Background: X-ray crystal structures of fluoroquinolone-gyrase-DNA complexes reveal a single drug-binding mode.
Results: A ciprofloxacin derivative with a chloroacetyl moiety at the C-7 end cross-linked with cysteine substitutions in both GyrA and GyrB that were 17 Å apart.
Conclusion: Cleaved complexes containing gyrase have two fluoroquinolone-binding modes.
Significance: The additional drug-binding mode provides new ways to investigate inhibitor-topoisomerase interactions.
Keywords: Antibiotic Action, Bacteria, DNA Topoisomerase, Enzyme Inhibitors, Escherichia coli, Mycobacterium smegmatis, Bacterium, Cleaved Complexes, Cross-linking, Resistance Mutations
Abstract
DNA gyrase and topoisomerase IV control bacterial DNA topology by breaking DNA, passing duplex DNA through the break, and then resealing the break. This process is subject to reversible corruption by fluoroquinolones, antibacterials that form drug-enzyme-DNA complexes in which the DNA is broken. The complexes, called cleaved complexes because of the presence of DNA breaks, have been crystallized and found to have the fluoroquinolone C-7 ring system facing the GyrB/ParE subunits. As expected from x-ray crystallography, a thiol-reactive, C-7-modified chloroacetyl derivative of ciprofloxacin (Cip-AcCl) formed cross-linked cleaved complexes with mutant GyrB-Cys466 gyrase as evidenced by resistance to reversal by both EDTA and thermal treatments. Surprisingly, cross-linking was also readily seen with complexes formed by mutant GyrA-G81C gyrase, thereby revealing a novel drug-gyrase interaction not observed in crystal structures. The cross-link between fluoroquinolone and GyrA-G81C gyrase correlated with exceptional bacteriostatic activity for Cip-AcCl with a quinolone-resistant GyrA-G81C variant of Escherichia coli and its Mycobacterium smegmatis equivalent (GyrA-G89C). Cip-AcCl-mediated, irreversible inhibition of DNA replication provided further evidence for a GyrA-drug cross-link. Collectively these data establish the existence of interactions between the fluoroquinolone C-7 ring and both GyrA and GyrB. Because the GyrA-Gly81 and GyrB-Glu466 residues are far apart (17 Å) in the crystal structure of cleaved complexes, two modes of quinolone binding must exist. The presence of two binding modes raises the possibility that multiple quinolone-enzyme-DNA complexes can form, a discovery that opens new avenues for exploring and exploiting relationships between drug structure and activity with type II DNA topoisomerases.
Introduction
DNA topoisomerases are ubiquitous enzymes whose central roles in DNA biology have made them popular targets for antibacterial and antitumor agents (1). The type II topoisomerases act by passing one region of duplex DNA through another, thereby introducing or removing DNA supercoils, catenanes, and knots (2, 3). As a part of the reaction mechanism, two single strand nicks, staggered by 4 base pairs, are introduced into DNA to provide a gate for double strand DNA passage (4, 5). Strand passage is followed by DNA resealing to prevent chromosome fragmentation when DNA is released through opening of a protein gate. The fluoroquinolones and some antitumor drugs reversibly trap type II topoisomerases on DNA as ternary complexes in which DNA is broken (6–10). Knowledge of these “cleaved complexes,” which block DNA replication and transcription, is central to establishing how quinolones and related antitopoisomerase agents act.
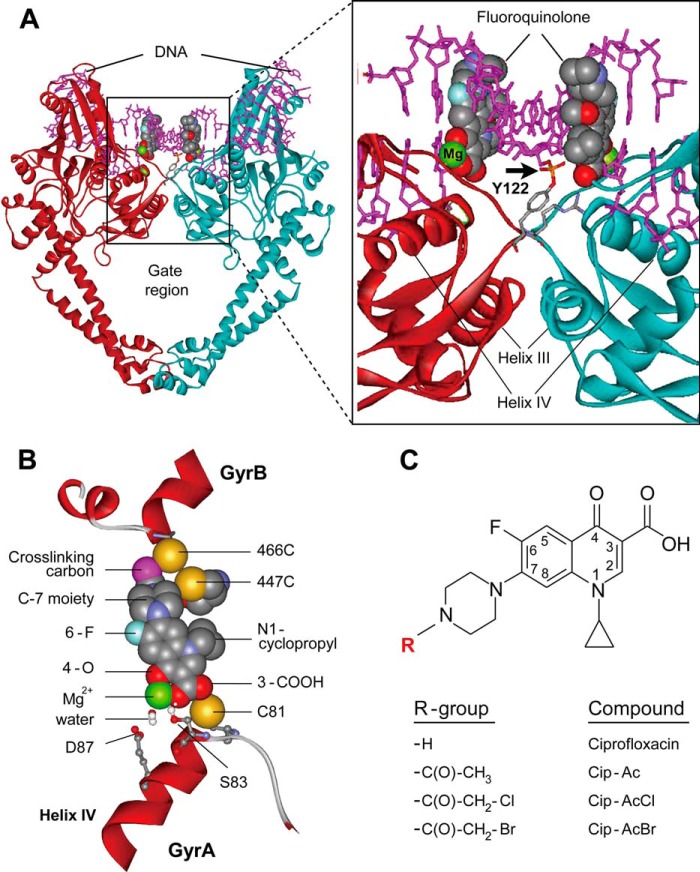
X-ray crystallography of gyrase and topoisomerase IV complexed with DNA and quinolone-class molecules provides a basis for understanding interactions between the inhibitors and their target enzymes (11–15). Gyrase is a tetramer composed of two GyrA and two GyrB subunits (the corresponding topoisomerase IV subunits are termed ParC and ParE (16)). Crystal structures of drug-enzyme-DNA complexes reveal that the drug intercalates into DNA at the nicks introduced by the topoisomerases and that the inhibitor C-7 ring system interacts with GyrB (ParE in topoisomerase IV; see Fig. 1, A and B). The distal portion of the C-7 ring system lies close to position 466 in Escherichia coli GyrB, whereas the 3-carboxyl end of the quinolone extends into GyrA (see Fig. 1B). The quinolone 3-carboxyl, along with an aspartic/glutamic acid and a serine in helix-IV of GyrA (ParC), participates in a magnesium-water bridge that stabilizes the drug-enzyme-DNA complex (17, 18). Although existing crystal structures explain many aspects of quinolone action, it is not clear why a spontaneous GyrA resistance substitution in Mycobacterium smegmatis (GyrA-Cys89) preferentially restricts the bacteriostatic action of fluoroquinolones that have bulky substitutions at the distal end of the fluoroquinolone C-7 piperazinyl ring (19): this region of the drug should be far from the GyrA-Cys89 residue in the cleaved complex. One possibility is that cleaved complexes contain additional drug-binding modes or sites not observed in currently available crystal structures.
FIGURE 1.
Structures of type II topoisomerase-DNA-quinolone complex and quinolones. A, cleaved complex. The side view of the three-dimensional structure of Acinetobacter baumannii topoisomerase IV bound to DNA and moxifloxacin (Protein Data Bank code 2XKK) is shown in which the DNA gate region is illustrated in an expanded view (right). Moxifloxacin is depicted in a space-filling representation; the arrow indicates the covalent bond between the DNA end and GyrA-Tyr122. One GyrA subunit is shown in maroon, the other is shown in turquoise, and DNA is pink. GyrB is omitted for clarity. B, enlargement of the quinolone-binding region. The proximity of GyrB cysteine substitutions to the cross-linking carbon of Cip-AcCl is shown. Other fluoroquinolone moieties and GyrA amino acids are present to provide orientation. C, structures of quinolones. Ciprofloxacin is shown along with the modified derivatives used in this work.
The present report describes a biochemical approach for mapping quinolone-protein contacts. For this work, we prepared derivatives of the fluoroquinolone ciprofloxacin (Cip)3 that contained a haloacetyl group located at the distal nitrogen of the C-7 piperazinyl ring (Cip-AcCl and Cip-AcBr; see Fig. 1C for fluoroquinolone structures). These derivatives were expected to react with cysteines in cleaved complexes if located within cross-linking distance of the bound drug. A strong interaction was observed between Cip-AcCl and GyrB-E466C gyrase, as predicted by crystal structures. Surprisingly, a strong interaction also occurred with GyrA-G81C gyrase, indicating the existence of a second binding mode. The GyrA-G81C variation is naturally occurring, which made intracellular tests straightforward: when living cells were treated with haloacetylated ciprofloxacin, a GyrA-G81C E. coli variant and the equivalent M. smegmatis variant exhibited effects consistent with GyrA-based cross-linking. Thus, two modes exist for binding of fluoroquinolone to gyrase-DNA complexes. This finding raises the possibility that an undiscovered type/conformation of cleaved complex exists. Such a new type of complex could be a key to understanding how quinolones rapidly kill bacterial cells.
EXPERIMENTAL PROCEDURES
Preparation of E. coli Gyrase
For most of the experiments described, GyrA and GyrB subunits of E. coli gyrase were expressed and purified separately as described by Schoeffler et al. (20). The subunits were combined to form gyrase. Briefly, full-length genes were cloned into an expression vector (pET His6 TEV LIC (1B), Addgene, Cambridge, MA) that was introduced into E. coli (BL21(DE3) RIL codon+) by bacterial transformation. Transformants were grown at 37 °C to midlog phase in 2× YT medium (Teknova, Hollister, CA), cultures were shifted to 18 °C, and expression was induced by addition of isopropyl 1-thio-β-d-galactopyranoside to 250 μm for 17 h. Cells were harvested by centrifugation, resuspended in A800 buffer (20 mm Tris-HCl, pH 7.9, 30 mm imidazole, pH 8, 800 mm NaCl, 10% (v/v) glycerol) plus protease inhibitors (1 mg/ml PMSF, 1 mm leukopeptin, 1 μg/ml pepstatin) and 2-mercaptoethanol (2 mm), and flash frozen in liquid nitrogen. Thawed cells were broken by sonication, debris was removed by centrifugation, and gyrase was purified by nickel affinity chromatography. The protein, eluted with a 30–300 mm imidazole gradient, was identified by absorption at 280 nm. Protein was concentrated, and the imidazole concentration was reduced using an Amicon Ultra centrifugal filter unit (Millipore, Billerica MA); overnight incubation at 4 °C with tobacco etch virus protease removed the His tag from the expressed protein. To remove unreacted protein, the preparation was passed over a His-Trap HP column (GE Healthcare) in A400 buffer (20 mm Tris-HCl, pH 7.9, 400 mm NaCl, 30 mm imidazole, pH 8, 10% (v/v) glycerol). Protein with the His tag removed was concentrated and applied to a Sephacryl S300 column in sizing buffer (50 mm HEPES, pH 7.5, 500 mm KCl, 1 mm EDTA, 10% (v/v) glycerol, 2 mm 2-mercaptoethanol) for final separation of protein. The protein peak having an appropriate molecular weight was pooled and concentrated with an Amicon centrifugal filter unit to about 200 μm GyrA or GyrB. Storage was in sizing buffer containing 33% (v/v) glycerol at −80 °C.
GyrA-G81C and GyrB-E466C were prepared by the same method used with wild-type protein. Expression vectors for these proteins were obtained using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) in which the Cys codon TGT was substituted into the nucleotide sequence of gyrA and gyrB, respectively. Both enzymes showed reduced sensitivity to ciprofloxacin with respect to complex-forming activity (9-fold for GyrB-E466C and 20-fold for GyrA-G81C). The supercoiling activity of GyrB-E466C gyrase was the same as that of the wild-type enzyme; GyrA-G81C gyrase was ∼20 times less active for supercoiling (data not shown).
For experiments shown in Fig. 3, A–C, the mutant gyrA G81C gene was constructed using the overlap extension PCR technique (21) and cloned into the pET-11c vector (22). The GyrA-G81C protein was expressed in E. coli BL21(DE3) (22). Mutant and wild-type proteins were purified as described (23, 24). Purified GyrA-G81C was mixed with wild-type GyrB to reconstitute GyrA-Cys81 gyrase. E. coli topoisomerase IV was prepared as described previously (25).
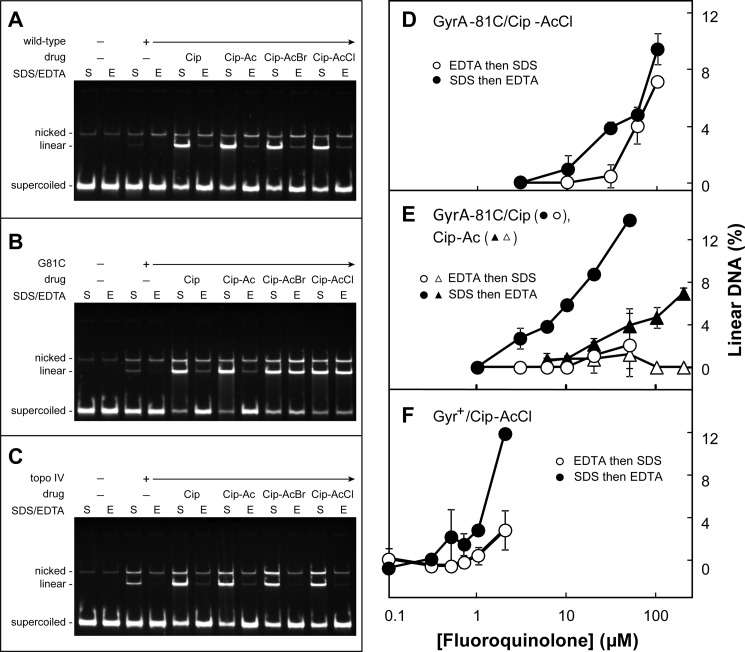
FIGURE 3.
Evidence for cross-linking of GyrA-G81C gyrase to haloacetyl derivatives of ciprofloxacin. Reaction mixtures containing gyrase or topoisomerase IV, supercoiled pBR322 DNA, and derivatives of ciprofloxacin were incubated at 37 °C as described under “Experimental Procedures.” A–C, display of plasmid DNA species following incubation with derivatives of ciprofloxacin and purified gyrase or topoisomerase IV. Concentrations of enzymes and drugs were chosen to obtain similar amounts of linear DNA. The DNA cleavage assay used 5 nm plasmid pBR322 DNA and 7.5 nm wild-type gyrase (A), 25 nm GyrA-G81C gyrase (B), or 7.5 nm topoisomerase IV (C). Ciprofloxacin was used at 1 μm for wild-type gyrase; it was 25 μm for GyrA-G81C gyrase and topoisomerase IV. Cip-Ac, Cip-AcBr, and Cip-AcCl were used at 5 μm for wild-type gyrase and 125 μm for GyrA-G81C gyrase and topoisomerase IV. S and E indicate DNA products from reaction mixtures without and with 50 mm EDTA-mediated reversal, respectively. The assay was repeated twice with identical results (less than 2% difference in the percentage of linear DNA). The effect of drug concentration on the recovery of linear DNA is shown in D–F; data were obtained as in Fig. 2 except that GyrA-G81C was substituted for GyrB-E466C. For the data shown, concentrations of enzyme and DNA were 20 and 7.4 nm, respectively; reactions were incubated for 30 min at 37 °C and then treated either with 50 mm EDTA and then SDS (empty symbols) or with SDS first and then EDTA (filled symbols). D shows the effect of the cross-linking agent Cip-AcCl with GyrA-G81C gyrase, E shows results for non-cross-linking ciprofloxacin with GyrA-G81C gyrase (circles) and for non-cross-linking Cip-Ac with GyrA-G81C gyrase (triangles), and F shows results for wild-type gyrase with the cross-linking agent Cip-AcCl. Examination of a variety of enzyme:DNA ratios and incubation times produced results similar to those shown. The number of total experiments and those used for the data shown (chosen so panels had the same incubation conditions), respectively, were 9 and 4 for the comparison of D, 10 and 2 for ciprofloxacin in E, 8 and 4 for Cip-Ac in E, and 7 and 2 for wild-type gyrase and the cross-linking agent Cip-AcCl in F. Total DNA recovery was unaffected by drug concentration, indicating that loss of specific bands was not due to preferential recovery. Error bars indicate S.D.
Fluoroquinolones
Cip and moxifloxacin (Mox) were obtained from Bayer Healthcare (Westhaven, CT). Synthesis of Cip-Ac, Cip-AcCl, Cip-AcBr, and Mox-AcCl followed methods reported previously (26) using modifications as described in the following paragraphs. Purity was determined by analytical HPLC (single >95% area peak at 285 and 210 nm).
The N-chloroacetyl derivative of ciprofloxacin (Cip-AcCl) (26) was synthesized by initially dissolving 35 mg (0.1 mmol) of ciprofloxacin hydrochloride in 1.5 ml of dimethylformamide (DMF) in the presence of 0.3 mmol of diisopropylethylamine. Chloroacetyl chloride (1.3 eq) was added followed by vigorous agitation. After 5-min incubation at ambient temperature, the mixture was poured into 10 ml of 0.1 m citric acid, and the precipitate was collected. The precipitate was washed with water, vacuum-dried, dissolved in DMF, and subjected to preparative silica gel thin layer chromatography (TLC) in a chloroform/ethanol (8:1) developing system. The product was eluted with methanol, and the solvent was removed by evaporation. The yield was 15 mg. An analytical HPLC (Restek Allure PFP propyl 5 μm, 150 × 4.6 mm) method was used as follows: 5–95% (v/v) acetonitrile, 0.1% (v/v) trifluoroacetic acid (TFA) in water over a 45-min gradient; the single product peak was at 22.73 min. Electrospray ionization MS-calculated [M + H+] was 408.11; the observed value was 408.10.
To prepare Cip-AcBr, 199 mg (0.60 mmol) of ciprofloxacin hydrochloride was stirred with 49.1 mg (1.23 mmol) of sodium hydroxide in 15 ml of water until clear. The solution was lyophilized, leaving as a white solid sodium salt of ciprofloxacin and NaCl (pretreatment to remove HCl was required to avoid halide exchange of the product during reaction, which otherwise resulted in significant quantities of Cip-AcCl). 43.6 mg (0.132 mmol) of the resulting residue was taken up in 1.5 ml of DMF and filtered through cotton to remove undissolved sodium chloride. To this solution was added 35.4 mg (0.132 mmol) of p-nitrophenylbromoacetate, the solution was stirred at 110 °C for 4 h, and then it was cooled to room temperature. The product was purified directly by semipreparative HPLC (Phenomenex Luna C18, 10 μm, 2.5 × 21.2 cm). Combined product fractions were concentrated by lyophilization to yield 16.4 mg (0.036 mmol; 28%). An analytical HPLC (Restek Allure PFP propyl 5 μm, 150 × 4.6 mm) method was used as follows: 5–95% (v/v) acetonitrile, 0.1% (v/v) TFA in water over a 45-min gradient; the single product peak was at 23.18 min. Electrospray ionization MS-calculated [M + H+] was 452.06; the observed value was 452.05.
For Cip-Ac (26), 175 mg (0.53 mmol) of ciprofloxacin hydrochloride was combined with 21.4 mg (0.54 mmol) of crushed sodium hydroxide and 4.2 mg (0.013 mmol) of tetrabutylammonium bromide and dissolved in 5 ml of DMF. The solution was stirred for 10 min and heated to 70 °C, and 199.6 mg (1.96 mmol) of acetic anhydride was added dropwise. The mixture was stirred for 50 min at 70 °C and then cooled to room temperature. The reaction mixture was diluted with water to precipitate the N-Ac-ciprofloxacin. Product was collected by filtration and then crystallized from methanol to yield 97.0 mg (0.260 mmol; 49%). An analytical HPLC (Restek Allure PFP propyl 5 μm, 150 × 4.6 mm) method was used as follows: 5–95% acetonitrile, 0.1% (v/v) TFA in water over a 45-min gradient; the single product peak was at 19.45 min. Electrospray ionization MS-calculated [M + H+] was 374.15; the observed value was 374.09.
Preparation of Mox-AcCl was initiated by supplementing a solution containing 43.8 mg (0.1 mmol) of moxifloxacin hydrochloride in 1 ml of anhydrous DMF with 30 μl (0.21 mmol) of triethylamine and 10 μl (0.125 mmol) of chloroacetyl chloride under vigorous agitation, and the resulting solution was allowed to stand at room temperature for 5 min. Then the reaction product was precipitated with 12 ml of 0.5% (w/v) aqueous citric acid. The precipitate was collected by filtration, washed several times with water, and dried in vacuo at room temperature. The yield was ∼40 mg or 80%. The product was homogenous as judged by TLC in a chloroform/ethanol (9:1) developing system. An analytical HPLC (Restek Allure PFP propyl 5 μm, 150 × 4.6 mm) method was used as follows: 5–95% acetonitrile, 0.1% (v/v) TFA in water over a 30-min gradient; the single product peak was at 20.25 min. Electrospray ionization MS-calculated [M + H+] was 478.15; the observed value was 478.14.
Cleaved Complex Formation and Detection
Reaction mixtures (20 μl) containing 2 mm MgCl2, 25 mm potassium glutamate, 15 mm Tris-HCl, pH 7.9, 2 mm ATP, 0.2 μg of supercoiled pBR322 DNA (4 nm; purified by preparative agarose gel electrophoresis), and quinolone were prepared on ice. Then gyrase (diluted in 50 mm Tris-HCl, pH 7.9, 30% (v/v) glycerol, 500 mm potassium glutamate) was added, usually at a protein:DNA ratio of 5:1, followed by incubation at 37 °C for either 30 or 45 min before mixtures were chilled on ice and sodium dodecyl sulfate (SDS) was added to 1% (w/v) to stop the reaction and denature the protein. Proteinase K was added to 100 μg/ml, and the mixtures were incubated at 37 °C for 15 min to remove protein covalently bound to linear and nicked DNA. EDTA (50 mm) was added for an additional 15-min incubation to minimize precipitation of SDS-magnesium complexes. Linear, nicked, and supercoiled plasmid species were separated by 1% (w/v) agarose gel electrophoresis using TAE buffer (0.04 m Tris acetate, 1 mm EDTA). Following electrophoresis, gels were stained with ethidium bromide (1 μg/ml) for 30 min at room temperature followed by destaining overnight in distilled water. Plasmid DNA bands were visualized using a High Performance Ultraviolet Transilluminator (354 nm; Ultraviolet Products Inc., Upland, CA), and image documentation was performed with a Kodak EDAS209 using Kodak Molecular Imaging Software (Eastman Kodak Co.). Relative fluorescence of the bands was measured, and the percentage of the linear band was determined for each lane at a series of five exposures to assure that the fluorescence was not saturating. The percentage of linear DNA was determined from an average determined with five exposures.
For experiments shown in Fig. 3, A–C, reaction mixtures (50 μl) contained 50 mm Tris-HCl, pH 8.0 at 23 °C, 10 mm MgCl2, 50 mg/liter bovine serum albumin, 1 mm ATP, 5 mg/liter tRNA, 0.3 μg of plasmid pBR322 DNA, the indicated amounts of either the wild-type gyrase, GyrA-G81C gyrase, or topoisomerase IV, and the indicated concentrations of fluoroquinolone. Mixtures were incubated at 37 °C for 15 min and then split into two samples. SDS was added at a concentration of 1% (w/v) to one half of the reaction mixtures, and the reaction mixtures were further incubated at 37 °C for 10 min. EDTA and proteinase K were then added to 50 mm and 100 mg/liter, respectively, and incubation was continued for an additional 15 min at 37 °C. The other half of the reaction mixtures was incubated for 10 min at 37 °C in the presence of 50 mm EDTA before the proteinase K treatment in the presence of SDS as described above. The DNA products were purified by extraction with phenol/chloroform/isoamyl alcohol and then analyzed by electrophoresis through vertical 1.2% (w/v) agarose gels at 2 V/cm for 12 h in TAE buffer that contained 0.5 mg/liter ethidium bromide. After destaining in water, gels were photographed and quantified using an Eagle Eye II system (Agilent Technologies).
Bacteriological Methods
E. coli K12 strains (Table 1) were grown in LB liquid medium or on LB agar plates (27). Growth was at 37 °C except for DNA synthesis experiments, which were carried out at 30 °C. M. smegmatis strains, also listed in Table 1, were grown at 37 °C in 7H9 liquid medium and on 7H10 agar plates, in both cases in the presence of 10% (v/v) albumin-dextrose-catalase (28). Mutant strains for both species were obtained as spontaneous fluoroquinolone-resistant mutants by single step selection on fluoroquinolone-containing agar. Bacteriophage P1-mediated transduction of mutant genes was used for construction of isogenic E. coli strains (29). Mutations were identified by nucleotide sequence analysis following amplification of gyrA by PCR.
TABLE 1.
Bacterial strains used in the study
| Strain number | Relevant genotype | Source or Ref. |
|---|---|---|
| E. coli | ||
| DM4100 | Wild type | 48 |
| KD1915 | DM4100 gyrA (G81C) | 49 |
| KD1973 | DM4100 gyrA (D82A) | 49 |
| KD2372 | DM4100 lon− | 34 |
| KD3052 | KD1915 lon− | This worka |
| KD3276 | DM4100 gyrA (G81D) | This workb |
| M. smegmatis | ||
| mc2155 | Wild type (KD1163) | S. Colec |
| KD1987 | mc2155 gyrA (A91V) | 19, 50 |
| KD2008 | mc2155 gyrA (G89C) | 19, 50 |
| KD2012 | mc2155 gyrA (G89A) | 19, 50 |
The minimal inhibitory concentration (MIC) was determined by broth dilution. Cells were grown to midexponential phase, diluted to about 105 CFU/ml in tubes containing fluoroquinolone, and incubated overnight at 37 °C. Growth was determined by visual inspection with the lowest fluoroquinolone concentration that blocked growth taken as the MIC.
DNA synthesis rate was measured as incorporation of [3H]thymidine into acid-precipitable radioactivity during a 2-min pulse of aliquots removed from cultures (9). DNA synthesis drops to a nadir rapidly (within minutes) and remains constant (9), thereby allowing reversal to be detected following removal of fluoroquinolone by rapid filtration of cultures and resuspension in drug-free growth medium.
Molecular Modeling
The structure of fluoroquinolone-gyrase cleaved complex was obtained from Protein Data Bank code 2XKK (14) using the C-8-H and C-7 ring structure of ciprofloxacin rather than moxifloxacin. Docking of ciprofloxacin and derivatives to E. coli gyrase in “GyrA-GyrA bridging pocket” and “inverted” modes was performed manually on the 2XKK structure using WebLab ViewerLight.
RESULTS
Cleaved Complexes Formed with Cip-AcCl and Purified GyrB-E466C Gyrase Resist EDTA-mediated Reversal
To study the immediate surroundings of fluoroquinolones within cleaved complexes, we prepared cysteine substitutions in the B subunit of gyrase near the drug binding site defined by crystallography (14) (Fig. 1B). We then attached a cysteine-reactive alkylating group to the distal nitrogen of the C-7 ring of ciprofloxacin, producing a compound designated as Cip-AcCl (Fig. 1C), to probe drug-enzyme contacts. According to crystal structures, the chloroacetyl group of Cip-AcCl is expected to be in close proximity to GyrB amino acid Glu466 such that a GyrB-E466C variant gyrase would cross-link with Cip-AcCl. Close proximity was also expected from our finding that cleaved complex formation with GyrB-E466C gyrase is sensitive to C-7 ring structure.4
To measure cleaved complex formation and reversal, plasmid DNA was incubated with gyrase and various concentrations of fluoroquinolone. After allowing DNA binding and DNA cleavage, one set of reaction mixtures received SDS to denature gyrase in the cleaved complexes and release double strand DNA breaks. The fraction of DNA in the linear form corresponds to the fraction of plasmids containing a cleaved complex. In these experiments, the fraction of plasmids with multiple complexes was kept low to avoid complications arising from multiple DNA cuts. A parallel set of reaction mixtures, also incubated to allow DNA binding and cleavage, was subsequently incubated for 15 min with 50 mm EDTA to reseal the paired DNA nicks and reverse cleaved complex formation. SDS was then added to release DNA breaks that had not been resealed, and proteinase K was added to all samples to remove gyrase covalently linked to DNA ends via GyrA-Tyr122 (Fig. 1A enlargement, heavy arrow). Gel electrophoresis of reaction products allowed the percentage of linear DNA to be determined.
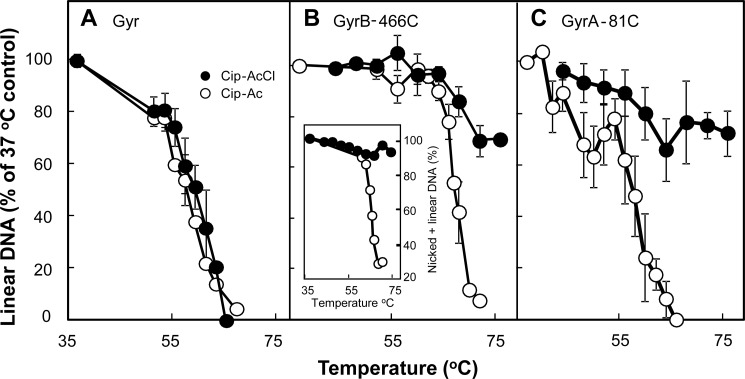
In an initial experiment, electrophoretic analysis of reaction mixtures showed increasing amounts of cleaved DNA with increasing fluoroquinolone concentration when reactions were stopped with SDS (Fig. 2, filled symbols). Increasing amounts of linear DNA were also observed when cleaved complexes were formed with GyrB-E466C gyrase and Cip-AcCl, and then reaction mixtures were treated with EDTA (Fig. 2A, empty circles). These data indicate the formation of cleaved complexes with GyrB-E466C gyrase and Cip-AcCl that were not reversed by EDTA. In contrast, little DNA escaped EDTA-mediated DNA resealing when reactions contained GyrB-E466C gyrase and either ciprofloxacin or Cip-Ac (Fig. 2B), a nonreactive structural analog of Cip-AcCl, or when reactions contained wild-type gyrase treated with Cip-AcCl (Fig. 2C).
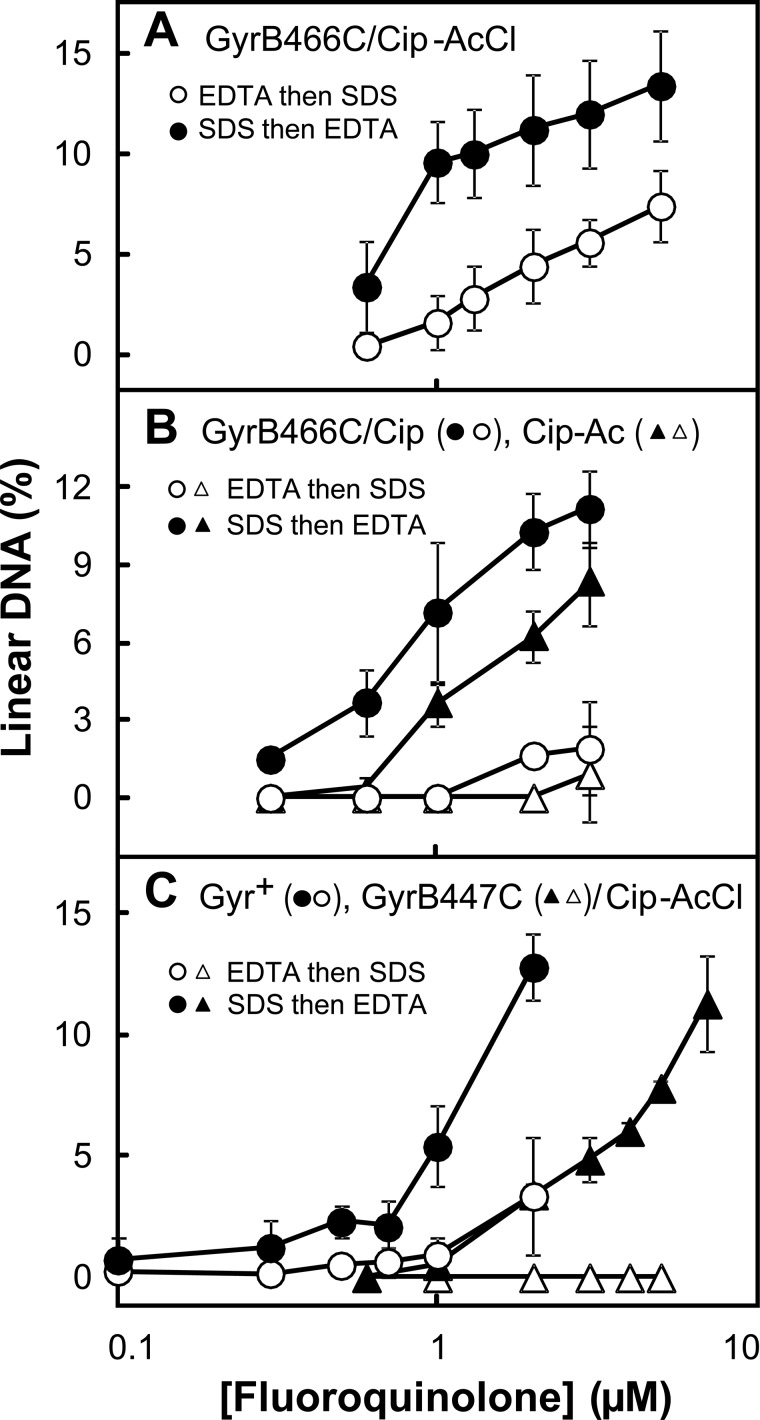
FIGURE 2.
Evidence for cross-linking of GyrB-E466C gyrase to Cip-AcCl. Mixtures containing gyrase, gel-purified supercoiled pBR322 DNA, and ciprofloxacin-based derivatives were incubated at 37 °C. Then reaction mixtures were treated with either SDS and proteinase K (filled symbols) to release DNA breaks generated by drug-gyrase action or with EDTA (empty symbols) to reseal DNA breaks. The SDS-treated samples were then treated with EDTA, and the EDTA samples were treated with SDS and proteinase K before samples were subjected to gel electrophoresis to separate the DNA species. The percentage of linear DNA within each sample was determined as a measure of DNA breaks generated by gyrase-drug action at the indicated drug concentrations. In the data shown, concentrations of gyrase and DNA were 20 and 7.4 nm, respectively; reactions were incubated for 15 min. Error bars indicate S.D. A shows GyrB-E466C gyrase incubated with the cross-linking agent Cip-AcCl; B shows results for GyrB-E466C gyrase with two non-cross-linking agents, ciprofloxacin (circles) and Cip-Ac (triangles); and C shows wild-type gyrase (circles) or GyrB-Cys447 gyrase (triangles) with the cross-linking agent Cip-AcCl. Examination of a variety of enzyme:DNA ratios and incubation times produced results similar to those shown. The number of total experiments and those used for the data shown (chosen so panels had the same conditions), respectively, were 14 and 5 for the comparison of A, 7 and 4 for ciprofloxacin in B, 4 and 4 for Cip-Ac in B, 3 and 3 for GyrB-Cys447 gyrase in C, and 11 and 4 for wild-type gyrase in C.
The specificity of the interaction between Cip-AcCl and GyrB-E466C gyrase was examined with gyrase having a GyrB-K447C substitution, which is located near the quinolone binding site but not as close as GyrB-E466C (Fig. 1B). Complexes formed with Cip-AcCl and GyrB-K447C gyrase were reversed by EDTA treatment (Fig. 2C). Collectively these data indicate that Cip-AcCl forms cross-links with a specific Cys-containing GyrB variant as predicted by x-ray crystallography of cleaved complexes. We next used Cip-AcCl to probe quinolone interactions with a Cys-containing variant of GyrA.
Cleaved Complexes Formed with Purified GyrA-G81C Gyrase and Cip-AcCl or Cip-AcBr Resist EDTA-mediated Reversal
In a preliminary characterization, we found that the E. coli GyrA-G81C substitution drastically reduced the quinolone sensitivity of gyrase: IC50 values for ciprofloxacin with wild-type and GyrA-G81C gyrase in a supercoiling reaction (30) were 0.45 and 28 μm, respectively. This difference was consistent with the loss of susceptibility observed with cultured cells (31), and it was used to adjust quinolone and enzyme concentrations to form similar amounts of cleaved complex in reaction mixtures containing wild-type or mutant gyrase. Electrophoretic analysis of reaction products showed that subsequent EDTA treatment resealed most of the linear plasmid DNA when cleaved complexes were formed with wild-type gyrase and ciprofloxacin, Cip-Ac, Cip-AcCl, or Cip-AcBr (Fig. 3A) or when formed with GyrA-G81C gyrase and either ciprofloxacin or Cip-Ac (Fig. 3B). However, the majority of cleaved complexes formed with GyrA-G81C gyrase and Cip-haloacetyl derivatives were not reversed by EDTA treatment (>90% for Cip-AcCl and >80% for Cip-AcBr; Fig. 3B and Table 2). As expected, irreversible complexes were not detected with wild-type topoisomerase IV (Fig. 3C), which lacks a cysteine at the position equivalent to GyrA81. A cysteine does occur naturally in topoisomerase IV at the ParC equivalent of GyrA85. Thus, the absence of cross-linking with this enzyme indicates that the location of the Cys residue is important for formation of irreversible complexes.
TABLE 2.
Reversibility of DNA cleavage
| Enzyme | Reversal of plasmid DNA cleavage |
|||
|---|---|---|---|---|
| Cip | Cip-Ac | Cip-AcCl | Cip-AcBr | |
| % | ||||
| Wild-type gyrase | 86.4 | 87.2 | 89.2 | 86.7 |
| GyrA-G81C gyrase | 82.5 | 85.3 | 4.5 | 11.9 |
| Topoisomerase IV | 88.7 | 87.3 | 96.4a | 94.5a |
a Note that with topoisomerase IV cleavage by Cip-AcCl and Cip-AcBr is completely reversible even though E. coli topoisomerase IV contains a cysteine at ParC position 82, which corresponds to GyrA position 85. Thus, cross-linkable fluoroquinolones have limited access to regions in helix-IV.
We also measured the effect of quinolone concentration on EDTA-mediated DNA resealing following cleaved complex formation. Reaction mixtures containing gyrase, plasmid DNA, and fluoroquinolone were incubated to form cleaved complexes, and as above, they were treated with EDTA before or after SDS-proteinase treatment. DNA resealing was then monitored by gel electrophoresis as a reduction in linear DNA. When GyrA-G81C gyrase was incubated with Cip-AcCl, the EDTA-resistant fraction appeared at the same fluoroquinolone concentration as total linear DNA (linear DNA detected when SDS was added before EDTA) once a threshold was passed (Fig. 3D). In contrast, treatment of wild-type gyrase with Cip-AcCl or mutant GyrA-G81C gyrase with Cip-Ac or ciprofloxacin produced much less EDTA-resistant linear DNA (Fig. 3, E and F). The latter data show that GyrA-G81C gyrase readily reseals DNA even though it has reduced supercoiling activity: the lack of resealing with Cip-AcCl is due to cross-linking, not weakened enzymatic activity of the mutant enzyme. In summary, EDTA-irreversible cleaved complexes are a large fraction of the total complexes only with the combination of GyrA-G81C gyrase and Cip-AcCl.
To determine whether the interaction between Cip-AcCl and GyrA-G81C gyrase is unique to derivatives of ciprofloxacin, we prepared a chloroacetyl derivative of moxifloxacin (Mox-AcCl) and incubated it with mutant gyrase and plasmid DNA. EDTA treatment failed to reverse ternary complexes formed with Mox-AcCl and GyrA-G81C gyrase (data not shown). In contrast, complexes were reversed by EDTA when moxifloxacin was incubated with GyrA-G81C gyrase or when Mox-AcCl was incubated with wild-type gyrase. We conclude that the ability to form EDTA-resistant complexes with GyrA-G81C gyrase extends from ciprofloxacin to other fluoroquinolones and that it applies to acetyl derivatives containing either chlorine or bromine.
Time Course of Cleaved Complex Formation with Cip-AcCl
The data described above suggest that fluoroquinolones have two binding modes in cleaved complexes. As an additional test, we examined the rate of cleaved complex formation under cross-linking conditions. Reaction mixtures containing plasmid DNA, Cip-AcCl, and one of the three gyrase variants were incubated for various times before reaction mixtures were subsequently incubated with either SDS and then EDTA or EDTA and then SDS. As above, reaction products were separated by gel electrophoresis, and linear DNA was quantified as an indicator of cleaved complex accumulation.
In an initial, baseline characterization, overall cleaved complex formation (SDS added before EDTA) was measured at the same concentration of Cip-AcCl (10 μm) for all three enzymes. At that concentration, complex formation was about 2 times faster with wild-type gyrase than with GyrB-E466C gyrase, which was in turn about 50 times faster than complex formation with GyrA-G81C gyrase (Fig. 4A). Slower complex formation by the GyrA-G81C enzyme is consistent with comparable supercoiling in the absence of drug requiring 20 times more mutant than wild-type enzyme for short (30-min) incubations.
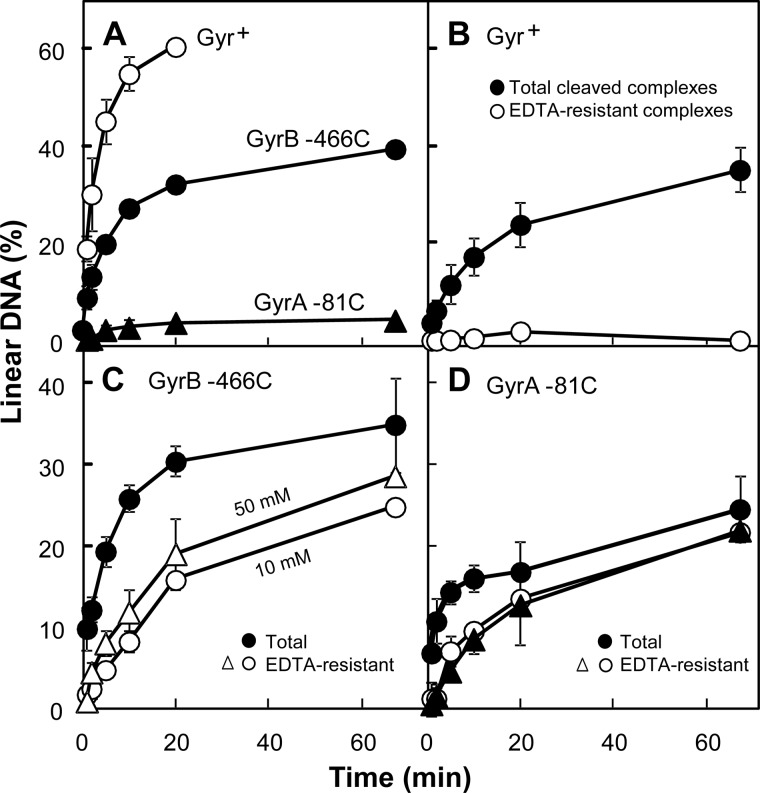
FIGURE 4.
Time course of formation of cleaved complexes containing Cip-AcCl. Reaction mixtures were prepared as in Fig. 2 using wild-type, GyrA-G81C, or GyrB-E466C gyrase, pBR322, and the cross-linking compound Cip-AcCl. Mixtures were then incubated at 37 °C for various times and processed to reveal the percentage of total linear DNA or the percentage of linear DNA remaining after reversal by EDTA. A, total cleaved complex formation. Gyrase (40 nm), supercoiled pBR322 DNA (3.7 nm), and Cip-AcCl (10 μm) were incubated for the indicated times followed by treatment with SDS and proteinase K for an additional 15 min. Empty circles, wild-type gyrase; filled circles, GyrB-E466C gyrase; filled triangles, GyrA-G81C gyrase. B, EDTA-resistant and total complexes with wild-type gyrase. Reaction mixtures, prepared as in A but with 2 μm Cip-AcCl, were incubated for the indicated times followed by an additional incubation with 10 mm EDTA for 15 min and then with SDS-proteinase K for another 15 min (empty circles). Parallel samples of reaction mixtures were incubated with SDS-proteinase before EDTA treatment (filled circles) as a measure of total complexes. C, EDTA-resistant and total complexes with GyrB-E466C gyrase. Reaction mixtures with GyrB-E466C gyrase, prepared as in A, were incubated for the indicated times followed by an additional incubation with 10 mm EDTA (empty circles) or 50 mm EDTA (empty triangles) for 15 min and then with SDS-proteinase K for another 15 min (filled circles). Parallel samples of reaction mixtures were incubated with SDS-proteinase before EDTA treatment (filled circles) to assess total complexes. D, EDTA-resistant and total complexes with GyrA-G81C gyrase. Conditions and treatments were as in C except for the use of 40 μm Cip-AcCl and 80 nm GyrA-G81C gyrase. Empty circles, 10 mm EDTA before SDS; filled triangles, 50 mm EDTA before SDS; filled circles, SDS before EDTA. Error bars indicate S.D. for four experiments.
To examine the rate of cross-linking, reaction mixtures containing Cip-AcCl were incubated for various times and then treated with 10 mm EDTA for 15 min rather than being stopped immediately with SDS. In this experiment, the concentration of Cip-AcCl was adjusted so that each enzyme would produce similar levels of total complex, defined as the percentage of DNA in the linear form when reactions were stopped immediately with SDS. With wild-type gyrase treated with 2 μm Cip-AcCl, almost all of the DNA breaks were resealed by EDTA treatment (Fig. 4B). As expected, EDTA-resistant complexes accumulated more rapidly and to a greater fraction of total cleaved complexes with GyrB-E466C gyrase than with wild-type enzyme (Fig. 4, compare B and C). However, the EDTA-resistant complexes formed with GyrB-E466C gyrase accumulated more slowly than total complexes (Fig. 4C, compare circles). With GyrA-G81C gyrase, we increased Cip-AcCl concentration to 40 μm and doubled the gyrase concentration because this gyrase construct is less active than the others. Under these conditions, formation of EDTA-resistant complexes exhibited a short lag and then reached the level seen for total complexes (Fig. 4D, circles). Thus, producing comparable levels of cleaved complexes requires more drug and enzyme with the GyrA than with the GyrB variant, but Cip-AcCl-Cys cross-linking appears to occur more quickly with GyrA-G81C gyrase once complexes form. This difference between the gyrase variants is consistent with Cip-AcCl forming distinct cross-linked complexes with GyrB-E466C and GyrA-G81C gyrases.
In separate work, we found that DNA resealing is more extensive at 10 mm EDTA than at 50 mm (DNA resealing displays an optimal response to EDTA concentration4), which provided another way to compare cleaved complexes formed with GyrB-E466C gyrase and GyrA-G81C gyrase. When we determined EDTA-resistant complex formation at the two EDTA concentrations, more cleaved complexes accumulated at 50 mm EDTA for the GyrB-E466C gyrase/Cip-AcCl combination (Fig. 4C). In contrast, little difference was observed for the GyrA-G81C gyrase/Cip-AcCl combination (Fig. 4D). These EDTA concentration effects are consistent with distinct cross-linked complexes forming with GyrA-G81C and GyrB-E466C gyrases. We speculate that cross-linking is slower with GyrB-E466C gyrase (relative to complex formation) and that 10 mm EDTA allows more competing DNA resealing to occur.
Cip-AcCl Forms Cleaved Complexes That Resist Thermal Reversal
Cleaved complexes formed with gyrase, DNA, and quinolone are also reversed by incubation at high temperature (17, 18) presumably due to drug dissociation. Thus, thermal DNA resealing provides an independent test for cross-linking between Cip-AcCl and Cys residues (GyrB E466C and GyrA G81C). When reversible complexes were formed at 37 °C with Cip-AcCl and wild-type gyrase, incubated at various temperatures for 5 min, and finally treated with SDS, DNA resealing was observed as the disappearance of linear plasmid DNA and the concomitant accumulation of supercoiled DNA (data not shown). Nicked DNA was a minor fraction that was largely unchanged by temperature. With wild-type gyrase, no difference was observed between Cip-AcCl and Cip-Ac for thermal DNA resealing when measured as a decrease in linear DNA (Fig. 5A).
FIGURE 5.
Effect of temperature on DNA resealing. Reaction mixtures containing gyrase, plasmid DNA, and derivatives of ciprofloxacin were incubated for 45 min at 37 °C followed by an additional 5-min incubation at the indicated temperature. Reactions were stopped by chilling on ice; additional SDS-proteinase K treatment and electrophoretic analysis were as described under “Experimental Procedures.” The percentage of DNA in the linear form was determined for each temperature; data were then normalized to the percentage of linear DNA observed at 37 °C to allow comparison of thermal resealing curves containing different levels of linear DNA prior to thermal treatment. A, wild-type gyrase (40 nm) and pBR322 (3.7 nm) were incubated with Cip-Ac (6 μm; empty circles) or Cip-AcCl (6 μm; filled circles). B, GyrB-E466C gyrase (40 nm) and pBR322 DNA (3.7 nm) were incubated with Cip-Ac (80 μm; empty circles) or Cip-AcCl (10 μm; filled circles). The inset shows the effect of thermal incubation on the sum of linear plus nicked DNA for complexes formed with Cip-Ac (empty circles) and Cip-AcCl (filled circles). C, GyrA-G81C gyrase (80 nm) and pBR322 (3.7 nm) were incubated with Cip-Ac (700 μm; empty circles) or Cip-AcCl (500 μm; filled circles). Elevated quinolone concentrations were required to obtain linear DNA at levels similar to those used in A and B (above 30%). Error bars indicate S.D. for the following number of determinations: A, Cip-AcCl, 4; Cip-Ac, 3; B, Cip-AcCl, 4; Cip-Ac, 3; C, Cip-AcCl, 8; Cip-Ac, 5.
When reaction mixtures containing GyrB-E466C gyrase were incubated at 37 °C and then at various temperatures for 5 min, DNA resealing was clearly observed with the non-cross-linking compound Cip-Ac, although the resealing temperature was about 5 °C higher than that seen with wild-type gyrase (Fig. 5B). With Cip-AcCl, little DNA resealing occurred at temperatures up to 70 °C. Above 70 °C, a moderate decrease (∼20%) in the linear DNA fraction was accompanied by an increase in nicked DNA (data not shown), indicating that the interaction between Cip-AcCl and GyrB-E466C is partially reversed by high temperature. Notably, the sum of linear and nicked DNA formed with GyrB-E466C gyrase and Cip-AcCl showed very little effect of temperature, whereas with the control compound Cip-Ac, the sum of linear plus nicked DNA dropped significantly (Fig. 5B, inset). Thus, consideration of either linear DNA or the sum of linear plus nicked DNA indicated that DNA resealing is restricted by Cip-AcCl. This finding is consistent with cross-linking, and it supports the conclusion that the distal end of the quinolone C-7 ring is near GyrB position 466 in cleaved complexes, in accordance with published crystal structures.
Cleaved complexes formed with GyrA-G81C gyrase and Cip-AcCl showed strong resistance to temperature; over the same temperature range, DNA resealing occurred readily with the control compound Cip-Ac (Fig. 5C). These data support the conclusion that cross-linking occurs between the C-7 ring system of fluoroquinolones and GyrA81. Because the result shown in Fig. 5C cannot be explained by crystal structures, we conclude that an additional, fluoroquinolone C7-GyrA81-mediated-binding mode occurs in cleaved complexes.
We note that DNA resealing in cleaved complexes formed with the control compound Cip-Ac and GyrA-G81C gyrase exhibited a complex response: at intermediate temperatures, a peak occurred in the resealing profile (Fig. 5C). The peak was not observed when the same experiment was carried out with ciprofloxacin (data not shown). This finding indicates that the acetyl moiety on the C-7 ring of Cip-Ac is responsible for the peak. Because the GyrB-E466C alteration increases the stability of the complexes formed with Cip-Ac, we examined thermal reversal with GyrB-E466C GyrA-G81C gyrase. The size of the peak was reduced but not eliminated, as would be expected if the GyrB-E466C subunit dominated the interaction (data not shown). A complex relationship appears to exist among the non-cross-linking compound Cip-Ac and the two mutant gyrases.
Enhanced Bacteriostatic Activity of Cip-AcCl and Cip-AcBr with Cysteine-containing GyrA Variants
To determine whether the strong interaction between GyrA-Gly81 and quinolone C-7 ring systems also occurs in living cells, we measured bacteriostatic activity (MIC) for Cip-AcCl and Cip-AcBr with several gyrA mutants of E. coli. The cysteine substitution at GyrA-Gly81 raised MIC by 16-fold for ciprofloxacin and Cip-Ac, but it had no effect on the MIC of Cip-AcCl and Cip-AcBr (Fig. 6). Several other amino acid substitutions in GyrA raised the MIC for all four compounds, showing no preferential effect with Cip-AcCl and Cip-AcBr (Fig. 6). These are the results expected for cross-linking occurring between the haloacetyl derivatives and GyrA-G81C.
FIGURE 6.
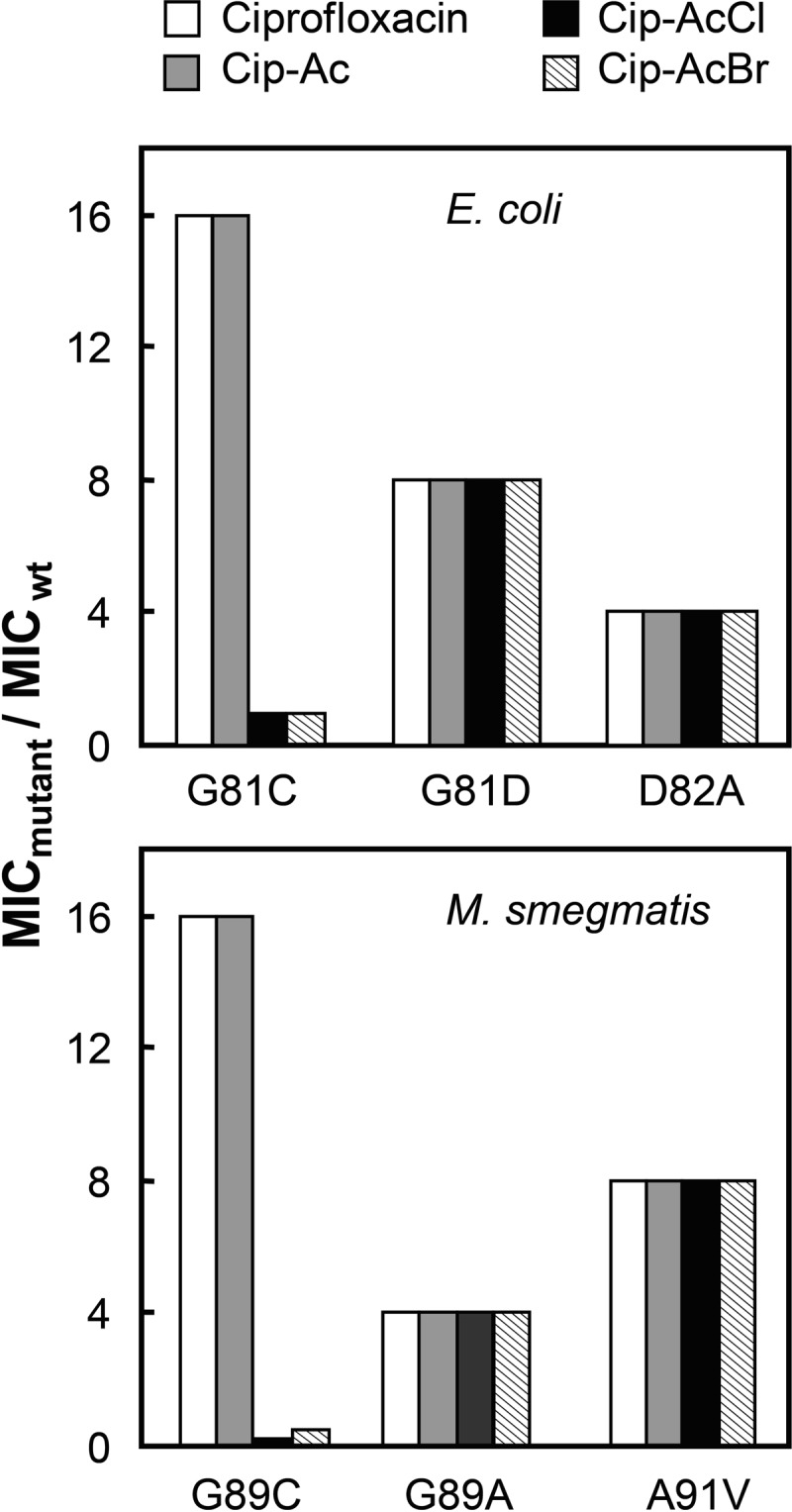
Effect of chloroacetyl and bromoacetyl substituents attached to ciprofloxacin on growth inhibition of GyrA Cys variants. The MIC was determined with the indicated mutants for ciprofloxacin derivatives (white bars, ciprofloxacin; gray bars, Cip-Ac; black bars, Cip-AcCl; striped bars, Cip-AcBr) with E. coli (upper panel) and M. smegmatis (lower panel). Data are expressed as multiples of values for MIC determined with wild-type E. coli strain DM4100 (MIC = 0.01 μg/ml for ciprofloxacin, 0.3 μg/ml for Cip-Ac, 0.25 μg/ml for Cip-AcCl, and 0.5 μg/ml for Cip-AcBr) or wild-type M. smegmatis strain mc2155 (0.1 μg/ml for ciprofloxacin, 0.16 μg/ml for Cip-Ac, 0.5 μg/ml for Cip-AcCl, and 0.5 μg/ml for Cip-AcBr). For strain numbers, see Table 1. Three independent experiments gave identical results, precluding determination of error bars.
Comparable results were obtained with M. smegmatis. Previous work with this organism indicated that substitution of Cys for Gly at position 89, the equivalent of position 81 in E. coli, increases ciprofloxacin and moxifloxacin MIC by 32-fold (19, 32). In the present work, the MIC was 16 times higher with the mutant for ciprofloxacin and the non-cross-linkable derivative Cip-Ac (Fig. 6), whereas the MIC for Cip-AcCl and Cip-AcBr with the GyrA-G89C variant dropped below that observed with wild-type cells (Fig. 6). Other naturally occurring resistance substitutions at position 89 (GyrA-G89A) or nearby at 91 (GyrA-A91V) were not expected to cross-link with either haloacetyl quinolone; their introduction caused substantial loss of susceptibility (increased MIC) to all four compounds (Fig. 6). These data extend our cross-linking conclusions beyond E. coli to mycobacteria.
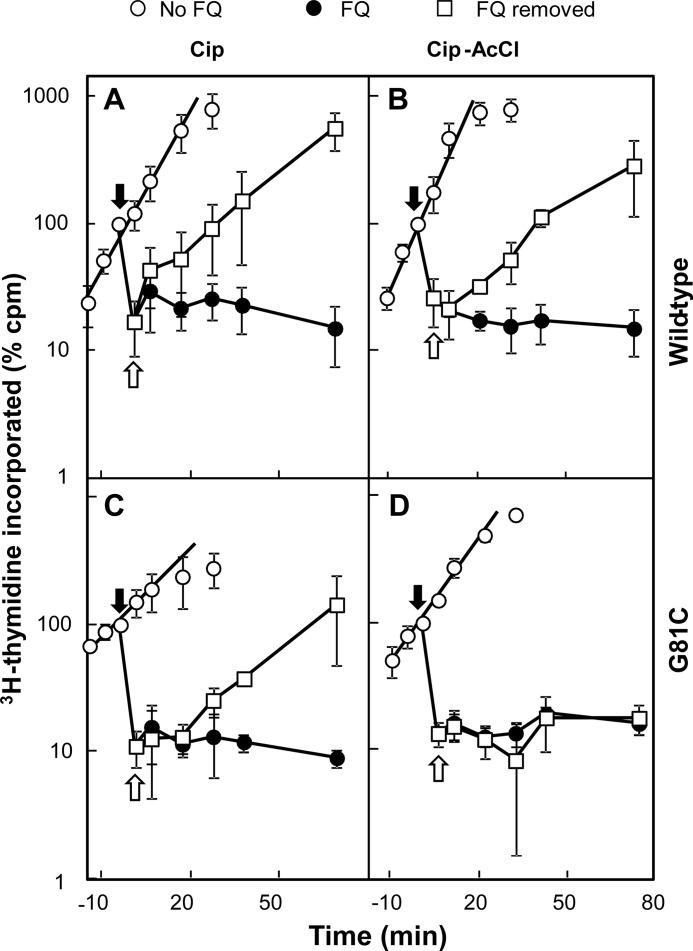
Irreversible Inhibition of Intracellular DNA Synthesis by Cip-AcCl with Cysteine-containing Variant of GyrA81
As an additional test for intracellular cross-linking, we examined the effect of Cip-AcCl on DNA synthesis. The addition of quinolone to growing cultures of E. coli immediately halts DNA synthesis (9, 33); however, such inhibition is reversible (33). Cross-linking of fluoroquinolone to gyrase was expected to form an irreversible block to DNA replication because cross-linked drug cannot be washed away. To test this prediction, we treated exponentially growing cultures of E. coli with ciprofloxacin or Cip-AcCl, and at various times, aliquots were withdrawn for determination of the DNA synthesis rate. As shown in Fig. 7, both compounds, added at time 0 (filled arrow) rapidly blocked DNA synthesis of either strain KD2372 (GyrA+) or strain KD3052 (GyrA-G81C), both of which were deficient in Lon protease to slow the repair of quinolone-gyrase-DNA complexes (34). After 5 min of drug treatment (open arrow), fluoroquinolones were removed by rapid filtration, cells were incubated in growth medium with shaking, and at various times the rate of recovery DNA synthesis was measured. Both GyrA+ and GyrA-G81C cells resumed DNA synthesis after removal of ciprofloxacin (Fig. 7, A and C) or the non-cross-linkable control compound Cip-Ac (data not shown). Resumption of DNA synthesis was also observed with GyrA+ cells following treatment and removal of Cip-AcCl (Fig. 7B). In contrast, DNA synthesis failed to resume following removal of Cip-AcCl from the growth medium of the GyrA-G81C mutant (Fig. 7D). Thus, cleaved complexes containing Cip-AcCl and GyrA-G81C gyrase are irreversible, unlike complexes formed by combinations of gyrase and quinolone that cannot form cross-links. We conclude that the fluoroquinolone C-7 ring can interact with residues at position GyrA81 inside living bacterial cells.
FIGURE 7.
Cip-AcCl preferentially blocks reversal of DNA synthesis inhibition with GyrA81 variant. E. coli strains KD2372 (gyrA+) and KD3052 (GyrA-G81C) were grown as liquid cultures that were treated at time 0 (filled arrow) with either ciprofloxacin (A and C) or Cip-AcCl (B and D) at 10× MIC. Five min later (empty arrow) an aliquot of treated cells was collected by filtration, and the cells were quickly resuspended in drug-free medium. At various times, aliquots were removed, and the DNA synthesis rate was measured as described under “Experimental Procedures.” Empty circles, untreated control; filled circles, treated with fluoroquinolone (FQ) at time 0; empty squares, treated with fluoroquinolone for 5 min, and then drug was removed by filtration. Error bars indicate S.D. from four determinations.
DISCUSSION
Formation of cleaved complexes containing drug, topoisomerase, and DNA is the central step in quinolone action as these complexes block transcription, DNA replication, and growth (for a review, see Ref. 35). A similar phenomenon likely occurs with several antitumor agents and human type II topoisomerases (36). Because cleaved complex formation can be reversed in several ways and assayed as resealing of plasmid DNA, exceptionally strong drug-enzyme interactions can be readily observed. The key finding in the present work was that a single type of fluoroquinolone, a C-7 chloroacetyl derivative of ciprofloxacin (Cip-AcCl), formed strong interactions, most likely cross-links, with two parts of gyrase that are far apart (17 Å according to crystal structures (14)). This observation indicates that two modes of drug binding occur in gyrase-containing cleaved complexes. To date, only one binding mode has been observed using x-ray crystallography (11–15); however, a second was anticipated from quinolone structure-activity relationships obtained with cultured M. smegmatis (19). Below we briefly discuss the methodology used to probe drug-topoisomerase interactions and present models for drug binding in gyrase-containing cleaved complexes.
Assays for Irreversible Cleaved Complexes Reveal Distinct Fluoroquinolone-binding Modes
The present study used both high (50 mm) and moderate (10 mm) concentrations of EDTA to examine irreversibility of interactions between Cip-AcCl and cysteine residues. These concentrations flanked the 35 mm concentration used previously to cause DNA resealing (37), and both were suitable for demonstrating irreversibility. A difference emerged between GyrB-E466C and GyrA-G81C gyrases: with the GyrB variant, accumulation of irreversible complexes, defined by the detection of linear DNA following treatment with EDTA, was slower than accumulation of total complexes (assayed by addition of SDS immediately after complex formation; Fig. 4C). This kinetic difference was smaller with the GyrA-G81C/Cip-AcCl combination (Fig. 4D). Raising the EDTA concentration from 10 to 50 mm had a small “accelerating” effect on irreversible complex accumulation with the GyrB-E466C/Cip-AcCl combination perhaps by retarding a competing DNA resealing reaction (at 50 mm, EDTA allows less resealing by gyrase than at 10 mm4). With the GyrA interaction, raising the EDTA concentration had little effect. Thus, the GyrA and GyrB sites differ with respect to cross-link accumulation.
Another difference between the two mutant enzymes was seen by comparing the fraction of complexes resistant to reversal by EDTA. For example, after 60-min incubation followed by treatment with 10 mm EDTA, the EDTA-resistant fraction remained at about half the total with GyrB-E466C gyrase (Fig. 4C). In contrast, with GyrA-G81C gyrase the fraction of EDTA-resistant complexes approached that of the total cleaved complexes (Fig. 4D). In both cases, accumulation of EDTA-resistant complexes lagged behind accumulation of total complexes. This behavior is consistent with an initial fluoroquinolone-stabilized DNA cleavage that is followed by cross-linking that occurs more rapidly with the GyrA variant. These differences add to the conclusion that GyrA-G81C and GyrB-E466C serve as distinct targets for irreversible, cleaved complex formation.
In the kinetic comparisons (Fig. 4, C and D), higher enzyme and drug concentrations were used with GyrA-G81C gyrase to make the yield similar to that with GyrB-E466C gyrase. Because the kinetic comparisons of cross-linking were then expressed relative to total cleavage, it is unlikely that concentration differences were responsible for the differences mentioned above.
We also noted for both mutant enzymes that binding kinetics were biphasic. The slow phase could reflect non-productive (non-cross-linking) binding that may interfere with productive binding or require a rearrangement to be productive (GyrB binding might be non-productive for GyrA cross-linking and vice versa; see discussion of models below).
Results from thermal DNA resealing measurements complemented those obtained with EDTA-mediated DNA resealing. With wild-type gyrase, both Cip-AcCl and the control compound Cip-Ac displayed temperature-stimulated DNA resealing (Fig. 5A). Complexes formed with GyrB-E466C and Cip-AcCl resisted resealing, although at high temperature, the accumulation of nicked DNA suggested partial resealing. Complexes formed with GyrA-G81C gyrase and Cip-AcCl also resisted thermal resealing. However, nicked DNA did not appear at high temperature with GyrA-G81C gyrase, further emphasizing that the binding modes are distinct.
Both variant gyrases produced surprising results with the non-cross-linkable compound Cip-Ac. GyrB-E466C gyrase increased the thermal stability of cleaved complexes formed with Cip-Ac (the temperature required for 50% resealing was 5 °C higher with GyrB-E466C than with wild-type gyrase). This increased stability may reflect an uncharacterized interaction between the C-7 ring of Cip-Ac and GyrB-E466C or the need to use higher inhibitor concentrations with mutant gyrase (we have found that resealing temperature is sensitive to inhibitor concentration4). More surprising was the peak in the thermal profile of Cip-Ac with GyrA-G81C gyrase (Fig. 5C). We speculate that during the 5-min treatment at moderate temperature a rearrangement occurs in which Cip-Ac shifts from one binding mode to the other such that stability increases.
Intracellular Cross-linking
Bacteria containing a Cys substitution at the position equivalent to E. coli GyrA81 are naturally occurring and have been associated with clinical resistance (38–40). Thus, the striking increase in susceptibility seen with haloacetyl derivatives of ciprofloxacin (Fig. 6) establishes that our biochemical observations reflect a relevant intracellular phenomenon. Obtaining the same result with two very different bacterial species (E. coli and M. smegmatis) and two cross-linking agents points to the generality of the novel quinolone-GyrA interaction. The reversibility of quinolone-mediated inhibition of DNA synthesis provided another way to observe intracellular cross-linking: the combination of Cip-AcCl and GyrA-G81C gyrase rendered inhibition of replication irreversible (Fig. 7). The effect was likely due to cleaved complexes because they are known to block DNA synthesis with purified topoisomerases (41, 42). Moreover, the presence of cleaved complexes in cultured cells correlates with inhibition of DNA synthesis (9, 43). These data solidify our conclusion that the C-7 portion of fluoroquinolones can interact with GyrA near position 81 in E. coli.
Models for Quinolone Binding
Published crystal structures of cleaved complexes reveal a drug-binding mode in which the C-7 ring of quinolone-like compounds interacts with amino acids in GyrB/ParC. Drug stoichiometry in the gyrase A2B2 tetramer is 2 (44), and in the crystal structure the two drug molecules are bound in a parallel orientation (Fig. 1A). The proximity of the distal end of the C-7 ring to GyrB-Glu466 was verified in the present work by a Cip-AcCl cross-link occurring with GyrB-E466C but not with GyrB-K447C gyrase, which is farther from the reactive chloroacetyl moiety (Fig. 1B). Other evidence for fluoroquinolone-GyrB/ParE interactions has been derived from the emergence of resistance substitutions in GyrB and ParE (37, 45). In the binding mode revealed by crystal structures (Fig. 1B), the carboxyl end of fluoroquinolones forms a stabilizing magnesium-water bridge with GyrA residues 83 and 87 (17, 18). This magnesium-mediated binding contributes to drug activity, and the loss of the interaction correlates with GyrA-mediated quinolone resistance (17, 18). Thus, substantial support exists for the binding mode shown in Fig. 1.
The second mode of drug binding, revealed in the present study, involves an interaction of the C-7 ring system with GyrA81. We envision two ways in which such an interaction might occur. One is an inversion of the drug orientation seen in crystal structures such that the drug remains bound to the same site (Fig. 8A). In the inverted binding mode, a Mg2+ coordination involving the 4-oxo-3-COOH chelate and the GyrB-Glu466 carboxylate potentially replaces the stabilizing fluoroquinolone-Mg2+-water-GyrA83/87 bridge. GyrB-Lys447 and the fluoroquinolone 3-carboxyl group can also form a salt bridge, which is functionally analogous to the interaction of the fluoroquinolone 3-carboxylate with GyrA-Arg121 seen in the orientation revealed by crystal structures. Thus, the drug-inverted mode fits with available data.
FIGURE 8.
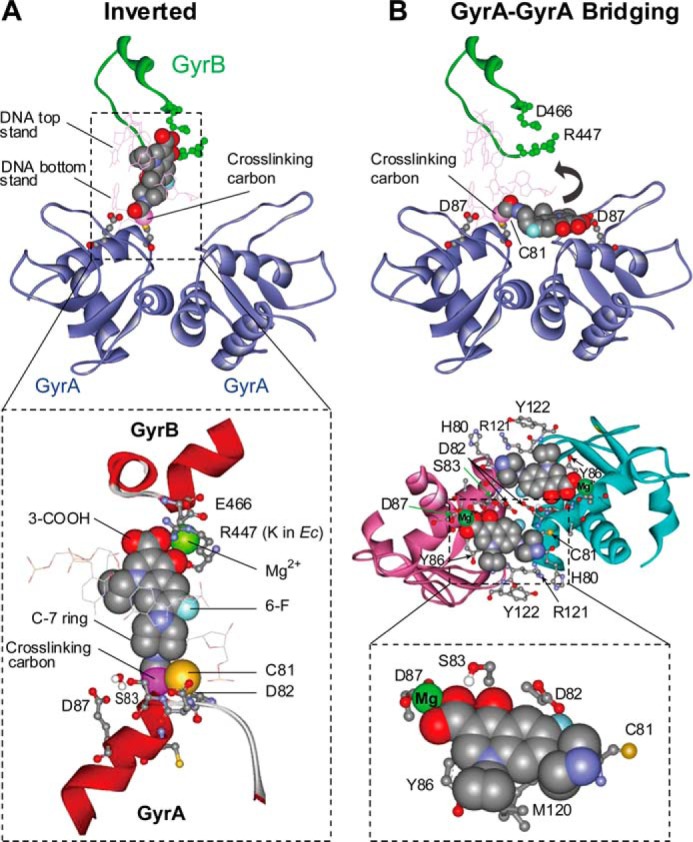
Models for binding of ciprofloxacin to gyrase-DNA complexes with interaction between fluoroquinolone C-7 ring and GyrA81. A, inverted configuration of the published x-ray crystal structure (Protein Data Bank code 2XKK). In the enlargement, GyrB447 is Arg in M. smegmatis and Lys in E. coli. B, GyrA-GyrA bridging model (from Protein Data Bank code 1AB4). The top portion shows one molecule of cross-linked ciprofloxacin in a bridging pocket. The curved arrow indicates the rotation needed to achieve the inverted structure shown in A. The center portion shows two antiparallel pockets filled with fluoroquinolone. The bottom portion shows a detail of nearby GyrA amino acid residues; GyrA87 and GyrA81 are on different GyrA subunits. In both panels, the cross-linking carbon is labeled.
The other possibility involves binding of quinolone to a different site. We have identified a potential binding pocket that bridges the GyrA-GyrA interface (Fig. 8B). A pair of antiparallel pockets, bounded by GyrA-Gly81 and GyrA-Asp87 on different GyrA subunits, is shown in the center of Fig. 8B. This GyrA-GyrA interface-bridging model explains the effects of amino acid substitutions that confer resistance, allows formation of a Mg2+-water coordination involving the quinolone carboxyl and Asp87, and places the quinolone C-7 ring near GyrA81. Comparison of the GyrA59 fragment and cleaved complexes (14, 46) indicates a better fit of drug to the fragment. It may be that the bridging model reflects binding to topoisomerase-DNA complexes formed prior to DNA cleavage. If so, it could represent the first binding step of a two-step process (47).
Concluding Remarks
Two additional comments are relevant to the work described above. First, our conclusion that cross-linking occurred in drug-gyrase-DNA complexes is based on indirect arguments: strong interactions occurred both with purified enzymes and with cultured bacteria of two species. Followup studies are in progress to directly identify cross-linking sites by mass spectroscopy and to define the second binding mode using x-ray crystallography. Second, cross-linking experiments can identify the existence of particular binding modes; however, they do not indicate the relative occupancy or allow determination of parameters based on equilibrium conditions because the cross-linked product accumulates. In the case of quinolones, minor binding modes can, in principle, be important because only a small fraction of the potential interactions must form to block replication. In the present case, we saw a very large effect of cross-linking on the MIC and DNA replication even though the low enzyme activity of GyrA-G81C gyrase likely allowed only a slow formation of cleaved complexes. Although additional work is required to assess the exact structure and relative usage of the binding modes, a deeper understanding of how the quinolone class inhibitors act on type II DNA topoisomerases is likely to emerge from finding multiple binding modes and from the ability to map intracellular drug-topoisomerase interactions by cross-linking.
Acknowledgments
We thank Y. Hong for assistance with preparing figures, K. Jude for technical advice, and M. Gennaro, M. Neiditch, R. Pine, and D. Wah for critical comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI073491 and R01AI87671 from the NIAID, R01GM-30717 from the NIGMS, and R01-CA077373 from the NCI.
K. Drlica, A. Mustaev, T. R. Towle, R. J. Kerns, and J. M. Berger, unpublished observations.
- Cip
- ciprofloxacin
- DMF
- dimethyl formamide
- MIC
- minimal inhibitory concentration
- Mox
- moxifloxacin.
REFERENCES
- 1. Nitiss J. L. (2009) Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 9, 338–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Champoux J. J. (2001) DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 [DOI] [PubMed] [Google Scholar]
- 3. Wang J. C. (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 [DOI] [PubMed] [Google Scholar]
- 4. Mizuuchi K., Fisher L. M., O'Dea M. H., Gellert M. (1980) DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc. Natl. Acad. Sci. U.S.A. 77, 1847–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kreuzer K. N., Cozzarelli N. R. (1980) Formation and resolution of DNA catenanes by DNA gyrase. Cell 20, 245–254 [DOI] [PubMed] [Google Scholar]
- 6. Liu L. F., Rowe T. C., Yang L., Tewey K. M., Chen G. L. (1983) Cleavage of DNA by mammalian DNA topoisomerase II. J. Biol. Chem. 258, 15365–15370 [PubMed] [Google Scholar]
- 7. Gellert M., Mizuuchi K., O'Dea M. H., Itoh T., Tomizawa J.-I. (1977) Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc. Natl. Acad. Sci. U.S.A. 74, 4772–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sugino A., Peebles C. L., Kreuzer K. N., Cozzarelli N. R. (1977) Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc. Natl. Acad. Sci. U.S.A. 74, 4767–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Snyder M., Drlica K. (1979) DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J. Mol. Biol. 131, 287–302 [DOI] [PubMed] [Google Scholar]
- 10. Pohlhaus J. R., Kreuzer K. N. (2005) Norfloxacin-induced DNA gyrase cleavage complexes block Escherichia coli replication forks, causing double-stranded breaks in vivo. Mol. Microbiol. 56, 1416–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dong K. C., Berger J. M. (2007) Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature 450, 1201–1205 [DOI] [PubMed] [Google Scholar]
- 12. Laponogov I., Sohi M. K., Veselkov D. A., Pan X. S., Sawhney R., Thompson A. W., McAuley K. E., Fisher L. M., Sanderson M. R. (2009) Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 16, 667–669 [DOI] [PubMed] [Google Scholar]
- 13. Bax B. D., Chan P. F., Eggleston D. S., Fosberry A., Gentry D. R., Gorrec F., Giordano I., Hann M. M., Hennessy A., Hibbs M., Huang J., Jones E., Jones J., Brown K. K., Lewis C. J., May E. W., Saunders M. R., Singh O., Spitzfaden C. E., Shen C., Shillings A., Theobald A. J., Wohlkonig A., Pearson N. D., Gwynn M. N. (2010) Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature 466, 935–940 [DOI] [PubMed] [Google Scholar]
- 14. Wohlkonig A., Chan P. F., Fosberry A. P., Homes P., Huang J., Kranz M., Leydon V. R., Miles T. J., Pearson N. D., Perera R. L., Shillings A. J., Gwynn M. N., Bax B. D. (2010) Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat. Struct. Mol. Biol. 17, 1152–1153 [DOI] [PubMed] [Google Scholar]
- 15. Laponogov I., Pan X. S., Veselkov D. A., McAuley K. E., Fisher L. M., Sanderson M. R. (2010) Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One 5, e11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kato J., Nishimura Y., Imamura R., Niki H., Hiraga S., Suzuki H. (1990) New topoisomerase essential for chromosome segregation in E. coli. Cell 63, 393–404 [DOI] [PubMed] [Google Scholar]
- 17. Aldred K. J., McPherson S. A., Wang P., Kerns R. J., Graves D. E., Turnbough C. L., Jr., Osheroff N. (2012) Drug interactions with Bacillus anthracis topoisomerase IV: biochemical basis for quinolone action and resistance. Biochemistry 51, 370–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aldred K. J., McPherson S. A., Turnbough C. L., Jr., Kerns R. J., Osheroff N. (2013) Topoisomerase IV-quinolone interactions are mediated through a water-metal ion bridge: mechanistic basis of quinolone resistance. Nucleic Acids Res. 41, 4628–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sindelar G., Zhao X., Liew A., Dong Y., Lu T., Zhou J., Domagala J., Drlica K. (2000) Mutant prevention concentration as a measure of fluoroquinolone potency against mycobacteria. Antimicrob. Agents Chemother. 44, 3337–3343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schoeffler A. J., May A. P., Berger J. M. (2010) A domain insertion in Escherichia coli GyrB adopts a novel fold that plays a critical role in gyrase function. Nucleic Acids Res. 38, 7830–7844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Horton R. M., Ho S. N., Pullen J. K., Hunt H. D., Cai Z., Pease L. R. (1993) Gene splicing by overlap extension. Methods Enzymol. 217, 270–279 [DOI] [PubMed] [Google Scholar]
- 22. Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. (1990) Use of T7 RNA polymerase to direct the expression of cloned genes. Methods Enzymol. 185, 60–89 [DOI] [PubMed] [Google Scholar]
- 23. Hiasa H., DiGate R. J., Marians K. J. (1994) Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J. Biol. Chem. 269, 2093–2099 [PubMed] [Google Scholar]
- 24. Hiasa H., Shea M. (2000) DNA gyrase-mediated wrapping of the DNA strand is required for the replication fork arrest by the DNA gyrase-quinolone-DNA ternary complex. J. Biol. Chem. 275, 34780–34786 [DOI] [PubMed] [Google Scholar]
- 25. Hiasa H. (2002) The Glu-84 of the ParC subunit plays critical roles in both topoisomerase IV-quinolone and topoisomerase IV-DNA interaction. Biochemistry 41, 11779–11785 [DOI] [PubMed] [Google Scholar]
- 26. Azéma J., Guidetti B., Dewelle J., Le Calve B., Mijatovic T., Korolyov A., Vaysse J., Malet-Martino M., Martino R., Kiss R. (2009) 7-((4-Substituted)piperazin-1-yl) derivatives of ciprofloxacin: synthesis and in vitro biological evaluation as potential antitumor agents. Bioorg. Med. Chem. 17, 5396–5407 [DOI] [PubMed] [Google Scholar]
- 27. Miller J. (1972) Experiments in Molecular Genetics, p. 433, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28. Jacobs W. R., Jr., Kalpana G. V., Cirillo J. D., Pascopella L., Snapper S. B., Udani R. A., Jones W., Barletta R. G., Bloom B. R. (1991) Genetic systems in mycobacteria. Methods Enzymol. 204, 537–555 [DOI] [PubMed] [Google Scholar]
- 29. Wall J. D., Harriman P. D. (1974) Phage P1 mutants with altered transducing abilities for Escherichia coli. Virology 59, 532–544 [DOI] [PubMed] [Google Scholar]
- 30. Oppegard L. M., Hamann B. L., Streck K. R., Ellis K. C., Fieldler H. P., Khodursky A. B., Hiasa H. (2009) In vivo and in vitro patterns of the activity of simocyclinone D8, an angucyclinone antibiotic from Streptomyces antibioticus. Antimicrob. Agents Chemother. 53, 2110–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yoshida H., Bogaki M., Nakamura M., Nakamura S. (1990) Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34, 1271–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Malik M., Marks K. R., Mustaev A., Zhao X., Chavda K., Kerns R. J., Drlica K. (2011) Fluoroquinolone and quinazolinedione activities against wild-type and gyrase mutant strains of Mycobacterium smegmatis. Antimicrob. Agents Chemother. 55, 2335–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goss W. A., Deitz W. H., Cook T. M. (1965) Mechanism of action of nalidixic acid on Escherichia coli. II. Inhibition of deoxyribonucleic acid synthesis. J. Bacteriol. 89, 1068–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malik M., Capecci J., Drlica K. (2009) Lon protease is essential for paradoxical survival of Escherichia coli when exposed to high concentrations of quinolone. Antimicrob. Agents Chemother. 53, 3103–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Drlica K., Malik M., Kerns R. J., Zhao X. (2008) Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 52, 385–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deweese J. E., Osheroff N. (2010) The use of divalent metal ions by type II topoisomerases. Metallomics 2, 450–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pan X. S., Gould K. A., Fisher L. M. (2009) Probing the differential interactions of quinazolinedione PD 0305970 and quinolones with gyrase and topoisomerase IV. Antimicrob. Agents Chemother. 53, 3822–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Golanbar G. D., Lam C. K., Chu Y. M., Cueva C., Tan S. W., Silva I., Xu H. H. (2011) Phenotypic and molecular characterization of Acinetobacter clinical isolates obtained from inmates of California correctional facilities. J. Clin. Microbiol. 49, 2121–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perlman D. C., El Sadr W. M., Heifets L. B., Nelson E. T., Matts J. P., Chirgwin K., Salomon N., Telzak E. E., Klein O., Kreiswirth B. N., Musser J. M., Hafner R. (1997) Susceptibility to levofloxacin of Mycobacterium tuberculosis isolates from patients with HIV-related tuberculosis and characterization of a strain with levofloxacin monoresistance. AIDS 11, 1473–1478 [DOI] [PubMed] [Google Scholar]
- 40. Weigel L. M., Steward C. D., Tenover F. C. (1998) gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob. Agents Chemother. 42, 2661–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wentzell L. M., Maxwell A. (2000) The complex of DNA gyrase and quinolone drugs on DNA forms a barrier to the T7 DNA polymerase replication complex. J. Mol. Biol. 304, 779–791 [DOI] [PubMed] [Google Scholar]
- 42. Shea M. E., Hiasa H. (1999) Interactions between DNA helicases and frozen topoisomerase IV-quinolone-DNA ternary complexes. J. Biol. Chem. 274, 22747–22754 [DOI] [PubMed] [Google Scholar]
- 43. Aedo S., Tse-Dinh Y. (2012) Isolation and quantitation of topoisomerase complexes accumulated on E. coli chromosomal DNA. Antimicrob. Agents Chemother. 56, 5458–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Critchlow S. E., Maxwell A. (1996) DNA cleavage is not required for the binding of quinolone drugs to the DNA gyrase-DNA complex. Biochemistry 35, 7387–7393 [DOI] [PubMed] [Google Scholar]
- 45. Yoshida H., Bogaki M., Nakamura M., Yamanaka L. M., Nakamura S. (1991) Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 35, 1647–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morais Cabral J. H., Jackson A. P., Smith C. V., Shikotra N., Maxwell A., Liddington R. C. (1997) Crystal structure of the breakage-reunion domain of DNA gyrase. Nature 388, 903–906 [DOI] [PubMed] [Google Scholar]
- 47. Kampranis S. C., Maxwell A. (1998) The DNA gyrase-quinolone complex, ATP hydrolysis and the mechanism of DNA cleavage. J. Biol. Chem. 273, 22615–22626 [DOI] [PubMed] [Google Scholar]
- 48. Sternglanz R., DiNardo S., Voelkel K. A., Nishimura Y., Hirota Y., Becherer K., Zumstein L., Wang J. C. (1981) Mutations in the gene coding for Escherichia coli DNA topoisomerase I affecting transcription and transposition. Proc. Natl. Acad. Sci. U.S.A. 78, 2747–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lu T., Zhao X., Drlica K. (1999) Gatifloxacin activity against quinolone-resistant gyrase: allele-specific enhancement of bacteriostatic and bactericidal activity by the C-8-methoxy group. Antimicrob. Agents Chemother. 43, 2969–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou J., Dong Y., Zhao X., Lee S., Amin A., Ramaswamy S., Domagala J., Musser J. M., Drlica K. (2000) Selection of antibiotic resistant bacterial mutants: allelic diversity among fluoroquinolone-resistant mutations. J. Infect. Dis. 182, 517–525 [DOI] [PubMed] [Google Scholar]
- 51. Trisler P., Gottesman S. (1984) lon transcriptional regulation of genes necessary for capsular polysaccharide synthesis in Escherichia coli K-12. J. Bacteriol. 160, 184–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Malik M., Hoatam G., Chavda K., Kerns R. J., Drlica K. (2010) Novel approach for comparing quinolones for emergence of resistant mutants during quinolone exposure. Antimicrob. Agents Chemother. 54, 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]