Background: Sulfur carrier proteins Rhd_2599, TusA, and DsrE2 occur in many sulfur oxidizing prokaryotes.
Results: Rhd_2599, TusA, and possibly DsrE2 are involved in cytoplasmic sulfur trafficking during dissimilatory sulfur oxidation.
Conclusion: Sulfur transfer from persulfide intermediates to dissimilatory sulfite reductase involves Rhd_2599, TusA, and possibly DsrE2.
Significance: Proteins involved in dissimilatory sulfur oxidation have been identified.
Keywords: Bacterial Metabolism, Intracellular Trafficking, Microbiology, Sulfur, Thiol, Allochromatium vinosum, Purple Sulfur Bacteria, Sulfur Oxidation, Sulfur Relay System, TusA
Abstract
The formation of periplasmic sulfur globules is an intermediate step during the oxidation of reduced sulfur compounds in various sulfur-oxidizing microorganisms. The mechanism of how this sulfur is activated and crosses the cytoplasmic membrane for further oxidation to sulfite by the dissimilatory reductase DsrAB is incompletely understood, but it has been well documented that the pathway involves sulfur trafficking mediated by sulfur-carrying proteins. So far sulfur transfer from DsrEFH to DsrC has been established. Persulfurated DsrC very probably serves as a direct substrate for DsrAB. Here, we introduce further important players in oxidative sulfur metabolism; the proteins Rhd_2599, TusA, and DsrE2 are strictly conserved in the Chromatiaceae, Chlorobiaceae, and Acidithiobacillaceae families of sulfur-oxidizing bacteria and are linked to genes encoding complexes involved in sulfur oxidation (Dsr or Hdr) in the latter two. Here we show via relative quantitative real-time PCR and microarray analysis an increase of mRNA levels under sulfur-oxidizing conditions for rhd_2599, tusA, and dsrE2 in Allochromatium vinosum. Transcriptomic patterns for the three genes match those of major genes for the sulfur-oxidizing machinery rather than those involved in biosynthesis of sulfur-containing biomolecules. TusA appears to be one of the major proteins in A. vinosum. A rhd_2599-tusA-dsrE2-deficient mutant strain, although not viable in liquid culture, was clearly sulfur oxidation negative upon growth on solid media containing sulfide. Rhd_2599, TusA, and DsrE2 bind sulfur atoms via conserved cysteine residues, and experimental evidence is provided for the transfer of sulfur between these proteins as well as to DsrEFH and DsrC.
Introduction
Reduced inorganic sulfur compounds serve as electron donors for carbon dioxide fixation during anoxygenic photosynthesis in purple and green sulfur bacteria. In the γ-proteobacterium Allochromatium vinosum periplasmic globules of zero-valent sulfur are formed as obligatory intermediates during the oxidation of sulfide, polysulfides, elemental sulfur, and thiosulfate to sulfate. The degradation of these globules involves essential steps in the cytoplasm and is catalyzed by soluble as well as membrane bound proteins of the Dsr system (1, 2). Despite its central importance, activation and transport of sulfur from the periplasm into the cytoplasm is not yet fully understood. The current model involves transport of sulfur into the cytoplasm via a persulfidic carrier molecule, possibly glutathione amide persulfide (3). It is well established that the Dsr mechanism includes the transfer of sulfur atoms from DsrEFH to DsrC, which in its persulfurated state then most likely serves as the substrate for dissimilatory sulfite reductase DsrAB (4). However, DsrEFH on its own is incapable of mobilizing sulfur from persulfidic carrier molecules (4) and, therefore, needs a donor protein. Intriguingly, DsrEFH and DsrC are homologs of Escherichia coli TusBCD and TusE, respectively (5, 6). These proteins are involved in the sulfur relay system leading to the synthesis of 2-thiouridine in the modified wobble base 5-methyl-aminomethyl-2-thiouridine ((mnm)5 s2U) in tRNA (7). The transferred sulfur originates from l-cysteine and is mobilized by the l-cysteine desulfurase IscS. The TusA protein is the first to accept sulfur from IscS and further transfers it to the TusD subunit of TusBCD. From here the transfer progresses to TusE and thiouridylase MnmA, which finally catalyzes 2-thiouridine formation.
A tusA gene also exists in A. vinosum opening the possibility that TusA may act as the cytoplasmic sulfur donor protein for DsrEFH in the purple sulfur bacterium. Bioinformatic and microarray analyses provided first hints that the role of TusA in bacteria not closely related to E. coli may not be limited to biosynthetic processes. In the acidophilic chemolithoautotrophic γ-proteobacterium Acidithiobacillus ferrooxidans, a tusA homologous gene exhibited a very conspicuous pattern of relative mRNA levels; significantly higher levels were observed under sulfur-oxidizing than under iron-oxidizing conditions (8). In A. ferrooxidans the tusA gene (AFE_2557) is flanked in the same direction of transcription by two genes encoding a rhodanese-like protein (rhd, AFE_2558) and a protein of the DsrE superfamily (AFE_2556, dsrE2). For the dsrE2 gene the relative mRNA level was also elevated under sulfur-oxidizing versus iron-oxidizing conditions. The rhd-tusA-dsrE2 arrangement is situated immediately upstream of genes encoding the putative heterodisulfide reductase-like complex HdrC1B1A1Orf2HdrC2B2. All of these genes followed the same transcription pattern. These findings led Quatrini et al. (8) to the following conclusions. 1) The Hdr-like complex was predicted to be responsible for the oxidation of organic persulfides that are formed as intermediates during the oxidation of externally available elemental sulfur to sulfite. 2) The rhodanese-like Rhd proteins, TusA, and DsrE2, were proposed to be involved in the transfer of sulfur from the persulfidic intermediates toward the Hdr complex.
The rhd-tusA-dsrE2 gene arrangement is not restricted to the genus Acidithiobacillus but is also present in A. vinosum (Alvin_2599–2601) and in a large number of further genome-sequenced chemotrophic and phototrophic sulfur-oxidizing prokaryotes (9). In most representatives of the phototrophic green sulfur bacteria Chlorobiaceae the complete set dsrE2-rhd-tusA is situated immediately upstream of the dsr gene cluster. Remarkably, the whole cluster is preceded by genes encoding the SoeABC-related PSRLC3 complex for which a role in the oxidation of sulfite in the cytoplasm is very probable (10, 11). Notably, the only Chlorobium species with incomplete rhd-tusA-dsrE2 clusters, Chlorobaculum parvum and Chlorobium ferrooxidans, are unable to convert accumulated sulfur globules to sulfate (12) and completely incapable of living on reduced sulfur compounds (13), respectively. Although the complete rhd-tusA-dsrE2 cluster is present in Chromatiaceae, an Alvin_2599 homolog is not present in phototrophic members of the Ectothiorhodospiraceae. In some of these organisms, tusA and dsrE2 are immediately linked with hdr genes; in others tusA resides close to the dsr and soeABC genes (9, 11, 14). The only phototrophic α-proteobacterium that contains tusA-dsrE2 is Rhodomicrobium vannielii, and in fact this is also the only genome-sequenced phototrophic member of this class that contains genes encoding the complete Dsr pathway. In this study we set out to investigate the roles of the rhodanese-like protein encoded next to tusA, TusA, and DsrE2 in dissimilatory sulfur metabolism and chose A. vinosum as a model organism.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
Table 1 lists the bacterial strains and plasmids that were used for this study. E. coli DH5α cells were employed for molecular cloning, whereas E. coli SM10 and E. coli S17-1 were used for conjugation with A. vinosum. E. coli BL21(DE3) was used for the overproduction of soluble proteins; for membrane-bound proteins E. coli strains C41 and C43 were employed. All E. coli strains were grown on LB medium. Wild type and mutant strains of A. vinosum DSM 180T were cultivated as described before (15). For photoorganoheterotrophic growth A. vinosum strains were cultivated on malate (16) supplemented with trace element solution SL12 (17). When cells were grown on plates the medium was solidified with 1% (w/v) Phytagel. 0.5% (w/v) NaCl was added to support gelling. For the characterization of phenotypes, A. vinosum strains were grown photolithoautotrophically on Pfennig's medium (18) lacking a sulfur source. The sulfur compound of interest was added directly to the freshly inoculated medium to start the experiment. Antibiotics were used at the following concentrations: for E. coli, 50 μg ml−1 kanamycin and 100 μg ml−1 ampicillin; for A. vinosum, 10 μg ml−1 kanamycin, 50 μg ml− rifampicin, 10 μg ml−1 ampicillin, and 10 μg ml−1 streptomycin.
TABLE 1.
Strains and plasmids used in this study
| Strains or plasmids | Genotype or phenotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| DH5α | F− ϕ80d lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1hsdR17 (rk− mk +) supE44 ë−thi-1 gyrA relA1 | (49) |
| S17-1 | 294 (recA pro res mod+) Tpr Smr (pRP4–2-Tc::Mu-Km::Tn7) | (50) |
| SM10 | Kmr supE44 thi-1 thr-1 recA leu B6 lacY1 ton A21 RP4–2-Tc::Mu-Km::Tn7 | (50) |
| BL21 (DE3) | F− ompT hsdSB (rB−mB−) gal dcm met(DE3) | Novagen |
| C41 (DE3) | Derived from BL21 (DE3), at least one uncharacterized mutation | (51) |
| C43 (DE3) | Derived from C41 (DE3), at least two uncharacterized mutations | (51) |
| A. vinosum strains | ||
| Rif50 | Rifr; spontaneous rifampicin-resistant mutant of A. vinosum DSM 180T | (31) |
| rhd_2599:: ΩSm | Rifr; insertion of ΩSm in rhd_2599 of Rif50 | This study |
| ΔtusA | Rifr; Rif50 with in-frame deletion of tusA | This study |
| Δrtd | Rifr; Rif50 with in-frame deletion of rhd_2599, tusA, and dsrE2 | This study |
| Δrtd::ΩKm | Rifr Kmr; Rif50 with in-frame deletion of rhd_2599, tusA, dsrE2, and ΩKm in center of deleted sequence | This study |
| Plasmids | ||
| pHP45-Ω | Apr Smr/Spcr | (52) |
| pHP45Ω-Km | Apr Kmr | (53) |
| pGEM-5Zf | Apr; lacZ f1 ori | Promega |
| pGEM5-tusA | Apr; rhd_2599-tusA-dsrE2 amplicon cloned into SalI restriction site of pGEM-5Zf | This study |
| pGEM5-rhd_2599::ΩSm | Apr Smr; SmaI fragment from pHP45-Ω ligated into Bsp68I restriction site of pGEM 5-tusA | This study |
| pK18mobsacB | Kmr, lacZ', sacB, Mob+ | (35) |
| pK18mobsacBΔtusA | Kmr; 1,21 kb amplicon with deleted tusA cloned into XbaI restriction site of pK18mobsacB | This study |
| pK18mobsacBΔrtd | Kmr; 1,21-kb amplicon with deleted rhd_2599-tusA-dsrE2 cloned into BamHI-restriction site of pK18mobsacB | This study |
| pSUP202 | Apr Cmr Tcr; Mob+ RP4 oriT | (50) |
| pSUP202-rhd_2599::ΩSm | SalI fragment from pGEM5-rhd_2599::ΩSm cloned into SalI of pSUP202 | This study |
| pSUP301 | Apr Kmr; RP4 oriT p15A ori Mob+ | (50) |
| pSUP301Δrtd | Apr; 1,21 kb amplicon with deleted rhd_2599-tusA-dsrE2 cloned into HindIII-restriction site of pSUP301 | This study |
| pSUP301Δrtd::ΩKm | Apr Kmr; pHP45Ω-Km kanamycin cassette inserted into pSUP301Δrtd using EcoRI restriction sites | This study |
| pET-15b | Apr, T7 promoter, His tag (N terminus) | Novagen |
| pET15bRhd | Apr, NdeI/XhoI fragment of amplified rhd_2599 in NdeI/XhoI of pET-15b | This study |
| pET15bRhd-C64S | Apr, NdeI/BamHI fragment of amplified rhd_2599-C64S in NdeI/BamHI of pET-15b | This study |
| pET15bRhd-C74S | Apr, NdeI/BamHI fragment of amplified rhd_2599-C74S in NdeI/BamHI of pET-15b | This study |
| pET15bTusA | Apr, NdeI/BamHI fragment of amplified tusA in NdeI/BamHI of pET-15b | This study |
| pET15bTusA-C15S | Apr; NdeI/BamHI fragment of amplified tusA-C15S in NdeI/BamHI of pET-15b | This study |
| pET15bEcTusA | Apr, NdeI/BamHI fragment of amplified E. coli tusA in NdeI/BamHI of pET-15b | This study |
| pASK-IBA5plus | Apr, Tet promoter, Strep-Tag (N-terminal) | IBA |
| pIBADsrE2 | Apr; BsaI fragment of amplified dsrE2 in BsaI of pASK-IBA5plus | This study |
| pIBADsrE2-C110S | Apr; BsaI fragment of amplified dsrE2-Cys110Ser in BsaI of pASK-IBA5plus | This study |
| pIBADsrE2-C120S | BsaI fragment of amplified dsrE2-C120S in BsaI of pASK-IBA5plus | This study |
| pIBADsrE2-C156S | Apr; BsaI fragment of amplified dsrE2-Cys156Ser in BsaI of pASK-IBA5plus | This study |
| pETEFH | Apr; NdeI/BamHI fragment of amplified dsrEFH in pET-15b | (54) |
| pETE78FH | Apr; NdeI/BamHI fragment of amplified dsrE78FH in pET-15b | (6) |
| pETCEX | Apr; NdeI/BamHI fragment of amplified dsrC in pET-15b | (5) |
| pETCEXSer-100 | Apr; NdeI/BamHI fragment of amplified dsrC100 in pET-15b | (5) |
| pETCEXSer-111 | Apr; NdeI/BamHI fragment of amplified dsrC111 in pET-15b | (5) |
RNA Preparation and cDNA Synthesis
Total RNA was extracted from A. vinosum wild type grown either photoorganoheterotrophically on malate (22 mm) or photolithoautotrophically on elemental sulfur (50 mm), sulfide (4 mm), or thiosulfate (5 mm) in thermostatted glass fermenters. The RNA extraction followed the protocol previously described by Weissgerber et al. (19). For cDNA synthesis the First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Schwerte, Germany) was used.
Analysis of Transcription by Quantitative Real-time PCR (qRT-PCR)
For qRT-PCR analysis 100 ng of RNA was used and analyzed via the QuantiTect SYBR green RT-PCR kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. Reactions without reverse transcriptase served as control for each sample. Alvin_0486 was used as a reference (19). The qRT-PCR conditions were as follows: 30 min at 50 °C, 15 s at 94 °C, 30 s at 58 °C, and 30 s at 72 °C. This was followed by a melting curve analysis in which the temperature was increased by 1 °C s−1 starting at 35 °C up to 95 °C. The qRT-PCR was performed in the iCycler iQ real-time detection system (Bio-Rad). The oligonucleotides used for this experiment are listed in Table 2. Analyses of melting curves and calculation of Ct (calculated threshold) values were automatically carried out by the iCycler iQ software. Ct values for each point in time were run in triplicate. Relative expression ratios (R) were calculated by the 2−ΔΔCt method (20) using Equations 1 and 2 with PLA referring to photolithotrophic growth and POH meaning photoorganoheterotroph growth.
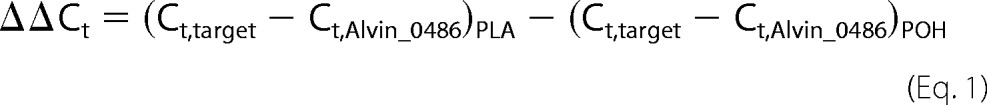 |
TABLE 2.
Oligonucleotides used in this study
Restriction sites and mutated nucleotides are printed in bold.
| Purpose | Oligonucleotide | Sequence 5′-3′ | Source or reference |
|---|---|---|---|
| Cloning of rhd_2599 | RhdTusA_for | GGGTAGGACCATATGGTGGTCAAC | This study |
| Rhd_rev | GTACTCGAGTCAGCCCTCGGG | This study | |
| Cloning of tusA into pET-15b | TusA_NdeI | GAGGCTCATATGGCTGATTTCGAT | This study |
| TusA_BamHI | GCCGTGCGGATCCATCAGGACTT | This study | |
| Cloning of EctusA into pET-15b | EcTusA_for | TGAAGCATATGCCCGATCTC | This study |
| EcTusA_rev | CATCAGGGATCCTCAACCGCC | This study | |
| Cloning of dsrE2 into pASK-IBA5+ | DE2_Strep_for | ATGGTAGGTCTCAGCGCCATGGAACAAAAGAAACTGGCGATC | This study |
| DE2_Strep_rev | ATGGTAGGTCTCATATCAGATGTAGAGACAGATGTCGCTCT | This study | |
| Cloning of rtd into pET-15b | RhdTusA_for | GGGTAGGACCATATGGTGGTCAAC | This study |
| DE2_rev | ACGTGCGGTCTCGAGACTTCAGATGTA | This study | |
| Cys-Ser exchange | DE_C110S_for | CGAGCTCTCACAGGAAGCC | This study |
| DE_C110S_rev | GGCTTCCTGTGAGAGCTCG | This study | |
| DE_C156S_for | AGCGACATCTCACTCTACATC | This study | |
| DE_C156S_rev | GATGTAGAGTGAGATGTCGCT | This study | |
| DE_C120S_rfor | TGATCGCCTCACAGATGACCG | This study | |
| DE_C120S_rev | CGGTCATCTGTGAGGCGATCA | This study | |
| IBA5_for | TGAGCTATGAGAAAGCGCC | This study | |
| IBA5_rev | GGCGACACGGAAATGTTGA | This study | |
| TusA_C15S_for | CCTGAACTCCCCGCTGC | This study | |
| TusA_C15S_rev | GCAGCGGGGAGTTCAGG | This study | |
| Rhd_C64S_for | GTGGTCATCTACTCCCGCAGC | This study | |
| Rhd_C64S_rev | GCTGCGGGAGTAGATGACCAC | This study | |
| Rhd_C74S_for | CAGGCTTCCGCCTATCTGATGC | This study | |
| Rhd_C74S_rev | GCATCAGATAGGCGGAAGCCTG | This study | |
| T7Prom | TAATACGACTCACTATAGGG | This study | |
| T7Term | GCTAGTTATTGCTCGCGG | This study | |
| Deletion of tusA | ΔTusA_for1 | GATCAAGAACTCGACTTCCACTTCCTGATC | This study |
| ΔTusA_rev1 | GATCAGGAAGTGGAAGTCGAGTTCTTGATC | This study | |
| ΔTusA_for2 | CAGCGCTCTAGACAGCTCAGGCGACAC | This study | |
| ΔTusA_rev2 | TTTGGCTCTAGAACGGTCATCTGACAG | This study | |
| Deletion of rtd | ΔRTD_F1 | CTTTGACACGGATCCGATAACG | This study |
| ΔRTD_R1 | GAGACTTCAGATGTAGACGTTGACCACACTATCG | This study | |
| ΔRTD_F2 | CGATAGTGTGGTCAACGTCTACATCTGAAGTCTC | This study | |
| ΔRTD_R2 | AAGTCGACGGATCCGCGATG | This study | |
| Interposon mutagenesis of rtd | RTD_Kan_F1 | CAGCGCAAGCTTCCGCGTG | This study |
| RTD_Kan_R1 | CTTCAGATGTAGGAATTCACCACACTATC | This study | |
| RTD_Kan_F2 | GATAGTGTGGTGAATTCCTACATCTGAAG | This study | |
| RTD_Kan_R2 | GACGCAAAGCTTGCACATTGG | This study | |
| qRT-PCR | RT-UROD-for | GTACCGCGCATCGAGGATT | (19) |
| RT-UROD-rev | GCATTACCGGCAGCGAGAA | (19) | |
| RT-Rhd-for | GTGTTGCTGGTGGACATCC | This study | |
| RT-Rhd-rev | GGCAGTAGATGACCACGTCG | This study | |
| RT-TusA-for | CGATCAAGAACTCGACGCAAGC | (19) | |
| RT-TusA-rev | GTTGCCGGTCTGCTTGGC | (19) | |
| RT-DsrE2-for | CGGTATGCAGGGCATGATGAC | (19) | |
| RT-DsrE2-rev | TTCGGCATATCGAAGAGGTCG | (19) |
Construction of A. vinosum Mutant Strains
For in-frame deletion of the rhd_2599-tusA-dsrE2 sequence and the single in-frame deletion of tusA, the plasmids pk18mobsacBΔrtd and pk18mobsacBΔtusA were created. PCR amplicons containing the deletions without disrupting the reading frame were obtained by gene SOEing (21) using the primers listed in Table 2. The amplicons were cloned into the respective restriction sites of pk18mobsacB. For replacement of the rhd_2599-tusA-dsrE2 cluster by a kanamycin resistance interposon, plasmid pSUP301Δrtd::ΩKm was generated; via gene SOEing a restriction site for EcoRI was created in the center of the remains of the gene cluster using pk18mobsacBΔrtd DNA as template. Before the Ω-kanamycin cassette from pHP45Ω-Km was cloned into the EcoRI site, the amplicon itself was inserted into the HindIII restriction site of pSUP301. For the generation of pSUP202-rhd_2599::ΩSm the Ω-Sm/Spc cassette from pHP45-Ω was isolated by digestion with SmaI and ligated into the Bsp68I site in the rhd_2599 sequence in pGEM-rtd (Table 1). The complete insert (rhd_2599::ΩSm-tusA-dsrE2) was then cut from the vector with SalI and cloned into the corresponding site in pSUP202. Plasmids based on pk18mobsacB and pSUP301 were transferred to E. coli S17-1, whereas pSUP202-rhd_2599::ΩSm was transferred to E. coli SM10 for conjugation with A. vinosum Rif50. The E. coli cells were mixed with stationary phase cells of A. vinosum in a ratio of 1:3 and incubated anaerobically on cellulose nitrate membranes (pore size 0.45 μm) on solid RCV medium (16). After 2 days the cells were washed from the filters with RCV medium. Cells were plated on RCV medium containing 1% (w/v) Phytagel for solidification. The plates were further supplemented with 0.5% (w/v) NaCl to support the gelling process and 0.02% (w/v) Na2S2O3 × 5 H2O, 2 mm sodium acetate, and 2.6 ml feeding solution (for 100 ml: 3.1 g of NaSH × H2O, 5.0 g of NaHCO3) per liter for growth enhancement. The selection process for in-frame mutants was as follows. Single-crossover mutants were identified via the resistance toward kanamycin provided by the pk18mobsacB-based plasmids and verified via colony PCR. After growth in non-selective RCV medium for three generations, double-crossover mutants were identified by plating cells on solid medium (see above) that contained 10% (w/v) sucrose. When plasmids of the pSUP series were used, double-crossover mutants were identified by their resistance to kanamycin or streptomycin and loss of the vector-encoded ampicillin resistance. Genotypes were verified via colony PCR and Southern blotting.
Characterization of A. vinosum Mutant Strain Phenotypes
To determine the turnover of reduced sulfur compounds under photolithoautotrophic conditions A. vinosum wild type and mutant strains were first grown photoorganoheterotrophically in liquid culture. Then 80 ml of the cultures were harvested, washed with Pfennig's medium without sulfur compounds (2500 × g; 10 min; room temperature), and used to inoculate 100 ml of Pfennig's medium in completely filled screw-capped bottles. Sulfide (4 mm), thiosulfate (5 mm), or sulfur (50 mm) was then added as sulfur substrates. The cultures were continuously illuminated and kept at 30 °C throughout the experiment. Elemental sulfur was determined via cyanolysis (22), and sulfate was measured by the method of Sörbo (23).
Cloning, Site-directed Mutagenesis, Overproduction, and Purification of Recombinant Proteins
For the amplification of the Alvin_2599 (rhd_2599), Alvin_2600 (tusA), and Alvin_2601 (dsrE2) genes, genomic DNA of A. vinosum served as the template. The oligonucleotides used for amplification and introduction of restriction sites for molecular cloning are listed in Table 2. After digestion with the respective restriction enzymes, the PCR products for rhd_2599 and tusA were ligated into the corresponding sites of pET-15b (Novagen) resulting in amino-terminally His-tagged proteins. The dsrE2 amplicon was ligated into pASK-IBA5plus (IBA, Göttingen, Germany) and thereby fused to an amino-terminal Strep tag. 500 ml of LB medium were inoculated with 5% (v/v) E. coli precultures hosting the respective plasmid and cultivated at 37 °C and 180 rpm until an A600 of 0.6–0.8 was reached. After induction with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside (for pET-15b-derived plasmids) or 50 μg ml−1 anhydrotetracycline (for the pASK-IBA5plus derived plasmid), the cells were cultivated for 2 more hours under the same conditions and harvested (14,000 × g; 20 min; 4 °C). Pellets were resuspended in a lysis buffer specific for the respective overproduction system, disrupted via sonication, and purified according to the manufacturer's instructions. DsrE2 and its mutant variants were isolated from the membrane fraction of E. coli. The membranes were separated from the soluble fraction after sonication via ultracentrifugation (145,000 rpm; 2 h; 4 °C). Solubilization was carried out with 1% Triton X-100 overnight on ice under gentle stirring. After another ultracentrifugation step the proteins were isolated from the supernatant. Buffers for purification and the storage buffer contained 0.1% (v/v) Triton X-100. Point mutations were introduced into rhd_2599, tusA, and dsrE2 via gene splicing by overlap extension (21) using standard PCR with Pfu polymerase (Thermo Fisher Scientific, Schwerte, Germany). Plasmids pET15bTusA, pET15Rhd, and pIBADsrE2 (Table 1), respectively, served as templates for PCR reactions using the primers listed in Table 2.
Preparation of Proteins for Electrophoretic Separation
For the electrophoretic separation of proteins by their molecular mass, the protocol by Laemmli was used. For native gel electrophoresis, 100 pmol of DsrEFH and 800 pmol of TusA from A. vinosum or E. coli were incubated as described previously (4). For the separation of native proteins, the Laemmli method was modified by preparing buffers without SDS. Gels were stained with 0.25% (w/v) Coomassie Brilliant Blue R250, 50% (v/v) methanol, and 10% (v/v) acetic acid. For destaining, 20% (v/v) methanol, 10% (v/v) acetic acid was used.
Thiosulfate-Cyanide Sulfurtransferase, Glutathionepersulfide:Cyanide Sulfurtransferase Activity
Thiosulfate:cyanide sulfurtransferase (rhodanese) activity was measured according to Ray et al. (24). The assay contained 100 mm glycine, pH 8.9, 50 mm sodium thiosulfate, 50 mm NaCN, and enzyme in a final volume of 500 μl. The reaction was started by adding NaCN, and the mixture was incubated for 1 min at 30 °C. 250 μl of 15% formaldehyde was used to stop the reaction before 750 μl of ferric nitrate reagent (25 g of Fe(NO3)3 × 9 H2O and 50 ml of 65% HNO3 per 375 ml) was added. Absorption was measured at 460 nm. Enzyme units are defined as the amount that catalyzed the production of 1 μmol of thiocyanate per min. As an alternative substrate, GSSH was tested in a concentration of 50 mm. GSSH was synthesized by incubating 500 mm oxidized glutathione with 500 mm sulfide for 30 min at 30 °C (25). Completion of the reaction was verified by the absence of sulfide in the product solution following the protocol for sulfide quantification published by Rethmeier et al. (26).
Sulfur Binding and Transfer Experiments with MALDI-TOF or N-(iodoacetyl)-N′-(5-sulfo-1-naphtyl)-ethylenediamine (1,5-IAEDANS)
Sulfur binding and transfer experiments with MALDI-TOF were performed exactly as described before (4). Under the applied experimental conditions, masses were detected for all tested proteins that agreed with the calculated masses within a range of a maximum of 3 Da. The basic principle of the identification of protein-bound persulfides by use of an 1,5-IAEDANS is documented in Zheng et al. (27). We applied this method based on the protocol of Thomé et al. (28). Proteins were always treated first with 2 mm dithiothreitol (DTT) for 30 min at room temperature to reduce thiol groups and to remove any preexisting persulfides. Excess DTT was removed by gel filtration on PD Mini-Trap columns (GE Healthcare) using 50 mm Tris-HCl, pH 7.5, 100 mm NaCl. When DsrE2 was analyzed, the buffer in addition contained 0.1% Triton X-100. For testing the principle reactivity of the reduced proteins with 1,5-IAEDANS, the proteins were concentrated using Vivaspin 500 centrifuge concentrators (5-kDa molecular weight cutoff, Sigma) to about 800 μg ml−1 at this stage. A volume containing 200 pmol of protein was then brought to a volume of 20 μl, and 1,5 IAEDANS was added. Conditions and further treatment were as described below for the sulfide-treated proteins. For persulfuration of proteins before treatment with 1,5-IAEDANS, the protein solutions eluted from PD Mini-Trap columns were immediately incubated with 4 mm NaHS or 4 mm sodium thiosulfate for 1 h at 30 °C followed by dialysis against the aforementioned Tris-HCl buffers to remove any excess of the reduced sulfur compound. The protein solutions were then concentrated to 300–700 μg ml−1. 100 pmol (in the case of DsrEFH) or 200 pmol (all other proteins) were incubated with 0.5 nmol (1 μl of a 0.5 mm stock solution) of 1,5-IAEDANS in a final volume of 21 μl for 1 h in the dark at 4 °C. Unbound 1,5-IAEDANS was allowed to react with 100 nmol l-cysteine (1 μl of a 100 mm stock solution were added) for 30 min at room temperature to prevent reaction of 1,5-IAEDANS in subsequent reaction steps. The reaction mixtures were now treated with DTT (2 μl of a 200 mm stock solution, i.e. 200 nmol) for 30 min at room temperature to reductively release bound persulfides as 1,5-AEDANS-sulfide conjugates. The samples were then mixed with 1 μl of native loading buffer (2 m sucrose, 1% (w/v) bromphenol blue), and the complete reaction mixture was applied to a 15% Laemmli gel that was run in the dark. Note that the pH of the resolving gel was adjusted to pH 9.5 to improve separation. The gels were then analyzed under UV light for visualization of 1,5-AEDANS-labeled proteins. The same gels were later stained with Coomassie Brilliant Blue to allow comparison of protein amounts in each lane. When sulfur transfer from one protein to another was studied, 100 pmol (in case of DsrEFH) or 200 pmol (all other proteins) of persulfurated donor protein was combined with the same amount of acceptor protein in a final volume of 20 μl of 50 mm Tris-HCl, pH 7.5, 100 mm NaCl for 1 h at 30 °C followed by the addition of 1,5 IAEDANS and further treatment as described above.
RESULTS
In A. vinosum Expression of rhd_2599, tusA, and dsrE2 Is Triggered under Sulfur Oxidizing Conditions
Recently, we assessed the genome wide transcriptional response of A. vinosum upon a shift from photoorganoheterotrophic to photolithoautotrophic growth on different inorganic sulfur compounds (19). In this work the cells were harvested at the time of maximum oxidation rate for the respective sulfur compound. In this experimental setting, relative mRNA levels for tusA and dsrE2 increased on sulfide and thiosulfate compared with malate, whereas the relative transcription of the rhodanese gene appeared unchanged. These results were verified via relative qRT-PCR of tusA and dsrE2 but not for rhd_2599 (19).
For assessment of a potential co-transcription of the three genes, we synthesized cDNA using the DE2_rev oligonucleotide from RNA that was isolated from sulfide-grown cells. Subsequent PCR yielded amplicons for the individual genes as well as for the rhd_2599-tusA and the tusA-dsrE2 combinations (Fig. 1, A and B), indicating that the three genes form a transcriptional unit. The bands obtained for rhd_2599 and the rhd_2599-tusA combinations (Fig. 1B, lanes 1 and 4) appear faint compared with those for tusA, dsrE2, and the tusA-dsrE2 combination (Fig. 1B, lanes 2, 3, and 5). This could be explained by two promoter sites of different strengths such that there is transcriptional linkage of rhd_2599 and its downstream genes but transcription of tusA/dsrE2 also occurs independently. Indeed, the promoter recognition program BPROM (29) predicts two promoter sites for the rhd_2599-tusA-dsrE2 region; the first is located upstream of rhd_2599, and the second in the intergenic space between rhd_2599 and tusA (Fig. 1A).
FIGURE 1.
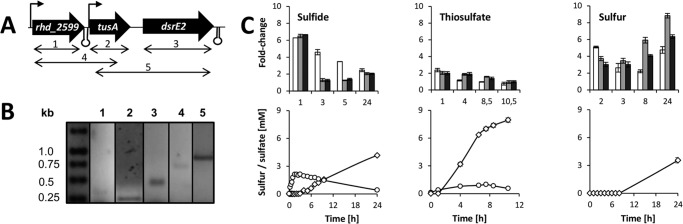
Transcription of rhd_2599, tusA, and dsrE2. A, genomic organization of the rhd_2599-tusA-dsrE2 gene cluster. Promoter and terminator sites predicted by BPROM (Softberry) (29) and TransTermHP (45), respectively, are indicated. Arrows below the genes indicate sequences amplified in PCR reactions 1–5 intended to evaluate (co)-transcription of the genes. B, agarose gel of the PCR products with the following expected sizes: 1, 330 bp; 2, 235 bp; 3, 495 bp; 4, 700 bp; 5, 860 bp. C, relative mRNA levels in A. vinosum during growth on sulfide, thiosulfate, and elemental sulfur compared with growth on malate. Results for rhd_2599 are shown as white bars, those for tusA as gray bars, and dsrE2 as black bars. mRNA levels for Alvin_0486 served as an endogenous reference (19). Concentrations of internal sulfur (○) and sulfate (♢) in the A. vinosum cultures are presented in the lower panels. Points represent means of three replicates. Error bars are too small to be visible in almost all cases.
Our initial whole genome transcriptional profiling study on A. vinosum covered only a single time point during the complex process of sulfur species conversion (19). We, therefore, reevaluated mRNA levels for tusA, dsrE2, and rhd_2599 by relative qRT-PCR at ample time points that were set such that they covered the phases of maximum oxidation rates for the respective sulfur compound, the point of maximum sulfur globule accumulation, and the period of sulfur globule conversion to sulfate (Fig. 1C). In the phases of maximum oxidation rates, our relative qRT-PCR results for tusA and dsrE2 exactly matched those of the previous study. qRT-PCR is more reliable and sensitive than genome-wide microarray analysis, and accordingly, we now also detected a significant positive transcriptional response for rhd_2599 in the presence of sulfide, thiosulfate, and elemental sulfur (Fig. 1C). The overall expression patterns for tusA and dsrE2 were very similar and differed by only a few percentage points with sulfide and thiosulfate as substrates. The effect exerted by thiosulfate was generally less prominent for all three genes than that observed on sulfide and elemental sulfur. Cells growing on thiosulfate contain lower concentration of sulfur stored in intracellular sulfur globules (Fig. 1C, lower panels) opening the possibility that the increased expression of rhd_2599, tusA, and dsrE2 is correlated with high concentrations of stored sulfur.
Transcriptome Evidence Toward the Physiological Function of Rhd, TusA, and DsrE2
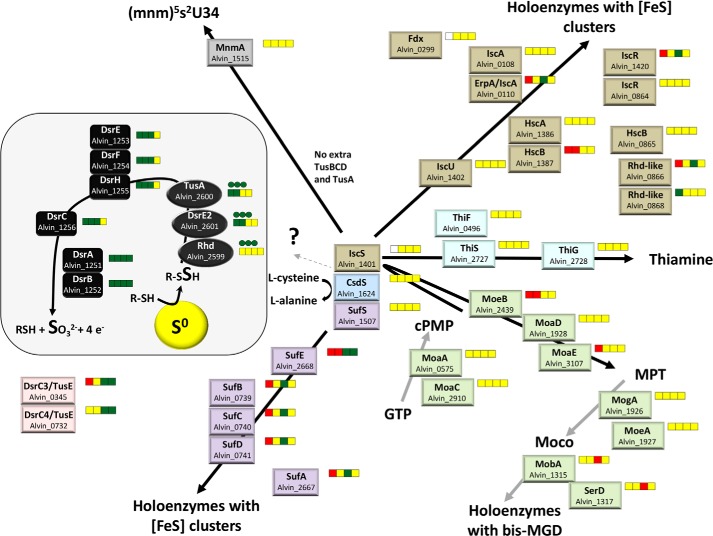
In E. coli, TusA affects not only the biosynthesis of 2-thiourdine but also the formation of FeS-clusters and the molybdenum cofactor (30). Besides proteins of central carbon metabolism, several important enzymes of the sulfur oxidation pathway in A. vinosum contain FeS-clusters, including DsrAB, DsrL, and the membrane complex DsrMKJOP (1, 2, 31, 32). Another example is the iron-sulfur molybdoenzyme SoeABC, the major player in the oxidation of the DsrAB product sulfite (11). It can, therefore, not a priori been excluded that the close genomic association of the rhd-tusA-dsrE2 genes with sulfur oxidation genes is related with a function of the former in biosynthesis of co-factors of the latter. If the function of A. vinosum TusA and also possibly Rhd and DsrE2 is indeed limited to biosynthesis, this should be reflected in similar transcription patterns of rhd-tusA-dsrE2 and the genes encoding proteins involved in biosynthetic machineries. Therefore, we compared the expression profiles of rhd-tusA-dsrE2 with those of the genes involved in biosynthesis of sulfur-containing co-factors. Data were taken from Weissgerber et al. (19) and were complemented by the qRT-PCR results described above (Fig. 2).
FIGURE 2.
Analysis of pathways for biosynthesis of FeS clusters, the molybdenum cofactor (Moco), thiamine, and 2-thiouridine in A. vinosum as predicted from the genome sequence and the transcription patterns of the respective genes. The transcriptomic profiles are depicted next to the respective protein and are color-coded; relative -fold changes in mRNA levels >2 (green) were considered significantly enhanced. Relative changes <0.5 (red) were considered as indicating significant decreases in mRNA levels. Relative -fold changes between 0.5 and 2 (yellow) indicated unchanged mRNA levels. For rhd_2599, tusA, and dsrE2, the results obtained via qRT-PCR in this study (values taken from the experiments presented in Fig. 1) are depicted in cycles. Administered sulfur compounds from left to right: sulfide, thiosulfate, elemental sulfur, and sulfite. In the figure DsrC/TusE proteins without any cysteine residues are omitted. cPMP, cyclic pyranopterin monophosphate; MPT, molybdopterin; bis-MGD, bis-molybdopterin-guanine-dinucleotide cofactor.
A. vinosum encodes the complete Isc and the Suf systems for the biosynthesis of FeS clusters. Genes crucial for the biosynthesis of the molybdenum cofactor and thiamine were also found (Fig. 2). Transcription of rhd-tusA-dsrE2 follows the same general pattern as that of the established sulfur-binding proteins of the Dsr pathway, DsrEFH, and DsrC as well as that of the dissimilatory sulfite reductase DsrAB itself. A very similar rise in relative mRNA levels in response to the presence of reduced sulfur compounds was seen for the genes encoding most other components of the Dsr system, genes for sulfur globule proteins, and for the cytoplasmic sulfite oxidation pathways (19). In contrast, similar patterns were not observed for any of the biosynthetic pathways (Fig. 2). Among the isc genes, only two (Alvin_0110 and Alvin_1420, encoding the iron-carrier IscA and the regulator IscR, respectively), responded positively under just single-growth conditions, i.e. in the presence of elemental sulfur. The same substrate triggered the expression of sufEBCDA, whereas the transcription of the cysteine desulfurase gene sufS remained unaltered under all tested conditions. Neither isc nor suf gene expression changed in the presence of sulfide and thiosulfate, the substrates that clearly exerted the strongest effect on rhd-tusA-dsrE2 and the established sulfur oxidation genes (Fig. 2 and Weissgerber et al. (19)). The levels for mRNAs of genes for molybdenum cofactor biosynthesis were either not affected or negatively affected by reduced sulfur compounds.
It is currently unclear which proteins might be involved in the synthesis of 2-thiouridine in A. vinosum. In E. coli, a multiprotein sulfur relay system involving TusA, TusBCD, TusE, and finally MnmA is responsible for this process. A. vinosum harbors a mnmA homolog the relative mRNA levels of which do not respond to reduced sulfur compounds. Extra copies of tusBCD apart from dsrEFH are not present. The tusA gene is also present in only a single copy. The tusE homolog dsrC is present with five genomic copies. All TusE/DsrC polypeptides functional in sulfur transfer contain a strictly conserved Cys residue (termed CysA) at the penultimate position in the carboxyl terminus (9) that serves as the site of sulfur binding (6). At a distance of 10 residues, DsrC proteins from sulfur oxidizers have an additional conserved cysteine (CysB). One A. vinosum DsrC homolog (Alvin_0345) has both conserved cysteine residues. This protein has recently been proposed to functionally operate as TusE (9). Neither CysA nor CysB is present in Alvin_0028 and Alvin_1508. These proteins are predicted to serve a regulatory function and have been termed regulatory sulfur-related proteins, RspA (9). Alvin_0732 contains only CysB that has no equivalent in E. coli TusE. Alvin_0732 also falls into the RspA group (6, 9). Increased mRNA levels were found on elemental sulfur and sulfite for Alvin_0732 and Alvin_0345.
Construction of rhd_2599-, tusA-, and dsrE2-deficient A. vinosum Mutants
To further assess the importance of the Rhd_2599, TusA, and DsrE2 proteins in A. vinosum, we set out to generate the mutant strains A. vinosum ΔtusA and A. vinosum Δrtd, carrying in-frame deletions of the tusA and the complete rhd-tusA-dsrE2 cluster, respectively. We have applied similar methodology successfully for construction of a whole range of A. vinosum deletion strains (33, 34). Amplified sequences carrying the deletion without disrupting the reading frame were cloned into the mobilizable suicide vector pk18mobsacB (35). Single crossover mutants, which were identified via the plasmid-encoded kanamycin resistance, were grown for several generations in non-selective RCV medium before they were plated on solid RCV medium containing 10% sucrose. Sucrose induces the gene expression of sacB encoding levansucrase leading to the death of cells still harboring the plasmid. Although single crossover mutants were verified, the next step yielded only revertants to the wild type genotype. Usually we obtain ∼50% deletion strains as would be theoretically expected. Thus this observation was a first hint toward a vital function of at least TusA for fitness of A. vinosum. We, therefore, changed our approach such that we now introduced a positive selection marker for the gene deletion; an Ω-kanamycin cassette was introduced into the remains of the rhd_2599-tusA-dsrE2 cluster to create A. vinosum Δrtd::ΩKm. The plates for selection contained kanamycin. Our selection plates routinely contain 0.5% yeast extract, 22 mm malate, 0.8 mm Na2S2O3, 2 mm sodium acetate, and 1.4 mm NaHS for growth. After 2 weeks of incubation under anoxic conditions in the light, we observed two colony types that differed with regard to diameter and color (Fig. 3). The first kind resembled typical A. vinosum colonies in radius, dark red color, and a shiny surface. The second kind was significantly smaller and had a non-shiny surface, and the color was a milky pink. Light microscopy showed that the milky appearance of the smaller colonies arose from massive accumulation of intercellular sulfur globules. Apparently, these cells were impaired in their ability to degrade the sulfur formed during the oxidation of sulfide and thiosulfate. A difference in the appearance of individual cells as reported for the E. coli TusA-deficient mutant (36) was not observed. Single colonies of both types were transferred to kanamycin/ampicillin and kanamycin-only plates to discriminate between single crossover recombinants that still contained the ampicillin resistance conferring shuttle plasmid and double crossover recombinants that had lost the vector-encoded ampicillin resistance. All of the regular colonies turned out to contain only single crossover recombinants, whereas the slowly growing cells of the smaller colonies were solely kanamycin-resistant. A considerable number of attempts to re-streak these colonies and transfer them into liquid media with and without reduced sulfur compounds remained unsuccessful. Inevitably, the cultures died after a few generations. It should be noted that we never obtained colonies of double-crossover recombinants on selection plates without sulfide and thiosulfate, indicating a requirement of A. vinosum for reduced sulfur in the absence of the rhd_2599-tusA-dsrE2 cluster. Yet it was possible to create A. vinosum rhd_2599::ΩSm, a mutant strain with a disruption in the reading frame of rhd_2599. Neither during photoorganoheterotroph nor during photolithoautotroph growth on sulfide, thiosulfate, or elemental sulfur did this strain behave differently from the wild type under the conditions applied. We conclude that Rhd_2599 can be either functionally replaced by another cytoplasmic rhodanese or the protein has a minor metabolic function. Although tusA and dsrE2 are present only once, the A. vinosum genome encodes three more cytoplasmic rhodaneses, Alvin_0866, Alvin_0868, and Alvin_1587. Two of these genes, Alvin_0866 and Alvin_1587, exhibit elevated relative mRNA levels under sulfur-oxidizing conditions and would thus be prime candidates for replacing Rhd_2599 (19). The different phenotypes for the rhd_2599-deficient mutant on the one hand and the rhd_2599-tusA-dsrE2-lacking mutant on the other hand indicated different promoter sites for rhd_2599 and tusA-dsrE2 and, therefore, supported the predictions by BPROM described above.
FIGURE 3.
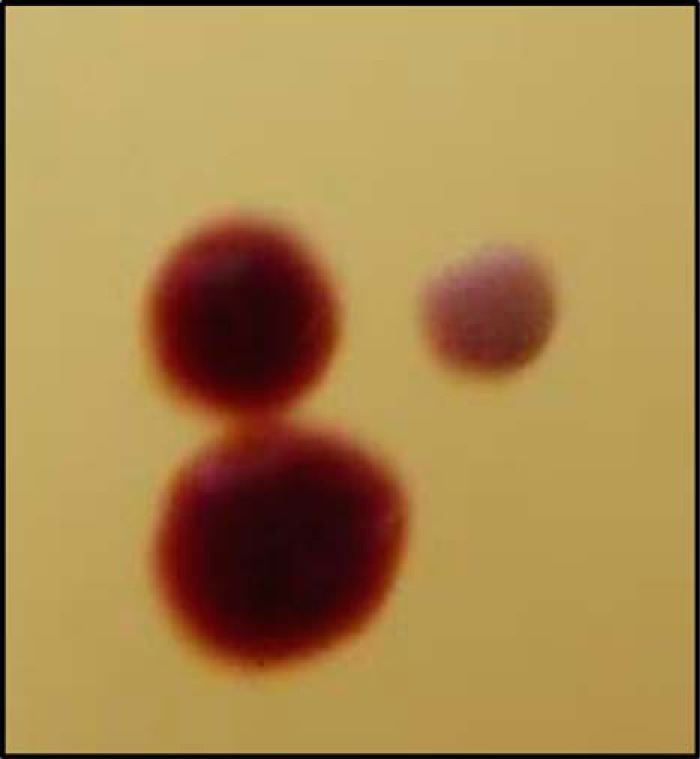
Different colony types of A. vinosum after transfer of plasmid pSUP301Δrtd::ΩKm.
Properties and Catalytic Activities of Rhd_2599, TusA, and DsrE2
With only one rhodanese domain and a molecular mass of 11.9 kDa, Rhd_2599 belongs in the group of single domain rhodaneses (pfam00581) together with GlpE and PspE from E. coli. Unlike the E. coli enzymes, Rhd_2599 has a second cysteine residue, Cys-74, in close proximity to the active site cysteine Cys-64. Both cysteines are strictly conserved among homologous rhodanese-like proteins of the Chromatiaceae and the Ectothiorhodospiraceae. The amino acid sequence does not contain predicted transmembrane helices. Rhd_2599 fused to an amino-terminal His tag was produced in and purified from E. coli BL21 (DE3). The protein eluted as a monomer from an analytical gel filtration column.
TusA (cd00291) from A. vinosum is a small 8.3-kDa protein. The characteristic active site motif for TusA-like proteins is Cys-19-Pro-X-Pro. In E. coli the motif contains a glutamate residue at the X position, and the residue contributes to the stabilization of the protein (37). In TusA from sulfur-oxidizing bacteria, glutamate is mostly replaced by a hydrophobic residue. For purification from E. coli, recombinant A. vinosum TusA was fused to a His tag at the amino terminus and isolated from the soluble fraction. From an analytical gel filtration column the protein eluted mostly as a monomer, and only a small portion eluted as a dimer. Note that A. vinosum TusA is not detectable at 280 nm due to the complete lack of tryptophan and tyrosine residues.
Alvin_2601 encodes a 159-amino acids, 17.5-kDa protein that is annotated as hypothetical protein of the DsrE/DsrF/DsrH superfamily (pfam13686). Two transmembrane helices are predicted that are arranged such that the carboxyl terminus is located in the cytoplasm. The A. vinosum protein harbors three cysteine residues located in the cytoplasmically oriented carboxyl terminus. Only Cys-120 is conserved in bacteria and archaea and corresponds to the active site cysteine of DsrE and TusD (6, 38). Recombinant DsrE2 was purified from the E. coli membrane fraction via affinity chromatography with an amino-terminal Strep tag. Cofactors were not identified for DsrE2 or for TusA and Rhd_2599.
All three recombinant proteins were subjected to thiosulfate:cyanide sulfurtransferase assays (24). Rhd_2599 proved to be active, mobilizing sulfur from thiosulfate and transferring it to cyanide (878 units mg−1). Because thiosulfate is already metabolized in the periplasm by the Sox proteins in A. vinosum (39), it is an unlikely in vivo substrate for cytoplasmic Rhd_2599. To date, persulfidic glutathione amide (GASSH) and glutathione (GSSH) are discussed as the organic carrier molecules for sulfane sulfur that is transported from the periplasmic sulfur globules to the cytoplasm (3, 4) and might, therefore, be substrates for rhodaneses in vivo. The unamidated GSSH was tested as the substrate for Rhd_2599, and indeed Rhd_2599 showed activity (25 units mg−1). In contrast, TusA and DsrE2 were inactive in assays with thiosulfate and glutathione persulfide.
Rhd_2599, TusA, and DsrE2 Are Sulfur Carriers
In our previous work on DsrC and DsrEFH we proved the sulfur binding capabilities of and sulfur transfer between these proteins via mass changes observed through MALDI-TOF mass spectrometry. However, we found this method not to be applicable to proteins exceeding a molecular mass of ∼18 kDa. Analysis of membrane proteins is also problematic due the detergents used during isolation and storage of the proteins. Therefore, we employed 1,5-IAEDANS as an additional or alternative method for visualization of persulfide-bound sulfur. 1,5-IAEDANS is a fluorescent reagent that can be coupled to proteins by displacement of its iodide group with the sulfur atom of a thiol group or sulfane sulfur of a persulfide. Excitation with UV light leads to the emission of light and allows the in-gel detection of proteins (27, 28). The method was validated by verification of sulfur binding of and sulfur transfer between DsrC and DsrEFH (not shown).
Above we showed that Rhd_2599 is able to mobilize sulfur from thiosulfate and GSSH, whereas TusA and DsrE2 cannot perform this reaction. We now verified the transfer of sulfane sulfur from thiosulfate and GSSH to the active site of Rhd_2599. This was done by mass spectrometry; exactly one sulfur atom (i.e. an additional mass of 32 Da for the single-charged and 16 Da for the double-charged protein molecules) was bound to the protein after incubation with each of the substrates (Fig. 4, A and B). When Rhd_2599 was incubated with sulfide, we found species carrying up to three sulfur atoms (not shown). We then incubated the Rhd_2599 mutant protein that carried a replacement of the probable active site cysteine Cys-64 to serine with thiosulfate. In this case a mass increase was not observed (Fig. 4C), clearly identifying Cys-64 as the sulfur binding residue and also showing that the second, partly conserved cysteine (Cys-74) is not able to mobilize sulfur from thiosulfate. This result was further verified by our finding that the Rhd_2559-C74S variant protein still exhibited the 32-Da mass addition after incubation with thiosulfate (not shown). The 1,5-IAEDANS method was not applicable to Rhd_2599. Neither the wild type nor the mutant proteins reacted with the reagent in the native or in their persulfurated states, which leads us to conclude that, both Cys-64 and Cys-74 are buried inside the enzyme and not surface-exposed.
FIGURE 4.
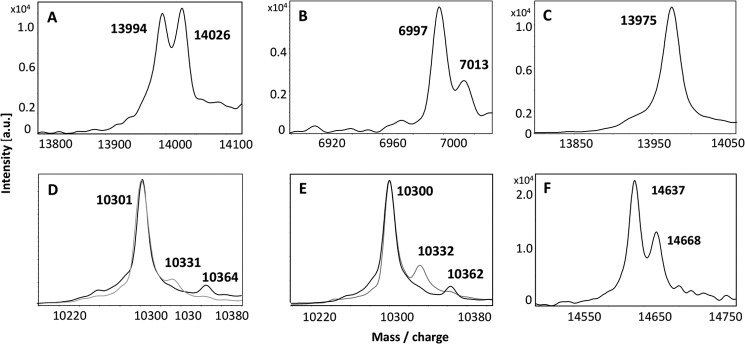
MALDI-TOF spectra of persulfurated Rhd_2599, TusA, and DsrC proteins. 30 μm protein solutions were incubated with a suitable sulfur substrate or sulfur-donating protein for 60 min at 30 °C. A, Rhd_2599 (mass 13,991 Da) after incubation with 2 mm thiosulfate. B, Rhd_2599 after incubation with 0.5 mm GSSH. C, the Rhd_2599-Cys-64Ser variant protein (mass 13,975 Da) is shown after incubation with 2 mm thiosulfate. D, TusA (mass 10,300 Da) after incubation with 2 mm sulfide. E, TusA after incubation with Rhd_2599 and thiosulfate. In D and E, two superimposed spectra obtained for two individually prepared samples are shown. F, persulfidic TusA was incubated with 30 μm DsrC (mass 14638 Da) as acceptor molecule. Binding of sulfur atoms is indicated by an additional mass of 32 Da. Note that in B the spectrum for the double-charged protein is shown. a.u., absorbance units.
Persulfuration of TusA was verified by additional masses of 32 and 64 Da after incubation with sulfide (Fig. 4D). In accordance with the enzyme assays described above, GSSH (0.5 mm) was not a suitable sulfur donor for the protein (not shown). The treatment of the variant protein TusA-C15S with sulfide did not lead to a mass change (not shown). Taken together these results unambiguously demonstrate the binding of sulfane sulfur to Cys-15 of TusA.
MALDI-TOF analysis could not be applied to the membrane protein DsrE2 because the presence of Triton X-100 in the storage buffer prevented its detection. However, the 1,5-IAEDANS method clearly demonstrated persulfuration of this protein with sulfide (Fig. 5A); DsrE2 reacted with the fluorescent reagent, and the protein was detected under UV light. Persulfurated DsrE2 lost the fluorescence upon reduction with DTT. Proteins variants carrying C110S or C156S replacements still showed the same reaction (not shown). The C120S variant of DsrE2 could not be tested because overproduction of this mutant protein resulted in massive formation of inclusion bodies, indicating that this cysteine is important for correct folding of the protein. It is therefore not yet possible to say which of the three cysteines present in the protein is responsible for sulfur binding.
FIGURE 5.
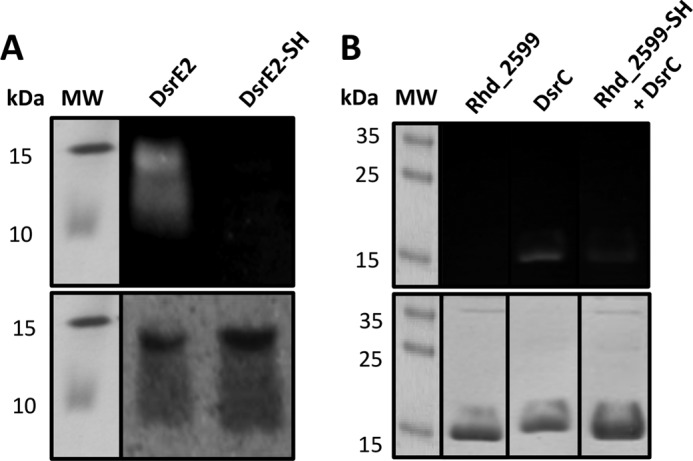
Identification of protein persulfides using 1,5-IAEDANS. A, persulfuration of DsrE2. The protein was reduced with DTT before incubation with sulfide, dialyzed, and treated with 1,5-IAEDANS and DTT as described under “Experimental Procedures.” Aliquots containing 200 pmol of protein were separated via SDS-PAGE (15%). B, persulfuration of DsrC after incubation with persulfidic Rhd_2599. 200 pmol of DsrC were added to 200 pmol of Rhd_2599, which had before been incubated with thiosulfate. After treatment with fluorescent 1,5-IAEDANS, the protein solution was analyzed via SDS-PAGE (15%). The upper images show the gels under UV light, and the lower images show the gels after staining with Coomassie. The molecular mass (MW) of the marker proteins is given in kDa.
In Vitro Sulfur Transfer Reactions
Our previous experiments established Rhd_2599 as capable of mobilizing sulfane sulfur from persulfides of low molecular weight compounds like glutathione persulfide. We now set out to identify possible acceptor proteins for Rhd_2599 and first tested TusA. To this end Rhd_2599 was incubated with 2 mm thiosulfate and TusA for 1 h. As a control TusA was incubated with thiosulfate under the same conditions as in the absence of Rhd_2599. TusA clearly remained unchanged in the control, whereas additional masses of 32 and 64 Da were observed when Rhd_2599 was present in the reaction mixtures (Fig. 4E). We then tested whether Rhd_2599 can also transfer sulfur to DsrE2 and possibly also directly to the sulfur-binding proteins of the Dsr system, DsrEFH and DsrC. The transfer to DsrE2 was assessed with 1,5-IAEDANS yielding a negative result. Rhd_2599 also proved unable to transfer sulfur to DsrEFH, which was verified with both mass spectrometry and the fluorescence method. A different result was obtained when DsrC served as the acceptor (Fig. 5B). When persulfurated Rhd_2559 was incubated with DsrC and 1,5-IAEDANS treatment was performed afterward, the fluorescence associated with DsrC was found to be noticeably decreased as compared with the control containing only DsrC. Please note that Rhd_2599 does not react with 1,5-IAEDANS (see above) and is, therefore, not visible under UV light (upper panel of Fig. 5B). DsrC and Rhd_2599 have similar molecular masses, 14.6 and 13.9 kDa, respectively, leading to similar migration patterns in the gel (Fig. 5B, lower panel).
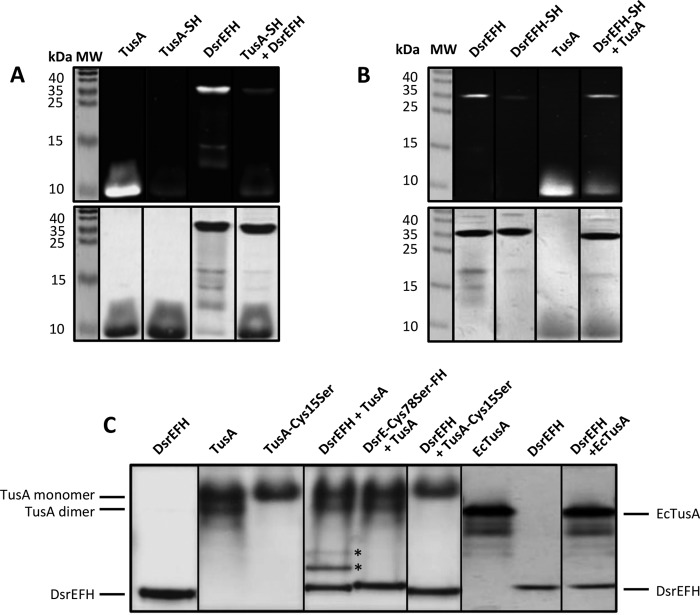
We now evaluated the sulfur transfer capabilities of TusA. Although the reaction with DsrE2 was negative, we found Rhd_2599 to be persulfurated upon incubation with persulfidic TusA (not shown). The reaction between TusA and DsrEFH was also tested (Fig. 6). It should be pointed out that DsrEFH migrated mainly in one band around 38 kDa in non-reducing SDS-PAGE, i.e. the three subunits were not separated from each other. Only small portions ran in the monomeric state. After incubation with persulfidic TusA, the fluorescence of DsrEFH was significantly reduced compared to the control sample, whereas that of TusA became more prominent again, thereby illustrating the transfer of sulfane sulfur from TusA to DsrEFH in vitro. Furthermore, the sulfur transfer was reversible (Fig. 6B); after TusA was incubated with persulfurated DsrEFH, the formation of a persulfide was detected in TusA. Although the fluorescence of TusA was reduced relative to the untreated sample, the fluorescence of DsrEFH was almost completely restored to the state before it had been incubated with sulfide. As a control, persulfurated DsrEFH was also incubated with the mutant TusA protein lacking Cys-15. In this case, the fluorescence of DsrEFH remained unchanged (not shown).
FIGURE 6.
Sulfur transfer between and interaction of TusA and DsrEFH. In panel A the capacity of TusA for sulfur transfer to DsrEFH was analyzed by the 1,5-IAEDANS method. In panel B the results for the reverse reaction, i.e. sulfur transfer from persulfurated DsrEFH to TusA, are presented. In both cases (A and B), the respective persulfurated donor protein was incubated with the acceptor protein for 60 min at 30 °C before the reaction mixtures were treated with 1,5-IAEDANS and DTT as described under “Experimental Procedures.” Samples were then electrophoresed on non-reducing 15% SDS-polyacrylamide gels. Gels were analyzed under UV-light (upper gels of panel A and B) and subsequently stained with Coomassie Brilliant Blue (lower gels of panels A and B). Part C shows the interaction of DsrEFH and TusA from A. vinosum (TusA) and E. coli (EcTusA), respectively. For the interaction experiments, 800 pmol of TusA/EcTusA were incubated with 100 pmol of DsrEFH (wild type and mutant proteins). Samples were then analyzed via native SDS-PAGE (7.5%). Bands arising from interaction complexes are marked with stars. Molecular mass (MW) of marker proteins is given in kDa.
In a further experiment persulfidic TusA was mixed with DsrC resulting in one sulfur atom bound to DsrC (Fig. 4F). Sulfur was clearly transferred to CysA (Cys-111 in A. vinosum DsrC) because the reaction did not occur with a mutant protein that contains only CysB (Cys-100 in A. vinosum DsrC) (not shown). A transfer of sulfur atoms from DsrC to TusA was not detected. Although DsrE2 was incubated with persulfidic TusA, Rhd_2599, DsrEFH, and DsrC, we did not detect persulfide formation for DsrE2 in any case. At this point it is not possible to say whether DsrE2 does not accept sulfane sulfur from any of these donor proteins or the failure to document sulfur transfer is due to the use of Triton X-100. The detergent is also the reason why DsrE2 could not be tested as the sulfur donor for other proteins. Dialysis of DsrE2 after incubation with sulfide in a buffer containing 0.1% Triton X-100 failed to remove excess sulfide.
Interaction of DsrEFH and TusA
In E. coli, an interaction between TusBCD and TusA was not detected, although sulfur transfer from TusA to TusBCD was evident (7). To evaluate a possible interaction between A. vinosum TusA and DsrEFH, 100 pmol of DsrEFH was incubated with 800 pmol of TusA for 30 min at 30 °C. Subsequently, the protein mix was applied to a native PAGE. Indeed, the migrating behavior of DsrEFH and TusA changed when incubated together. Two extra bands appeared (Fig. 6C). The presence of DsrE-Cys-78 and TusA-Cys-15 proved to be crucial for this reaction as these additional bands were missing when mutant proteins were used. Interestingly, the use of TusA from E. coli did not result in altered migration patterns. E. coli TusA did not interact with DsrEFH. The native gel also revealed that TusA forms dimers via formation of intermolecular disulfide bonds. For the mutated protein, only the monomeric form was observed. Together, these data strongly resemble the results found for the interaction of DsrEFH with DsrC (4–6).
DISCUSSION
In this work we combined several independent lines of evidence that all strongly indicate a function of the rhd_2599-tusA-dsrE2-encoded proteins in oxidative dissimilatory sulfur metabolism in A. vinosum and probably also in other sulfur oxidizing phototrophs.
The frequent occurrence of the rhd-tusA-dsrE2 genes in phototrophic sulfur-oxidizing bacteria as well as their conspicuous co-localization with genes encoding established (Dsr) or very probable (Hdr) major components of dissimilatory sulfur oxidation pathways point at the general and major importance of the encoded proteins for the oxidation of reduced sulfur compounds. Transcriptomic patterns for rhd_2599, tusA, and dsrE2 match those of major genes of the sulfur-oxidizing system (here the dsr genes) rather than those of genes encoding components of machineries for biosynthesis of sulfur-containing biomolecules like iron-sulfur clusters, thionucleosides, thiamine, or molybdopterin (Fig. 2). Further hints toward the function of TusA come from proteomic data (40). TusA is one of the major proteins in A. vinosum cells grown on reduced sulfur compounds. Its abundance is comparable with that for proteins involved in central carbon and energy metabolism as well as in photosynthesis. TusA even outnumbers the most prominent proteins of the Dsr system, DsrC, and DsrAB (40). Taken together, these observations strongly support the idea that A. vinosum TusA is not restricted to biosynthesis of sulfur-containing co-factors.
However, TusA cannot be exclusively dedicated to sulfur oxidation, which is indicated by the strongly decreased vitality on solid media and the incapability to grow in liquid media of the rhd_2599-tusA-dsrE2-deficient A. vinosum mutant strain even in the presence of the organic substrate malate. Concerning a possible important function at least for TusA in 2-thiouridine biosynthesis, it should be emphasized that 5-methyl-2-thiouridine tRNA derivatives are universally present (41); however, the Tus proteins are not conserved in all domains of life (7). TusB, TusC, TusD, and TusE are not ubiquitous even in bacteria. A. vinosum harbors no extra copies for tusA or tusBCD apart from Alvin_2600 and dsrEFH, respectively. The tusE homolog dsrC is present with five genomic copies. This implies that either the role of TusA and TusBCD is exerted by completely different sulfur transferases or 2-thiouridine is formed via IscS and MnmA alone as has originally been proposed for E. coli (42). A role of TusA in molybdopterin biosynthesis and the impact of the protein on the iron-sulfur cluster formation as described for E. coli (30) is also possible in A. vinosum and would result in a much stronger phenotype than that described for the E. coli tusA mutant (43). A balanced synthesis of iron-sulfur clusters would be expected to be of essential importance for a photolithotrophic organism. The biosynthesis of a high potential iron-sulfur protein, the primary electron donor to the reaction center in A. vinosum, may serve as an example.
As demonstrated via the bandshift assays with DsrEFH, A. vinosum TusA and E. coli TusA behave differently in vitro. The incubation of DsrEFH with AvTusA led to altered migration patterns in native PAGE gels, whereas EcTusA had no effect. Because we also showed the dependence of the interaction of the cysteine residues DsrE-Cys-78 and AvTusA-Cys-15, it is tempting to conclude that the entire Cys-Pro-X-Pro sequence and not only the conserved sulfur binding cysteine residue is essential for interaction with other proteins. With a few exceptions, TusA proteins encoded next to rhd_2599 and dsrE2 homologs harbor a hydrophobic non-aromatic amino acid at the X position in the motif. Within Chromatiaceae, the E. coli motif Cys-Pro-Glu-Pro is only found in Rheinheimera strains, which lack both sulfur oxidizing systems and DsrE2. The two TusA homologs of E. coli, YeeD and YedF, both harbor hydrophobic aromatic amino acids in the motif: Cys-Pro-Phe-Pro and Cys-Pro-Tyr-Pro, respectively. Neither YeeD nor YedF were able to complement a tusA-deficient E. coli mutant (30), which further points at the non-conserved amino acid in the motif as the decisive element for protein-protein interaction.
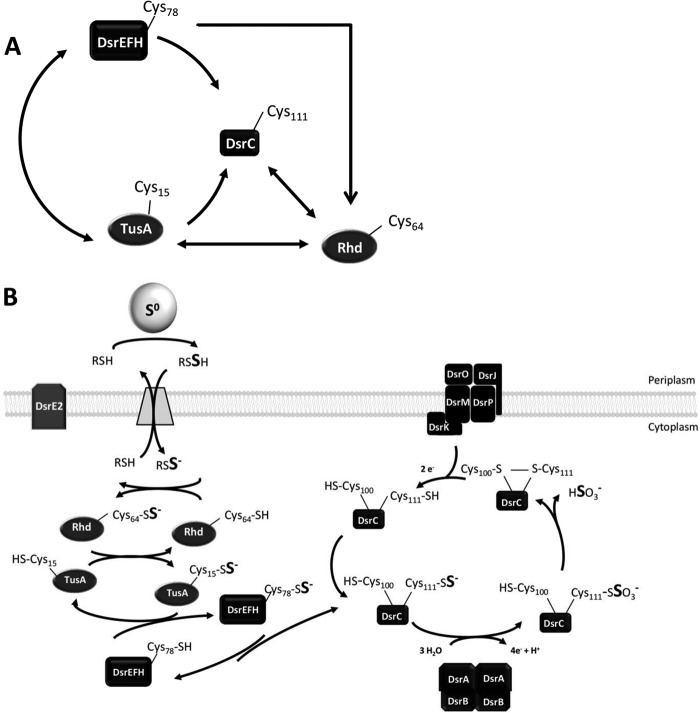
The successful overproduction and purification of Rhd_2599, TusA, and DsrE2 allowed characterization of the proteins as sulfur carriers and sulfur transferases. A schematic summary of the sulfur transfer reactions detected in vitro is given in Fig. 7A. Rhd_2599 was the only enzyme showing thiosulfate and GSSH:cyanide sulfurtransferase activity. Thus, cytoplasmic sulfur trafficking in the course of dissimilatory sulfur oxidation in A. vinosum most probably starts with mobilization of sulfane sulfur from the low molecular weight persulfide serving as a carrier for sulfur from the periplasmic sulfur globules to the cytoplasm by Rhd_2599. Persulfidic proteins also serve as substrates for this enzyme; Rhd_2599 accepted sulfane sulfur from all protein persulfides tested in this study. More importantly, the rhodanese served as an efficient sulfur donor for TusA and DsrC in vitro. However, a direct sulfur transfer from Rhd_2599 to DsrC in vivo is unlikely because the presence of the sulfurtransferase DsrEFH is essential for the degradation of sulfur globules in A. vinosum (4). Thus, sulfur transfer from Rhd_2599 to TusA is strongly indicated as the relevant reaction in vivo. TusA was the only protein that was capable of transferring sulfur to DsrEFH in vitro. Taken together with the tight in vitro interaction of the TusA and DsrEFH proteins (Fig. 6), a direct sulfur transfer from TusA to DsrEFH is also the most likely scenario in vivo.
FIGURE 7.
Cytoplasmic sulfur trafficking during sulfur globule oxidation in A. vinosum. A, summary of sulfur transfer reactions detected in vitro. The cysteines responsible for sulfur binding are depicted. B, model of sulfur transfer reactions occurring in vivo. Rhd_2599 and TusA are introduced as sulfur-donating proteins for the Dsr system. Thiol groups and persulfides are shown in the ionized or protonated state according to their supposed pKa values of around 8.5 (46) and 6.2 (47), respectively. Because persulfides are 1–2 pKa units more acidic than their thiol equivalents, we calculated the pKa of the assumed carrier molecule glutathione amide persulfide on the basis of the pKa for glutathione (48) to be ∼7.2. DsrC-Cys-111 and Cys-100 correspond to CysA and CysB, respectively (9). See “Discussion” for detailed explanation.
From DsrEFH, sulfur is further transferred to DsrC (4). The proposed sulfur relay system involving transfer of sulfur atoms from TusA via DsrEFH to DsrC (Fig. 7B) agrees with that derived for the related proteins from E. coli, TusA, TusBCD, and TusE (7). DsrC in its persulfidic state appears to be very stable as we did not detect any reverse transfer reaction from DsrC to TusA or DsrEFH. Persulfurated DsrC very probably serves as a direct substrate for DsrAB in vivo (4, 44), and it is, therefore, feasible that DsrC acts as a sulfur trap. This would ensure a constant flow of sulfur atoms necessary for a high turnover rate of DsrAB.
This work was supported by Deutsche Forschungsgemeinschaft Grant Da 351/6-1 (to C. D.).
- mnm
- 5-methyl-aminomethyl
- qRT
- quantitative real-time
- 1,5-IAEDANS
- N-(iodoacetyl)-N′-(5-sulfo-1-naphtyl)-ethylenediamine.
REFERENCES
- 1. Pott A. S., Dahl C. (1998) Sirohaem-sulfite reductase and other proteins encoded in the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology 144, 1881–1894 [DOI] [PubMed] [Google Scholar]
- 2. Dahl C., Engels S., Pott-Sperling A. S., Schulte A., Sander J., Lübbe Y., Deuster O., Brune D. C. (2005) Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J. Bacteriol. 187, 1392–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Frigaard N.-U., Dahl C. (2009) Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 54, 103–200 [DOI] [PubMed] [Google Scholar]
- 4. Stockdreher Y., Venceslau S. S., Josten M., Sahl H. G., Pereira I. A., Dahl C. (2012) Cytoplasmic sulfurtransferases in the purple sulfur bacterium Allochromatium vinosum: evidence for sulfur transfer from DsrEFH to DsrC. PLoS ONE 7, e40785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cort J. R., Selan U., Schulte A., Grimm F., Kennedy M. A., Dahl C. (2008) Allochromatium vinosum DsrC: solution-state NMR structure, redox properties, and interaction with DsrEFH, a protein essential for purple sulfur bacterial sulfur oxidation. J. Mol. Biol. 382, 692–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dahl C., Schulte A., Stockdreher Y., Hong C., Grimm F., Sander J., Kim R., Kim S.-H., Shin D. H. (2008) Structural and molecular genetic insight into a wide-spread bacterial sulfur oxidation pathway. J. Mol. Biol. 384, 1287–1300 [DOI] [PubMed] [Google Scholar]
- 7. Ikeuchi Y., Shigi N., Kato J., Nishimura A., Suzuki T. (2006) Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell 21, 97–108 [DOI] [PubMed] [Google Scholar]
- 8. Quatrini R., Appia-Ayme C., Denis Y., Jedlicki E., Holmes D. S., Bonnefoy V. (2009) Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Venceslau S. S., Stockdreher Y., Dahl C., Pereira I. A. C. (2014) The “bacterial heterodisulfide” DsrC is a key protein in dissimilatory sulfur metabolism. Biochim. Biophys. Acta 10.1016/j.bbabio.2014.03.007 [DOI] [PubMed] [Google Scholar]
- 10. Frigaard N.-U., Bryant D. A. (2008) Genomic insights into the sulfur metabolism of phototrophic green sulfur bacteria. In (Hell R., Dahl C., Knaff D. B., Leustek T., eds) pp. 337–355, Sulfur metabolism in phototrophic organisms, Springer, Dordrecht, The Netherlands [Google Scholar]
- 11. Dahl C., Franz B., Hensen D., Kesselheim A., Zigann R. (2013) Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology 159, 2626–2638 [DOI] [PubMed] [Google Scholar]
- 12. Kelly D. P. (2008) Stable sulfur isotope fractionation by the green bacterium Chlorobaculum parvum during photolithoautotrophic growth on sulfide. Pol. J. Microbiol. 57, 275–279 [PubMed] [Google Scholar]
- 13. Heising S., Richter L., Ludwig W., Schink B. (1999) Chlorobium ferrooxidans sp. nov., a phototrophic green sulfur bacterium that oxidizes ferrous iron in coculture with a “Geospirillum” sp. strain. Arch. Microbiol. 172, 116–124 [DOI] [PubMed] [Google Scholar]
- 14. Dahl C. (2008) Inorganic sulfur compounds as electron donors in purple sulfur bacteria. In Sulfur in Phototrophic Organisms (Hell R., Dahl C., Knaff D. B., Leustek T., eds) pp. 289–317, Springer, Dordrecht, The Netherlands [Google Scholar]
- 15. Pattaragulwanit K., Dahl C. (1995) Development of a genetic system for a purple sulfur bacterium: conjugative plasmid transfer in Chromatium vinosum. Arch. Microbiol. 164, 217–222 [Google Scholar]
- 16. Weaver P. F., Wall J. D., Gest H. (1975) Characterization of Rhodopseudomonas capsulata. Arch. Microbiol. 105, 207–216 [DOI] [PubMed] [Google Scholar]
- 17. Overmann J., Fischer U., Pfennig N. (1992) A new purple sulfur bacterium from saline littoral sediments, Thiorhodovibrio winogradskyi gen. nov. and sp. nov. Arch. Microbiol. 157, 329–335 [Google Scholar]
- 18. Pfennig N., Trüper H. G. (1992) The family Chromatiaceae. In The Prokaryotes. A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications (Balows A., Trüper H. G., Dworkin M., Harder W., Schleifer K.-H., eds) pp. 3200–3221, Springer-Verlag, New York [Google Scholar]
- 19. Weissgerber T., Dobler N., Polen T., Latus J., Stockdreher Y., Dahl C. (2013) Genome-wide transcriptional profiling of the purple sulfur bacterium Allochromatium vinosum DSM 180T during growth on different reduced sulfur compounds. J. Bacteriol. 195, 4231–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 21. Horton R. M. (1995) PCR mediated recombination and mutagenesis: SOEing together tailor-made genes. Mol. Biotechnol. 3, 93–99 [DOI] [PubMed] [Google Scholar]
- 22. Kelly D. P., Chambers L. A., Trudinger P. A. (1969) Cyanolysis and spectrophotometric estimation of trithionate in mixture with thiosulfate and tetrathionate. Anal. Chem. 41, 898–901 [Google Scholar]
- 23. Sörbo B. (1987) Sulfate: turbidometric and nephelometric methods. Methods Enzymol. 143, 3–6 [DOI] [PubMed] [Google Scholar]
- 24. Ray W. K., Zeng G., Potters M. B., Mansuri A. M., Larson T. J. (2000) Characterization of a 12-kilodalton rhodanese encoded by glpE of Escherichia coli and its interaction with thioredoxin. J. Bacteriol. 182, 2277–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rohwerder T., Sand W. (2003) The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 149, 1699–1710 [DOI] [PubMed] [Google Scholar]
- 26. Rethmeier J., Rabenstein A., Langer M., Fischer U. (1997) Detection of traces of oxidized and reduced sulfur compounds in small samples by combination of different high- performance liquid chromatography methods. J. Chromatogr. A 760, 295–302 [Google Scholar]
- 27. Zheng L., White R. H., Cash V. L., Dean D. R. (1994) Mechanism for the desulfurization of l-cysteine catalyzed by the nifS gene product. Biochemistry 33, 4714–4720 [DOI] [PubMed] [Google Scholar]
- 28. Thomé R., Gust A., Toci R., Mendel R., Bittner F., Magalon A., Walburger A. (2012) A sulfurtransferase is essential for activity of formate dehydrogenases in Escherichia coli. J. Biol. Chem. 287, 4671–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Solovyev V., Salamov A. (2010) Automatic annotation of microbial genomes and metagenomic sequences. In Metagenomics and Its Applications in Agriculture, Biomedicine, and Environmental Studies (Li R. W., ed.) pp. 71–78, Nova Science Publishers, Hauppage, NY [Google Scholar]
- 30. Dahl J. U., Radon C., Bühning M., Nimtz M., Leichert L. I., Denis Y., Jourlin-Castelli C., Iobbi-Nivol C., Méjean V., Leimkühler S. (2013) The sulfur carrier protein TusA has a pleiotropic role in Escherichia coli that also affects molybdenum cofactor biosynthesis. J. Biol. Chem. 288, 5426–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lübbe Y. J., Youn H.-S., Timkovich R., Dahl C. (2006) Siro(haem)amide in Allochromatium vinosum and relevance of DsrL and DsrN, a homolog of cobyrinic acid a,c diamide synthase for sulphur oxidation. FEMS Microbiol. Lett. 261, 194–202 [DOI] [PubMed] [Google Scholar]
- 32. Grein F., Pereira I. A., Dahl C. (2010) The Allochromatium vinsosum DsrMKJOP transmembrane complex: biochemical characterization of individual components aids understanding of complex function in vivo. J. Bacteriol. 192, 6369–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Grimm F., Cort J. R., Dahl C. (2010) DsrR, a novel IscA-like protein lacking iron- and FeS-binding function involved in the regulation of sulfur oxidation in Allochromatium vinosum. J. Bacteriol. 192, 1652–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sander J., Engels-Schwarzlose S., Dahl C. (2006) Importance of the DsrMKJOP complex for sulfur oxidation in Allochromatium vinosum and phylogenetic analysis of related complexes in other prokaryotes. Arch. Microbiol. 186, 357–366 [DOI] [PubMed] [Google Scholar]
- 35. Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73 [DOI] [PubMed] [Google Scholar]
- 36. Yamashino T., Isomura M., Ueguchi C., Mizuno T. (1998) The yhhP gene encoding a small ubiquitous protein is fundamental for normal cell growth of Escherichia coli. J. Bacteriol. 180, 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Katoh E., Hatta T., Shindo H., Ishii Y., Yamada H., Mizuno T., Yamazaki T. (2000) High precision NMR structure of YhhP, a novel Escherichia coli protein implicated in cell division. J. Mol. Biol. 304, 219–229 [DOI] [PubMed] [Google Scholar]
- 38. Numata T., Fukai S., Ikeuchi Y., Suzuki T., Nureki O. (2006) Structural basis for sulfur relay to RNA mediated by heterohexameric TusBCD complex. Structure 14, 357–366 [DOI] [PubMed] [Google Scholar]
- 39. Hensen D., Sperling D., Trüper H. G., Brune D. C., Dahl C. (2006) Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol. Microbiol. 62, 794–810 [DOI] [PubMed] [Google Scholar]
- 40. Weissgerber T., Sylvester M., Kröninger L., Dahl C. (2014) A comparative quantitative proteome study identifies new proteins relevant for sulfur oxidation in the purple sulfur bacterium Allochromatium vinosum. Appl. Environ. Microbiol. 80, 2279–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suzuki T. (2005) Biosynthesis and function of tRNA wobble modifications. In Topics in current genetics (Grosjean H., ed.) pp. 24–69, Springer-Verlag, New York [Google Scholar]
- 42. Kambampati R., Lauhon C. T. (2003) MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry 42, 1109–1117 [DOI] [PubMed] [Google Scholar]
- 43. Ishii Y., Yamada H., Yamashino T., Ohashi K., Katoh E., Shindo H., Yamazaki T., Mizuno T. (2000) Deletion of the yhhP gene results in filamentous cell morphology in Escherichia coli. Biosci. Biotechnol. Biochem. 64, 799–807 [DOI] [PubMed] [Google Scholar]
- 44. Oliveira T. F., Vonrhein C., Matia P. M., Venceslau S. S., Pereira I. A. C., Archer M. (2008) The crystal structure of Desulfovibrio vulgaris dissimilatory sulfite reductase bound to DsrC provides novel insights into the mechanism of sulfite respiration. J. Biol. Chem 283, 34141–34149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kingsford C. L., Ayanbule K., Salzberg S. L. (2007) Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biology 8, R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Riederer B. M. (2009) Oxidation proteomics: the role of thiol modifications. Curr. Proteomics 6, 51–62 [Google Scholar]
- 47. Everett S. A., Folkes L. K., Wardman P., Asmus K. D. (1994) Free radical repair by a novel perthiol: reversible hydrogen-transfer and perthiyl radical formation. Free Radic. Res. 20, 387–400 [DOI] [PubMed] [Google Scholar]
- 48. Tajc S. G., Tolbert B. S., Basavappa R., Miller B. L. (2004) Direct determination of thiol pKa by isothermal titration microcalorimetry. J. Am. Chem. Soc. 126, 10508–10509 [DOI] [PubMed] [Google Scholar]
- 49. Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]
- 50. Simon R., Priefer U., Pühler A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1, 784–791 [Google Scholar]
- 51. Miroux B., Walker J. E. (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 [DOI] [PubMed] [Google Scholar]
- 52. Prentki P., Krisch H. M. (1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29, 303–313 [DOI] [PubMed] [Google Scholar]
- 53. Fellay R., Frey J., Krisch H. (1987) Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vivo insertional mutagenesis of Gram-negative bacteria. Gene 52, 147–154 [DOI] [PubMed] [Google Scholar]
- 54. Dahl C., Schulte A., Shin D. H. (2007) Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of DsrEFH from Allochromatium vinosum. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 890–892 [DOI] [PMC free article] [PubMed] [Google Scholar]