Background: Once DNA repair is completed, the DNA damage checkpoint is terminated, and the cell cycle is resumed.
Results: Src inhibition induced a delay in G2 checkpoint recovery and persistent ATR-Chk1 activation.
Conclusion: Src inhibits ATR signaling to promote recovery from G2 checkpoint arrest.
Significance: Src sends a termination signal between the completion of DNA repair and the initiation of checkpoint termination.
Keywords: Cell Cycle, Checkpoint Control, DNA Damage, DNA Damage Response, Src, Checkpoint Recovery
Abstract
The DNA damage checkpoint arrests cell cycle progression to allow time for repair. Once DNA repair is completed, checkpoint signaling is terminated. Currently little is known about the mechanism by which checkpoint signaling is terminated, and the disappearance of DNA lesions is considered to induce the end of checkpoint signaling; however, here we show that the termination of checkpoint signaling is an active process promoted by Src family tyrosine kinases. Inhibition of Src activity delays recovery from the G2 phase DNA damage checkpoint following DNA repair. Src activity is required for the termination of checkpoint signaling, and inhibition of Src activity induces persistent activation of ataxia telangiectasia mutated (ATM)- and Rad3-related (ATR) and Chk1 kinases. Src-dependent nuclear protein tyrosine phosphorylation and v-Src expression suppress the ATR-mediated Chk1 and Rad17 phosphorylation induced by DNA double strand breaks or DNA replication stress. Thus, Src family kinases promote checkpoint recovery through termination of ATR- and Chk1-dependent G2 DNA damage checkpoint. These results suggest a model according to which Src family kinases send a termination signal between the completion of DNA repair and the initiation of checkpoint termination.
Introduction
Genotoxic stress activates DNA damage checkpoint responses and arrests cell cycle progression to allow time for DNA repair. After completion of DNA damage repair, checkpoint signaling must be terminated so that checkpoint-arrested cells can resume cell cycle progression. The initiation of checkpoint signaling has been studied in depth, but little is known about the termination of checkpoint signaling and the resumption of cell cycle progression. Several reports have shown that the recovery from DNA damage checkpoint is not a passive or spontaneous process following the completion of DNA repair. Cell cycle resumption requires the active involvement of Polo-like and Aurora A kinases to reactivate the Cdk-cyclin complex, indicating that cell cycle resumption is not a direct result of the termination of checkpoint signaling. The termination or silencing of checkpoint signaling is also an active process that involves dephosphorylation and proteasomal degradation of checkpoint proteins; however, except for the direct reversal of phosphorylation or removal of the activated checkpoint proteins, very little is known about the signal transduction pathway that is triggered by the completion of DNA repair to initiate the termination of checkpoint signaling. Currently the disappearance of DNA lesions is considered to bring about the end of the activation of the most upstream components of the DNA damage checkpoint, namely the sensors that detect DNA lesions (1–3).
As part of the sensor mechanism that detects DNA lesions or stalled replication forks, ataxia telangiectasia mutated (ATM)6 and ATM- and Rad3-related (ATR) protein kinases are crucial regulatory proteins in the DNA damage checkpoint and maintenance of genomic stability. ATM is activated in response to DNA double strand breaks (DSBs) induced by ionizing radiation and radiomimetic anticancer agents. ATR is activated in response to replication protein A (RPA)-coated single-stranded DNA, which is formed during DNA replication and repair. The checkpoint signals retain the Cdk1-cyclin B1 complex in an inactive state, resulting in cell cycle blockage at the G2/M transition, namely G2 DNA damage checkpoint arrest (3–6).
In the S and G2 phases of the cell cycle, DSB ends are processed by nucleases and resected in an ATM-dependent manner to generate the RPA-coated single-stranded DNA that activates ATR and induces subsequent phosphorylation and activation of Chk1 (7–11). Despite the similarity in substrate specificity between ATM and ATR, it is believed that Chk1 is a direct substrate of ATR but not ATM and that Chk2 functions exclusively downstream of ATM in vivo (4). Chk1 activation is essential for the maintenance of G2 checkpoint arrest in response to DSB induction, and inhibition of Chk1 activity during G2 checkpoint arrest induces premature mitotic entry even though DNA repair has not been completed (12–16). Rad17 is another phosphorylation substrate of ATR, and the phosphorylation of Rad17 is required for its interaction with Claspin and Chk1 activation (17–19). Claspin mediates the ATR-dependent phosphorylation of Chk1 to activate the ATR-Chk1 signaling pathway (20). Following checkpoint activation, several protein phosphatases and ubiquitin ligases target the activated checkpoint proteins including Rad17 (21), Claspin (22–24), and Chk1 (25–30). The direct dephosphorylation and degradation of checkpoint proteins promote the termination of checkpoint signaling (1–3).
Src family kinases (SFKs) are the largest family of non-receptor tyrosine kinases. Activated SFKs phosphorylate a number of substrates and play important roles in the intracellular signal transduction that regulates cell proliferation, differentiation, migration, and morphological changes. SFK kinase activity is autoinhibited through the intramolecular interaction between the SH2 domain and a C-terminal phosphotyrosine residue (31, 32). SFKs are mainly located on the cytoplasmic side of the plasma membrane but are also found in late endosomes/lysosomes, secretory granules/phagosomes, and Golgi membranes (33–38). Intriguingly, cell fractionation and confocal microscopy showed that a fraction of the SFKs are expressed in the nucleus (39–43). Lyn, one of the SFK members, is activated and translocated into the nucleus upon DNA damage induction (44, 45). In DNA damage responses, Lyn plays positive and negative roles in apoptosis induction (46–50). Fyn is also translocated to the nucleus upon UV-B irradiation (51). These results indicate that SFKs are engaged in DNA damage responses; however, little is known about the involvement of the nuclear SFKs in the ATM/ATR-regulated checkpoint pathways.
The present study shows that the termination of checkpoint signaling is an active process promoted by Src family tyrosine kinases. Inhibition of SFK activity delays recovery from G2 DNA damage checkpoint following DNA DSB repair. Src activity is required for termination of checkpoint signaling but is dispensable for the resumption of the cell cycle that follows. SFKs are involved in the silencing of the ATR-Chk1 signaling pathway, and inhibition of SFK activity leads to persistent checkpoint activation and prolonged cell cycle arrest. SFKs also suppress ATR-Chk1 signaling activated by replication stress. These results suggest a model according to which SFKs play a crucial role in the signal transduction pathway that terminates DNA damage checkpoint signaling and suggest that SFKs send a termination signal between completion of DNA repair and initiation of checkpoint termination to promote checkpoint recovery.
EXPERIMENTAL PROCEDURES
Plasmids, Cell Lines, and Cell Culture
The cDNA encoding human wild-type Lyn was provided by Tadashi Yamamoto (The University of Tokyo) (52). Chicken v-Src was provided by Hiroshi Ohnishi (Gunma University) (53). Human c-Src was provided by Donald J. Fujita (University of Calgary) (54). cDNAs were subcloned into the pcDNA4-TO vector (Invitrogen). Wild-type Lyn was tagged with FLAG-HA (FH) epitopes and a nuclear localization signal (NLS) at its N terminus (55). FH-NLS-Lyn retains the inhibitory tyrosine phosphorylation site at the C-terminal tail. The constitutively active mutants LynΔC-HA (deleted of residues 507–512) and NLS-LynΔC-HA were described previously (36, 43). To generate HeLa S3 cells with an inducible v-Src allele (HeLa S3-TR/v-Src), HeLa S3 cells were transfected with the tetracycline repressor (TR) and selected with hygromycin (HeLa S3-TR). HeLa S3-TR cells were subsequently transfected with pcDNA4-TO-neo/v-Src and selected with G418. The neomycin-resistant pcDNA4-TO-neo vector was described previously (56). The expression of v-Src was induced with 1 μg/ml doxycycline. Gene transfection was performed using acidified polyethylenimine (38, 57) or Lipofectamine 2000 (Invitrogen). Parental HeLa S3 and HeLa S3 cells stably expressing NLS-Lyn or TR were cultured in Iscove's modified Dulbecco's medium supplemented with 5% bovine serum.
Checkpoint Recovery and Mitotic Entry after Adriamycin Exposure
Cells were synchronized at the G1/S boundary with 4 mm thymidine for 24 h. For G2 synchronization, cells were released from thymidine block and cultured for an additional 12 h with 9 μm RO-3306, a Cdk1 inhibitor (EMD Biosciences, 217699). The cells were exposed for 1 h to 110 nm Adriamycin (Sigma-Aldrich, D1515) except where noted. Then cells were released from G1/S or G2 synchronization and cultured in the presence of 40 ng/ml nocodazole to determine the mitotic index. The numbers of mitotic cells with a round shape were counted under a phase-contrast microscope. To inhibit the activity of SFKs, 5 μm SU6656 (Sigma-Aldrich, S9692) (58) or 20 μm PP2 (Sigma-Aldrich, P0042) was added 1 h before or after Adriamycin and included in the medium until the end of the culture period.
Mitotic Entry after Checkpoint Abrogation
Cells were synchronized in G1/S or G2 phase, exposed to 500 or 250 nm Adriamycin for 1 h, and allowed to recover. To inactivate checkpoint signaling, 5 mm caffeine, 0.1 mm Gö6976 (LC Laboratories, G-6203), 10 μm KU-55933 (Chemdea, CD0191), 10 μm Chk2 inhibitor II (Sigma-Aldrich, C3742), or VE-821 (Tinib-Tools, V134) was added to the culture. One hour before checkpoint inactivation, 5 μm SU6656 or 100 ng/ml BI2536 (Selleck Chemicals, S1109) was added to the culture medium and included until the end of the culture period.
For analysis with shRNA, HeLa S3 cells were transfected with pENTR4-H1/shLyn together with pcDNA4-TO/EGFP, cultured for 8 h, and cultured with fresh medium for an additional 16 h. Then cells were exposed to 4 mm thymidine for 24 h. The cell were exposed to 250 nm Adriamycin for 1 h and allowed to recover for 16 h. To inactivate checkpoint signaling, 0.1 mm Gö6976 was added to the culture, and cells were cultured in the presence of 40 ng/ml nocodazole. BI2536 was added at 100 ng/ml when Gö6976 was added.
Analysis of Checkpoint Proteins
For analysis of checkpoint responses in G2 phase, cells were treated as follows. After release from G1/S synchronization, cells were cultured for 6 h to reach the late S/G2 phase. Then cells were exposed to 110 nm Adriamycin for 1 h and allowed to recover. One hour before adding Adriamycin or after the end of Adriamycin exposure, 5 μm SU6656, 20 μm PP2, or 100 ng/ml BI2536 was added to the culture medium and included until the end of the culture period. Cells were solubilized in SDS-PAGE sample buffer by boiling and sonication or lysed in high salt buffer (50 mm HEPES-NaOH (pH 7.8), 300 mm KCl, 1% Triton X-100, 20% glycerol, and inhibitors of proteases and phosphatases) (59). The intensity of each protein band was quantitated using a ChemiDoc XRS-Plus image analyzer (Bio-Rad) and calculated as -fold increase ± standard deviation (S.D.) over that obtained with DMSO-treated cells.
For analysis of checkpoint responses activated by replication stress, HeLa S3 cells were treated with 4 mm thymidine for 24 h and fractionated to obtain the chromatin-enriched fractions. Six hours before harvest, 10 μm SU6656 was added. The chromatin fractions were prepared as follows. HeLa S3 cells were collected and lysed in low salt buffer (10 mm HEPES-NaOH (pH 7.8), 10 mm KCl, 0.1% Triton X-100, 10% glycerol, 0.34 m sucrose, and 1.5 mm MgCl2) (60). The resulting insoluble materials were washed once with low salt buffer, suspended in low salt buffer, and solubilized by sonication. The soluble fraction was cleared by centrifugation and used as the solubilized chromatin fraction. Alternatively, HeLa S3 cells were transfected with shRNA expression vector, cultured for 72 h, and solubilized in SDS-PAGE sample buffer. Cells were exposed to 4 mm thymidine for 24 h before harvest. In these experiments, cells were not exposed to Adriamycin.
Binucleation Assay
HeLa S3 cells were synchronized at the G1/S boundary and exposed to 110 nm Adriamycin for 1 h. Then cells were released from synchronization and cultured in the presence of 0.2 μg/ml cytochalasin D. After 23 h of recovery, cells were fixed with 4% paraformaldehyde and stained with propidium iodide as described previously (42, 43). Binucleated cells were counted under a fluorescence microscope (Zeiss Axiovert S100). In parallel, cells were cultured in the presence of 40 ng/ml nocodazole for mitotic index determination.
Flow Cytometry
Flow cytometric analyses were performed as described previously (56, 61). For analysis of Chk1-Ser317 phosphorylation, HeLa S3 cells were transfected 24 h after seeding. At 8 h after transfection, cell culture medium was changed with fresh medium, and the cells were cultured for an additional 16 h. Then the cells were exposed to 4 mm thymidine for 16 h before harvest. The cells were fixed with 1.5% paraformaldehyde, stained with the indicated antibodies, and analyzed on a MoFlo cell sorter (Beckman Coulter) or BD FACSCanto II (BD Biosciences).
For analysis of recovery from G2 checkpoint arrest, HeLa S3 cells were transfected with pENTR4-H1/shLyn together with pcDNA4-TO/EGFP and cultured for 24 h. Then cells were exposed to 4 mm thymidine for 24 h. The cells were exposed to 110 nm Adriamycin for 1 h, released from thymidine block, and cultured in the presence of 40 ng/ml nocodazole. At 30–36 h after the end of Adriamycin exposure, cells were fixed for flow cytometry.
Immunofluorescence Microscopy
For detection of γ-H2AX and RPA foci, cells were extracted with cytoskeleton buffer supplemented with 0.5% Triton X-100 and fixed with 4% paraformaldehyde. For detection of nuclear localization of LynΔC, the transfected cells were fixed with 4% paraformaldehyde. Subsequently, the cells were permeabilized with 0.5% Triton X-100 for 5 min and stained as described previously (42, 43). DNA was stained with propidium iodide.
Antibodies
The following antibodies were used: actin (Chemicon, MAB1501), Cdk1 (Santa Cruz Biotechnology, sc-54), Cdk1-Tyr(P)15 (Cell Signaling Technology, 4539S), Chk1 (MBL International, K0086-3), Chk1-Ser(P)317 (Cell Signaling Technology, 2344 or 8191), Chk1-Ser(P)345 (Cell Signaling Technology, 2348), Chk2 (MBL International, K0088-3), Chk2-Thr(P)68 (Cell Signaling Technology, 2661), Claspin (Sigma-Aldrich, C7867), cyclin A (Sigma-Aldrich, C4710), cyclin B1 (Cell Signaling Technology, 4135; Santa Cruz Biotechnology, sc-752), Fyn (BD Transduction Laboratories, 610163), histone H3-Ser(P)10 (Cell Signaling Technology, 9706), Hsc70 (Santa Cruz Biotechnology, sc-7298), Lyn (44, Santa Cruz Biotechnology, sc-15; H-6, Santa Cruz Biotechnology, sc-7274), Rad17 (MBL International, K0120-3), Rad17-Ser(P)645 (Bethyl Laboratories, A300-153A), Src (Santa Cruz Biotechnology, sc-19; Calbiochem, OP07), and Yes (BD Transduction Laboratories, 610375).
Oligo DNA Sets Used for shRNA
Oligo DNA sets used for shRNAs targeting SFKs are as follows: shLyn sense, 5′-GAT CTC TGG ACA GAG GTT TCA AAC TAT TCA AGA GAT AGT TTG AAA CCT CTG TCC TTT TTG CGC AT-3′; shLyn antisense, 5′-CTA GAT GCG CAA AAA GGA CAG AGG TTT CAA ACT ATC TCT TGA ATA GTT TGA AAC CTC TGT CCA GA-3′; shYes sense, 5′-GAT CTC TGG TTA ATT GAA GAC AAT GAT TCA AGA GAT CAT TGT CTT CAA TTA ACC TTT TT-3′; shYes antisense, 5′-CTA GAA AAA GGT TAA TTG AAG ACA ATG ATC TCT TGA ATC ATT GTC TTC AAT TAA CCA GA-3′; shFyn sense, 5′-GAT CTC TGG AAG AGC TCT GAA ATT ACT TCA AGA GAG TAA TTT CAG AGC TCT TCC TTT TT-3′; shFyn antisense, 5′-CTA GAA AAA GGA AGA GCT CTG AAA TTA CTC TCT TGA AGT AAT TTC AGA GCT CTT CCA GA-3′. An shRNA targeting luciferase (shLuci) was described previously (56).
RESULTS
SFK Activity Is Required for G2 DNA Damage Checkpoint Recovery
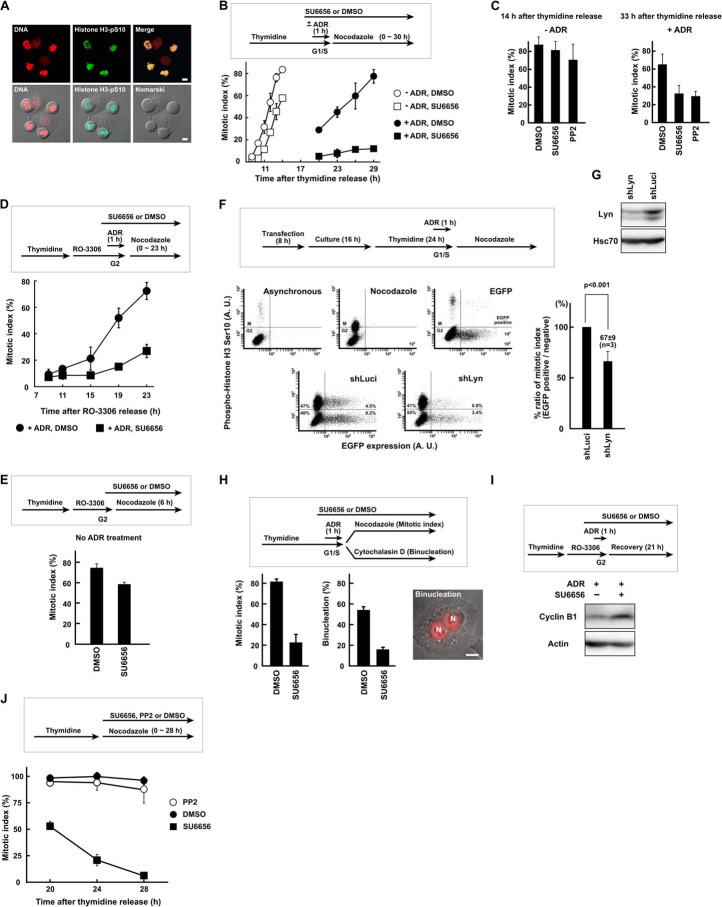
Exposure of cells to Adriamycin induces DSBs, which activate the DNA damage checkpoint and arrest cell cycle progression at the G2/M phase boundary. After completion of DNA repair, cells recover from the DNA damage checkpoint arrest and enter mitosis. Mitotic HeLa S3 cells exhibited a round cell shape and phosphorylation of histone H3 at Ser10 (Fig. 1A). Exposure to SU6656, an SFK inhibitor, brought about a severe delay in mitotic entry when cells were exposed to Adriamycin (Fig. 1B). Without Adriamycin, SU6656-treated cells slightly delayed mitotic entry, but the mitotic index curve had the same slope as mock-treated cells. Likewise, exposure to PP2, another SFK inhibitor, severely delayed mitotic entry after Adriamycin exposure (+ADR), but PP2 alone did not appear to affect mitotic entry (−ADR) (Fig. 1C). To exclude the effect of S phase checkpoint on mitotic entry, cells were synchronized in G2 phase with the Cdk1 inhibitor RO-3306 and exposed to Adriamycin. Cells treated with RO-3306 complete DNA replication and arrest in G2 phase (62, 63). Again, SU6656-treated cells showed a decreased mitotic index after Adriamycin exposure (Fig. 1, D and E). To exclude the possibility that SU6656 and PP2 may nonspecifically inhibit other kinases to delay checkpoint recovery, the involvement of SFKs was addressed by RNAi-mediated gene knockdown. In HeLa S3 cells, at least three family members, Lyn, c-Yes, and Fyn, are abundantly expressed and might have considerable functional redundancy (64). Because previous studies reported the involvement of Lyn in DNA damage responses (44, 45), we examined the role of Lyn. shRNA-mediated knockdown of Lyn induced a delay in mitotic entry after Adriamycin exposure (Fig. 1F). These results suggest that SFK inhibition leads to prolonged G2 DNA damage checkpoint arrest and that SFK activity is required for G2 checkpoint recovery and mitotic entry.
FIGURE 1.
SFK activity is required for G2 DNA damage checkpoint recovery. A, HeLa S3 cells with a round shape were stained with anti-phosphohistone H3 (Ser10) antibody and propidium iodide and confirmed as mitotic cells. B, HeLa S3 cells were synchronized at the G1/S phase boundary by thymidine block, exposed to 110 nm Adriamycin (ADR) for 1 h, and allowed to recover. Cells were exposed to 5 μm SU6656 or DMSO. Results without Adriamycin are also shown. C, HeLa S3 cells were treated as in B except that 20 μm PP2 was used. The mitotic index was determined at 14 and 33 h after release from thymidine synchronization for untreated (−ADR) and Adriamycin-treated (+ADR) cells, respectively. D and E, cells were treated as in B but synchronized in G2 phase with RO-3306. Results without Adriamycin are shown in E. F, HeLa S3 cells were transfected with shRNA expression vector targeting Lyn (shLyn). EGFP expression vector was co-transfected. Recovery from G2 checkpoint arrest was analyzed by flow cytometry as described under “Experimental Procedures.” An shRNA targeting luciferase (shLuci) was used as a control. p values were calculated using t test. A. U., arbitrary units. G, HeLa S3 cells were transfected with shRNA expression vector and cultured for 72 h. SDS lysates were prepared and probed with the indicated antibodies. H, Adriamycin-treated cells were cultured for 23 h in the presence of cytochalasin D to inhibit cytokinesis. The numbers of binucleated cells were counted, and the mitotic index was determined in parallel. N, nucleus. I, cells were exposed to Adriamycin as in D and harvested 21 h later. SDS lysates were prepared and probed for cyclin B1 and actin. J, HeLa S3 cells were synchronized at the G1/S phase boundary by thymidine block. Cells were released from thymidine and exposed to 5 μm SU6656, 20 μm PP2, or DMSO. The mitotic index was determined to monitor slippage from nocodazole-induced mitotic arrest. Scale bar, 10 μm for all panels. Error bars represent S.D.
Data confirmed that the decreased mitotic index was not due to slippage from mitotic arrest. Although SU6656 often induces mitotic slippage (65), PP2 does not, a result confirmed in the present study (Fig. 1J). In addition, checkpoint recovery was examined without monitoring mitotic arrest. In the presence of cytochalasin D, mitotic cells can complete chromosome segregation but fail cytokinesis, resulting in binucleation (Fig. 1H) (66). After Adriamycin exposure, SU6656-treated cells showed a lower number of binucleated cells, which correlated with the decreased mitotic index (Fig. 1H). In addition, degradation of cyclin B1 was also inhibited by SU6656 after Adriamycin exposure (Fig. 1I), which is reminiscent of results showing that cyclin B1 accumulates during DNA damage-induced G2 checkpoint arrest and is degraded in mitosis (see Fig. 3A) (61). By contrast, cyclin B1 was found to be degraded during SU6656-induced mitotic slippage.7 These results suggest that the decreased mitotic index upon SFK inhibition is not due to mitotic slippage but to cell cycle arrest.
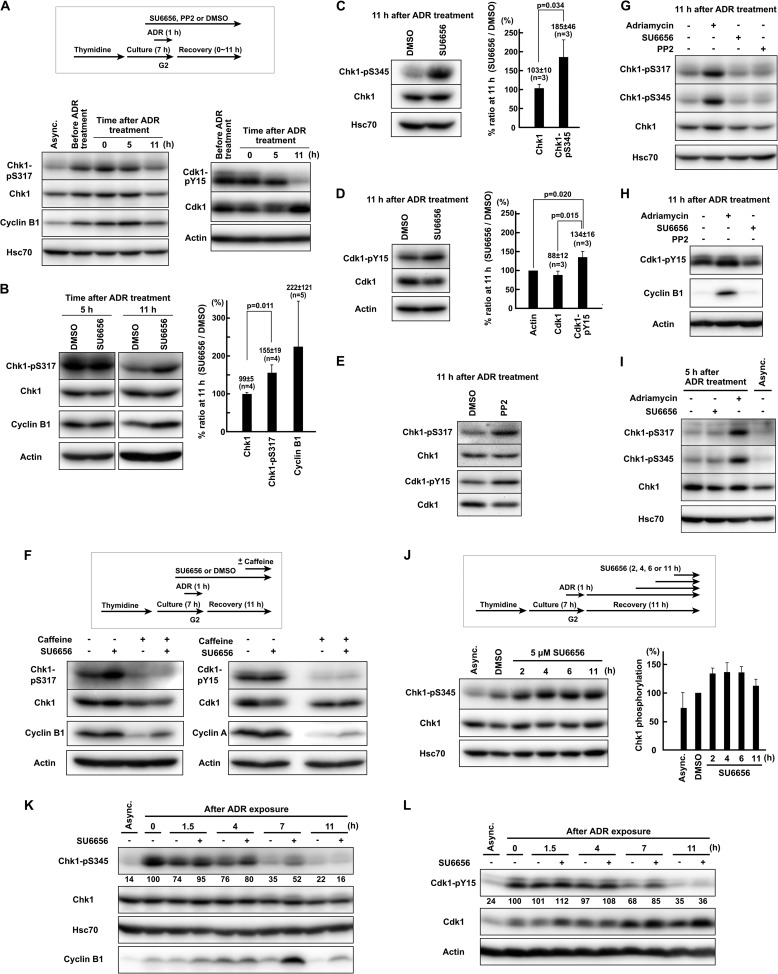
FIGURE 3.
SFKs promote down-regulation of ATR-mediated Chk1 phosphorylation during G2 checkpoint recovery. A, HeLa S3 cells were exposed to 110 nm Adriamycin (ADR) in G2 phase as described under “Experimental Procedures.” Cells were allowed to recover for the indicated times, and SDS lysates were prepared for Western blotting. B, C, and D, cells were treated as in A except that 5 μm SU6656 was added 1 h before Adriamycin exposure. -Fold increases in band intensity induced by SU6656 are shown for samples with 11 h of recovery. Results are expressed as means ± S.D. E, instead of SU6656, 20 μm PP2 was used. F, HeLa S3 cells were treated as in B. One hour before harvest, 5 mm caffeine was added. G, H, and I, the same experiment as in B–E. J, HeLa S3 cells were synchronized by thymidine block for 16 h. Cells were exposed to 110 nm Adriamycin for 1 h from 6 h after release from thymidine block. At 11 h after the end of Adriamycin exposure, cells were harvested, and SDS lysates were prepared. For the indicated times before harvesting, cells were treated with 5 μm SU6656. The graph represents results obtained from two independent experiments. K and L, HeLa S3 cells were synchronized by double thymidine block. From 6 h after release from thymidine block, cells were exposed to 110 nm Adriamycin for 1 h. Cells were allowed to recover for the indicated times, and SDS lysates were prepared. Representative data from two independent experiments are shown. p values were calculated using t test. Error bars represent S.D. Async., asynchronously growing cells.
SFK Activity Is Involved in Silencing of ATR- and Chk1-dependent G2 DNA Damage Checkpoint
Next, the mechanism by which SFK kinase activity is involved in the recovery from G2 DNA damage checkpoint was assessed. For this purpose, maintenance of G2 DNA damage checkpoint was abrogated by caffeine, an ATM/ATR inhibitor, and checkpoint-arrested cells were allowed to enter mitosis in the presence of kinase inhibitors (Fig. 2, A and B). The design of this protocol mimics cell cycle restart after completion of DNA repair and termination of ATM/ATR signaling (67) and enables the differentiation of kinases involved in checkpoint silencing from those involved in cell cycle resumption. Plk1 is required for cell cycle resumption after checkpoint termination (67). Indeed, exposure to the Plk1 inhibitor BI2536 blocked entry into mitosis after checkpoint abrogation by caffeine (Fig. 2A) as shown previously (68). In contrast to BI2536, SU6656 was incapable of blocking mitotic entry when Adriamycin-treated cells were exposed to caffeine (Fig. 2, A and B). With a prolonged exposure to SU6656 from G1/S phase, Adriamycin-treated cells were also allowed to enter mitosis upon exposure to caffeine despite a slight delay (Fig. 2C). These results suggest that SFK activity is dispensable for cell cycle resumption but required for the silencing of ATM/ATR-dependent G2 DNA damage checkpoint signaling.
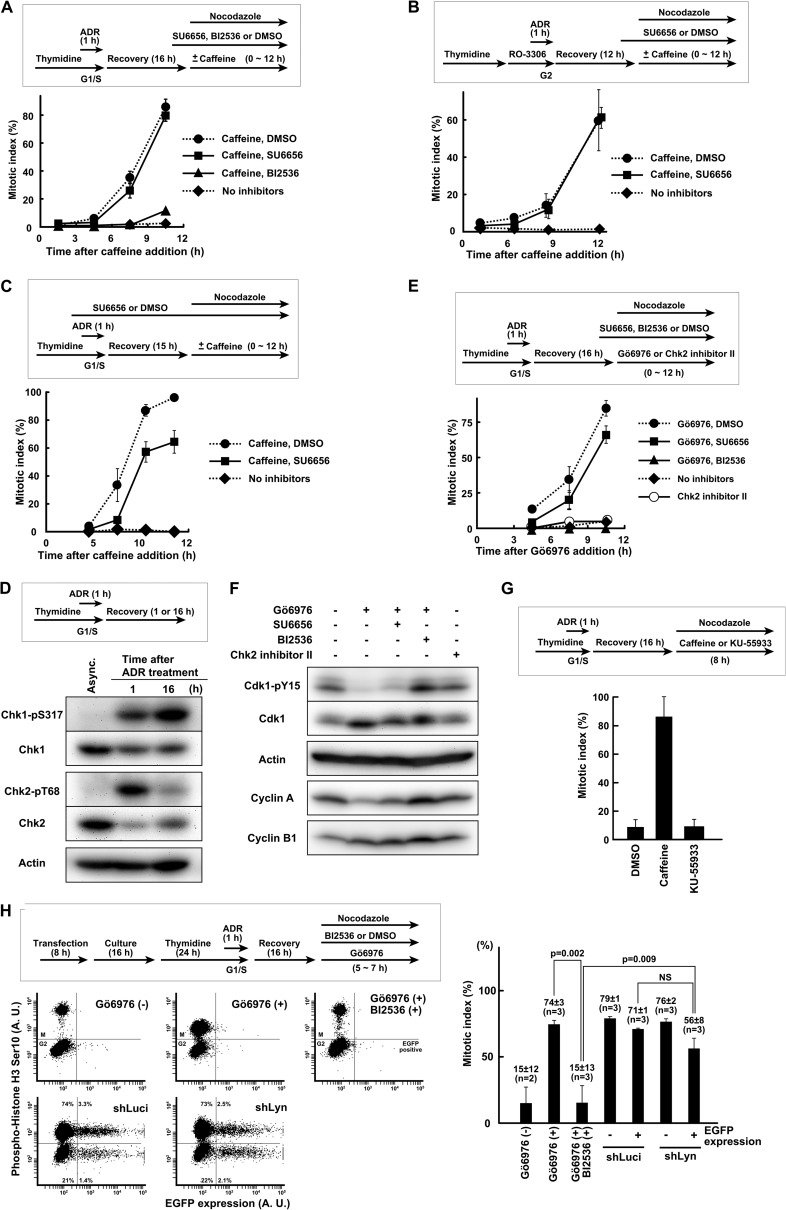
FIGURE 2.
SFK activity is involved in silencing of ATR- and Chk1-dependent G2 DNA damage checkpoint. A, HeLa S3 cells were synchronized at the G1/S boundary and exposed to 500 nm Adriamycin (ADR). After 16 h of recovery, cells were exposed to caffeine and allowed to enter mitosis. SU6656 or BI2536 was added 1 h before caffeine. B, cells were synchronized in G2 phase and exposed to 250 nm Adriamycin. After recovery, cells were exposed to caffeine. C, cells were treated as in A except that SU6656 was added 1 h before Adriamycin. D, cells were synchronized at the G1/S boundary, exposed to 250 nm Adriamycin for 1 h, and allowed to recover for the indicated times. SDS lysates were prepared and probed with the indicated antibodies. Asynchronously growing cells (Async.) were used as controls. E, cells were treated as in D. After 16 h of recovery, Gö6976 or Chk2 inhibitor II was added. F, cells were treated as in E except that nocodazole was not used. At 10 h after Gö6976 addition, cells were harvested for Western blotting. G, cells were treated as in A. After recovery, caffeine or KU-55933 was added. H, HeLa S3 cells were transfected with shLyn together with EGFP expression vector. After synchronization, cells were exposed to 250 nm Adriamycin for 1 h and allowed to recover for 16 h. Mitotic entry was induced by Gö6976 addition, and cells were fixed for flow cytometric analysis. p values were calculated using t test. NS, not significant. Error bars represent S.D. A. U., arbitrary units.
Because previous works showed that G2 checkpoint maintenance is largely dependent on Chk1 (12–16), the possibility that SFK activity is required for the silencing of ATR-Chk1 signaling was examined. At 16 h after the end of Adriamycin exposure, ATM-mediated Chk2 phosphorylation was drastically decreased, but ATR-mediated Chk1 phosphorylation was increased (Fig. 2D), suggesting that ATM and Chk2 are almost inactivated but that ATR and Chk1 are still active and responsible for the maintenance of G2 checkpoint arrest at this time point. The reduction of Chk1 and Chk2 protein after Adriamycin exposure should be due to an electrophoretic mobility shift and proteasomal degradation (28–30, 69–71). Because caffeine has a pleiotropic effect, we used Gö6976, which selectively inhibits the activity of Chk1 but not Chk2 (14, 72). In fact, exposure to Gö6976 released cells from G2 checkpoint arrest and induced mitotic entry, whereas exposure to the Chk2 inhibitor II (73) or specific ATM inhibitor KU-55933 did not (Fig. 2, E and G). Exposure to VE-821, a specific ATR inhibitor, released cells from G2 checkpoint arrest as reported previously (data not shown) (74–76). Furthermore, Gö6976 induced dephosphorylation of the inhibitory Tyr15 on Cdk1 (Cdk1-Tyr15) (Fig. 2F). These results suggest that the maintenance of G2 DNA damage checkpoint arrest is mediated by ATR-Chk1 signaling, which is consistent with previous studies (12, 14, 16). Exposure to BI2536 strongly inhibited Gö6976-induced mitotic entry and dephosphorylation of Cdk1-Tyr15 (Fig. 2, E, F, and H). In sharp contrast to this, SU6656 affected neither Gö6976-induced mitotic entry nor the dephosphorylation of Cdk1-Tyr15, although it caused a slight delay. Similarly, Gö6976 induced mitotic entry in cells transfected with shLyn (Fig. 2H). These results suggest that SFK activity is required for the silencing of G2 checkpoint signaling mediated by ATR and Chk1.
SFKs Promote Down-regulation of ATR-mediated Chk1 Phosphorylation during G2 Checkpoint Recovery
To understand how SFK activity exerts checkpoint silencing, ATR-mediated phosphorylation of Chk1 during checkpoint recovery was examined because phosphorylation of Chk1 at Ser317 and Ser345 is a reliable indicator of activation of ATR and Chk1 in vivo (4, 13, 77, 78). Upon Adriamycin exposure in G2 phase, the accumulation of cyclin B1 peaked at 5 h after the end of Adriamycin exposure, and the phosphorylation of Cdk1-Tyr15 was strikingly decreased at 11 h (Fig. 3A), indicating that the recovery from G2 DNA damage checkpoint occurred between 5 and 11 h after Adriamycin exposure. Before Adriamycin exposure, Chk1 was already phosphorylated and activated because of DNA replication stress in S phase. Upon Adriamycin exposure, Chk1 was up-regulated and then down-regulated gradually (Fig. 3A). In this experiment, cells were not treated with SFK inhibitors.
To examine the involvement of SFKs in Chk1 inactivation, cells were treated with SU6656. At 5 h after Adriamycin exposure, SU6656 treatment affected neither phosphorylation of Chk1-Ser317 nor degradation of cyclin B1 (Fig. 3B), indicating that SFK inhibition does not affect Chk1 phosphorylation during G2 checkpoint activation. By contrast, at 11 h after Adriamycin exposure, SU6656 increased phosphorylation of Chk1 at Ser317 and Ser345 (Fig. 3, B and C), suggesting that the down-regulation of Chk1 is prevented. SU6656 also inhibited both the degradation of cyclin B1 and the dephosphorylation of Cdk1-Tyr15 (Fig. 3, B and D), indicating that the recovery from G2 checkpoint arrest is delayed. Similarly, PP2 increased the phosphorylation of Chk1 (Fig. 3E). The phosphorylation of Chk1 at Ser317 and Ser345 is mediated by ATR, but not ATM, in vivo (4). Caffeine effectively blocked SU6656-induced phosphorylation of Chk1-Ser317 (Fig. 3F), which is consistent with the finding that SU6656 did not affect caffeine-induced abrogation of G2 DNA damage checkpoint (Fig. 2, A and B). Along with the dephosphorylation of Chk1-Ser317, caffeine induced dephosphorylation of Cdk1-Tyr15 and degradation of cyclin A and cyclin B1 (Fig. 3F). Without exposure to Adriamycin, SU6656 or PP2 did not increase Chk1 phosphorylation, Cdk1 phosphorylation, or the amount of cyclin B1 (Fig. 3, G and H). These results indicate that SFK activity is critical for the down-regulation of ATR-dependent Chk1 phosphorylation to promote G2 checkpoint recovery.
To further examine the involvement of SFKs in Chk1 down-regulation, SU6656 was added at various time points after the end of Adriamycin exposure instead of treating the cells with the inhibitor before and during Adriamycin exposure. Adding SU6656 at 2–6 h before harvesting also increased Chk1 phosphorylation (Fig. 3J). These data suggest that SFK inhibition increases Chk1 phosphorylation not through the inhibition of DSB repair. In this experiment, cells were synchronized by single thymidine block.
The effect of SFK inhibition on Chk1 phosphorylation was also examined by a time course experiment. SFK inhibition increased phosphorylation of Chk1, accumulation of cyclin B1, and phosphorylation of Cdk1-Tyr15 at 7 h after the end of Adriamycin exposure (Fig. 3, K and L). At 11 h after Adriamycin exposure, Chk1 phosphorylation had returned to the basal level and was not affected by SU6656 treatment (Fig. 3K), indicating that Chk1 is eventually dephosphorylated in the presence of SU6656. The discrepancy between Fig. 3, B, C, and D, and Fig. 3, K and L, should be due to the fact that we used double thymidine block in the time course experiment.
Although Chk1 was substantially phosphorylated by replication stress before Adriamycin exposure (Fig. 3A), Chk1 phosphorylation induced by replication stress became much weaker than that induced by Adriamycin exposure even in the presence of SU6656 at 5 or 11 h after the end of Adriamycin exposure (Fig. 3, G and I). Moreover, without Adriamycin exposure, SFK inhibition slightly delayed mitotic entry, but cells entered mitosis even in the presence of SU6656 (Fig. 1B). These data indicate that the observed checkpoint inactivation defect is not due to the replication stress induced before Adriamycin exposure.
DSB Repair Is Largely Unaffected by SFK Inhibition
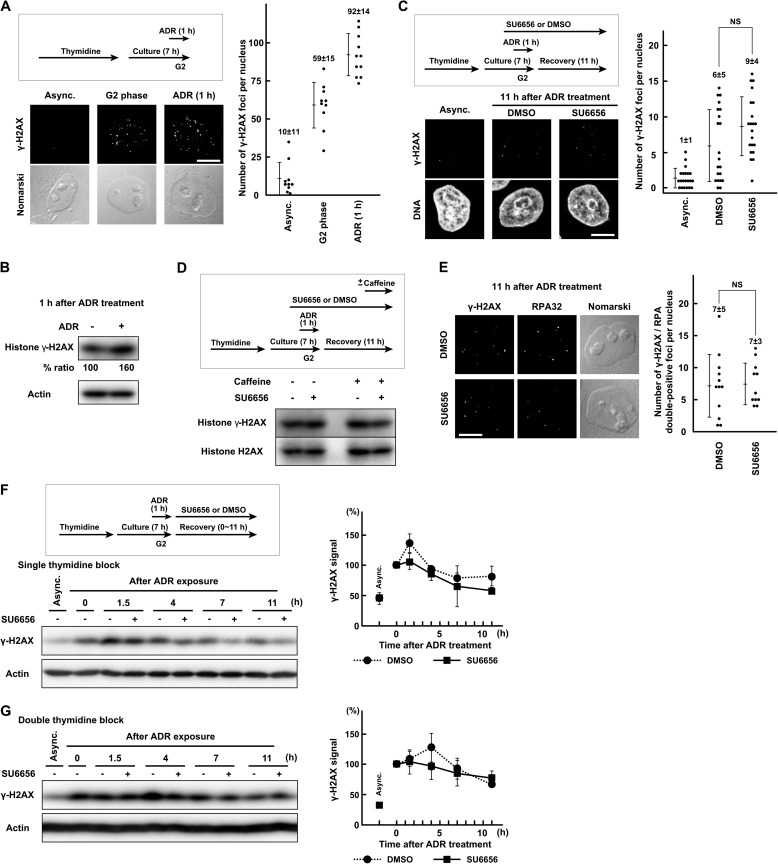
G2 checkpoint recovery is preceded by DSB repair. To understand how SFKs regulate Chk1 phosphorylation, the possibility that the SFK inhibition may increase Chk1 phosphorylation by the inhibition of DSB repair was assessed. ATM-mediated phosphorylation of histone H2AX at Ser139 (γ-H2AX) is widely accepted as an indicator of DSBs after ionizing radiation and exposure to radiomimetic agents (4, 5). The γ-H2AX signal was already up-regulated before Adriamycin exposure by DNA replication stress (Fig. 4, A and B) (79), which was confirmed by the Chk1-Ser(P)317 blot in Fig. 3A. At 11 h after the end of Adriamycin exposure, most of the γ-H2AX foci had disappeared (Fig. 4C). The number of γ-H2AX foci was slightly increased in SU6656-treated cells, but the difference was not statistically significant. In Western blotting, exposure to SU6656 and/or caffeine did not affect the γ-H2AX signal at 11 h after Adriamycin exposure (Fig. 4D). The effect of SFK inhibition on γ-H2AX signal was also examined by a time course experiment with single or double thymidine block. SU6656 treatment did not increase but rather inhibited the γ-H2AX signal (Fig. 4, F and G). γ-H2AX/RPA-double positive foci were also counted because DSB-induced ATR activation is mediated by DSB end resection and generation of RPA-coated single-stranded DNA (7–11). The number of γ-H2AX/RPA-double positive foci was not affected by SU6656 treatment (Fig. 4E). These results indicate that the majority of DSBs had already been repaired after 11 h of recovery and that the efficiency of DSB repair is largely unaffected by SFK inhibition. These results suggest that SFK inhibition increases Chk1 phosphorylation not through the inhibition of DSB repair and that SFKs regulate checkpoint signaling independently of DSB repair.
FIGURE 4.
DSB repair is largely unaffected by SFK inhibition. A, HeLa S3 cells were exposed to 110 nm Adriamycin (ADR) in G2 phase as described under “Experimental Procedures” and fixed for immunofluorescence microscopy. After fixation, the cells were stained with anti-γ-H2AX antibody. B, cells were treated as in A. SDS lysates were prepared and probed with the indicated antibodies. C and E, cells were treated as in A and fixed 11 h after the end of Adriamycin exposure. One hour before Adriamycin exposure, 5 μm SU6656 was added. D, the same experiment as in Fig. 3F. Cells were treated as in C and E, and SDS lysates were prepared. One hour before harvest, 5 mm caffeine was added. F and G, HeLa S3 cells were exposed to 110 nm Adriamycin for 1 h from 6 h after release from thymidine block and allowed to recover for the indicated times. SDS lysates were prepared and probed with the indicated antibodies. Cells were synchronized by single thymidine block for 16 h (F) or double thymidine block (G). Each graph represents results obtained from two independent experiments. NS, not significant. Error bars represent S.D. Scale bar, 10 μm in all panels. Async., asynchronously growing cells.
SFKs Regulate ATR Signaling Independently from Regulation of ATM Activity
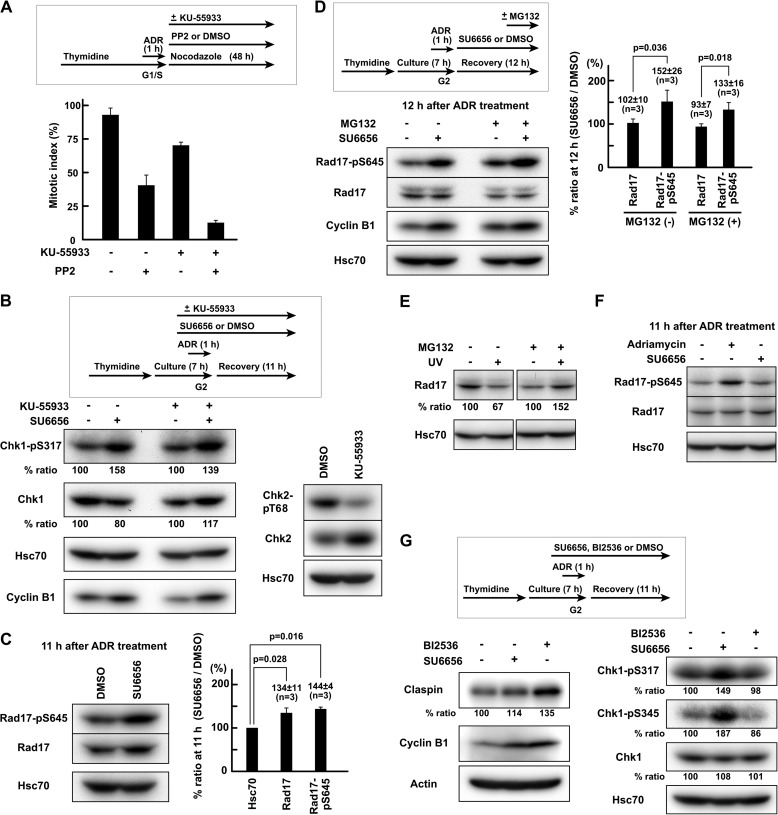
ATM-dependent DSB end resection mediates the activation of ATR by DSBs in the G2 phase (7–11). To understand how SFKs regulate checkpoint signaling, the possibility that the regulation of ATR activity by SFKs may be exerted through the regulation of ATM activity was assessed. In the presence of the ATM inhibitor KU-55933, SFK inhibition still delayed mitotic entry after Adriamycin exposure (Fig. 5A). In the presence of KU-55933, SU6656 consistently induced an increase in both the phosphorylation of Chk1-Ser317 and the amount of cyclin B1 (Fig. 5B). These results suggest that SFKs regulate checkpoint signaling independently from regulation of ATM activity and that SFKs regulate ATR signaling to Chk1.
FIGURE 5.
SFKs regulate Rad17 phosphorylation independently of proteasomal degradation of Rad17. A, HeLa S3 cells were treated as in Fig. 1B except that 20 μm PP2 and/or 10 μm KU-55933 was used. The mitotic index was determined after 48 h of recovery. B, cells were exposed to 110 nm Adriamycin (ADR) in G2 phase as described under “Experimental Procedures.” One hour before Adriamycin exposure, 10 μm KU-55933 was added. SDS lysates were prepared. C, the same experiment as in Fig. 3, B–E. Cells were exposed to 110 nm Adriamycin in G2 phase as described under “Experimental Procedures,” and SDS lysates were prepared 11 h after the end of Adriamycin exposure. D, HeLa S3 cells were treated as in C except that 5 μm SU6656 was added after Adriamycin exposure. Two hours before harvest, 10 μm MG132 was added. E, HeLa S3 cells were UV-irradiated (10 J/m2), allowed to recover for 4 h, and solubilized in SDS-PAGE sample buffer. One hour before UV irradiation, 10 μm MG132 was added. The data were obtained from the same blot. F, the same experiment as in C. G, HeLa S3 cells were treated as in C. Cells were exposed to SU6656 or BI2536. p values were calculated using t test. Error bars represent S.D.
SFKs Regulate Rad17 Phosphorylation Independently of Proteasomal Degradation of Rad17
To understand how SFKs regulate ATR signaling to Chk1, the possibility that SFKs regulate proteins involved in ATR-Chk1 signaling was assessed. ATR-dependent checkpoint signaling is mediated by protein complex formation on damaged chromatin (4–6). Rad17 is phosphorylated by ATR, and the Rad17 phosphorylation is required for interaction with Claspin and activation of Chk1 (17–19). Therefore, phosphorylation of Rad17 at Ser645 (Rad17-Ser645) was examined. It is of note that SU6656 significantly increased the phosphorylation of Rad17-Ser645 (Fig. 5C). Without exposure to Adriamycin, SU6656 did not increase Rad17 phosphorylation (Fig. 5F). These results suggest that SFK inhibition increased Rad17 phosphorylation and hence resulted in the elevated Chk1 phosphorylation.
Because the stability of Rad17 is regulated by proteasome-dependent protein degradation during checkpoint recovery (1–3, 21), the involvement of proteasomal protein degradation was examined. Exposure to the proteasome inhibitor MG132 did not affect the increase in Rad17 phosphorylation induced by SU6656 (Fig. 5D). We confirmed that MG132 inhibits UV irradiation-induced Rad17 degradation as reported previously (Fig. 5E) (21). The amount of Rad17 was increased by SFK inhibition in Fig. 5C but not in Fig. 5D. In Fig. 5D, SU6656 was added after Adriamycin exposure. These results indicate that proteasomal degradation of Rad17 is not involved in the regulation of Rad17 phosphorylation by SFKs.
SFKs Regulate ATR Signaling via a Mechanism Other than Claspin Degradation
To examine whether SFKs regulate downstream components of Rad17 in addition to the Rad17 phosphorylation, the possibility that SFKs may regulate proteasomal degradation of Claspin was assessed. Claspin interacts with Chk1, recruits Chk1 to phosphorylated Rad17, and mediates ATR-dependent Chk1 phosphorylation (19, 20). During checkpoint recovery, proteasome-dependent degradation of Claspin, which is promoted by Plk1, contributes to the down-regulation of ATR-mediated Chk1 phosphorylation (22–24). Indeed, exposure to the Plk1 inhibitor BI2536 increased the amount of Claspin (Fig. 5G). By contrast, SU6656 did not increase the amount of Claspin, but it induced a much higher phosphorylation of Chk1 than did BI2536. These results suggest that SFKs are capable of suppressing ATR-mediated Chk1 phosphorylation via a mechanism other than Claspin degradation. Although we did not investigate whether SFK inhibition perturbs Chk1 dephosphorylation or degradation (25–30), these data suggest that SFKs regulate ATR signaling via ATR-dependent Rad17 phosphorylation.
v-Src Suppresses ATR-Chk1 Signaling
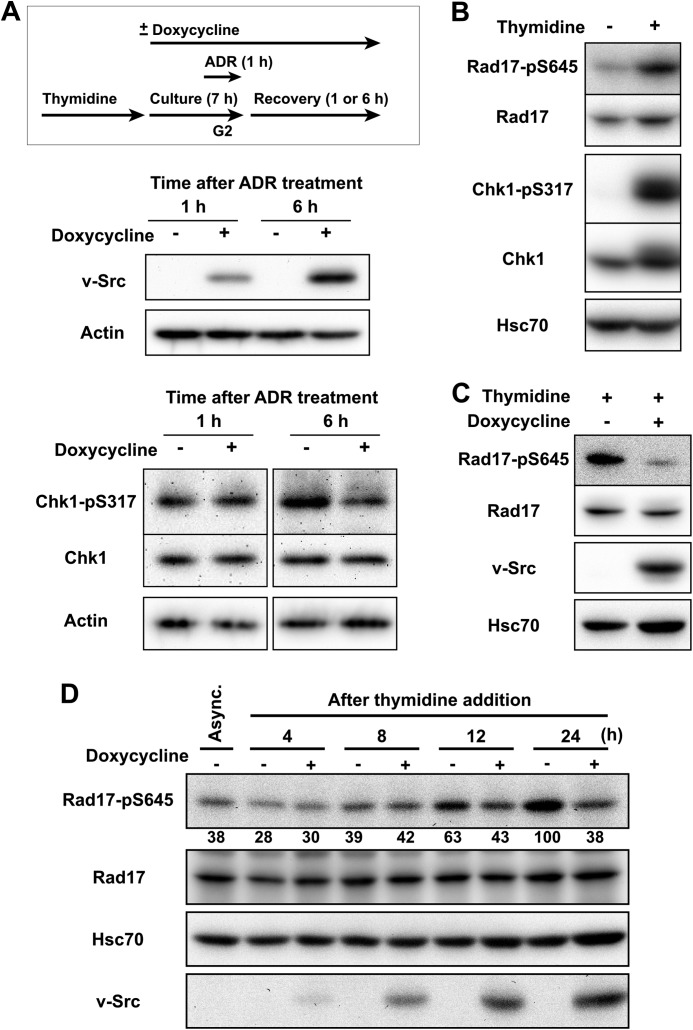
The data described above indicate that SFKs inhibit ATR-dependent Rad17 phosphorylation. To confirm the role of SFKs in ATR-Chk1 signaling, the impact of oncogenic v-Src on the ATR-dependent checkpoint was examined. For this purpose, we constructed HeLa S3 cells carrying an inducible v-Src allele (HeLa S3-TR/v-Src cells). Consistent with the data obtained with SFK inhibition, v-Src expression suppressed Chk1-Ser317 phosphorylation during recovery from Adriamycin-induced G2 checkpoint activation (Fig. 6A). This supports the above conclusion.
FIGURE 6.
v-Src suppresses ATR-Chk1 signaling. A, HeLa S3-TR/v-Src cells were synchronized at the G1/S boundary by exposure to thymidine. After release from thymidine block, doxycycline was added, and the cells were cultured for 6 h. Then the cells were exposed to 110 nm Adriamycin (ADR) and allowed to recover for the indicated times. SDS lysates were prepared and probed with the indicated antibodies. B, asynchronously growing HeLa S3 cells were exposed to 4 mm thymidine for 24 h. SDS lysates were prepared and probed with the indicated antibodies. C, HeLa S3-TR/v-Src cells were exposed to 4 mm thymidine for 24 h. Expression of v-Src was induced with doxycycline for 24 h. Cells were lysed in high salt buffer, and the lysates were probed with the indicated antibodies. D, the same experiment as in C. Cells were harvested at the indicate times after addition of thymidine and/or doxycycline. SDS lysates were prepared and probed with the indicated antibodies. Representative data from two independent experiments are shown. Async., asynchronously growing cells.
ATR-dependent DNA damage checkpoint is also activated by replication stress because replication arrest induces the generation of RPA-coated single-stranded DNA by helicase and polymerase uncoupling (4, 6, 80). To further confirm that SFKs regulate ATR-Chk1 signaling not through regulation of DSB repair, ATR-Chk1 signaling was activated by thymidine exposure, and the effect of v-Src expression on Rad17 phosphorylation was examined. Thymidine treatment induced Chk1-Ser317 and Rad17-Ser645 phosphorylation (Fig. 6B). In addition to the Chk1 phosphorylation induced by DSBs, v-Src also suppressed the Rad17 phosphorylation induced by replication stress upon exposure to thymidine (Fig. 6, C and D). This supports the conclusion that SFKs regulate Rad17 phosphorylation but not through the regulation of DSB repair.
SFKs Negatively Regulate ATR-Chk1 Signaling Activated by Replication Stress
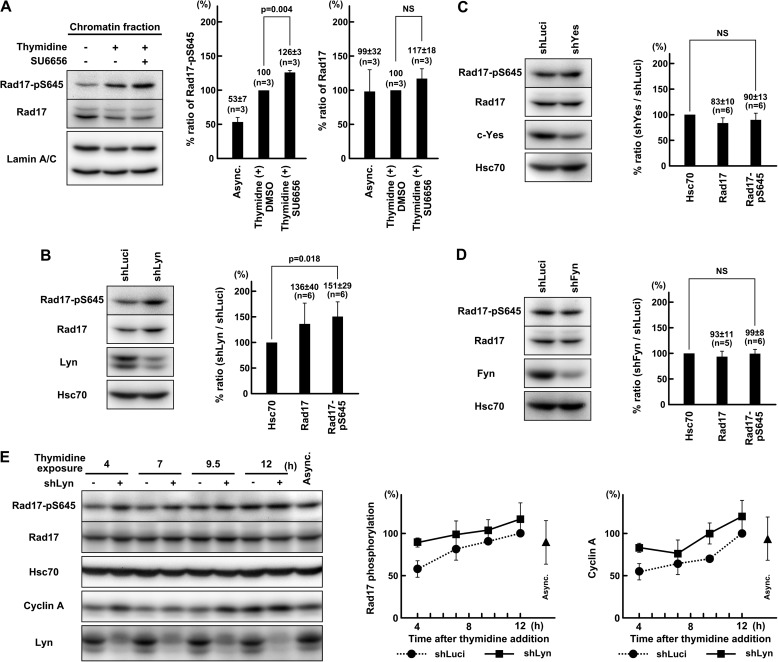
In the above experiments, v-Src suppressed ATR-Chk1 signaling activated by replication stress. To confirm that endogenous SFKs regulate ATR-Chk1 signaling independently of DSB repair, we examined whether endogenous SFKs also regulate ATR-Chk1 signaling activated by replication stress. For this purpose, HeLa S3 cells were treated with thymidine, and SFKs were inhibited. SU6656 increased Rad17 phosphorylation induced by thymidine (Fig. 7A). shRNA-mediated knockdown of Lyn, but not c-Yes or Fyn, increased Rad17-Ser645 phosphorylation (Fig. 7, B, C, D, and E). The effect of these shRNAs on Chk1 activation was marginal (data not shown); however, shLyn increased cyclin A accumulation (Fig. 7E). These data indicate that SFKs regulate ATR-Chk1 signaling activated by replication stress as well as DSBs. In these experiments, cells were not exposed to Adriamycin. These data further confirm that endogenous SFKs regulate ATR-Chk1 signaling in a manner that is independent of regulation of DSB repair.
FIGURE 7.
SFKs negatively regulate ATR-Chk1 signaling activated by replication stress. A, HeLa S3 cells were exposed to 4 mm thymidine for 24 h. Chromatin fractions were prepared as described under “Experimental Procedures” and probed with the indicated antibodies. Six hours before harvest, 10 μm SU6656 was added. B, C, and D, HeLa S3 cells were transfected with shRNA expression vectors targeting Lyn (shLyn), c-Yes (shYes), or Fyn (shFyn), and SDS lysates were prepared 72 h after transfection. Cells were exposed to 4 mm thymidine for 24 h before harvest. E, HeLa S3 cells were transfected with shLyn and inoculated 24 h after transfection to reduce confluence. At 48 h after transfection, the medium was changed with fresh medium containing 4 mm thymidine. Cells were harvested, and SDS lysates were prepared at the indicated times. The asynchronous control (Async.) was harvested 24 h after inoculation. The graph represents results obtained from two independent experiments. p values were calculated using t test. NS, not significant. Error bars represent S.D. shLuci, shRNA targeting luciferase.
SFKs Promote Checkpoint Silencing through Nuclear Protein Tyrosine Phosphorylation
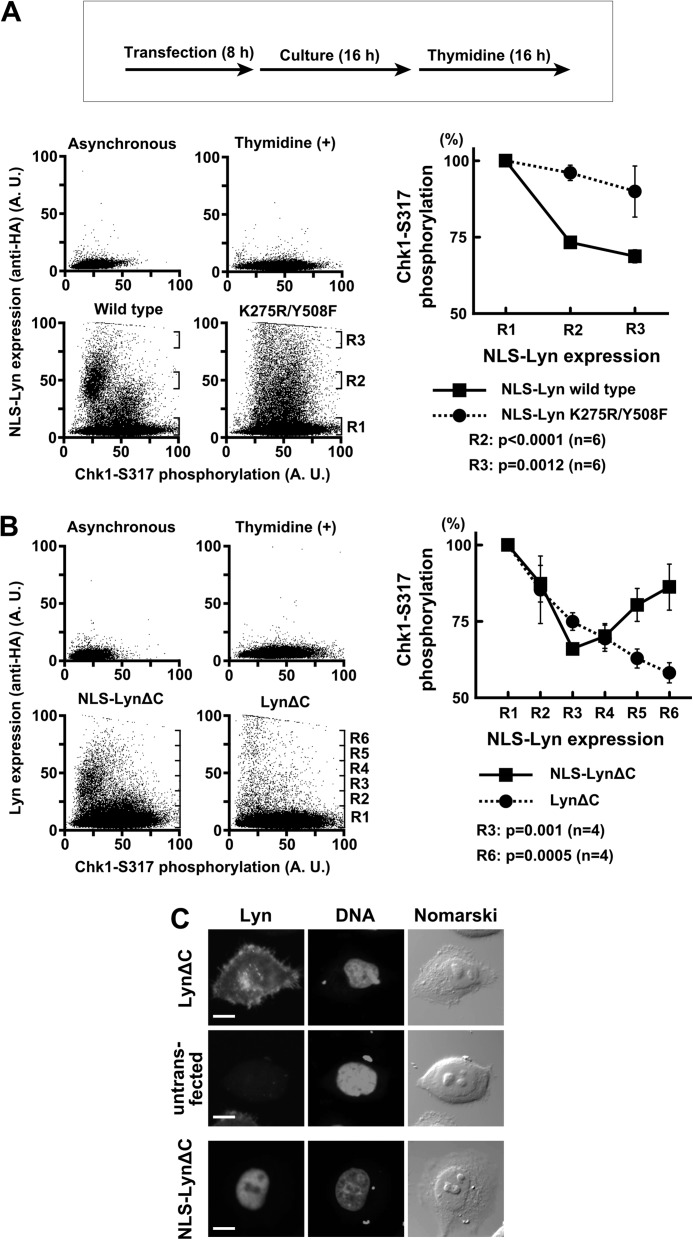
Several reports showed the nuclear localization of SFKs (39–45, 51). Therefore, we examined whether SFKs exert silencing of ATR-Chk1 signaling in the nucleus. For this purpose, the SFK member Lyn was tagged with a nuclear localization signal (NLS-Lyn) and transiently expressed in thymidine-treated cells. Transient expression of NLS-Lyn promotes protein tyrosine phosphorylation in the nucleus (43, 55). Chk1-Ser317 phosphorylation was induced by thymidine treatment and detected in flow cytometry (Fig. 8A). NLS-Lyn, but not the kinase-inactive mutant (NLS-Lyn-K275R/Y508F), suppressed Chk1-Ser317 phosphorylation, which is consistent with the above data.
FIGURE 8.
SFKs promote checkpoint silencing through nuclear protein tyrosine phosphorylation. A and B, HeLa S3 cells were transfected and subsequently exposed to thymidine for 16 h. The cells were fixed, stained with the indicated antibodies, and analyzed by flow cytometry. The intensity of the Chk1-Ser(P)317 signal was quantitated in each region (Rn) and expressed relative to that obtained with untransfected cells (R1). p values were calculated using t test. Error bars represent S.D. C, the cells treated as in B were fixed for immunofluorescence and stained as indicated. An untransfected cell on the same coverslip as LynΔC-transfected cells is shown as a control. Scale bar, 10 μm in all panels. A. U., arbitrary units.
NLS-Lyn and wild-type Lyn were compared in their ability to suppress Chk1 phosphorylation. Previously we showed that the N-terminal lipid modification of SFKs is critical for proper subcellular localization (37, 42). A constitutively active mutant of Lyn with the lipid modification (LynΔC) was localized in the nucleus as well as the cell membrane and the perinuclear cytoplasm (Fig. 8C), which is consistent with our previous reports (16, 50). By contrast, NLS-LynΔC accumulated in the nucleus. LynΔC also suppressed Chk1-Ser317 phosphorylation (Fig. 8B). However, as indicated in Fig. 8B (R3), when NLS-LynΔC and LynΔC were expressed in equal amounts, NLS-LynΔC suppressed Chk1 phosphorylation more effectively than did LynΔC. These results suggest that SFKs promote silencing of ATR-Chk1 signaling through protein tyrosine phosphorylation in the nucleus. Curiously, high level expression of NLS-LynΔC reincreased Chk1-Ser317 phosphorylation (Fig. 8B). Because inhibition of Chk1 activity increases ATR-dependent Chk1 phosphorylation (27, 81, 82), one of the possible explanations is that higher expression of NLS-LynΔC reincreases ATR-dependent Chk1 phosphorylation through inhibition of Chk1 activity. Conversely, another tyrosine kinase is involved in ATR activation (83, 84). Therefore, another possibility is that excessive protein tyrosine phosphorylation by NLS-LynΔC in the nucleus might stimulate other tyrosine phosphorylation-dependent checkpoint regulation in addition to ATR-Chk1 down-regulation. These data raise the possibility that SFKs regulate ATR-Chk1 signaling in the nucleus.
DISCUSSION
The present study reveals that SFKs promote recovery from G2 DNA damage checkpoint arrest by inhibiting ATR-Chk1 signaling. DNA damage checkpoint recovery involves at least two events: checkpoint silencing and cell cycle resumption (1–3). Plk1 and Aurora A are essential for cell cycle resumption (67, 68). By contrast, SFK activity is dispensable for cell cycle resumption but is required for checkpoint silencing (Fig. 2, A, B, and E). Thus, our findings shed light on the upstream signaling for inactivation of the ATR-dependent DNA damage checkpoint.
G2 checkpoint recovery is preceded by DSB repair; however, the delay in G2 checkpoint recovery induced by SFK inhibition is not due to a delay in DSB repair. When SFK inhibition resulted in increased phosphorylation of Chk1 and Rad17, the γ-H2AX signal had returned to basal levels and was not affected significantly by SFK inhibition or caffeine (Fig. 4, C, D, and E). Over the time course, SFK inhibition decreased the γ-H2AX signal (Fig. 4, F and G). Moreover, adding SU6656 later in the time course also increased Chk1 phosphorylation (Fig. 3J). Furthermore, SFKs regulate ATR-Chk1 signaling activated by replication stress (Figs. 6, C and D; 7, A, B, and E; and 8, A and B). SFKs also suppress ATR-Chk1 signaling induced by UV irradiation.8 Thus, regulation of ATR-Chk1 signaling by SFKs does not involve regulation of DSB repair, and the delay in G2 checkpoint recovery induced by SFK inhibition is due to prolonged activation of ATR-Chk1 signaling after completion of DSB repair.
Although activation of ATR by DSBs is mediated by ATM-dependent DSB end resection (7–11), the regulation of ATR-Chk1 signaling by SFKs is mostly independent of the regulation of ATM for the following reasons. First, SFKs regulate maintenance of G2 checkpoint arrest (Fig. 1, B, C, and D) that is dependent on ATR and Chk1 but is independent of ATM and Chk2 (Fig. 2, D, E, F, and G). Second, in the presence of the ATM inhibitor KU-55933, SFK inhibition still induced a delay in G2 checkpoint recovery and an increase in ATR-mediated Chk1 phosphorylation (Fig. 5, A and B). Consistent with these observations, ATM inhibition was shown not to affect ATR and Chk1 activity once ATM-dependent DSB end resection occurs and ATR is activated (16). Third, SFKs regulate the ATR-mediated Chk1 phosphorylation that is induced by replication stress in an ATM-independent manner (Figs. 6C; 7, A and B; and 8, A and B). Previous work reported the presence of at least two molecularly distinct G2 checkpoint arrests (10, 16, 85): early and transient ATM-dependent G2 checkpoint arrest and late and prolonged G2 checkpoint arrest whose maintenance is dependent on ATR and Chk1 activity but independent of ATM. Our results indicate that SFKs promote recovery from the late and prolonged G2 checkpoint arrest through negative regulation of ATR-Chk1 signaling.
Chk1 is dephosphorylated by PP1, PP2A, and PPM1D (25–27) and is also subjected to proteasomal degradation upon genotoxic stress (28–30). Although we cannot exclude the possibility that SFK inhibition perturbs Chk1 dephosphorylation or degradation, SFK inhibition increased and v-Src suppressed Rad17-Ser645 phosphorylation (Figs. 5, C and D; 6, C and D; and 7, A, B, and E), and the Rad17 phosphorylation regulates ATR-dependent Chk1 phosphorylation (17–19). Therefore, regulation of ATR-dependent Rad17 phosphorylation is involved in the mechanism by which SFKs regulate Chk1 phosphorylation and activation.
Currently the mechanism by which SFKs regulate Rad17 phosphorylation is unclear. Proteasomal degradation of Rad17 was reported (21), but SFKs regulate Rad17 phosphorylation independently of Rad17 degradation (Fig. 5D). Most likely, SFKs would affect upstream components of the ATR signaling pathway. Because ectopic expression of NLS-Lyn suppressed Chk1 phosphorylation (Fig. 8, A and B), SFKs would exert this role in the nucleus. We have not examined the effect of SFK inhibition on Claspin, RPA chromatin loading, and other checkpoint proteins.
Multiple SFK members have redundant function in inhibition of ATR-Chk1 signaling because overexpression of v-Src and Lyn inhibited ATR-Chk1 signaling (Figs. 6, A and C, and 8, A and B). However, shRNA targeted to Lyn, but not c-Yes or Fyn, increased Chk1 phosphorylation in thymidine-arrested cells (Fig. 7, B, C, and D). This should be due to the differential regulation of expression level, activity, and subcellular localization of each SFK member (31, 36–38).
The effect of v-Src on Rad17 phosphorylation is not due to a possible alteration in thymidine-induced cell cycle arrest for the following reasons. v-Src suppressed Adriamycin-induced Chk1 phosphorylation at 6 h, but not 1 h, after the end of Adriamycin exposure (Fig. 6A). v-Src suppressed Rad17 phosphorylation (Fig. 6C) but not ATR autophosphorylation at Thr1989 (86) induced by thymidine (data not shown), indicating that replication stress is induced. Furthermore, we confirmed that v-Src suppresses Chk1 phosphorylation without affecting thymidine arrest.9
SFK activation is found in a large number of common tumors; however, the role of SFKs in human cancer development and progression is still not fully understood (32). Disruptions in ATR-dependent checkpoint pathways cause genomic instability because ATR-Chk1 signaling is essential for the stability of DNA replication forks and recovery of stalled forks (4, 6). In addition, we and others showed that inhibition of ATR-Chk1 signaling induces premature mitotic entry and mitotic catastrophe (Fig. 2, A, B, C, and E) (12–16). In the present study, we showed that v-Src, an activated form of cellular SFKs, suppressed ATR-Chk1 signaling during checkpoint maintenance (Fig. 5, A and C). Therefore, the ATR suppression induced by activated SFKs should result in genomic instability through aberrant DNA replication and premature mitotic entry, which provides another molecular basis for the carcinogenesis promoted by SFK activation.
Conversely, given the oncogenic nature of v-Src, inhibition of ATR signaling by v-Src is a very surprising result because most tumor cells rely on the ATR-Chk1 pathway to survive genotoxic stress (87–90). Because certain tumor cells express high levels of Chk1 and its upstream regulators (89, 91–93), the negative regulation by activated SFKs would be counteracted in these cell types. SFKs are highly expressed and activated in colorectal and other cancer cell lines (94–96), and it may be fruitful to examine the role of endogenous SFKs in regulating ATR signaling in these cell lines.
In response to DNA damage caused by chemo- and radiotherapy, many tumor cells, especially those lacking p53 function, rely on the ATR-dependent DNA damage checkpoint for survival (87–90), and DNA damage checkpoint recovery should be an important chemotherapeutic target. Several SFK inhibitors are already available for clinical use or clinical trials. Therefore, our findings raise the possibility of chemical modulation of chemo- and radiotherapy by inhibition of SFKs through modulation of DNA damage checkpoint recovery.
The results of this study demonstrate that SFKs are important negative regulators of ATR-Chk1 signaling in DNA damage checkpoint recovery. Currently, the disappearance of DNA lesions is considered to bring about the end of checkpoint signaling (1–3); however, the present results proved that the termination of checkpoint signaling is an active process promoted by SFK activity, and our finding reveals a new layer of regulation between the completion of DNA repair and the initiation of checkpoint termination. Further study will reveal how the DNA repair process is monitored and how its completion sends a termination signal to DNA damage checkpoints for checkpoint recovery.
Acknowledgments
We thank Tadashi Yamamoto, Hiroshi Ohnishi, and Donald J. Fujita for plasmids.
This work was supported in part by grants-in-aid for scientific research, the Global Centers of Excellence (COE) Program (Global Center for Education and Research in Immune Regulation and Treatment), and Special Funds for Education and Research (Development of single photon emission computed tomography (SPECT) Probes for Pharmaceutical Innovation) from the Japanese Ministry of Education, Culture, Sports, Science and Technology and a research grant from the Mochida Memorial Foundation for Medical and Pharmaceutical Research.
Y. Fukumoto, unpublished data.
Y. Fukumoto, unpublished observations.
T. Miura and Y. Fukumoto, unpublished observations.
- ATM
- ataxia telangiectasia mutated
- ATR
- ATM- and Rad3-related
- DSB
- double strand break
- RPA
- replication protein A
- SFK
- Src family kinase
- SH
- Src homology
- NLS
- nuclear localization signal
- TR
- tetracycline repressor
- EGFP
- enhanced GFP
- γ-H2AX
- phosphorylated H2AX.
REFERENCES
- 1. Bartek J., Lukas J. (2007) DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19, 238–245 [DOI] [PubMed] [Google Scholar]
- 2. Calonge T. M., O'Connell M. J. (2008) Turning off the G2 DNA damage checkpoint. DNA Repair 7, 136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clémenson C., Marsolier-Kergoat M. C. (2009) DNA damage checkpoint inactivation: adaptation and recovery. DNA Repair 8, 1101–1109 [DOI] [PubMed] [Google Scholar]
- 4. Cimprich K. A., Cortez D. (2008) ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thompson L. H. (2012) Recognition, signaling, and repair of DNA double-strand breaks produced by ionizing radiation in mammalian cells: the molecular choreography. Mutat. Res. 751, 158–246 [DOI] [PubMed] [Google Scholar]
- 6. Flynn R. L., Zou L. (2011) ATR: a master conductor of cellular responses to DNA replication stress. Trends Biochem. Sci. 36, 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gatei M., Sloper K., Sorensen C., Syljuäsen R., Falck J., Hobson K., Savage K., Lukas J., Zhou B. B., Bartek J., Khanna K. K. (2003) Ataxia-telangiectasia-mutated (ATM) and NBS1-dependent phosphorylation of Chk1 on Ser-317 in response to ionizing radiation. J. Biol. Chem. 278, 14806–14811 [DOI] [PubMed] [Google Scholar]
- 8. Adams K. E., Medhurst A. L., Dart D. A., Lakin N. D. (2006) Recruitment of ATR to sites of ionizing radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene 25, 3894–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cuadrado M., Martinez-Pastor B., Murga M., Toledo L. I., Gutierrez-Martinez P., Lopez E., Fernandez-Capetillo O. (2006) ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J. Exp. Med. 203, 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jazayeri A., Falck J., Lukas C., Bartek J., Smith G. C., Lukas J., Jackson S. P. (2006) ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8, 37–45 [DOI] [PubMed] [Google Scholar]
- 11. Myers J. S., Cortez D. (2006) Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J. Biol. Chem. 281, 9346–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Graves P. R., Yu L., Schwarz J. K., Gales J., Sausville E. A., O'Connor P. M., Piwnica-Worms H. (2000) The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J. Biol. Chem. 275, 5600–5605 [DOI] [PubMed] [Google Scholar]
- 13. Liu Q., Guntuku S., Cui X. S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., DeMayo F., Bradley A., Donehower L. A., Elledge S. J. (2000) Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 14, 1448–1459 [PMC free article] [PubMed] [Google Scholar]
- 14. Kohn E. A., Yoo C. J., Eastman A. (2003) The protein kinase C inhibitor Gö6976 is a potent inhibitor of DNA damage-induces S and G2 cell cycle checkpoints. Cancer Res. 63, 31–35 [PubMed] [Google Scholar]
- 15. Latif C., den Elzen N. R., O'Connell M. J. (2004) DNA damage checkpoint maintenance through sustained Chk1 activity. J. Cell Sci. 117, 3489–3498 [DOI] [PubMed] [Google Scholar]
- 16. Shibata A., Barton O., Noon A. T., Dahm K., Deckbar D., Goodarzi A. A., Löbrich M., Jeggo P. A. (2010) Role of ATM and the damage response mediator proteins 53BP1 and MDC1 in the maintenance of G2/M checkpoint arrest. Mol. Cell. Biol. 30, 3371–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bao S., Tibbetts R. S., Brumbaugh K. M., Fang Y., Richardson D. A., Ali A., Chen S. M., Abraham R. T., Wang X. F. (2001) ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress response. Nature 411, 969–974 [DOI] [PubMed] [Google Scholar]
- 18. Post S., Weng Y. C., Cimprich K., Chen L. B., Xu Y., Lee E. Y. (2001) Phosphorylation of serines 635 and 645 of human Rad17 is cell cycle regulated and is required for G1/S checkpoint activation in response to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 98, 13102–13107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang X., Zou L., Lu T., Bao S., Hurov K. E., Hittelman W. N., Elledge S. J., Li L. (2006) Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol. Cell 23, 331–341 [DOI] [PubMed] [Google Scholar]
- 20. Kumagai A., Dunphy W. G. (2000) Claspin, a novel protein required for the activation of Chk1 during DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell 6, 839–849 [DOI] [PubMed] [Google Scholar]
- 21. Zhang L., Park C. H., Wu J., Kim H., Liu W., Fujita T., Balasubramani M., Schreiber E. M., Wang X. F., Wan Y. (2010) Proteolysis of Rad17 by Cdh1/APC regulates checkpoint termination and recovery from genotoxic stress. EMBO J. 29, 1726–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mailand N., Bekker-Jensen S., Bartek J., Lukas J. (2006) Destruction of Claspin by SCFβTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell 23, 307–318 [DOI] [PubMed] [Google Scholar]
- 23. Mamely I., van Vugt M. A., Smits V. A., Semple J. I., Lemmens B., Perrakis A., Medema R. H., Freire R. (2006) Polo-like kinase-1 controls proteasome-dependent degradation of claspin during checkpoint recovery. Curr. Biol. 16, 1950–1955 [DOI] [PubMed] [Google Scholar]
- 24. Peschiaroli A., Dorrello N. V., Guardavaccaro D., Venere M., Halazonetis T., Sherman N. E., Pagano M. (2006) SCFβTrCP-mediated degradation of claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell 23, 319–329 [DOI] [PubMed] [Google Scholar]
- 25. den Elzen N. R., O'Connell M. J. (2004) Recovery from DNA damage checkpoint arrest by PP1-mediated inhibition of Chk1. EMBO J. 23, 908–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu X., Nannenga B., Donehower L. A. (2005) PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes Dev. 19, 1162–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leung-Pineda V., Ryan C. E., Piwnica-Worms H. (2006) Phosphorylation of Chk1 by ATR is antagonized by a Chk1-regulated protein phosphatase 2A circuit. Mol. Cell. Biol. 26, 7529–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y. W., Otterness D. M., Chiang G. G., Xie W., Liu Y. C., Mercurio F., Abraham R. T. (2005) Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol. Cell 19, 607–618 [DOI] [PubMed] [Google Scholar]
- 29. Leung-Pineda V., Huh J., Piwnica-Worms H. (2009) DDB1 targets Chk1 to the Cul4 E3 ligase complex in normal cycling cells and in cells experiencing replication stress. Cancer Res. 69, 2630–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y. W., Brognard J., Coughlin C., You Z., Dolled-Filhart M., Aslanian A., Manning G., Abraham R. T., Hunter T. (2009) The F box protein Fbx6 regulates Chk1 stability and cellular sensitivity to replication stress. Mol. Cell 35, 442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thomas S. M., Brugge J. S. (1997) Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol. 13, 513–609 [DOI] [PubMed] [Google Scholar]
- 32. Yeatman T. J. (2004) A renaissance for SRC. Nat. Rev. Cancer 4, 470–780 [DOI] [PubMed] [Google Scholar]
- 33. Möhn H., Le Cabec V., Fischer S., Maridonneau-Parini I. (1995) The src-family protein-tyrosine kinase p59hck is located on the secretory granules in human neutrophils and translocates towards the phagosome during cell activation. Biochem. J. 309, 657–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bijlmakers M. J., Marsh M. (1999) Trafficking of an acylated cytosolic protein: newly synthesized p56lck travels to the plasma membrane via the exocytic pathway. J. Cell Biol. 145, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaplan K. B., Swedlow J. R., Varmus H. E., Morgan D. O. (1992) Association of p60c-src with endosomal membranes fibroblasts. J. Cell Biol. 118, 321–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kasahara K., Nakayama Y., Ikeda K., Fukushima Y., Matsuda D., Horimoto S., Yamaguchi N. (2004) Trafficking of Lyn through the Golgi caveolin involves the charged residues on αE and αI helices in the kinase domain. J. Cell Biol. 165, 641–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sato I., Obata Y., Kasahara K., Nakayama Y., Fukumoto Y., Yamasaki T., Yokoyama K. K., Saito T., Yamaguchi N. (2009) Differential trafficking of Src, Lyn, Yes and Fyn is specified by the state of palmitoylation in the SH4 domain. J. Cell Sci. 122, 965–975 [DOI] [PubMed] [Google Scholar]
- 38. Obata Y., Fukumoto Y., Nakayama Y., Kuga T., Dohmae N., Yamaguchi N. (2010) The Lyn kinase C-lobe mediates Golgi export of Lyn through conformation-dependent ACSL3 association. J. Cell Sci. 123, 2649–2662 [DOI] [PubMed] [Google Scholar]
- 39. Radha V., Nambirajan S., Swarup G. (1996) Association of Lyn tyrosine kinase with the nuclear matrix and cell-cycle-dependent changes in matrix-associated tyrosine kinase activity. Eur. J. Biochem. 236, 352–359 [DOI] [PubMed] [Google Scholar]
- 40. Jain A. K., Jaiswal A. K. (2007) GSK-3β acts upstream of Fyn kinase in regulation of nuclear export and degradation of NF-E2 related factor 2. J. Biol. Chem. 282, 16502–16510 [DOI] [PubMed] [Google Scholar]
- 41. Paliwal P., Radha V., Swarup G. (2007) Regulation of p73 by Hck through kinase-dependent and independent mechanisms. BMC Mol. Biol. 8, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ikeda K., Nakayama Y., Togashi Y., Obata Y., Kuga T., Kasahara K., Fukumoto Y., Yamaguchi N. (2008) Nuclear localization of Lyn tyrosine kinase mediated by inhibition of its kinase activity. Exp. Cell Res. 314, 3392–3404 [DOI] [PubMed] [Google Scholar]
- 43. Takahashi A., Obata Y., Fukumoto Y., Nakayama Y., Kasahara K., Kuga T., Higashiyama Y., Saito T., Yokoyama K. K., Yamaguchi N. (2009) Nuclear localization of Src-family tyrosine kinases is required for growth factor-induced euchromatinization. Exp. Cell Res. 315, 1117–1141 [DOI] [PubMed] [Google Scholar]
- 44. Kharbanda S., Yuan Z. M., Rubin E., Weichselbaum R., Kufe D. (1994) Activation of Src-like p56/p53Lyn tyrosine kinase by ionizing radiation. J. Biol. Chem. 269, 20739–20743 [PubMed] [Google Scholar]
- 45. Kharbanda S., Saleem A., Yuan Z. M., Kraeft S., Weichselbaum R., Chen L. B., Kufe D. (1996) Nuclear signaling induced by ionizing radiation involves colocalization of the activated p56/p53Lyn tyrosine kinase with p34cdc2. Cancer Res. 56, 3617–3621 [PubMed] [Google Scholar]
- 46. Qin S., Minami Y., Kurosaki T., Yamamura H. (1997) Distinctive functions of Syk and Lyn in mediating osmotic stress- and ultraviolet C irradiation-induced apoptosis in chicken B cells. J. Biol. Chem. 272, 17994–117999 [DOI] [PubMed] [Google Scholar]
- 47. Maruo A., Oishi I., Sada K., Nomi M., Kurosaki T., Minami Y., Yamamura H. (1999) Protein tyrosine kinase Lyn mediates apoptosis induced by topoisomerase II inhibitors in DT40 cells. Int. Immunol. 11, 1371–1380 [DOI] [PubMed] [Google Scholar]
- 48. Yoshida K., Weichselbaum R., Kharbanda S., Kufe D. (2000) Role for Lyn tyrosine kinase as a regulator of stress-activated protein kinase activity in response to DNA damage. Mol. Cell. Biol. 20, 5370–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Grishin A. V., Azhipa O., Semenov I., Corey S. J. (2001) Interaction between growth arrest-DNA damage protein 34 and Src kinase Lyn negatively regulates genotoxic apoptosis. Proc. Natl. Acad. Sci. U.S.A. 98, 10172–10177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shangary S., Lerner E. C., Zhan Q., Corey S. J., Smithgall T. E., Baskaran R. (2003) Lyn regulates the cell death response to ultraviolet radiation through c-Jun N terminal kinase-dependent Fas ligand activation. Exp. Cell Res. 289, 67–76 [DOI] [PubMed] [Google Scholar]
- 51. He Z., Cho Y. Y., Ma W. Y., Choi H. S., Bode A. M., Dong Z. (2005) Regulation of ultraviolet B-induced phosphorylation of histone H3 at serine 10 by Fyn kinase. J. Biol. Chem. 280, 2446–2454 [DOI] [PubMed] [Google Scholar]
- 52. Yamanashi Y., Fukushige S., Semba K., Sukegawa J., Miyajima N., Matsubara K., Yamamoto T., Toyoshima K. (1987) The yes-related cellular gene lyn encodes a possible tyrosine kinase similar to p56lck. Mol. Cell. Biol. 7, 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ohnishi H., Yamamori S., Ono K., Aoyagi K., Kondo S., Takahashi M. (2001) A src family tyrosine kinase inhibits neurotransmitter release from neuronal cells. Proc. Natl. Acad. Sci. U.S.A. 98, 10930–10935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bjorge J. D., Bellagamba C., Cheng H. C., Tanaka A., Wang J. H., Fujita D. J. (1995) Characterization of two activated mutants of human pp60c-src that escape c-Src kinase regulation by distinct mechanisms. J. Biol. Chem. 270, 24222–24228 [DOI] [PubMed] [Google Scholar]
- 55. Kubota S., Fukumoto Y., Aoyama K., Ishibashi K., Yuki R., Morinaga T., Honda T., Yamaguchi N., Kuga T., Tomonaga T., Yamaguchi N. (2013) Phosphorylation of KRAB-associated protein 1 (KAP1) at Tyr-449, Tyr-458, and Tyr-517 by nuclear tyrosine kinases inhibits the association of KAP1 and heterochromatin protein 1α (HP1α) with heterochromatin. J. Biol. Chem. 288, 17871–17883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nakayama Y., Igarashi A., Kikuchi I., Obata Y., Fukumoto Y., Yamaguchi N. (2009) Bleomycin-induced over-replication involves sustained inhibition of mitotic entry through the ATM/ATR pathway. Exp. Cell Res. 315, 2515–2528 [DOI] [PubMed] [Google Scholar]
- 57. Fukumoto Y., Obata Y., Ishibashi K., Tamura N., Kikuchi I., Aoyama K., Hattori Y., Tsuda K., Nakayama Y., Yamaguchi N. (2010) Cost-effective gene transfection by DNA compaction at pH 4.0 using acidified, long shelf-life polyethylenimine. Cytotechnology 62, 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Blake R. A., Broome M. A., Liu X., Wu J., Gishizky M., Sun L., Courtneidge S. A. (2000) SU6656, a selective Src family kinase inhibitor, used to probe the growth factor signaling. Mol. Cell. Biol. 20, 9018–9027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fukumoto Y., Dohmae N., Hanaoka F. (2008) Schizosaccharomyces pombe Ddb1 recruits substrate-specific adaptor proteins through a novel protein motif, the DDB-box. Mol. Cell. Biol. 28, 6746–6756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Méndez J., Stillman B. (2000) Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kikuchi I., Nakayama Y., Morinaga T., Fukumoto Y., Yamaguchi N. (2010) A decrease in cyclin B1 levels leads to polyploidization in DNA damage-induced senescence. Cell Biol. Int. 34, 645–653 [DOI] [PubMed] [Google Scholar]
- 62. Vassilev L. T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H., Heimbrook D. C., Chen L. (2006) Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl. Acad. Sci. U.S.A. 103, 10660–10665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nakayama Y., Matsui Y., Takeda Y., Okamoto M., Abe K., Fukumoto Y., Yamaguchi N. (2012) c-Src but not Fyn promotes proper spindle orientation in early prometaphase. J. Biol. Chem. 287, 24905–24915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Matsui Y., Nakayama Y., Okamoto M., Fukumoto Y., Yamaguchi N. (2012) Enrichment of cell populations in metaphase, anaphase, and telophase by synchronization using nocodazole and blebbistatin: a novel method suitable for examining dynamic changes in proteins during mitotic progression. Eur. J. Cell Biol. 91, 413–419 [DOI] [PubMed] [Google Scholar]
- 65. Riffell J. L., Zimmerman C., Khong A., McHardy L. M., Roberge M. (2009) Effects of chemical manipulation of mitotic arrest and slippage on cancer cell survival and proliferation. Cell Cycle 8, 3025–3038 [PubMed] [Google Scholar]
- 66. Fenech M. (2007) Cytokinesis-block micronucleus cytome assay. Nat. Protoc. 2, 1084–1104 [DOI] [PubMed] [Google Scholar]
- 67. van Vugt M. A., Brás A., Medema R. H. (2004) Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol. Cell 15, 799–811 [DOI] [PubMed] [Google Scholar]
- 68. Macůrek L., Lindqvist A., Lim D., Lampson M. A., Klompmaker R., Freire R., Clouin C., Taylor S. S., Yaffe M. B., Medema R. H. (2008) Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 455, 119–123 [DOI] [PubMed] [Google Scholar]
- 69. Zhang D., Zaugg K., Mak T. W., Elledge S. J. (2006) A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 126, 529–542 [DOI] [PubMed] [Google Scholar]
- 70. Kass E. M., Ahn J., Tanaka T., Freed-Pastor W. A., Keezer S., Prives C. (2007) Stability of checkpoint kinase 2 is regulated via phosphorylation at serine 456. J. Biol. Chem. 282, 30311–30321 [DOI] [PubMed] [Google Scholar]
- 71. Lovly C. M., Yan L., Ryan C. E., Takada S., Piwnica-Worms H. (2008) Regulation of Chk2 ubiquitylation and signaling through autophosphorylation of serine 379. Mol. Cell. Biol. 28, 5874–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ishimi Y., Komamura-Kohno Y., Kwon H. J., Yamada K., Nakanishi M. (2003) Identification of MCM4 as a target of the DNA replication block checkpoint system. J. Biol. Chem. 278, 24644–24650 [DOI] [PubMed] [Google Scholar]
- 73. Arienti K. L., Brunmark A., Axe F. U., McClure K., Lee A., Blevitt J., Neff D. K., Huang L., Crawford S., Pandit C. R., Karlsson L., Breitenbucher J. G. (2005) Checkpoint kinase inhibitors: SAR and radioprotective properties of a series of 2-arylbenzimidazoles. J. Med. Chem. 48, 1873–1885 [DOI] [PubMed] [Google Scholar]
- 74. Charrier J. D., Durrant S. J., Golec J. M., Kay D. P., Knegtel R. M., MacCormick S., Mortimore M., O'Donnell M. E., Pinder J. L., Reaper P. M., Rutherford A. P., Wang P. S., Young S. C., Pollard J. R. (2011) Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J. Med. Chem. 54, 2320–2330 [DOI] [PubMed] [Google Scholar]
- 75. Prevo R., Fokas E., Reaper P. M., Charlton P. A., Pollard J. R., McKenna W. G., Muschel R. J., Brunner T. B. (2012) The novel ATR inhibitor VE-821 increases sensitivity of pancreatic cancer cells to radiation and chemotherapy. Cancer Biol. Ther. 13, 1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vávrová J., Zárybnická L., Lukášová E., Řezáčová M., Novotná E., Sinkorová Z., Tichý A., Pejchal J., Durišová K. (2013) Inhibition of ATR kinase with the selective inhibitor VE-821 results in radiosensitization of cells of promyelocytic leukemia (HL-60). Radiat. Environ. Biophys. 52, 471–479 [DOI] [PubMed] [Google Scholar]
- 77. Katsuragi Y., Sagata N. (2004) Regulation of Chk1 kinase by autoinhibition and ATR-mediated phosphorylation. Mol. Biol. Cell 15, 1680–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ng C. P., Lee H. C., Ho C. W., Arooz T., Siu W. Y., Lau A., Poon R. Y. (2004) Differential mode of regulation of the checkpoint kinases CHK1 and CHK2 by their regulatory domains. J. Biol. Chem. 279, 8808–8819 [DOI] [PubMed] [Google Scholar]
- 79. Ward I. M., Chen J. (2001) Histone H2AX is phosphorylated in an ATR-dependent manner in response to replication stress. J. Biol. Chem. 276, 47759–47762 [DOI] [PubMed] [Google Scholar]
- 80. Byun T. S., Pacek M., Yee M. C., Walter J. C., Cimprich K. A. (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19, 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Syljuåsen R. G., Sørensen C. S., Hansen L. T., Fugger K., Lundin C., Johansson F., Helleday T., Sehested M., Lukas J., Bartek J. (2005) Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol. Cell. Biol. 25, 3553–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Morgan M. A., Parsels L. A., Zhao L., Parsels J. D., Davis M. A., Hassan M. C., Arumugarajah S., Hylander-Gans L., Morosini D., Simeone D. M., Canman C. E., Normolle D. P., Zabludoff S. D., Maybaum J., Lawrence T. S. (2010) Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 Involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res. 70, 4972–4981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wagner M. W., Li L. S., Morales J. C., Galindo C. L., Garner H. R., Bornmann W. G., Boothman D. A. (2008) Role of c-Abl kinase in DNA mismatch repair-dependent G2 cell cycle checkpoint arrest responses. J. Biol. Chem. 283, 21382–21393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang X., Zeng L., Wang J., Chau J. F., Lai K. P., Jia D., Poonepalli A., Hande M. P., Liu H., He G., He L., Li B. (2011) A positive role for c-Abl in Atm and Atr activation in DNA damage response. Cell Death Differ. 18, 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Xu B., Kim S. T., Lim D. S., Kastan M. B. (2002) Two molecularly distinct G2/M checkpoints are induced by ionizing irradiation. Mol. Cell. Biol. 22, 1049–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liu S., Shiotani B., Lahiri M., Maréchal A., Tse A., Leung C. C., Glover J. N., Yang X. H., Zou L. (2011) ATR autophosphorylation as a molecular switch for checkpoint activation. Mol. Cell 43, 192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhou B. B., Bartek J. (2004) Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat. Rev. Cancer 4, 216–225 [DOI] [PubMed] [Google Scholar]
- 88. Smith J., Tho L. M., Xu N., Gillespie D. A. (2010) The ATM-Chk2 and ATR-Chk1 pathways in DNA damage silencing and cancer. Adv. Cancer Res. 108, 73–112 [DOI] [PubMed] [Google Scholar]
- 89. Ma C. X., Janetka J. W., Piwnica-Worms H. (2011) Death by releasing the breaks: Chk1 inhibitors as cancer therapeutics. Trends Mol. Med. 17, 88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fokas E., Prevo R., Hammond E. M., Brunner T. B., McKenna W. G., Muschel R. J. (2014) Targeting ATR in DNA damage response and cancer therapeutics. Cancer Treat. Rev. 40, 109–117 [DOI] [PubMed] [Google Scholar]
- 91. Madoz-Gúrpide J., Cañamero M., Sanchez L., Solano J., Alfonso P., Casal J. I. (2007) A proteomics analysis of cell signaling alterations in colorectal cancer. Mol. Cell. Proteomics 6, 2150–2164 [DOI] [PubMed] [Google Scholar]
- 92. Verlinden L., Vanden Bempt I., Eelen G., Drijkoningen M., Verlinden I., Marchal K., De Wolf-Peeters C., Christiaens M. R., Michiels L., Bouillon R., Verstuyf A. (2007) The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor−/progesterone receptor−/HER-2− breast carcinomas. Cancer Res. 67, 6574–6581 [DOI] [PubMed] [Google Scholar]
- 93. Speers C., Tsimelzon A., Sexton K., Herrick A. M., Gutierrez C., Culhane A., Quackenbush J., Hilsenbeck S., Chang J., Brown P. (2009) Identification of novel kinase targets for the treatment of estrogen receptor-negative breast cancer. Clin. Cancer Res. 15, 6327–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Irby R. B., Yeatman T. J. (2000) Role of Src expression and activation in human cancer. Oncogene 19, 5636–5642 [DOI] [PubMed] [Google Scholar]
- 95. Martin G. S. (2001) The hunting of the Src. Nat. Rev. Mol. Cell Biol. 2, 467–475 [DOI] [PubMed] [Google Scholar]
- 96. Yeatman T. J. (2004) A renaissance for SRC. Nat. Rev. Cancer 4, 470–480 [DOI] [PubMed] [Google Scholar]