate a Gib2 interactive network, the S. cerevisiae proteins with which proteins of C. neoformans var. grubii and var. neoformans share homologies were identified. Proteins with an E-value of <1e−5 are considered acceptable. The S. cerevisiae proteins (Saccharomyces Genome Database) were compiled using the Search Tool for the Interacting Genes/Proteins (STRING) database v9.0 (45) with a STRING confidence score over 0.7 (high confidence) to generate an Asc1-protein interactive network. The Gib2 interactive network was then established based on the S. cerevisiae Asc1 model.
We used pairwise cluster analysis of functional families, Y2H screening, and co-IP to validate selected Gib2-protein interactions. In the process of establishing the Gib2 interactome, we used blastp to search S. cerevisiae proteins that are homologous (E-value <1e−5) to the C. neoformans proteins identified through GST pulldown. Through the STRING database, we then predicted proteins that interact with Asc1 with each protein having a STRING confidence score ranging from 0.31 to 0.99 (>0.7, high confidence; 0.4–0.7, medium confidence; <0.4, low confidence). We clustered both C. neoformans and S. cerevisiae by OrthoMCL and identified C. neoformans proteins that could be clustered with at least one S. cerevisiae protein in each cluster, respectively. We also used Y2H screening and co-IP as another means of data validation to examine the association between Gib2 and eIF4A.
RESULTS
Gib2 Is Required for Cellular Viability and Virulence
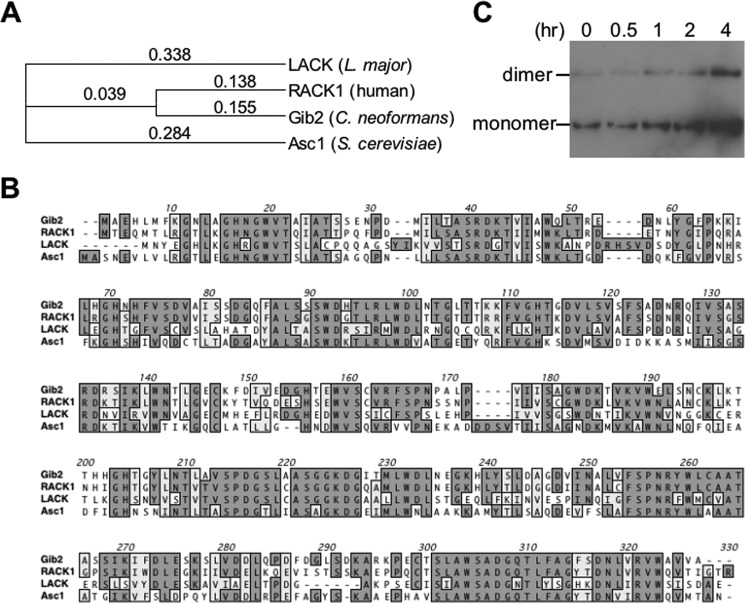
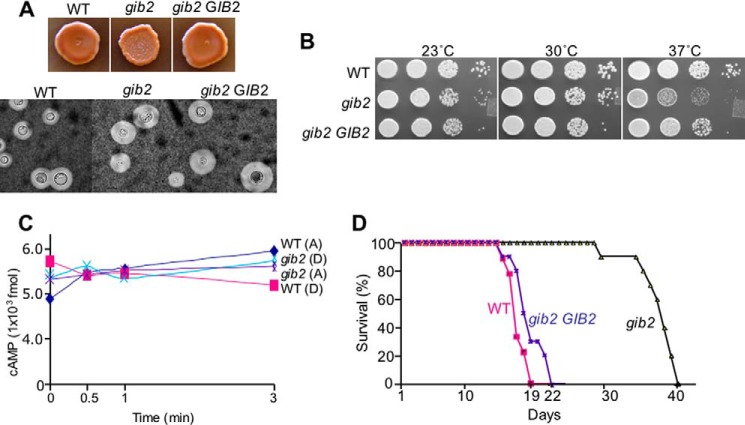
Gib2 was previously discovered in Y2H screening using Gα Gpa1 as protein bait, and it was shown to also promote cAMP levels when overexpressed in cells lacking Gpa1 (17). Gib2 adopts a seven-bladed β-propeller fold and shares high conservation in amino acid sequences with mammalian RACK1, parasitic Leishmania major LACK1, and S. cerevisiae Asc1 (Fig. 1, A and B). In addition, Western blotting analysis using the previously obtained Gib2-specific antibody showed that Gib2 occurs as monomeric and dimeric complexes (Fig. 1C). To characterize Gib2 function, we had previously tried but failed to obtain a gib2 knock-out mutant using the auxotrophic URA5 marker (17). However, when switching to dominant selective marker genes such as the nourseothricin resistance NAT and neomycin resistance NEO genes and selecting putative mutants at ambient temperature (25 °C) instead of 30 °C, we were able to generate gib2 knock-out mutants in both var. grubii (serotype A) and var. neoformans (serotype D) strains. Disruption of the GIB2 gene results in no apparent reduction in formation of the melanin pigment or capsules under the laboratory conditions (Fig. 2A). However, the gib2 mutant strains exhibited reduced growth in comparison with the wild-type and complemented (gib2 GIB2) strains at 37 °C (Fig. 2B). No statistically significant differences in cAMP levels were found between the gib2 mutants of var. grubii (serotype A) and var. neoformans (serotype D) and the wild type H99 and Jec21 strains (Fig. 2C).
FIGURE 1.
Gib2 shares high homology with human Gβ-like/RACK1, parasitic L. major LACK1, and S. cerevisiae Asc1 proteins and exists as monomer and dimer. A, multiple sequence alignment was made using ClustalW (v1.83). B, Gib2 is highly homologous to RACK1/LACK1/Asc1 proteins sharing, respectively, 84, 69, and 70% in amino acid sequence similarities. C, Western blotting showing that Gib2 is capable of forming dimeric complexes under normal physiological conditions. hr, time in yeast nitrogen base minimal medium after switching from yeast extract-peptone-dextrose.
FIGURE 2.
Gib2 is involved in growth and pathogenicity of C. neoformans. A, Gib2 is not directly involved in melanin and capsule formation. The upper panel shows the gib2::NAT mutant (var. grubii) and the wild-type strain H99 grown on Niger seed medium agar for 2 days at 30 °C. The lower panel shows cells recovered from mouse brain tissues exhibiting normal sized capsules following India ink staining. B, the growth of the gib2 mutant is reduced at 37 °C in comparison with H99. Partial complementation is achieved in the complemented mutant strain (gib2 GIB2). C, Gib2 has no apparent direct roles in cAMP levels in a transient cAMP assay using the Amersham Biosciences cAMP enzyme immunoassay system (see text). Each cAMP level represents the value estimated from 1 × 106 cells. D, Gib2 is required for full virulence expression. Ten A/JCr mice were infected with the gib2 mutant, H99, and the complemented (gib2 GIB2) strains, respectively. The difference in survival between gib2 and H99 and between gib2 and the complemented strain was statistically significant (p < 0.05), whereas there was no statistically significant difference between H99 and the complemented strain. A, serotype A; D, serotype D.
We also evaluated the impact of GIB2 disruption on its ability to cause diseases through an experimental murine model of cryptococcosis. Using the var. grubii gib2::NAT mutant and A/JCr mice, we showed that disruption of the GIB2 gene results in severe attenuation of virulence with the mice surviving nearly twice as long as the mice infected with the wild type H99 strain (Fig. 2D). The mean survival for the gib2 mutant is 39 days postinfection in comparison with 17.5 days by the wild-type strain and 21 days by the complemented gib2 GIB2 strain (p < 0.01). India ink staining of the brain smear collected from the moribund mice showed the presence of the gib2 mutant cells with capsules comparable with those of the wild-type strain (Fig. 2A).
Gib2 Binding of Gpa1 Is Not Mediated by S. cerevisiae Gβ Ste4
Gib2 was identified through its interaction with Gα Gpa1 through Y2H screening and protein pulldown (17). Gib2 contains a seven-WD domain motif and was modeled to show the β-transducin structure. Binding of Gib2 to Gpa1, which lacks a traditional Gβ, and Gpg1/Gpg2 suggests that Gib2 may provide a structural component for the canonical heterotrimeric Gpa1 protein complex. Because mammalian RACK1 is known to interact with a subset of heterotrimeric Gαβγ and heterodimeric Gβγ proteins (24–26), we deleted the STE4 gene from S. cerevisiae AH109 and PJ69-4 strains to test whether Gib2 remains interactive with Gpa1 and Gpg1/Gpg2.
The S. cerevisiae STE4 gene was disrupted in both PJ69-4a and AH109 host strains using a ste4::NEO knock-out allele. The ste4::NEO allele was generated through PCR amplification with primers containing an ∼75-oligonucleotide sequence of S. cerevisiae STE4 and ∼18 oligonucleotides for linking the neomycin resistance gene (46). Both yeast ste4 mutant strains were sterile as expected and exhibited no other defects (data not shown). Interactions between Gib2 and Gpa1 and between Gib2 and Gpg2 were again demonstrated (Fig. 3A), confirming that Gib2 indeed binds directly with Gpa1 and Gpg1/Gpg2 and not through a complex with the host G protein β subunit.
FIGURE 3.
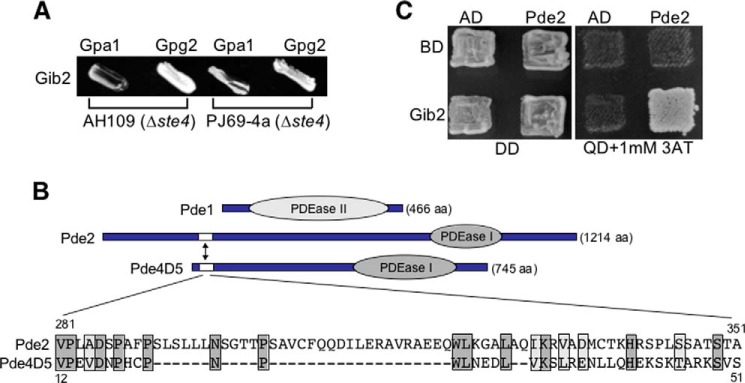
Gib2 directly interacts with Gpa1 and Gpg2 subunits and with Pde2 through the RAID. A, Gib2 interacts with Gpa1, Gpg1, and Gpg2 in the absence of yeast Ste4. B, schematic representations of C. neoformans PDEs Pde1 and Pde2. Pde2 is 1214 amino acids (aa) long and, similar to Pde4D5, contains a putative RAID between amino acids 280 and 350 (marked by an arrow and enlarged below). Darkly shaded areas indicate identical amino acid residues, and lightly shaded areas show conserved residues. PDEase, phosphodiesterase. C, Gib2 interacts with Pde2 through the putative RAID in a yeast two-hybrid assay. DD indicates dropout medium lacking leucine and tryptophan, and QD indicates dropout medium lacking leucine, tryptophan, histidine, and adenine. 3AT, 3-amino-1,2,4-triazole. 1 mm was used. AD, pGADT7. BD, pGBKT7.
Gib2 Interacts with Phosphodiesterase Pde2
cAMP is a ubiquitous second messenger produced by cells in response to external stimuli that activates cAMP-dependent protein kinase A and in turn transcription factors to promote cellular growth and differentiation. The levels of the cellular cAMP depend on the activity of adenylyl cyclase that synthesizes cAMP as well as PDEs that degrade cAMP. In humans, RACK1 modulates the cAMP levels through inhibition of the Pde4D5 PDE activity, and the inhibition is mediated by an interaction at the conserved RACK1 interaction domain (RAID) (27, 47). Binding of RACK1 to Pde4D5 blocks the site on which Pde4D5 normally has access to cAMP, thus preventing degradation of cAMP. Therefore, we hypothesized and tested the possibility of a similar operating mechanism involved in cAMP regulation by Gib2.
In C. neoformans var. grubii (serotype A) strains, the low affinity PDE Pde1 is a prominent member of the Gpa1-cAMP signaling pathway, whereas the high affinity Pde2 has a minor or negligible impact (15). Because Gib2 was initially found to regulate cAMP levels in the highly related but divergent var. neoformans (serotype D) strains, we characterized Pde1 and Pde2 functions by searching for the presence of the RAID and by generating pde1, pde2, and pde1 pde2 in the wild-type JEC20 (MATa) and the mutant BAC21 (MATα gpa1) strain backgrounds.
No putative RAID was found in Pde1 of either serotype that shares high homology. A discrepancy was found, however, in the sequences of Pde2. Rapid amplification of cDNA ends and RT-PCR revealed that var. neoformans Pde2 (Cnb01170) consists of 1214 amino acids in contrast to 555 amino acids as deposited (GenBankTM accession number AAX73258 (15)) (Fig. 3B). Interestingly, a region at the N terminus (amino acid residues 281–351) of Pde2 exhibits certain sequence homology (27%; 18% identity and 9% similarity) with the RAID of human Pde4D5 (Fig. 3B). In addition, a Y2H assay showed that this region interacts with Gib2 (Fig. 3C), suggesting that Gib2 associates with Pde2 through this putative RAID to potentially modulate its function.
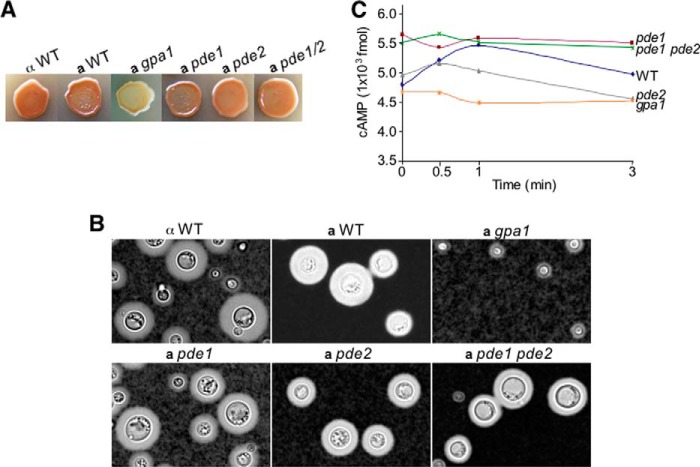
Characterization of pde1 and pde2 mutant strains through melanin and capsule formation revealed little useful information as the pde1, pde2, and pde1 pde2 mutant strains showed no detectable differences when compared with wild-type control strains (Fig. 4, A and B). Therefore, we resorted to measuring the basal and transient levels of intracellular cAMP following nutrient deprivation and glucose triggering as this method provides an accurate assessment of cAMP signaling status as demonstrated in several previous studies (17, 48, 49). Higher basal levels of cAMP were found in the pde1 and pde1 pde2 mutant strains as well as transient levels when the cells were starved and stimulated with glucose in comparison with the other strains (p < 0.05; Fig. 4C). The pde1 and pde1 pde2 mutant strains showed similar profiles with cAMP levels of 5.5–5.7, 5.4–5.7, 5.5–5.6, and 5.4–5.5 × 103 fmol at 0, 0.5, 1, and 3 min postglucose stimulation. The wild-type strain (JEC20) and pde2 mutant strain showed similar basal levels of 4.8–4.9 × 103 fmol, whereas the gpa1 mutant showed the overall lowest cAMP levels at all sampling points (Fig. 4C) as expected. This result suggests that Pde1, but not Pde2, has a measurable role in cAMP degradation. This finding is consistent with that of var. grubii PDEs, and it argues against a potential role of Pde2 in promoting cAMP degradation governed by Gib2.
FIGURE 4.
Pde1, but not Pde2, is required for regulation of cAMP levels. A and B, impacts of C. neoformans var. neoformans PDE1 and PDE2 gene disruptions on melanin and capsule formation (cAMP signaling). Cells were spotted on Niger seed agar for melanin and inoculated in DMEM for capsule induction for 3 days at 30 °C, respectively. C, cAMP assay suggests that Pde1, but Pde2, affects cAMP accumulation. Each cAMP level represents the value estimated from 1 × 106 cells.
Ras1 Interacts with Adenylyl Cyclase Cac1 and Gib2
Because Pde2 not having an apparent measurable role in cAMP regulation is inconsistent with our hypothesis that Gib2 might promote cAMP levels by inhibiting cAMP degradation mediated by Pde2, we next examined the effect of Gib2 on other conserved proteins known to be involved in the synthesis of cAMP. In S. cerevisiae and other fungi, cAMP is synthesized from ATP by adenylyl cyclase Cyr1, and loss of Cyr1 activity results in a decrease in intracellular cAMP levels (50). Cyr1 is activated through interactions with proteins, such as cyclase-associated Srv2 and Ras GTPases Ras1 and/or Ras2 (51, 52). The presence of an RA domain in Cyr1 is critical for binding to and activation by Ras proteins (53).
C. neoformans contains two Ras proteins, Ras1 and Ras2, and previous studies found that they play distinct as well as shared roles in promoting fungal growth at high temperature as well as in filamentation and mating (16, 54). The ras1 mutant strain had a defect in growth at 37 °C, and overexpression of Ras2 could suppress the defect of Ras1 in thermal resistance (16). In addition, overexpression of Ras1 in a gpa1 mutant partially suppressed the defects in melanin and capsule formation, which are the traits regulated by cAMP signaling (54). However, whether or not Ras1 or Ras2 interacts with Cac1 and whether they have any impact on cAMP levels remain unknown.
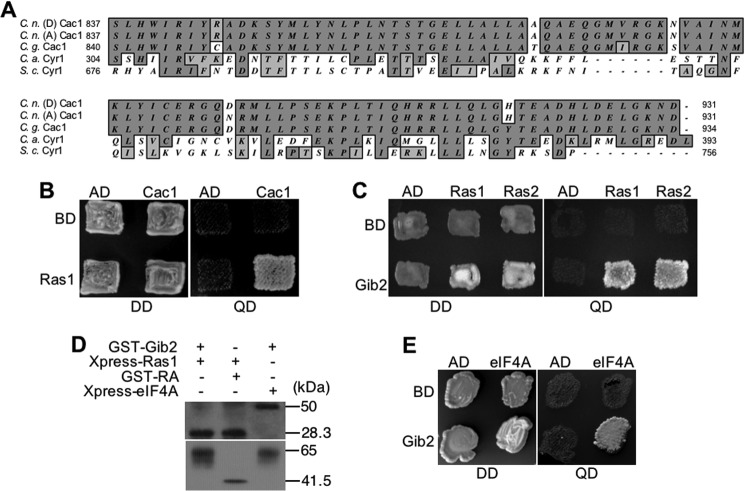
C. neoformans var. grubii encodes an adenylyl cyclase Cac1 protein (13). Cac1 of the var. neoformans strain is similar to var. grubii in that it contains 2272 amino acids. In Cac1 identified from several sources (C. neoformans var. grubii, var. neoformans, and var. gattii), there is a region sharing relatively high homology with the RA domains of other fungi: 38% similarity with RA domain of S. cerevisiae Cyr1 and 51% similarity with RA domain of Candida albicans Cyr1 (Fig. 5A). This suggests that Cac1 of C. neoformans could interact with Ras1 and function as a Ras1 effector molecule in cAMP signaling in addition to Gα Gpa1. To investigate whether or not Gib2-mediated cAMP up-regulation involves the function of Ras-Cac1 interactions, we first tested the interactions between Ras1 and Cac1 and between Ras1 and Gib2 through Y2H screening. We then examined whether or not cAMP signaling by Gib2 is dependent on Ras1 and/or Cac1.
FIGURE 5.
Ras1 interacts with Cac1 and Gib2. A, C. neoformans var. neoformans (serotype D) (C. n. (D)), var. grubii Cac1 (serotype A) (C. n. (A)), and C. gattii (C. g.) contain putative RA domains that are homologous to those of S. cerevisiae (S. c.) and C. albicans (C. a.). Numbers mark the amino acid locations of the RA domains. B, Ras1 interacts with Cac1 through the conserved RA domain in a yeast two-hybrid assay. C, Ras1 and paralog Ras2 interact with Gib2. D, co-immunoprecipitation confirms the interactions between Gib2 and Ras1, Ras1 and Cac1 (RA domain), and Gib2 and eIF4A. Anti-GST and Anti-Xpress antibodies were used to detect the respective fusion proteins following PAGE and Western blot analysis. E, Gib2 interacts with eukaryotic initiation factor eIF4A homolog. The Y2H assay conditions and media used were the same as that described in Fig. 3. DD indicates dropout medium lacking leucine and tryptophan, and QD indicates dropout medium lacking leucine, tryptophan, histidine, and adenine. co-IP was performed following the previously published method (17). AD, pGADT7; BD, pGBKT7.
Ras1 as well as paralog Ras2 indeed associates with Cac1, and this interaction is mediated through the putative RA domain, and it also interacts with Gib2 through Y2H screening (Fig. 5, B and C). These interactions were subsequently confirmed by co-IP (Fig. 5E). Although these findings are in contrast to S. cerevisiae Asc1 that inhibits Cyr1 cAMP production through a direct contact (30), they support a proposition that Gib2 could regulate cAMP signaling through functions of Ras1 and Cac1.
Gib2 Promotes cAMP Levels through Modulating the Ras1-Cac1 Interaction
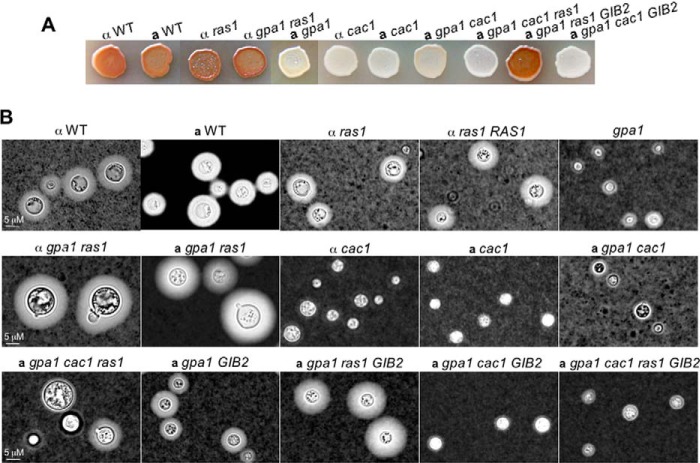
In C. neoformans var. grubii, Ras1 is known to play a major cellular role, whereas the role of Ras2 is minimal (16) in a manner similar to Pde2. To establish functional connections between Ras1 and Cac1, we disrupted the RAS1 and/or CAC1 genes in various var. neoformans strains and performed epistasis analysis. Consistent with previous studies, the ras1 mutant exhibited reduced thermal resistance (55). The ras1 mutant strain also did not show any detectable differences in cAMP-regulated traits such as melanin or capsule formation (Fig. 6, A and B); however, the ras1 gpa1 mutant colony showed a very mucus-like appearance in yeast extract-peptone-dextrose medium, a non-inductive condition for capsule formation, which suggests enhanced capsule formation (high cAMP levels). Indeed, the ras1 gpa1 mutant cells showed large capsules following induction in comparison with ras1, ras1 RAS1 complemented, and the wild-type JEC21 strains (Fig. 6B). The gpa1 ras1 Pgpd1GAL7-GIB2 strains showed capsule formation consistent with that of the gpa1 ras1 strain (Fig. 6B). These results suggest that Ras1 negatively regulates cAMP signaling in cells lacking Gpa1.
FIGURE 6.
Genetic evidence suggests that Gib2 promotes cAMP levels through the Ras1-Cac1 interaction. A, Ras1 negatively regulates melanin formation, whereas Cac1 is required for melanin formation (cAMP signaling) in all of the strains including F4 (gpa1 GAL7-GIB2). B, Ras1 negatively regulates capsule formation in the absence of normal Gpa1 signaling. Induction conditions for melanin and capsule were as the same as those described in Fig. 2.
Moreover, disruption of the CAC1 gene in all strain backgrounds (gpa1, gpa1 ras1, gpa1 ras1 Pgpd1GAL7-GIB2, and wild types) resulted in defects in melanin and capsule formation, indicating a near total defect in cAMP production (Fig. 6, A and B). Although this result is consistent with previous studies in which the var. grubii cac1 mutant was defective in capsule and melanin formation (13), it suggests that Cac1 is the downstream target of both Gib2 and Ras1. As controls, the gpa1 mutant exhibited defects in melanin and capsule, whereas the gpa1 mutant with the GAL7-GIB2 (gpa1 GIB2) construct showed partially complemented capsule formation due to the leaky nature of the GAL7 promoter (Fig. 6, A and B) (17).
Based on these studies, we propose that Ras1 has a previously uncharacterized negative role in regulating Cac1 activity and that Gib2 promotes cAMP levels through the inhibition of Ras1 function on Cac1. An updated version of the cAMP signaling pathway is illustrated in Fig. 7.
FIGURE 7.
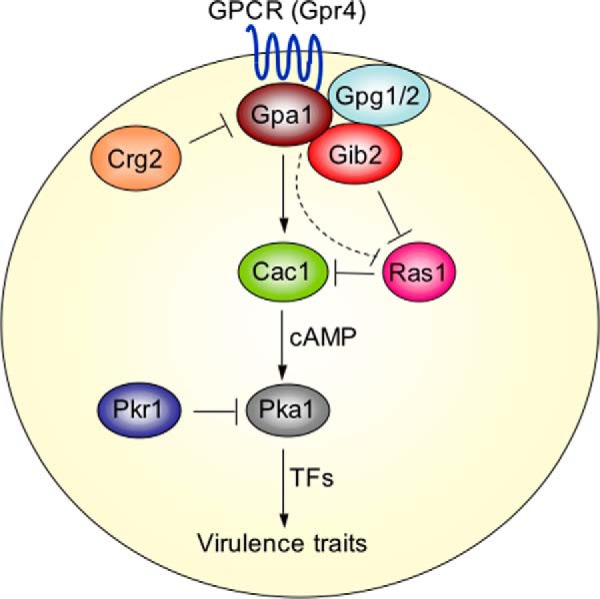
Ras1 regulates the Cac1-cAMP signaling pathway in C. neoformans. The Ras1 function opposes that of Gpr4/Gpa1. Gpa1, G protein α subunit (8, 62); Gpg1/2, G protein γ subunits 1 and 2 (11); Crg2, cryptococcal regulator of G protein signaling 2 (39, 63); Pkr1 and Pka1, cAMP-dependent protein kinase regulatory and catalytic subunit, respectively (14). TFs denote Nrg1 (60) and other unknown transcriptional factors.
Gib2 Is a Scaffolding Protein
Genetic analysis has provided a molecular basis by which Gib2 promotes cAMP levels in the absence of Gpa1. Because Gib2 was previously shown to also interact with Gpa1, Pkc1, Cin1, and other proteins (17, 39), the newly established interactions provided further support for the hypothesis that Gib2 is a scaffolding protein capable of interconnecting various cellular processes and pathways through multiple protein interactions.
Mammalian RACK1 has as many as 80 protein partners (56). To identify additional proteins to which Gib2 might bind, we used a proteomic approach of GST affinity purification combined with LC-MS/MS analysis. To do this, the GST gene was fused to the N terminus of the GIB2 gene, generating a GST-Gib2 fusion protein. Using the crude protein extracts prepared from cells grown first in nutrient-rich yeast extract-peptone-dextrose medium and then grown for 2 h in nutrient poor yeast nitrogen base, we identified 47 proteins (Table 3). These proteins are highly homologous to those of other model systems with diverse functions including intracellular trafficking (Ykt6), stress responses (response to stress-related protein and flavohemoglobin), metabolism (sn-glycerol-3-phosphate dehydrogenase, UDP-glucose dehydrogenase, and glucose-6-phosphate 1-dehydrogenase), and ribosomal assembly and translation (40 S ribosomal proteins S7 and s3ae and 60 S ribosomal proteins L4, L13, and L19). These findings strongly support the hypothesis that Gib2 is a signal-transducing scaffolding protein that interacts with various proteins to integrate many cellular pathways necessary for growth, differentiation, and virulence.
TABLE 3.
Gib2-interacting proteins identified through GST-Gib2 affinity purification combined with mass spectrometry
Please see “Experimental Procedures” for detailed description. Data shown were the unique proteins from combined extracts of C. neoformans var. grubii (H99) and var. neoformans (gpa1).
| CCPR_CRYNE; cytochrome c peroxidase, mitochondrial; C. neoformans |
| EF1A_CRYNE; elongation factor 1-α; C. neoformans |
| IF4A_CRYNE; ATP-dependent RNA helicase eIF4A; C. neoformans |
| Q2XPZ3_CRYNV; eukaryotic ADP/ATP carrier; C. neoformans |
| Q5KQ18_CRYNJ CRYNE; putative prohibitin PHB1; C. neoformans |
| Q55IY2_CRYNE; putative ribosomal protein L4; C. neoformans |
| Q55KS9_CRYNE; putative uncharacterized protein (pyruvate decarboxylase); C. neoformans |
| Q55MH3_CRYNE; UDP-glucose dehydrogenase; C. neoformans |
| Q55NI0_CRYNE; 40 S ribosomal protein S7; C. neoformans |
| Q55SW7_CRYNE; ATP synthase γ chain; C. neoformans |
| Q55W79_CRYNE; conserved hypothetical protein (mucin-like glycoprotein); C. neoformans |
| Q55Y63_CRYNE; endothelin-converting enzyme 1; C. neoformans |
| Q55ZV5_CRYNE; putative uncharacterized protein (elongation factor 1-γ); C. neoformans |
| Q561E6_CRYNE; T-complex protein 1 ϵ subunit; C. neoformans |
| Q561J1_CRYNE; putative proteolysis- and peptidolysis-related protein; C. neoformans |
| Q5K7I1_CRYNE; expressed protein (Pfam exo-endo-phos); C. neoformans |
| Q5K7W8_CRYNE; 60 S ribosomal protein L13; C. neoformans |
| Q5K9S1_CRYNE; hypothetical protein (SNARE protein YKT6); C. neoformans |
| Q5KAF2_CRYNE; 60 S ribosomal protein L4e; C. neoformans |
| Q5KBW9_CRYNE; 60 S ribosomal protein L19, putative; C. neoformans |
| Q5KDP3_CRYNE; glucose-6-phosphate 1-dehydrogenase; C. neoformans |
| Q5KE76_CRYNE; hypothetical protein; C. neoformans |
| Q5KEJ6_CRYNE; 60 S acidic ribosomal protein P0; C. neoformans |
| Q5KFA0_CRYNE; Putative ketol-acid reductoisomerase; C. neoformans |
| Q5KFT4_CRYNE; electron transporter, transferring electrons within CoQH2-c; C. neoformans |
| Q5KFU0_CRYNE; ATP synthase subunit β; C. neoformans |
| Q5KGN7_CRYNE; putative Ran small monomeric GTPase; C. neoformans |
| Q5KJB0_CRYNE; putative response to stress-related protein; C. neoformans |
| Q5KJK1_CRYNE; putative zinc-binding dehydrogenase; C. neoformans |
| Q5KKA2_CRYNE; hypothetical protein; C. neoformans |
| Q5KKL4_CRYNE; hypothetical protein (putative sugar kinase); C. neoformans |
| Q5KL26_CRYNE; ATP synthase, putative; C. neoformans |
| Q5KLL1_CRYNE; 40 S ribosomal protein s3ae-a; C. neoformans |
| Q5KLS4_CRYNE; cytoplasm protein, putative; C. neoformans |
| Q5KN57_CRYNE; putative NADH dehydrogenase; C. neoformans |
| Q5KNI6_CRYNE; putative 60 S ribosomal protein I7; C. neoformans |
| Q5KP06_CRYNE; actin; C. neoformans |
| Q5KP62_CRYNE; S-adenosylmethionine synthetase; C. neoformans |
| Q5KPZ1_CRYNE; putative ribosomal protein S3; C. neoformans |
| Q6XX21_CRYNV; flavohemoglobin; C. neoformans |
| Q7Z875_CRYNV; elongation factor 1-α; C. neoformans |
| Q85SZ4_CRYNV; cytochrome c oxidase subunit 2; C. neoformans |
| Q8NKG5_CRYNE; sn-glycerol-3-phosphate dehydrogenase NAD+; C. neoformans |
| Q5KGM8_CRYNE; 60 S ribosomal protein L6; C. neoformans |
| Q5KAP9_CRYNE; putative uncharacterized protein; C. neoformans |
| Q5KJP2_CRYNE; putative voltage-dependent ion-selective channel; C. neoformans |
| Q560D1_CRYNE; putative uncharacterized protein; C. neoformans |
Establishment of a Gib2-Protein Interactome
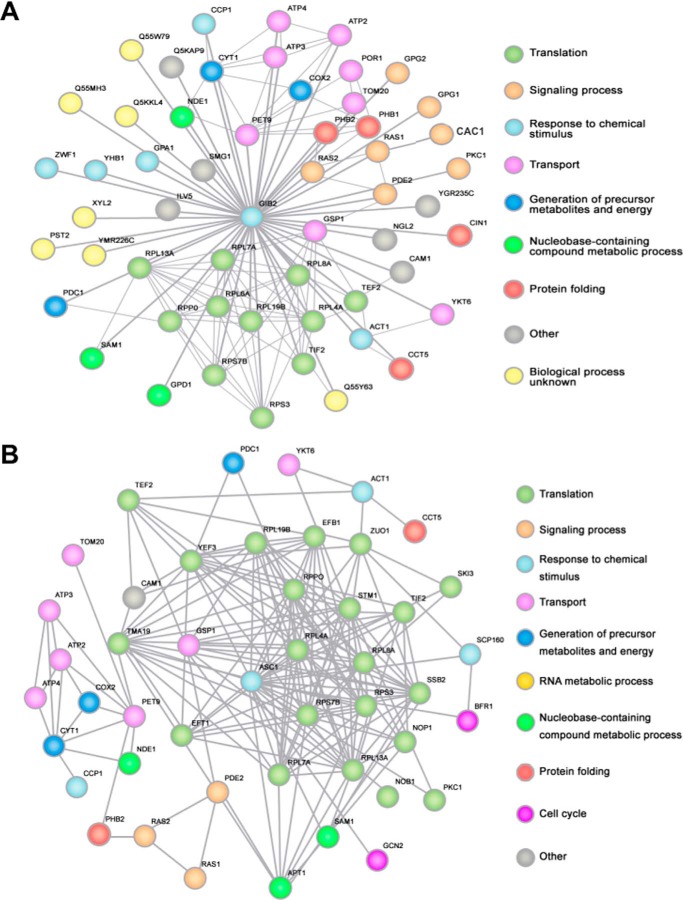
A genomic catalog of protein-protein interactions is a rich source of information, particularly for exploring the genetic relationships between various proteins and elucidating their functions. Given the large number of proteins with which Gib2 interacts, we sought to generate a Gib2 interactive protein network to better illustrate the complexity of Gib2 networking in events such as cAMP signaling, Pkc1-mediated stress responses, Cin1-mediated intracellular trafficking, and others. To do this, we first identified the respective homologous proteins in the S. cerevisiae genome (Saccharomyces Genome Database) and compiled yeast proteins with an E-value of <1e−5. Using the STRING database v9.0 (45) for protein-protein interactions and a STRING confidence score over 0.7 (high confidence), we created a S. cerevisiae Asc1 interactive network as reference followed by a Gib2 interactive network using C. neoformans proteins.
Approximately 55 proteins are shown in the interactive network centered by Gib2 (Fig. 8A). Surprisingly, proteins involved in cellular signaling rank third in the number of targets by Gib2 (6 of 53, 11.3%). The preferential targets of Gib2 appear to be various proteins involved in ribosomal function and protein translation (12 of 53, 22.6%) as well as intracellular transport (8 of 53, 15.1%). This gradient of targeting is, not surprisingly, also seen in the reference network for S. cerevisiae Asc1 (Fig. 8B) with ribosomal targets accounting for 26.7% (12 of 45), transport proteins accounting for 15.6% (7 of 45), and signaling proteins accounting for 6.6% (3 of 45). Taking into consideration that these interactions are not exhaustive as several proteins that interact with Asc1 through traditional experimental approaches (30) are not included, the percentage of Asc1 that modulates signal transduction could be higher than 6.6%. The functions of 11 proteins with which Gib2 interacts (22.9%, 11 of 48) remain undefined compared with S. cerevisiae where only the function of Cam1 is unclear. In addition, several cryptococcal proteins do not appear to have any corresponding homologs in S. cerevisiae or vice versa, which could be due to the evolutionary divergence existing between the two organisms. As a result, these proteins were not included in the interactive network.
FIGURE 8.
A Gib2 interactive network demonstrates that Gib2 functions as a signal-transducing adaptor protein interconnecting many pathways in C. neoformans. Respective S. cerevisiae proteins homologous to cryptococcal proteins (E-value <1e−5) we found were first identified in the genome of S. cerevisiae (Saccharomyces Genome Database). The S. cerevisiae proteins were then compiled using the STRING database v9.0 for protein-protein interactions using a STRING confidence score over 0.7 to generate an Asc1-protein interactive network (see text). Two interactive networks were generated accordingly: one for C. neoformans Gib2 (A) and another for S. cerevisiae Asc1 (B). Thick lines in A indicate that the connections were experimentally established by us, whereas thin lines indicate the connections derived from those of S. cerevisiae.
Validation of the Gib2-protein Interactome
Our analysis so far indicates that Gib2 has evolved to play a major adaptor role regulating various cellular functions by recruiting and chaperoning multiple proteins. To further substantiate the Gib2-protein interaction network we established through affinity pulldown, we adopted the following two methods for data validation: 1) pairwise cluster analysis of functional families (57, 58) and 2) Y2H screening and co-IP of selected proteins.
In the process of establishing a Gib2 interactome, we used blastp to search S. cerevisiae proteins that are homologous (E-value <1e−5) to the 47 C. neoformans proteins identified through GST pulldown and identified 42 protein homologs. Through the STRING database, approximately half of these proteins (22) were predicted to interact with Asc1 with each protein having a STRING confidence score ranging from 0.31 to 0.99 (>0.7, high confidence; 0.4–0.7, medium confidence; <0.4, low confidence). We clustered all of these proteins (both C. neoformans and S. cerevisiae) by OrthoMCL and found that 42 of the 47 C. neoformans proteins could be clustered with at least one S. cerevisiae protein in each cluster, respectively. This is consistent with the results of blastp. We assume that Asc1 interacts with RPL4A with high confidence (S. cerevisiae), and likewise, Gib2 (Asc1 homolog) and Q5KAF2 (RPL4e, a RPL4A homolog) should also interact with high confidence. Twenty-two C. neoformans proteins were thus predicted to interact with Gib2 with various degrees of confidence (Table 4).
TABLE 4.
Pairwise cluster analysis of Gib2-interacting proteins

We also validated Gib2-protein interactions through a classical approach. Specifically, we examined the interaction between Gib2 and eIF4A, a eukaryotic initiation factor homolog of C. neoformans, through Y2H screening and co-IP. Cryptococcal eIF4A is composed of 401 amino acids (GenBank accession number AFR92916), and it shares high amino acid sequence homology with S. cerevisiae Tif2 (65% identity and similarity 14%). The latter is a DEAD (Asp-Glu-Ala-Asp) box-containing RNA helicase that confers essential functions in S. cerevisiae (59). Consistent with GST tag pulldown and comparative cluster analysis results, yeast cells expressing Gib2 (pGADT7-Gib2) and eIF4A (pGBKT7-eIF4A) were able to grow in selective medium, and eIF4A (expressed in pRSET-B) could also be precipitated by Gib2 (expressed in pET41a(+)) in co-IP analysis (Fig. 5, D and E). Although the function of cryptococcal eIF4A remains to be defined, its high sequence homology with Tif2 and the confirmed interaction with Gib2 are consistent with Gib2 functioning as a RACK1-like protein adaptor.
DISCUSSION
C. neoformans infects mammalian hosts with weakened immune systems, causing severe clinical manifestations that include meningoencephalitis. As a soil-borne pathogen, C. neoformans has acquired the ability to resist harsh environmental conditions. The transition into a human host poses another drastic change for the fungus, involving further adaptation to even harsher conditions. Signal transduction pathways such as those mediated by G proteins are of paramount importance in allowing for such adaptations. Previous studies have shown that C. neoformans maintains two parallel G protein signaling pathways: Gpa2/Gpa3/Gpb1/Gpg1/Gpg2 for pheromone-responsive mating and Gpa1/Cac1 for cAMP signaling. Although recombination through mating is arguably the most conserved approach to evolution and adaptation, mating does not play a significant role in virulence (9–11). By contrast, Gpa1 adapts a conserved cAMP signaling pathway that senses nutrients and presumably amino acids such as methionine to regulate growth characteristics and virulence. Gpa1 functions through a conserved mechanism that likely involves the activation of Cac1 upon stimulation by G protein-coupled receptor Gpr4 with the resulting increased cAMP levels through the activation of protein A kinase Pka1 (8, 12, 13, 48). Although the definitive target of cAMP signaling remains elusive, it is generally believed that the main target of the Gpa1-cAMP pathway is a network of transcription factors (60). A canonical Gβ for Gpa1 was not found, suggesting that Gpa1 may function through a “noncanonical” fashion. Our previous identification of Gib2 as an atypical Gβ binding to Gpa1 sheds light on the possible functional mechanism of Gpa1 under such “dire” circumstances.
Gib2 binding to Gα Gpa1 and Gγs Gpg1 and Gpg2 suggests that Gib2 couples with Gγ to function as a βγ dimeric complex and to maintain the activation/inactivation cycle of Gpa1. Further studies that include measuring the guanine nucleotide dissociation inhibitor activity could perhaps provide additional supporting evidence. Intriguingly, Gib2 promoted cAMP levels when overexpressed in strains lacking normal Gpa1-cAMP signaling (17). This finding suggests that Gib2 has an additional role in the Gpa1-cAMP pathway. To illustrate this role, we focused on the proteins involved in the biogenesis of cAMP, namely Pde1/Pde2, Ras1, and Cac1 of C. neoformans. We found that Gib2 associates with Pde2 and Ras1, but only Ras1 has a meaningful role because it also associates with Cac1.
Although Gib2 interacts with Pde2 through the putative RAID, the significance of Pde2 in cAMP regulation appears negligible under normal testing conditions, raising the question of whether or not the interaction has any biological significance. C. neoformans Cac1 positively regulates cAMP levels, but an interaction between Gib2 and Cac1 was not found. We then resorted to examining the role of Ras1 in cAMP signaling as Ras proteins are known to regulate adenylyl cyclase activities in fungi including S. cerevisiae. Interactions between Gib2 and Ras and Ras1-Cac1 were sequentially established and substantiated. Finally, we provide evidence demonstrating that Ras1 indeed functions in cAMP regulation by inhibiting the activity of Cac1, a novel role not yet established in C. neoformans, and that Gib2 promotes cAMP levels by recruiting Ras1. It is worth noting that these genetic interactions occur in cells lacking normal Gpa1 signaling. Our findings suggest that Gib2 regulates cAMP signaling in C. neoformans through a mechanism distinct from that of S. cerevisiae Asc1, which is in accord with the opposing impact of each protein on cAMP levels. Whether or not the presence of Gpa1 limits the Ras1-Cac1 interaction and how Gpa1 and Ras1 interact with Cac1 in normal cells remain important but undefined research subjects. Moreover, our findings also do not rule out the possibility of additional regulatory elements of Cac1-cAMP signaling and that there may be more than one target in cAMP accumulation by Gib2. Altogether, our study highlights the complexity of fungal regulatory mechanisms and illustrates the importance of secondary regulatory pathways whose function may often be masked by the major regulatory pathways. This theme is very reminiscent of that of the latest study demonstrating the dual functions of S. pombe Sck1/Sch9 (61).
Adaptor proteins do not possess any catalytic domains but rather function to chaperone and tether other proteins to provide spatial and temporal regulations. The fact that Gib2 interacts with several proteins including Gpa1, Gpg1/Gpg2, Smg1, Cin1, and Pkc1 strongly suggests that Gib2 is an adaptor protein. It is also hypothesized that the β-transducin structure of Gib2 provides multiple rigid surfaces allowing for its interactions with multiple proteins. The dynamic and complex processes may be enhanced by its ability to oligomerize, forming dimeric and/or multimeric complexes. Here, we reason that Gib2 tethers with Gpa1 and Gpg1/Gpg2 to foster a heterotrimeric complex. Gib2 also recruits Ras1 to function in cAMP regulation. We anticipate that more functions can be revealed through careful investigation of various regions of Gib2 by approaches such as residue- and domain-specific mutagenesis studies.
Proteins often physically interact to carry out their functions within living cells. The human RACK1 protein is estimated to interact with as many as 80 protein partners (56). Given the multitude of RACK1 and Asc1 functions, we aim to search for evidence of more proteins that interact with Gib2. A proteomic approach using GST affinity purification combined with mass spectrometry allows us to expand the list of binding partners of Gib2 to ∼50, more than half that of RACK1 and approximately the same as S. cerevisiae Asc1. We were also able to validate approximately half of the interactive partners identified through GST affinity purification. By examining orthologs of Asc1-interacting proteins and predicting possible interlogs, we generated a protein-protein interaction network of C. neoformans Gib2 that will provide much insight into functions of Gib2.
Finally, given that C. neoformans is evolutionarily divergent from the classic model yeast S. cerevisiae, our finding represents a novel perspective on the global scale of the Gib2 interactive network. We believe that the ability of Gib2 to interact with multiple protein partners underlies the capability of the fungus to cope with changes in its living environment, particularly that of the mammalian host. Gib2 provides a means to maintain the activation/inactivation cycle of Gpa1 and promotes cAMP signaling in the absence of Gpa1. Gib2 likely influences Pkc1 signaling through conserved mechanisms to respond to stresses. Gib2 tethers Cin1, which is involved in intracellular trafficking. Given the fact that Gib2 is not an essential protein and that the gib2 mutant was able to eventually disseminate to the brain of infected AJCR mice despite slowed growth and reduced virulence, the regulatory networks underlying the growth, differentiation, and pathogenicity of C. neoformans are likely to be very complex.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI054958 and R01AI074001 (to P. W.). This work was also supported by a fund from The Research Institute for Children, Children's Hospital of New Orleans, Louisiana (to P. W.). Research in the Zhang laboratory was supported in part by an especially appointed professorship (Jiangsu, China).
- RA
- Ras association
- Y2H
- yeast two-hybrid
- PDE
- phosphodiesterase
- NAT
- nourseothricin resistance gene
- NEO
- neomycin resistance gene
- RAID
- RACK1 interaction domain
- RACK1
- receptor for activated protein C kinase 1
- IP
- immunoprecipitation
- STRING
- Search Tool for the Interacting Genes/Proteins.
REFERENCES
- 1. Park B. J., Wannemuehler K. A., Marston B. J., Govender N., Pappas P. G., Chiller T. M. (2009) Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23, 525–530 [DOI] [PubMed] [Google Scholar]
- 2. Kozel T. R. (1995) Virulence factors of Cryptococcus neoformans. Trends Microbiol. 3, 295–299 [DOI] [PubMed] [Google Scholar]
- 3. Buchanan K. L., Murphy J. W. (1998) What makes Cryptococcus neoformans a pathogen? Emerg. Infect. Dis. 4, 71–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lengeler K. B., Davidson R. C., D'souza C., Harashima T., Shen W.-C., Wang P., Pan X., Waugh M., Heitman J. (2000) Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64, 746–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kays A. M., Borkovich K. A. (2004) Signal Transduction Pathways Mediated by Heterotrimeric G Proteins, pp. 175–207, Springer-Verlag, Berlin and Heidelberg [Google Scholar]
- 6. Hoffman C. S. (2005) Except in every detail: comparing and contrasting G-protein signaling in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Eukaryot. Cell 4, 495–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gilman A. G. (1987) G-proteins: transducers of receptor-generated signals. Annu. Rev. Biochem. 56, 615–649 [DOI] [PubMed] [Google Scholar]
- 8. Alspaugh J. A., Perfect J. R., Heitman J. (1997) Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes Dev. 11, 3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang P., Perfect J. R., Heitman J. (2000) The G-protein β subunit GPB1 is required for mating and haploid fruiting in Cryptococcus neoformans. Mol. Cell. Biol. 20, 352–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hsueh Y. P., Xue C., Heitman J. (2007) G protein signaling governing cell fate decisions involves opposing Gα subunits in Cryptococcus neoformans. Mol. Biol. Cell 18, 3237–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li L., Shen G., Zhang Z. G., Wang Y. L., Thompson J. K., Wang P. (2007) Canonical heterotrimeric G proteins regulating mating and virulence of Cryptococcus neoformans. Mol. Biol. Cell 18, 4201–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xue C., Bahn Y. S., Cox G. M., Heitman J. (2006) G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol. Biol. Cell 17, 667–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alspaugh J. A., Pukkila-Worley R., Harashima T., Cavallo L. M., Funnell D., Cox G. M., Perfect J. R., Kronstad J. W., Heitman J. (2002) Adenylyl cyclase functions downstream of the Gα protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot. Cell 1, 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. D'Souza C. A., Alspaugh J. A., Yue C., Harashima T., Cox G. M., Perfect J. R., Heitman J. (2001) Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell. Biol. 21, 3179–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hicks J. K., Bahn Y. S., Heitman J. (2005) Pde1 phosphodiesterase modulates cyclic AMP levels through a protein kinase A-mediated negative feedback loop in Cryptococcus neoformans. Eukaryot. Cell 4, 1971–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Waugh M. S., Nichols C. B., DeCesare C. M., Cox G. M., Heitman J., Alspaugh J. A. (2002) Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148, 191–201 [DOI] [PubMed] [Google Scholar]
- 17. Palmer D. A., Thompson J. K., Li L., Prat A., Wang P. (2006) Gib2, a novel Gβ-like/RACK1 homolog, functions as a Gβ subunit in cAMP signaling and is essential in Cryptococcus neoformans. J. Biol. Chem. 281, 32596–32605 [DOI] [PubMed] [Google Scholar]
- 18. Guillemot F., Billault A., Auffray C. (1989) Physical linkage of a guanine nucleotide-binding protein-related gene to the chicken major histocompatibility complex. Proc. Natl. Acad. Sci. U.S.A. 86, 4594–4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ron D., Chen C. H., Caldwell J., Jamieson L., Orr E., Mochly-Rosen D. (1994) Cloning of an intracellular receptor for protein kinase C: a homolog of the β subunit of G proteins. Proc. Natl. Acad. Sci. U.S.A. 91, 839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ron D., Jiang Z., Yao L., Vagts A., Diamond I., Gordon A. (1999) Coordinated movement of RACK1 with activated βIIPKC. J. Biol. Chem. 274, 27039–27046 [DOI] [PubMed] [Google Scholar]
- 21. Liliental J., Chang D. D. (1998) Rack1, a receptor for activated protein kinase C, interacts with integrin β subunit. J. Biol. Chem. 273, 2379–2383 [DOI] [PubMed] [Google Scholar]
- 22. Chang B. Y., Conroy K. B., Machleder E. M., Cartwright C. A. (1998) RACK1, a receptor for activated C kinase and a homolog of the β subunit of G proteins, inhibits activity of src tyrosine kinases and growth of NIH 3T3 cells. Mol. Cell. Biol. 18, 3245–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chang B. Y., Harte R. A., Cartwright C. A. (2002) RACK1: a novel substrate for the Src protein-tyrosine kinase. Oncogene 21, 7619–7629 [DOI] [PubMed] [Google Scholar]
- 24. Dell E. J., Connor J., Chen S., Stebbins E. G., Skiba N. P., Mochly-Rosen D., Hamm H. E. (2002) The βγ subunit of heterotrimeric G proteins interacts with RACK1 and two other WD repeat proteins. J. Biol. Chem. 277, 49888–49895 [DOI] [PubMed] [Google Scholar]
- 25. Chen S., Dell E. J., Lin F., Sai J., Hamm H. E. (2004) RACK1 regulates specific functions of Gβγ. J. Biol. Chem. 279, 17861–17868 [DOI] [PubMed] [Google Scholar]
- 26. Chen S., Lin F., Hamm H. E. (2005) RACK1 binds to a signal transfer region of Gβγ and inhibits phospholipase C β2 activation. J. Biol. Chem. 280, 33445–33452 [DOI] [PubMed] [Google Scholar]
- 27. Yarwood S. J., Steele M. R., Scotland G., Houslay M. D., Bolger G. B. (1999) The RACK1 signaling scaffold protein selectively interacts with the cAMP-specific phosphodiesterase PDE4D5 isoform. J. Biol. Chem. 274, 14909–14917 [DOI] [PubMed] [Google Scholar]
- 28. Ceci M., Gaviraghi C., Gorrini C., Sala L. A., Offenhäuser N., Marchisio P. C., Biffo S. (2003) Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature 426, 579–584 [DOI] [PubMed] [Google Scholar]
- 29. Nilsson J., Sengupta J., Frank J., Nissen P. (2004) Regulation of eukaryotic translation by the RACK1 protein: a platform for signalling molecules on the ribosome. EMBO Rep. 5, 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zeller C. E., Parnell S. C., Dohlman H. G. (2007) The RACK1 ortholog Asc1 functions as a G-protein β subunit coupled to glucose responsiveness in yeast. J. Biol. Chem. 282, 25168–25176 [DOI] [PubMed] [Google Scholar]
- 31. Gerbasi V. R., Weaver C. M., Hill S., Friedman D. B., Link A. J. (2004) Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol. Cell. Biol. 24, 8276–8287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Valerius O., Kleinschmidt M., Rachfall N., Schulze F., López Marín S., Hoppert M., Streckfuss-Bömeke K., Fischer C., Braus G. H. (2007) The Saccharomyces homolog of mammalian RACK1, Cpc2/Asc1p, is required for FLO11-dependent adhesive growth and dimorphism. Mol. Cell. Proteomics 6, 1968–1979 [DOI] [PubMed] [Google Scholar]
- 33. McLeod M., Shor B., Caporaso A., Wang W., Chen H., Hu L. (2000) Cpc2, a fission yeast homologue of mammalian RACK1 protein, interacts with Ran1 (Pat1) kinase to regulate cell cycle progression and meiotic development. Mol. Cell. Biol. 20, 4016–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang L., Berndt P., Xia X., Kahnt J., Kahmann R. (2011) A seven-WD40 protein related to human RACK1 regulates mating and virulence in Ustilago maydis. Mol. Microbiol. 81, 1484–1498 [DOI] [PubMed] [Google Scholar]
- 35. Kwon-Chung K. J., Edman J. C., Wickes B. L. (1992) Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60, 602–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Perfect J. R., Toffaletti D. L., Rude T. H. (1993) The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect. Immun. 61, 4446–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang P., Cutler J., King J., Palmer D. (2004) Mutation of the regulator of G protein signaling Crg1 increases virulence in Cryptococcus neoformans. Eukaryot. Cell 3, 1028–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sambrook J., Russell D. G. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39. Shen G., Whittington A., Song K., Wang P. (2010) Pleiotropic function of intersectin homologue Cin1 in Cryptococcus neoformans. Mol. Microbiol. 76, 662–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Edman J. C. (1992) Isolation of telomerelike sequences from Cryptococcus neoformans and their use in high-efficiency transformation. Mol. Cell. Biol. 12, 2777–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McDade H. C., Cox G. M. (2001) A new dominant selectable marker for use in Cryptococcus neoformans. Med. Mycol. 39, 151–154 [DOI] [PubMed] [Google Scholar]
- 42. Wang P., Cox G. M., Heitman J. (2004) A Sch9 protein kinase homologue controlling virulence independently of the cAMP pathway in Cryptococcus neoformans. Curr. Genet. 46, 247–255 [DOI] [PubMed] [Google Scholar]
- 43. James P., Halladay J., Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Qian J., Cole R. B., Cai Y. (2011) Synthesis and characterization of a ‘fluorous’ (fluorinated alkyl) affinity reagent that labels primary amine groups in proteins/peptides. J. Mass Spectrom. 46, 1–11 [DOI] [PubMed] [Google Scholar]
- 45. Jensen L. J., Kuhn M., Stark M., Chaffron S., Creevey C., Muller J., Doerks T., Julien P., Roth A., Simonovic M., Bork P., von Mering C. (2009) STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dolinski K., Muir S., Cardenas M., Heitman J. (1997) All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 94, 13093–13098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bolger G. B., McCahill A., Yarwood S. J., Steele M. R., Warwicker J., Houslay M. D. (2002) Delineation of RAID1, the RACK1 interaction domain located within the unique N-terminal region of the cAMP-specific phosphodiesterase, PDE4D5. BMC Biochem. 3, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Davidson R. C., Blankenship J. R., Kraus P. R., de Jesus Berrios M., Hull C. M., D'Souza C., Wang P., Heitman J. (2002) A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148, 2607–2615 [DOI] [PubMed] [Google Scholar]
- 49. Shen G., Wang Y. L., Whittington A., Li L., Wang P. (2008) The RGS protein Crg2 regulates pheromone and cyclic AMP signaling in Cryptococcus neoformans. Eukaryot. Cell 7, 1540–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kataoka T., Broek D., Wigler M. (1985) DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell 43, 493–505 [DOI] [PubMed] [Google Scholar]
- 51. Toda T., Uno I., Ishikawa T., Powers S., Kataoka T., Broek D., Cameron S., Broach J., Matsumoto K., Wigler M. (1985) In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40, 27–36 [DOI] [PubMed] [Google Scholar]
- 52. Mintzer K. A., Field J. (1994) Interactions between adenylyl cyclase, CAP and RAS from Saccharomyces cerevisiae. Cell. Signal. 6, 681–694 [DOI] [PubMed] [Google Scholar]
- 53. Kido M., Shima F., Satoh T., Asato T., Kariya K., Kataoka T. (2002) Critical function of the Ras-associating domain as a primary Ras-binding site for regulation of Saccharomyces cerevisiae adenylyl cyclase. J. Biol. Chem. 277, 3117–3123 [DOI] [PubMed] [Google Scholar]
- 54. Alspaugh J. A., Cavallo L. M., Perfect J. R., Heitman J. (2000) RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol. Microbiol. 36, 352–365 [DOI] [PubMed] [Google Scholar]
- 55. Shen G., Zhou E., Alspaugh J. A., Wang P. (2012) Wsp1 is downstream of Cin1 and regulates vesicle transport and actin cytoskeleton as an effector of Cdc42 and Rac1 in Cryptococcus neoformans. Eukaryot. Cell 11, 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Adams D. R., Ron D., Kiely P. A. (2011) RACK1, A multifaceted scaffolding protein: structure and function. Cell Commun. Signal. 9, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Marcotte E. M., Pellegrini M., Thompson M. J., Yeates T. O., Eisenberg D. (1999) A combined algorithm for genome-wide prediction of protein function. Nature 402, 83–86 [DOI] [PubMed] [Google Scholar]
- 58. Mohseni-Zadeh S., Camonis J., Wojcik J., Barillot E. (2004) in Genome Informatics 2004, Presented at the 15th International Conference on Genome InformaticsDecember 16–18, 2004, Yokohama Pacifico, Japan, Abstr. P120, Japanese Society for Bioinformatics, Tokyo [Google Scholar]
- 59. Linder P., Slonimski P. P. (1989) An essential yeast protein, encoded by duplicated genes TIF1 and TIF2 and homologous to the mammalian translation initiation factor eIF-4A, can suppress a mitochondrial missense mutation. Proc. Natl. Acad. Sci. U.S.A. 86, 2286–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Cramer K. L., Gerrald Q. D., Nichols C. B., Price M. S., Alspaugh J. A. (2006) Transcription factor Nrg1 mediates capsule formation, stress response, and pathogenesis in Cryptococcus neoformans. Eukaryot. Cell 5, 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mudge D. K., Yang F., Currie B. M., Kim J. M., Yeda K., Bashyakarla V. K., Ivey F. D., Hoffman C. S. (2014) Sck1 negatively-regulates Gpa2-mediated glucose signaling in Schizosaccharomyces pombe. Eukaryot. Cell 13, 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tolkacheva T., McNamara P., Piekarz E., Courchesne W. (1994) Cloning of a Cryptococcus neoformans gene, GPA1, encoding a G-protein α-subunit homolog. Infect. Immun. 62, 2849–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xue C., Hsueh Y. P., Chen L., Heitman J. (2008) The RGS protein Crg2 regulates both pheromone and cAMP signalling in Cryptococcus neoformans. Mol. Microbiol. 70, 379–395 [DOI] [PMC free article] [PubMed] [Google Scholar]