Background: ARVCF occurs both at adherens junctions and in the nucleus, but little is known about its nuclear function.
Results: ARVCF interacts with RNA-binding proteins involved in splicing and contributes to alternative splicing of specific transcripts.
Conclusion: ARVCF influences splicing of specific pre-mRNAs.
Significance: We propose a new role of nuclear ARVCF in post-transcriptional regulation of gene expression.
Keywords: Adherens Junction, Alternative Splicing, Catenin, Cell Junctions, Molecular Cell Biology, Ribonuclear Protein (RNP), Post-transcriptional Gene Expression Regulation, Arm Repeat Protein
Abstract
The armadillo repeat protein ARVCF is a component of adherens junctions. Similar to related proteins, such as p120-catenin and β-catenin, with known signaling functions, localization studies indicate a cytoplasmic and a nuclear pool of ARVCF. We find that ARVCF interacts with different proteins involved in mRNA-processing: the splicing factor SRSF1 (SF2/ASF), the RNA helicase p68 (DDX5), and the heterogeneous nuclear ribonucleoprotein hnRNP H2. All three proteins bind to ARVCF in an RNA-independent manner. Furthermore, ARVCF occurs in large RNA-containing complexes that contain both spliced and unspliced mRNAs of housekeeping genes. By domain analysis, we show that interactions occur via the ARVCF C terminus. Overexpression of ARVCF, p68, SRSF1, and hnRNP H2 induces a significant increase in splicing activity of a reporter mRNA. Upon depletion of ARVCF followed by RNA sequence analysis, several alternatively spliced transcripts are significantly changed. Therefore, we conclude that nuclear ARVCF influences splicing of pre-mRNAs. We hypothesize that ARVCF is involved in alternative splicing, generating proteomic diversity, and its deregulation may contribute to diseased states, such as cancer and neurological disorders.
Introduction
In multicellular organisms, the establishment and regulation of cell adhesion is fundamental to the development and maintenance of tissues. In cell-cell adhesion, key structural elements are adherens junctions. Transmembrane proteins mediate the interaction to neighboring cells and are coupled to the cytoskeleton through protein-protein interactions (1). Besides stabilizing tissue architecture, adherens junctions also act as signaling platforms via catenins (2). These proteins not only bind to cadherins but have numerous additional roles in various cellular compartments.
The family of catenins with the exception of α-catenin possess central armadillo (Arm)2 repeat domains and consists of β-catenin and plakoglobin and the p120-related proteins. Members of the p120-related proteins are p120-catenin, ARVCF (armadillo repeat gene deleted in velocardiofacial syndrome), p0071-catenin (also known as PKP4), δ-catenin (also known as neurojungin or neural plakophilin-related protein (NRAP)), and plakophilin 1, 2, and 3 (3–5). ARVCF, identified by Sirotkin et al. (6), is one of the gene products deleted in velocardiofacial syndrome. It is the closest relative to p120-catenin and contains an N-terminal coiled-coil domain, nine central Arm repeats, and a nuclear localization signal (2, 3, 6, 7). Four isoforms of human ARVCF are known that contain either the coiled-coil domain or an extended nuclear localization signal due to alternative splicing (6). Specifically, all ARVCF isoforms together with p0071-catenin and δ-catenin have a C-terminal PDZ-binding motif known to mediate the binding to the scaffolding protein Erbin (8), the tight junction proteins ZO-1 and ZO-2 (9), and the protein FRMPD2 (10).
Catenins in their junction-bound and non-junctional forms may modulate cadherin stability by influencing the endocytosis of cadherins (5, 11, 12) and may regulate the cytoskeleton by interacting with small GTPases, such as RhoA and Rac (13–15). In addition, some catenins enter the nucleus to interact with transcription factors and therefore have an impact on gene expression and developmental decisions. β-Catenin is a central component of the Wnt signaling pathway and triggers transcription of Wnt-specific genes (16). In a comparable manner, nuclear p120-catenin binds to Kaiso and relieves Kaiso-mediated repression (17). Besides their direct effect on transcription, catenins may also influence gene expression post-transcriptionally. We showed that plakophilin 1 and 3 are associated with RNA-binding proteins and are part of mRNA-ribonucleoprotein particles (RNPs) (18). RNPs play critical roles not only in pre-mRNA processing, including splicing and polyadenylation, but also in stability, transport, and cellular localization of mRNAs (19). Although ARVCF was described to localize to the nucleus more than a decade ago (14), little is known about its associations and functions in this compartment.
To explore the nuclear function of ARVCF, we performed a yeast two-hybrid screen and identified proteins involved in mRNA processing. The heterogeneous nuclear ribonucleoprotein (hnRNP) H2, the RNA helicase p68, and the splicing factor SRSF1 are found in ARVCF-containing RNPs and bind to ARVCF via protein-protein interactions. Upon overexpression of ARVCF, p68, SRSF1, or hnRNP H2, splicing activity was increased. Moreover, in ARVCF-depleted cells, several alternatively spliced transcripts were significantly changed. Our findings demonstrate for the first time that a member of the p120-catenin family, ARVCF, influences splicing of pre-mRNA.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfections
Human colon carcinoma (Caco-2) cells were cultured in minimum Eagle's medium with 1% glutamine, 10% FCS, and 1% sodium pyruvate. Human embryonic kidney (HEK 293) cells were cultured in DMEM with 1% glutamine and 10% FCS. Transient plasmid transfections were performed with X-tremeGene9 (Roche Applied Science) according to the manufacturer's protocol in 6- and 12-well cell culture plates. Cells were analyzed 48 h after transfection. siRNA transfections were performed with Dharmafect 1 (Thermo Scientific, Pittsburgh, PA) according to the manufacturer's protocol in 12-well cell culture plates. RNA isolation was performed 72 h after transfection.
Antibodies
Primary antibodies to ARVCF (h2ARVCF gp1 affinity-purified and h2ARVCF gp2, mARVCF gp, and mouse monoclonal ARVCF-1056) were used as described previously (20). Antibodies to β-catenin (C 2206, Sigma), E-cadherin (EP700Y, Biomol, Hamburg, Germany), FLAG (F3165, Sigma), GAPDH (ab8245, Abcam (Cambridge, UK)), GFP (ab290, Abcam), hnRNP A1 (ab4791, Abcam), hnRNP H2 (HPA001359, Sigma), lamin (X67, 65147A, Progen (Heidelberg, Germany)), nucleolin (ab22758, Abcam), p0071 (hp0071 gp, as described previously (21)), p68 (ab21696, Abcam), PKP2 (651101, Progen), SRSF1 (SF2/ASF, 32–4600, Invitrogen), and symplekin (610644, BD Biosciences) were used. Secondary HRP-coupled antibodies anti-mouse, anti-rabbit and anti-guinea pig (Jackson ImmunoResearch, West Grove, PA) were used for Western blot analysis, and Alexa 488-coupled antibodies (MoBiTec, Göttingen, Germany) or Cy3-coupled antibodies (BioTrend, Köln, Germany) were used for immunofluorescence.
cDNA Clones and Constructs
ARVCF (NM_001670; SC315001) and Myc-DDK-tagged hnRNP H2 (NM_019597; RC221204) cDNAs were purchased from OriGene (via VWR, Bruchsal, Germany). GFP-tagged ARVCF was generated via amplification with the primers 5′-gcggtcgacgctctggtcatggaggactg-3′ and 5′-gcgggatcccatgcaagcgagacccaggaa-3′ with deletion of the ARVCF stop codon and cloning into the pEGFP-N1 vector with SalI and BamHI restriction enzymes (New England Biolabs, Ipswich, MA). ARVCF constructs ARVCF-NA, -A, and -AC were amplified with the primers 5′-gcggtcgacgctctggtcatggaggactg-3′ and 5′-gcgggatccacgatttcgtggatggtgtt-3′, 5′-gcggaattcgaggacacagcagatgatgg-3′ and 5′-gcgggatccacgatttcgtggatggtgtt-3′, and 5′-gcggaattcgaggacacagcagatgatgg-3′ and 5′-gcgggatccacccaggaatcaacgggctgagg-3′, respectively, and cloned into the pEGFP-N1 vector. ARVCF construct ARVCF-ΔP was generated using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies, Waldbronn, Germany) with the primers 5′-ctaagcctcagcccgtggatccaccggtc-3′ and 5′-gaccggtggatccacgggctgaggcttag-3′. Isoform sARVCF was cloned with the primers 5′-gcggtcgacgctgtacaagtccggaatgc-3′ and 5′-gcgggatccacccaggaatcaacgggctgagg-3′. The NLS insert was generated using the QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies) with the primers 5′-ggaggcaagaaggccaaagaggagtggttccaccaaggaaagaaggatgg-3′ and 5′-ccatccttctttccttggtggaaccactcctctttggccttcttgcctcc-3′. p0071 (NM_001005476, IRATp970F0662D, RZPD, Berlin, Germany) was cloned with the primers 5′-gcgagatcttgcctaagctggaaagagga-3′ and 5′-gcgaccggttgggcatcttgatgctacac-3′ into the pEGFP-N1 vector (without GFP tag) with restriction enzymes BglIII and AgeI (New England Biolabs, Ipswich, MA). p68 (NM_004396, clone 191513512, DDX5) and SRSF1 (BC010264, clone 146755482) cDNAs were purchased in pDONR vectors (Genomics and Proteomics Core Facility, DKFZ (Heidelberg, Germany)) and cloned into the destination vector pDEST-N-FLAG (Genomics and Proteomics Core Facility, DKFZ) to generate an N-terminal FLAG tag via clonase reaction (Invitrogen) according to the manufacturer's protocol. mARVCF constructs (NM_033474.3) containing the N-terminal and Arm repeat domains (amino acids 1–780; mARVCF-NA) (22) or the C-terminal domain only (C11 isoform, amino acids 782–976; mARVCF-CT) (23) were used for rescue experiments.
Immunofluorescence and Microscopy
Caco-2 cells grown on glass slides for 2 days were fixed in 2% formaldehyde for 5 min, rinsed in NH4Cl, permeabilized in 0.2% Triton X-100 for 3 min, and washed twice in PBS. After a 1-h incubation with the primary antibodies, cells were washed again in PBS and incubated with the secondary antibodies for 30 min. After the addition of DAPI for another 5 min, cells were washed again in PBS and water and mounted with Fluoromount-G (Southern Biotech, Birmingham, AL) on coverslips. A block of transcription was achieved by incubation with actinomycin D (AppliChem, Darmstadt, Germany) for 3 h in a final concentration of 5 μg/ml.
Samples were examined on either a motorized inverted wide field microscope (Axio Observer.Z1, Zeiss; equipped with ×40/1.3 numerical aperture oil differential interference contrast objective EC PlnN and a dual camera setup 2× AxioCam MRm) or a motorized inverted laser-scanning confocal microscope (DMI6000 CFS Sp5, Leica (Wetzlar, Germany); equipped with ×40/1.25 numerical aperture oil objective HCX PL APO Lambda Blue and tunable spectral photomultipliers). The images were taken with and exported from Zeiss AxioVision or Leica LAS AF software, respectively, and digital images were prepared using Adobe Photoshop software.
Yeast Two-hybrid Screen
A yeast two-hybrid screen was performed by the Genomics and Proteomics Core Facility (DKFZ) using the bait vector pGBKT7 (Gateway, via the Genomics and Proteomics Core Facility, DKFZ) containing full-length ARVCF (NM_001670). For screening, two different human libraries were used, a library of more than 10,000 full-length open reading frames and a library from kidney tissue. The protocol has been described in detail (24).
Nuclear Extraction
Caco-2 cells were harvested in buffer (10 mm HEPES, 1.5 mm MgCl2, 10 mm KCl, 1 mm DTT, and 0.5 mm PMSF), incubated for 15 min on ice, mechanically disrupted with a 23-gauge needle, and centrifuged. The cytoplasmic supernatant was collected, and nucleoplasm was extracted in high salt buffer (20 mm HEPES, 1.5 mm MgCl2, 25% glycerol, 420 mm NaCl, 0.2 mm EDTA, 1 mm DTT, and 0.5 mm PMSF), followed by 5 min of centrifugation to pellet the cellular debris (25). Equal volumes of cytoplasmic, nuclear, and pellet fraction were loaded onto SDS gels and analyzed by Western blot.
Immunoprecipitation and RNA Immunoprecipitation
For immunoprecipitation, HEK 293 cells were grown for 3 days in 10-cm plates or in 6-well plates and, if applicable, transfected with plasmids on day 2. Cells were lysed in immunoprecipitation buffer (140 mm NaCl, 20 mm HEPES, 5 mm EDTA, 1% Nonidet P-40, pH 7.5) for 10 min on ice, followed by 10 min of centrifugation. The supernatant (load) was used for the immunoprecipitation. In some experiments, RNase A (final concentration 0.2 μg/μl; Roche Applied Science) was added to immunoprecipitation buffer prior to cell lysis.
Immunoprecipitation was performed with magnetic protein A and pan-mouse-coupled DYNAL beads (Invitrogen). Beads were loaded with the primary antibodies for 2 h at 4 °C. In parallel, the cell extract (load) was precleaned with unloaded beads (control). Immunoprecipitation was performed for 2 h at 4 °C. Proteins of the load, precleaning, and immunoprecipitation were lysed in Laemmli buffer.
RNA immunoprecipitation was performed overnight at 4 °C. Bound RNA was isolated via TRIzol extraction (Ambion, Carlsbad, CA). An additional sample of the lysate after RNA immunoprecipitation (post in Figs. 4, 6, and 7) was taken to control RNA stability during the experiment. The RNA was transcribed into cDNA with the High Capacity cDNA reverse transcription kit (Applied Biosystems, Carlsbad, CA) and analyzed via PCR using the following primers: GAPDH, 5′-tgttccaatatgattccaccc-3′ and 5′-cttctccatggtggtgaaga-3′; actin, 5′-cacggccgagcgggaaat-3′ and 5′-tgcatcctgtcggcaatgc-3′; GAPDH pre-mRNA, 5′-tgttccaatatgattccaccc-3′ and 5′-aagggagccacaccatcct-3′; actin pre-mRNA, 5′-cacggccgagcgggaaat-3′ and 5′-cgggagacagtctccac-3′; PKP2, 5′-ttctgggtggcctgaaggagact-3′ and 5′-cacttccggccgtgaggttc-3′; ACA44 small nucleolar RNA, 5′-gtttccaagggctgtggct-3′ and 5′-ccacatgccatataccagattacaac-3′; U2 snRNA, 5′-ttctcggccttttggctaag-3′ and 5′-ctccctgctccaaaaatcca-3′. For MYL6, NDUFV3, and Tau, the primers listed under “RT-PCR and Capillary Electrophoresis” were used.
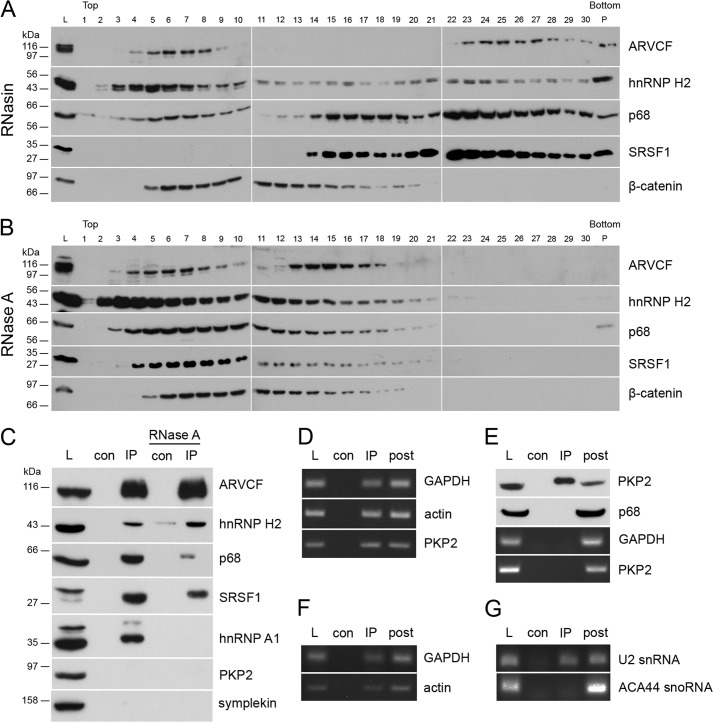
FIGURE 4.
ARVCF is part of an RNP containing classical mRNAs and other RNA-binding proteins. Sucrose gradient centrifugation with lysates from HEK 293 cells was done with the addition of RNasin (A) or RNase A (B). Western blot was performed with antibodies against ARVCF, hnRNP H2, p68, SRSF1, and β-catenin. Top, 10% sucrose; bottom, 40% sucrose; L, load; P, pellet. C, immunoprecipitates using ARVCF antibody with or without the addition of RNase A were analyzed by Western blotting with antibodies against ARVCF, hnRNP H2, p68, SRSF1, hnRNP A1, PKP2, and symplekin. D–F, samples from RNA immunoprecipitations of ARVCF were analyzed by RT-PCR with primers against GAPDH, actin, or PKP2 mRNA (D) and GAPDH or actin pre-mRNA (E) and U2 snRNA or ACA44 small nucleolar RNA (F). G, immunoprecipitates using PKP2 antibody were analyzed by Western blotting with antibodies against PKP2 and p68, and corresponding samples from RNA immunoprecipitations were analyzed by RT-PCR with primers against GAPDH or PKP2 mRNA. L, load; con, control; IP, immunoprecipitation; post, supernatant after RNA immunoprecipitation.
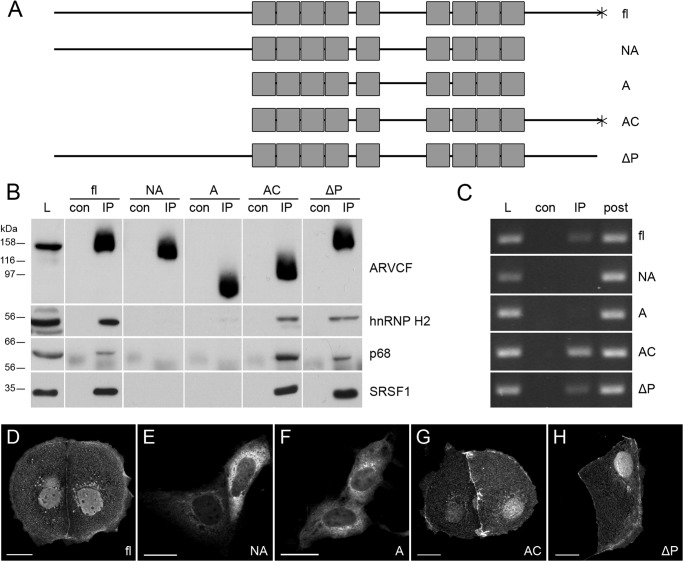
FIGURE 6.
ARVCF C terminus is necessary for the interaction with splicing factors. A, schematic representation of full-length (fl) ARVCF and ARVCF partial constructs. Gray boxes, Arm repeats; *, PDZ-binding motif. Mutants are shown as follows. NA, N-terminal domain and Arm repeats; A, Arm repeats only; AC, Arm repeats and the C-terminal domain; ΔP, deletion of the PDZ-binding domain. B, HEK 293 cells were co-transfected with C-terminally GFP-tagged full-length ARVCF or GFP-tagged ARVCF constructs together with FLAG-tagged hnRNP H2, p68, or SRSF1, as indicated. Immunoprecipitation was performed with GFP antibody, and Western blot was performed with FLAG antibody or p68 antibody. L, load; con, control; IP, immunoprecipitation. C, RNA immunoprecipitation using GFP antibody in HEK 293 cells transfected with full-length ARVCF or ARVCF partial constructs. PCR using GAPDH primers is shown. post, supernatant after RNA immunoprecipitation. D–H, fluorescence staining of Caco-2 cells transfected with GFP-tagged ARVCF constructs, as indicated. Scale bars, 20 μm.
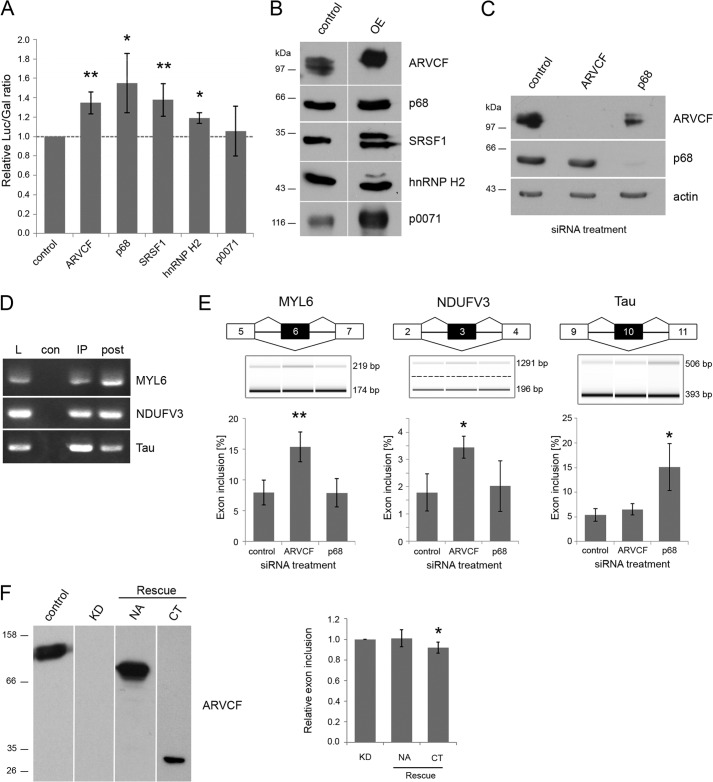
FIGURE 7.
ARVCF regulates pre-mRNA splicing. A, double reporter splicing assay in HEK 293 cells co-transfected with the reporter plasmid pTN23 and the cDNAs coding for ARVCF, p68, SRSF1, hnRNP H2, p0071, or empty vector as control. The ratio of luciferase to β-galactosidase activity is shown. B and C, verification of protein overexpression and knockdown via Western blot analysis. Laemmli lysates of HEK 293 cells transfected with the plasmids (B) or siRNAs (C) indicated at the top were separated on SDS gels and analyzed via Western blot with the antibodies indicated on the right. Equal amounts of lysates were loaded, except for ARVCF overexpression less lysate was used. OE, overexpressing HEK 293 cells. D, samples from RNA immunoprecipitations of ARVCF were analyzed by RT-PCR with primers against MYL6, NDUFV3, and Tau. E, capillary gel electrophoresis of RT-PCR products from HEK 293 cells treated with siRNA as indicated. Primers against the targets MYL6, NDUFV3, and Tau are used. The percentage of exon inclusion for these gene products is calculated. F, mARVCF constructs containing the N-terminal and Arm repeat domain (NA) or the C-terminal domain (CT) only were expressed in siRNA-treated HEK 293 cells verified by Western blotting using antibodies, as indicated. The percentage of exon inclusion for MYL6 was measured and normalized to siRNA-treated cells transfected with empty vector (KD). n ≥ 3 biological replicates. Error bars, S.D.; *, p < 0.05; **, p < 0.01.
Sucrose Gradient Centrifugation
HEK 293 cells were lysed with immunoprecipitation buffer containing either 0.05% RNasin (Promega) or 0.2 μg/μl RNase A (Roche Applied Science), incubated for 10 min on ice, and centrifuged for 10 min. The supernatant was loaded on top of a 10–40% sucrose gradient, and centrifugation was performed for 16 h at 23,000 rpm and 4 °C in an ultracentrifuge (Sorvall WX ultra 100, Thermo Scientific). 400-μl fractions were collected beginning at the top of the gradient, and proteins were analyzed by Western blot.
Splicing Reporter Assay
HEK 293 cells were transfected in 12-well cell culture plates with reporter plasmid pTN23 (26) and the cDNA of interest in parallel. After 48 h, cells were collected in TEN buffer (40 mm Tris, 1 mm EDTA, 150 mm NaCl, pH 7.5) and pelleted, and luciferase and galactosidase activity was assayed using the Dual-light reporter system (Applied Biosystems) according to the manufacturer's protocol. Light emission was measured using an Infinite 200 reader (Tecan, Crailsheim, Germany).
RNA Extraction and RNA Sequencing
RNA was isolated from siRNA-treated HEK 293 cells with the RNeasy minikit (Qiagen, Hilden, Germany) according to the manufacturer's protocol, including on-column DNase treatment with the RNase-free DNase set (Qiagen). RNA samples were analyzed by the Genomics and Proteomics Core Facility (DKFZ) in triplicates with high throughput sequencing using the Illumina HiSeq 2000 system. Mapping to the reference genome hg19 was done with TopHat (27). We allowed for uniquely mapped reads only and provided TopHat with known transcripts based on the consensus CDS annotation (28) to support the detection of junction reads. Subsequent data analysis was done with the programs SplicingCompass (29) and DEXSeq (30).
RT-PCR and Capillary Electrophoresis
Reverse transcription was performed on 2 μg of total RNA using the High Capacity cDNA reverse transcription kit (Applied Biosystems). For verification of RNA sequencing, PCR analysis was done using 25 ng of cDNA, DreamTaq (Fermentas, St. Leon-Rot, Germany), and the following primers: MYL6, 5′-acttcgggtgtttgacaagg-3′ (exon 5) and 5′-gaaggtcctcagccattcag-3′ (exon 7); NDUFV3, 5′-gccacagaattccaagaagc-3′ (exon 2) and 5′-ccctcagtgtcgaggtgact-3′ (exon 4); Tau, 5′-caacgccaccaggattccagcaaa-3′ (exon 9) and 5′-atgttgcctaatgagccacacttg-3′ (exon 11). PCR products were purified with the QIAquick PCR purification kit (Qiagen, Hilden, Germany), and 1 μl was used for capillary electrophoresis using the Agilent DNA 1000 kit (Agilent Technologies). Data generation was done with the 2100 Bioanalyzer (Agilent Technologies) and the Agilent 2100 Expert software. The percentage of exon inclusion was calculated as follows, [exon included]/([exon included] + [exon skipped]).
RESULTS
Cellular Localization of ARVCF
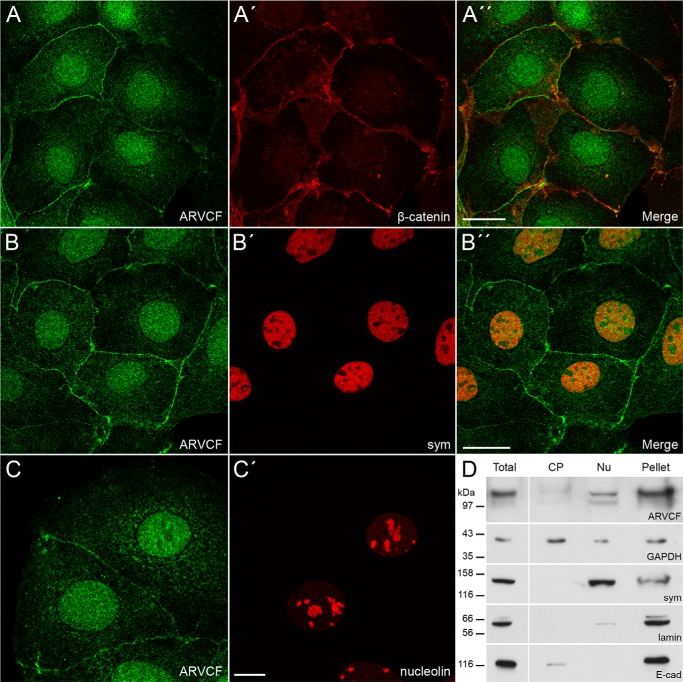
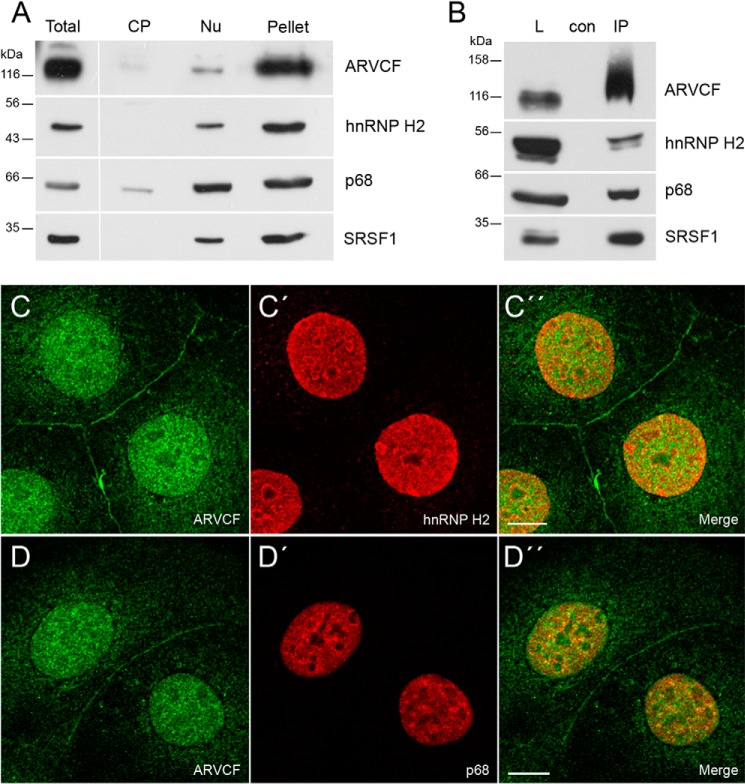
Using our novel monoclonal and polyclonal antibodies against ARVCF (20) for localization studies, we noticed that ARVCF occurred in junction-bound and non-junction-bound forms in Caco-2 cells (Fig. 1) and the renal cell line HK-2 (data not shown). The junction-bound form co-localized with β-catenin, a marker protein for adherens junctions (Fig. 1, A–A″). On the other hand, ARVCF was enriched in the nucleus. Here, ARVCF showed a distribution similar to that of nuclear symplekin (31) (Fig. 1, B–B″) but was only weakly detectable in nucleoli, as indicated by the localization of nucleolin (Fig. 1, C and C′). In addition to the nuclear localization, a weak cytoplasmic staining was also observed for ARVCF. To characterize the distribution of ARVCF in more detail, we generated a cytoplasmic and nuclear fraction (25). Additionally, the remaining pellet consisting of components of the nucleocytoskeleton and membranes was also analyzed. In the cytoplasmic extract, GAPDH was enriched, and the nuclear extract showed a strong signal for symplekin (31); in the pellet, lamin A/C, a marker for the nucleocytoskeleton, and E-cadherin, a marker for adherens junctions, were detectable. Endogenous ARVCF was found in the nuclear fraction but was also observed in the pellet fraction, indicating that a considerable amount of the ARVCF pool occurred in the nucleus.
FIGURE 1.
Cellular localization of ARVCF in Caco-2 cells. Shown is double immunofluorescence of ARVCF (A–C) with β-catenin (A′), symplekin (B′, sym), and nucleolin (C′) analyzed by confocal microscopy. A″ and B″, merge pictures. Scale bars, 20 μm (A and B) or 10 μm (C). D, in total, cytoplasmic (CP), and nuclear (Nu) cellular extracts and in the residual pellet of Caco-2 cells, the distribution of ARVCF was compared with marker proteins by Western blots (cytoplasmic, GAPDH; nuclear, symplekin (sym); pellet, lamin, E-cadherin (E-cad)).
ARVCF Interacts with mRNA-processing Proteins
To identify novel interaction partners, we performed a yeast two-hybrid analysis and obtained 79 positive clones, of which notably 67 clones encoded kazrin (NM_001017999). The identification of kazrin as ARVCF-interacting protein coincides with earlier findings (32) and confirms our experimental setup. The other genes identified were grouped into the subcategories mRNA-processing, DNA-associated, and cytoskeleton-associated (Table 1). Two independent clones corresponded to the HNRNPH2 gene that is involved in mRNA processing (33, 34). Similarly, RNA helicase p68/DDX5 has been shown to contribute to mRNA processing (35, 36). Both proteins had not yet been reported to interact with ARVCF.
TABLE 1.
Results of the yeast two-hybrid screen using full-length ARVCF as bait
| Prey | Description | Accession no. |
|---|---|---|
| mRNA-processing | ||
| HNRNPH2 | Heterogeneous nuclear ribonucleoprotein H2 (H') | NM_019597 |
| p68/DDX5 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | NM_004396 |
| DNA-associated | ||
| CHD4 | Chromodomain helicase DNA binding protein 4 | NM_001273 |
| ZNF189 | Zinc finger protein 189 | NM_003452 |
| Cytoskeleton-associated | ||
| ACTN4 | Actinin, α4 | NM_004924 |
| EPS8L2 | EPS8-like 2 | XM_001129221 |
| KAZ | Kazrin | NM_001017999 |
| Others | ||
| BEX4 | BEX family member 4 | XM_043653 |
| CD14 | CD14 molecule | NM_000591 |
| FAM46C | Family with sequence similarity 46, member C | NM_017709 |
| UBE2S | Ubiquitin-conjugating enzyme E2S | NM_014501 |
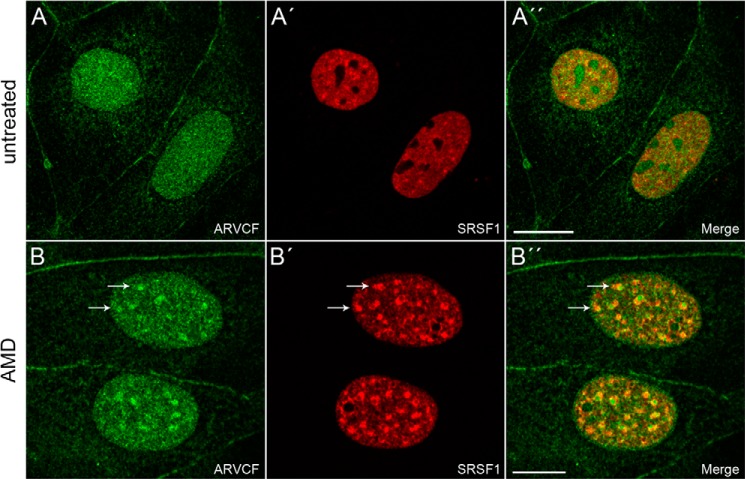
To verify whether the interaction of ARVCF with hnRNP H2 and p68 also occurs in cultured cells, we checked whether the two proteins co-fractionated with ARVCF in nuclear extracts. As a control, we included the known splicing factor SRSF1 (SF2/ASF) (37) in our analysis. All three proteins occurred in the nuclear extract and the pellet fraction and had a distribution similar to that of ARVCF (Fig. 2A). In addition, p68 was detectable in the cytoplasmic extract, although here the signal was much weaker than in the nuclear extract. To further explore the interaction with the RNA-binding proteins, we performed immunoprecipitation experiments (Fig. 2B). Using ARCVF-specific antibodies, the proteins hnRNP H2, p68, and SRSF1 were enriched in the immunoprecipitate, indicating that the three proteins formed a complex with ARVCF. Moreover, upon double immunolocalization, hnRNP H2, p68, and SRSF1 occurred in nuclei showing a co-distribution with ARVCF in the nucleoplasm (Figs. 2 (C–D″) and 3 (A–A″)). The distribution in nuclear speckles that function as storage, assembly, or modification compartments (38) was less obvious for ARVCF (Figs. 2 (C and D) and 3A) than for the proteins SRSF1 (Fig. 3A′) and p68 (Fig. 2D′). However, when Caco-2 cells were treated with actinomycin D that inhibits transcription and thereby leads to a redistribution of nuclear proteins (38), the occurrence in nuclear speckles of SRSF1 but also of ARVCF was enhanced (Fig. 3, B–B″).
FIGURE 2.
ARVCF interacts with the splicing factors hnRNP H2, p68, and SRSF1. A, distribution of ARVCF in total, cytoplasmic (CP), and nuclear (Nu) extracts and the residual pellet of Caco-2 cells was compared with the proteins hnRNP H2, p68, and SRSF1. B, co-immunoprecipitation of endogenous ARVCF was verified by Western blotting using the appropriate antibodies. L, load; con, negative control; IP, immunoprecipitation. C–D″, double immunofluorescence of ARVCF (C and D) with hnRNP H2 (C′) and p68 (D′) on Caco-2 cells analyzed by confocal microscopy. C″–E″) merge pictures. Scale bars, 10 μm.
FIGURE 3.
Cellular localization of ARVCF in Caco-2 cells treated with actinomycin D. Double immunofluorescence staining of ARVCF (A and B) with SRSF1 (A′ and B′) on untreated (A–A″) Caco-2 cells or cells treated (B–B″) with actinomycin D (AMD). A″ and B″, merge pictures. Scale bars, 20 μm. Note that arrows point to a partial co-distribution in interchromatin granule clusters upon actinomycin D treatment.
ARVCF Is Part of a Ribonucleoprotein Complex
Because ARVCF was co-distributed and co-precipitated with RNA-binding proteins (Fig. 2), we wondered whether ARVCF might also be part of an RNP. To test this, we separated complexes according to size by density gradient centrifugation and analyzed the fractionation in high molecular weight complexes (Fig. 4A). ARVCF was found in fractions 4–9 and fractions 23–30, with maxima at fraction 5 and 25, implying that ARVCF occurred in smaller and larger migrating complexes. The RNA-binding proteins hnRNP H2, p68, and SRSF1 showed partial co-fractionation with large ARVCF-containing complexes. In contrast, β-catenin was found in fractions 5–20, indicating that the large complexes are specific for ARVCF.
To determine whether the occurrence of these high molecular weight complexes depends on RNA, we added RNase A to the lysates prior to density gradient centrifugation (Fig. 4B). Upon RNase treatment, ARVCF and the proteins hnRNP H2, p68, and SRSF1 were no longer found in fractions 22–30, indicating that larger complexes contained RNA also. The distribution of β-catenin was, however, unchanged, emphasizing that β-catenin is not part of an RNP.
The interactions within an RNP may be mediated by protein-protein or protein-RNA interactions. To test this, we again added RNase A to lysates and immunoprecipitated ARVCF. The efficiency of the ARVCF immunoprecipitation was not affected by RNase A treatment, and hnRNP H2, p68, and SRSF1 were found to associate with ARVCF (Fig. 4C). In contrast, the RNA-binding protein hnRNP A1 remained no longer associated with ARVCF after RNase A treatment. Because the ARVCF-related protein PKP2 in its non-junctional form occurred also in nuclei (39), we tested whether PKP2 was enriched in ARVCF-containing complexes, and we could not observe a co-precipitation (Fig. 4C). Moreover, symplekin that acts as a scaffold factor of the multiprotein complex involved in cleavage and polyadenylation of pre-mRNA (31) was also not observed in ARVCF-containing complexes (Fig. 4C). Because the larger ARVCF-containing complexes were sensitive to RNase A treatment, we analyzed whether these complexes contain mRNA. We performed an immunoprecipitation experiment with ARVCF-specific antibodies, isolated RNA, and tested for the presence of mRNAs and pre-mRNAs by RT-PCR (Fig. 4, D and E). By this approach, the mRNA and also the pre-mRNA of the housekeeping genes GAPDH and actin and the mRNA of a less abundant gene, PKP2, were detected. As a control, we checked whether small nuclear RNAs were also enriched (40–42). The U2 snRNA, a spliceosomal RNA component, but not ACA44, a small nucleolar RNA, was detected (Fig. 4F). In addition, we did not observe that the ARVCF-related protein PKP2 bound p68 or the mRNAs of GAPDH or PKP2 (Fig. 4G). Taken together, our data provide evidence for the association of ARVCF with hnRNP H2, p68, and SRSF1 and demonstrate that these interactions are RNA-independent and that ARVCF-containing complexes include mRNA.
The C Terminus of ARVCF Mediates the Interactions
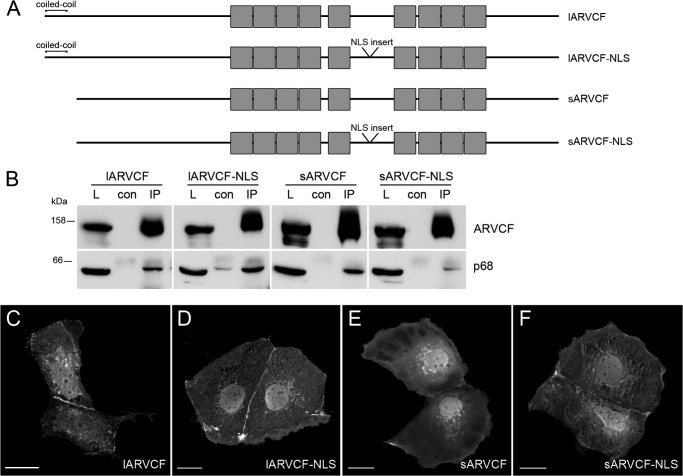
Because human ARVCF occurs in four different isoforms, we wondered whether they differ in their association with the RNA-binding proteins. All four ARVCF isoforms were transfected in HEK cells (Fig. 5) and tested for co-precipitation with p68. Indeed, all four isoforms were detectable in a complex with p68 (Fig. 5B). Moreover, all proteins occurred upon forced expression in Caco-2 cells in cell-cell contacts, in the nucleus, and to a lesser extent in the cytoplasm (Fig. 5, C–F). This indicates that neither the N-terminal coiled-coil domain nor the internal NLS motif specify the interaction with the RNA-binding proteins and the localization within a cell.
FIGURE 5.
All four ARVCF isoforms interact with RNA-binding proteins. A, schematic representation of the four human ARVCF isoforms containing an N-terminal extension of a coiled-coil domain or the insertion into the NLS motif. Gray boxes, Arm repeats. B, HEK 293 cells were transfected with C-terminally GFP-tagged ARVCF isoforms. Immunoprecipitation was performed with GFP antibody, and Western blot was performed with p68 antibody. L, load; con, control; IP, immunoprecipitation. C–F, fluorescence staining of Caco-2 cells transfected with GFP-tagged ARVCF isoform constructs as indicated. Scale bars, 20 μm.
To resolve ARVCF domains necessary for the interaction with the three RNA-binding proteins, four ARVCF GFP-tagged constructs (Fig. 6A) were expressed in HEK 293 cells and used for immunoprecipitation (Fig. 6B). Deletion of the C-terminal 176 amino acid residues abolished ARVCF association with hnRNP H2, p68, and SRSF1. However, a deletion of the C-terminal PDZ-binding motif DSWV did not disrupt the ARVCF interaction with the RNA-binding proteins. Moreover, deletion of the C-terminal 176 amino acid residues abolished ARVCF recruitment to GAPDH mRNA (Fig. 6C). Similarly, ARVCF-GFP tagged constructs containing the C-terminal portion of ARVCF accumulated in the nuclear compartment upon transient transfection to Caco-2 cells (Fig. 6, D–H). Taken together, these data indicate that the region of ARVCF necessary for the interaction with the RNA-binding proteins resides between amino acid residues 776 and 952 and that the C-terminal domain of ARVCF is also necessary for the mRNA recruitment. Because this domain also mediates the interaction with the RNA-binding proteins, this implies that ARVCF is recruited to the RNP via protein-protein interactions.
Altered ARVCF Expression Leads to Changes in Pre-mRNA Splicing
All three ARVCF-interacting proteins were reported to be involved in splicing of pre-mRNA (33, 36, 37). Therefore, we tested whether ARVCF also influences splicing. A double reporter assay was used to detect changes in the ratio of spliced and unspliced mRNA (26). Overexpression of ARVCF, hnRNP H2, p68, SRSF1, or p0071 was verified by immunoblotting (Fig. 7B). To quantify the splicing efficiency, luciferase and galactosidase activities were measured. In cells overexpressing the proteins p68, SRFS1, and hnRNP H2, which are well known modulators of splicing efficiency, the relative luciferase/galactosidase ratio was significantly higher (Fig. 7A). Similarly, overexpression of ARVCF also led to a higher luciferase/galactosidase ratio, whereas the overexpression of the ARVCF-related protein p0071 showed no influence on splicing efficiency of the reporter mRNA. The data indicate that ARVCF is a novel factor that affects mammalian splicing.
To further validate the influence of ARVCF on splicing processes, we knocked down the protein in HEK 293 cells, which was verified via immunoblotting (Fig. 7C). The total RNA was isolated and analyzed by RNA sequencing, and differential splicing events were identified using the programs SplicingCompass (29) and DEXSeq (30). Together, 30 differentially spliced genes were predicted (adjusted p value < 0.1) (Table 2). Two of them were chosen for validation: the myosin light chain protein MYL6 and NDUFV3, a subunit of the ubiquinone oxidoreductase complex. Both of these proteins exist in two isoforms, differing in the inclusion or exclusion of one exon (Fig. 7E). In MYL6, exon 6 is alternatively included (43), and in NDUFV3, exon 3 is changed (44). As an experimental control, we chose a splicing target of p68, Tau, which shows p68-dependent changes of exon 10 inclusion (45). First, we verified whether the mRNAs of the three different genes were detectable in immunoprecipitates using ARVCF-specific antibodies from lysates of untreated HEK cells (Fig. 7D). All three mRNAs were detectable in ARVCF-containing complexes and might therefore be splicing targets of ARVCF. To test this, we used RT-PCR from HEK 293 cells treated with siRNA against ARVCF, p68, or control with primers flanking the alternatively spliced exons. Via capillary electrophoresis, we could show an increase of exon inclusion for MYL6 and NDUFV3 in cells treated with ARVCF siRNA (Fig. 7E). In contrast, Tau, which shows an increase of exon 10 inclusion in p68 knockdown cells, is not changed after ARVCF knockdown. To rule out off-target effects, we performed rescue experiments using RNAi-resistant ARVCF constructs (Fig. 7F). Upon re-expression of a C-terminal ARVCF fragment but not an N-terminal and Arm repeat domain, the exon inclusion for MYL6 was significantly reduced. Therefore, we conclude that ARVCF is involved in alternative splicing of specific targets.
TABLE 2.
Genes predicted to be differentially spliced between HEK 293 cells treated with ARVCF siRNA and control siRNA
Results from SplicingCompass and DEXSeq are combined, and the smallest p value is shown. Genes chosen for validation are shown in boldface type.
| Gene ID | Description | p value |
|---|---|---|
| FAM89B | Family with sequence similarity 89, member B | 1.33E-05 |
| RPS28 | 40 S ribosomal protein S28 | 4.04E-05 |
| ATP5D | ATP synthase, H+-transporting, mitochondrial F1 complex, δ subunit | 7.44E-05 |
| ZDHHC12 | Zinc finger, DHHC-type containing 12 | 1.25E-04 |
| GPX1 | Glutathione peroxidase 1 | 1.25E-04 |
| TMBIM1 | Transmembrane BAX inhibitor motif containing 1 | 6.89E-04 |
| RPS17 | 40 S ribosomal protein S17 | 1.01E-03 |
| PIAS4 | Protein inhibitor of activated STAT, 4 | 1.03E-03 |
| ACTR5 | ARP5 actin-related protein 5 homolog (yeast) | 4.00E-03 |
| ZNF275 | Zinc finger protein 275 | 6.79E-03 |
| SRP19 | Signal recognition particle 19 kDa | 6.90E-03 |
| STXBP2 | Syntaxin-binding protein 2 | 1.17E-02 |
| CYB5R1 | Cytochrome b5 reductase 1 | 1.78E-02 |
| NOP14 | NOP14 nucleolar protein homolog (yeast) | 2.65E-02 |
| FAM128A | Mitotic spindle-organizing protein 2A | 3.06E-02 |
| HSP90AA1 | Heat shock protein 90 kDa α (cytosolic), class A member 1 | 3.48E-02 |
| C14orf153 | Apoptogenic 1, mitochondrial | 3.64E-02 |
| C16orf70 | Chromosome 16 open reading frame 70 | 4.00E-02 |
| RCN2 | Reticulocalbin 2, EF-hand calcium binding domain | 4.90E-02 |
| RAP1GAP | RAP1 GTPase-activating protein | 5.44E-02 |
| MYL6 | Myosin, light chain 6, alkali, smooth muscle, and non-muscle | 5.63E-02 |
| NDUFV3 | NADH dehydrogenase (ubiquinone) flavoprotein 3, 10 kDa | 6.09E-02 |
| NAT14 | N-Acetyltransferase 14 (GCN5-related, putative) | 6.62E-02 |
| ZNHIT6 | Zinc finger, HIT-type containing 6 | 6.93E-02 |
| CSTB | Cystatin B (stefin B) | 7.54E-02 |
| TRPT1 | tRNA phosphotransferase 1 | 8.05E-02 |
| TMEM170B | Transmembrane protein 170B | 8.22E-02 |
| HOXC13 | Homeobox C13 | 8.38E-02 |
| C6orf35 | Transmembrane protein 242 | 9.60E-02 |
| PITX1 | Paired-like homeodomain 1 | 9.76E-02 |
DISCUSSION
ARVCF, an Arm repeat protein of the p120-catenin family, is located in adherens junctions but is also found in the nucleus and the cytoplasm. The present study reveals nuclear ARVCF as a component of RNPs in complex with SRSF1, p68, and hnRNPH2. Furthermore, ARVCF-containing RNPs show an enrichment of mRNA and pre-mRNA, and overexpression and knockdown experiments suggest an involvement of ARVCF in pre-mRNA splicing.
By biochemical fractionation and localization studies, we identified ARVCF in the nucleus. Our findings correlate with earlier observations using forced expression approaches (14, 23) or endogenous protein localization studies (22). In the past, several studies focused on the identification of motifs necessary for nuclear enrichment of ARVCF (9, 14, 22). Mariner et al. (14) performed forced expression using a fibroblast cell line and noticed by domain swapping between ARVCF and p120-catenin that the N-terminal portion of ARVCF fused to p120-catenin mediates nuclear enrichment of this construct. Kaufmann et al. (22) observed that, upon forced expression, a construct consisting of Arm repeats and the C-terminal portion was recruited to the nucleus, and this is in line with our findings. Kausalya et al. (9) focused on the PDZ-binding domain of ARVCF and its interaction with ZO-2. Upon introducing point mutations into this motif, the in vitro binding was strongly reduced, and the ARVCF mutant was no longer enriched in the nucleus upon forced expression. This observation seems to contradict our findings but may be explained by the different experimental setup. In our study, none of the four human ARVCF isoforms was preferentially recruited to the nucleus. The insertion of a basic amino acid stretch into the putative NLS signal within the Arm repeat domain seemed not to influence its cellular localization as it was supposed in earlier reports (6). Similar to ARVCF, several p120-catenin isoforms can be expressed due to alternative splicing and multiple translation initiation codons (46). On the contrary, nuclear trafficking of p120-catenin depends on NLS and NES signals as well as on the Arm repeat domain (7, 47–50), and only p120-catenin isoforms containing these signals will be recruited to the nucleus (48, 51). For ARVCF, our findings suggest that the interaction with other cellular components and not internal sequences define the localization of ARVCF within a cell.
To elucidate the function of ARVCF in the nucleus, we identified nuclear interaction partners. ARVCF is a component of RNPs containing mRNA and interacts with the RNA-binding proteins SRSF1, p68, and hnRNP H2. Our observation that the interaction with the three proteins was resistant to RNase treatment indicates that ARVCF is bound to RNPs by protein-protein interactions. Moreover, our domain analysis data suggest that the region of ARVCF necessary for the interaction with the RNA-binding proteins resides between amino acid residues 776 and 952. In contrast, the recruitment of ARVCF to adherens junctions by binding to the juxtamembrane domain of cadherins is mediated by the Arm repeats of ARVCF (22, 23, 52). The PDZ binding domain consisting of the last C-terminal amino acids was not essential for binding to the RNA-binding proteins but mediates binding to PDZ domain-containing proteins, such as erbin, ZO-1, and ZO-2 (8, 9).
Because all three proteins, SRSF1, p68, and hnRNP H2, that were identified as ARVCF-interacting proteins are involved in pre-mRNA splicing, it was tempting to speculate that nuclear ARVCF also contributes to this process. Indeed, ARVCF-containing complexes were associated with pre-mRNA, and upon overexpression, ARVCF had an impact on splicing of a reporter construct. Splicing of pre-mRNA is catalyzed by the spliceosome, a dynamic multimegadalton RNP. In the past, core spliceosomal components and spliceosomal dynamics have been studied intensively (40, 53, 54). Both proteins, SRSF1 and p68, are ubiquitous components of the spliceosome and are essential for constitutive splicing (36, 55). In contrast, neither hnRNP H2 nor ARVCF were identified as core spliceosomal components (40). However, in higher eukaryotes, alternative splicing expands the complexity of the proteome in different tissues (56, 57), and this is achieved through complex networks of protein-RNA interactions (58–60). All three ARVCF-interacting proteins were reported to be involved in alternative splicing of specific genes (37, 45, 59, 61). Upon knockdown of ARVCF, alternative splicing of several genes was significantly altered in our study. Specifically, we noticed a differential inclusion of exon 6 in MYL6, one of the myosin light chain proteins. In ARVCF-depleted cells, inclusion of exon 6 occurred more frequently. Besides, the exon 6-containing MYL6 isoform is preferentially expressed in smooth muscle cells (43), suggesting that ARVCF might have an impact on cellular integrity via alternative splicing events of regulatory structural components (62).
Surprisingly, all mRNAs tested so far were detectable in ARVCF-containing RNPs, but only a subset of these mRNAs was alternatively spliced. Recent findings show that the regulation of alternative splicing events is characterized by an extremely high complexity that relies on cis-acting intronic and exonic elements and trans-acting splicing factors with overlapping affinities (59, 60). Differences in alternative splicing can be mediated by changes in the stochiometry of RNA-binding proteins or by post-translational mechanisms, including protein modification or subcellular sequestration in response to external stimuli (65). Here, the regulated exchange of the junctional and non-junctional form of ARVCF by its modification (14) may couple extra- and intercellular signaling to splicing regulation. Similarly, the ARVCF-related proteins, PKP1 and PKP3, also associate in their non-junctional form with RNA-binding proteins and are components of RNPs. Therefore, a role in translation and/or mRNA decay has been suggested (18, 63, 64).
In this study, we focused on the function of ARVCF in epithelial cells, but ARVCF is broadly expressed not only in a variety of epithelia but also in non-epithelial tissues, such as skeletal muscle and brain in the adult and in developmental stages (6, 20, 66). It is noteworthy that missense mutations in the ARVCF gene specifically in an exon coding for the C terminus have been linked to the risk of schizophrenia (67, 68). Aberrant RNA processing by alternative splicing of genes related to schizophrenia had been frequently observed in the past (69, 70), and this was correlated with the pathophysiology of the disorder. In future studies, it will be clarified whether ARVCF contributes to malfunctions in alternative splicing that may lead to aberrant pre-mRNA splicing events giving rise to neurological disorders, such as schizophrenia, or other human diseases, such as myotonic dystrophy or cancer (33, 61, 71, 72).
In conclusion, our finding that ARVCF binds to hnRNP H2, p68, and SRSF1 and influences splicing opens a new research direction. We show for the first time that the non-junctional form of a member of the p120-catenin family has an impact on pre-mRNA splicing and influences the abundance of alternatively spliced mRNAs. Future studies will clarify the exact function of ARVCF in this highly regulated process. We hypothesize that ARVCF is involved in alternative splicing, and its deregulation may contribute to diseased states, such as cancer.
Acknowledgments
We thank Dr. I. C. Eperon (University of Leicester, UK) for providing the splicing reporter vector pTN23 and Prof. Dr. A. Starzinski-Powitz (University of Frankfurt, Germany) for providing the mouse ARVCF constructs. We thank both the DKFZ Genomics and Proteomics Core Facility unit Yeast Two-Hybrid Screening Service and the unit for High Throughput Sequencing led by Dr. Stephan Wolf for providing the sequencing data used in this project. We gratefully acknowledge helpful comments and critical reading by Dr. Barbara Janssens (DKFZ).
This work was supported by Grant 108978 from the German Cancer Aid (to I. H.).
- Arm
- armadillo
- RNP
- ribonucleoprotein particle
- hnRNP
- heterogeneous nuclear ribonucleoprotein particle
- NLS
- nuclear localization signal
- DKFZ
- Deutsches Krebsforschungszentrum (German Cancer Research Center).
REFERENCES
- 1. Stepniak E., Radice G. L., Vasioukhin V. (2009) Adhesive and signaling functions of cadherins and catenins in vertebrate development. Cold Spring Harb. Perspect. Biol. 1, a002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McCrea P. D., Gu D., Balda M. S. (2009) Junctional music that the nucleus hears: cell-cell contact signaling and the modulation of gene activity. Cold Spring Harb. Perspect. Biol. 1, a002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carnahan R. H., Rokas A., Gaucher E. A., Reynolds A. B. (2010) The molecular evolution of the p120-catenin subfamily and its functional associations. PLoS One 5, e15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McCrea P. D., Gu D. (2010) The catenin family at a glance. J. Cell Sci. 123, 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reynolds A. B. (2007) p120-catenin: past and present. Biochim. Biophys. Acta 1773, 2–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sirotkin H., O'Donnell H., DasGupta R., Halford S., St Jore B., Puech A., Parimoo S., Morrow B., Skoultchi A., Weissman S. M., Scambler P., Kucherlapati R. (1997) Identification of a new human catenin gene family member (ARVCF) from the region deleted in velo-cardio-facial syndrome. Genomics 41, 75–83 [DOI] [PubMed] [Google Scholar]
- 7. van Hengel J., Vanhoenacker P., Staes K., van Roy F. (1999) Nuclear localization of the p120ctn Armadillo-like catenin is counteracted by a nuclear export signal and by E-cadherin expression. Proc. Natl. Acad. Sci. U.S.A. 96, 7980–7985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laura R. P., Witt A. S., Held H. A., Gerstner R., Deshayes K., Koehler M. F., Kosik K. S., Sidhu S. S., Lasky L. A. (2002) The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of δ-catenin and ARVCF. J. Biol. Chem. 277, 12906–12914 [DOI] [PubMed] [Google Scholar]
- 9. Kausalya P. J., Phua D. C., Hunziker W. (2004) Association of ARVCF with zonula occludens (ZO)-1 and ZO-2: binding to PDZ-domain proteins and cell-cell adhesion regulate plasma membrane and nuclear localization of ARVCF. Mol. Biol. Cell 15, 5503–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stenzel N., Fetzer C. P., Heumann R., Erdmann K. S. (2009) PDZ-domain-directed basolateral targeting of the peripheral membrane protein FRMPD2 in epithelial cells. J. Cell Sci. 122, 3374–3384 [DOI] [PubMed] [Google Scholar]
- 11. Kowalczyk A. P., Nanes B. A. (2012) Adherens junction turnover: regulating adhesion through cadherin endocytosis, degradation, and recycling. Subcell. Biochem. 60, 197–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nanes B. A., Chiasson-MacKenzie C., Lowery A. M., Ishiyama N., Faundez V., Ikura M., Vincent P. A., Kowalczyk A. P. (2012) p120-catenin binding masks an endocytic signal conserved in classical cadherins. J. Cell Biol. 199, 365–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fang X., Ji H., Kim S. W., Park J. I., Vaught T. G., Anastasiadis P. Z., Ciesiolka M., McCrea P. D. (2004) Vertebrate development requires ARVCF and p120 catenins and their interplay with RhoA and Rac. J. Cell Biol. 165, 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mariner D. J., Wang J., Reynolds A. B. (2000) ARVCF localizes to the nucleus and adherens junction and is mutually exclusive with p120ctn in E-cadherin complexes. J. Cell Sci. 113, 1481–1490 [DOI] [PubMed] [Google Scholar]
- 15. Anastasiadis P. Z. (2007) p120-ctn: A nexus for contextual signaling via Rho GTPases. Biochim. Biophys. Acta 1773, 34–46 [DOI] [PubMed] [Google Scholar]
- 16. Valenta T., Hausmann G., Basler K. (2012) The many faces and functions of β-catenin. EMBO J. 31, 2714–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Roy F. M., McCrea P. D. (2005) A role for Kaiso-p120ctn complexes in cancer? Nat. Rev. Cancer 5, 956–964 [DOI] [PubMed] [Google Scholar]
- 18. Hofmann I., Casella M., Schnölzer M., Schlechter T., Spring H., Franke W. W. (2006) Identification of the junctional plaque protein plakophilin 3 in cytoplasmic particles containing RNA-binding proteins and the recruitment of plakophilins 1 and 3 to stress granules. Mol. Biol. Cell 17, 1388–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Glisovic T., Bachorik J. L., Yong J., Dreyfuss G. (2008) RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582, 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Walter B., Schlechter T., Hergt M., Berger I., Hofmann I. (2008) Differential expression pattern of protein ARVCF in nephron segments of human and mouse kidney. Histochem. Cell Biol. 130, 943–956 [DOI] [PubMed] [Google Scholar]
- 21. Hofmann I., Kuhn C., Franke W. W. (2008) Protein p0071, a major plaque protein of non-desmosomal adhering junctions, is a selective cell-type marker. Cell Tissue Res. 334, 381–399 [DOI] [PubMed] [Google Scholar]
- 22. Kaufmann U., Zuppinger C., Waibler Z., Rudiger M., Urbich C., Martin B., Jockusch B. M., Eppenberger H., Starzinski-Powitz A. (2000) The armadillo repeat region targets ARVCF to cadherin-based cellular junctions. J. Cell Sci. 113, 4121–4135 [DOI] [PubMed] [Google Scholar]
- 23. Waibler Z., Schäfer A., Starzinski-Powitz A. (2001) mARVCF cellular localisation and binding to cadherins is influenced by the cellular context but not by alternative splicing. J. Cell Sci. 114, 3873–3884 [DOI] [PubMed] [Google Scholar]
- 24. Mohr K., Koegl M. (2012) High-throughput yeast two-hybrid screening of complex cDNA libraries. Methods Mol. Biol. 812, 89–102 [DOI] [PubMed] [Google Scholar]
- 25. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nasim M. T., Chowdhury H. M., Eperon I. C. (2002) A double reporter assay for detecting changes in the ratio of spliced and unspliced mRNA in mammalian cells. Nucleic Acids Res. 30, e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trapnell C., Pachter L., Salzberg S. L. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pruitt K. D., Harrow J., Harte R. A., Wallin C., Diekhans M., Maglott D. R., Searle S., Farrell C. M., Loveland J. E., Ruef B. J., Hart E., Suner M. M., Landrum M. J., Aken B., Ayling S., Baertsch R., Fernandez-Banet J., Cherry J. L., Curwen V., Dicuccio M., Kellis M., Lee J., Lin M. F., Schuster M., Shkeda A., Amid C., Brown G., Dukhanina O., Frankish A., Hart J., Maidak B. L., Mudge J., Murphy M. R., Murphy T., Rajan J., Rajput B., Riddick L. D., Snow C., Steward C., Webb D., Weber J. A., Wilming L., Wu W., Birney E., Haussler D., Hubbard T., Ostell J., Durbin R., Lipman D. (2009) The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 19, 1316–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aschoff M., Hotz-Wagenblatt A., Glatting K. H., Fischer M., Eils R., König R. (2013) SplicingCompass: differential splicing detection using RNA-Seq data. Bioinformatics 29, 1141–1148 [DOI] [PubMed] [Google Scholar]
- 30. Anders S., Reyes A., Huber W. (2012) Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hofmann I., Schnölzer M., Kaufmann I., Franke W. W. (2002) Symplekin, a constitutive protein of karyo- and cytoplasmic particles involved in mRNA biogenesis in Xenopus laevis oocytes. Mol. Biol. Cell 13, 1665–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cho K., Vaught T. G., Ji H., Gu D., Papasakelariou-Yared C., Horstmann N., Jennings J. M., Lee M., Sevilla L. M., Kloc M., Reynolds A. B., Watt F. M., Brennan R. G., Kowalczyk A. P., McCrea P. D. (2010) Xenopus Kazrin interacts with ARVCF-catenin, spectrin and p190B RhoGAP, and modulates RhoA activity and epithelial integrity. J. Cell Sci. 123, 4128–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stark M., Bram E. E., Akerman M., Mandel-Gutfreund Y., Assaraf Y. G. (2011) Heterogeneous nuclear ribonucleoprotein H1/H2-dependent unsplicing of thymidine phosphorylase results in anticancer drug resistance. J. Biol. Chem. 286, 3741–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Honoré B., Rasmussen H. H., Vorum H., Dejgaard K., Liu X., Gromov P., Madsen P., Gesser B., Tommerup N., Celis J. E. (1995) Heterogeneous nuclear ribonucleoproteins H, H′, and F are members of a ubiquitously expressed subfamily of related but distinct proteins encoded by genes mapping to different chromosomes. J. Biol. Chem. 270, 28780–28789 [DOI] [PubMed] [Google Scholar]
- 35. Liew C. C., Smith H. C. (1989) Immunological evidence for the role of phosphoprotein p68/pI = 7.3 in premessenger RNA splicing. FEBS Lett. 248, 101–104 [DOI] [PubMed] [Google Scholar]
- 36. Lin C., Yang L., Yang J. J., Huang Y., Liu Z. R. (2005) ATPase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol. Cell Biol. 25, 7484–7493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krainer A. R., Conway G. C., Kozak D. (1990) The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell 62, 35–42 [DOI] [PubMed] [Google Scholar]
- 38. Lamond A. I., Spector D. L. (2003) Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4, 605–612 [DOI] [PubMed] [Google Scholar]
- 39. Mertens C., Hofmann I., Wang Z., Teichmann M., Sepehri Chong S., Schnölzer M., Franke W. W. (2001) Nuclear particles containing RNA polymerase III complexes associated with the junctional plaque protein plakophilin 2. Proc. Natl. Acad. Sci. U.S.A. 98, 7795–7800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wahl M. C., Will C. L., Lührmann R. (2009) The spliceosome: design principles of a dynamic RNP machine. Cell 136, 701–718 [DOI] [PubMed] [Google Scholar]
- 41. Kiss T., Fayet-Lebaron E., Jády B. E. (2010) Box H/ACA small ribonucleoproteins. Mol. Cell 37, 597–606 [DOI] [PubMed] [Google Scholar]
- 42. Galiveti C. R., Rozhdestvensky T. S., Brosius J., Lehrach H., Konthur Z. (2010) Application of housekeeping npcRNAs for quantitative expression analysis of human transcriptome by real-time PCR. RNA 16, 450–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lenz S., Lohse P., Seidel U., Arnold H. H. (1989) The alkali light chains of human smooth and nonmuscle myosins are encoded by a single gene. Tissue-specific expression by alternative splicing pathways. J. Biol. Chem. 264, 9009–9015 [PubMed] [Google Scholar]
- 44. Rajan P., Dalgliesh C., Carling P. J., Buist T., Zhang C., Grellscheid S. N., Armstrong K., Stockley J., Simillion C., Gaughan L., Kalna G., Zhang M. Q., Robson C. N., Leung H. Y., Elliott D. J. (2011) Identification of novel androgen-regulated pathways and mRNA isoforms through genome-wide exon-specific profiling of the LNCaP transcriptome. PLoS One 6, e29088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kar A., Fushimi K., Zhou X., Ray P., Shi C., Chen X., Liu Z., Chen S., Wu J. Y. (2011) RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5′ splice site. Mol. Cell Biol. 31, 1812–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Keirsebilck A., Bonné S., Staes K., van Hengel J., Nollet F., Reynolds A., van Roy F. (1998) Molecular cloning of the human p120ctn catenin gene (CTNND1): expression of multiple alternatively spliced isoforms. Genomics 50, 129–146 [DOI] [PubMed] [Google Scholar]
- 47. Kelly K. F., Spring C. M., Otchere A. A., Daniel J. M. (2004) NLS-dependent nuclear localization of p120ctn is necessary to relieve Kaiso-mediated transcriptional repression. J. Cell Sci. 117, 2675–2686 [DOI] [PubMed] [Google Scholar]
- 48. Aho S., Levänsuo L., Montonen O., Kari C., Rodeck U., Uitto J. (2002) Specific sequences in p120ctn determine subcellular distribution of its multiple isoforms involved in cellular adhesion of normal and malignant epithelial cells. J. Cell Sci. 115, 1391–1402 [DOI] [PubMed] [Google Scholar]
- 49. Daniel J. M. (2007) Dancing in and out of the nucleus: p120ctn and the transcription factor Kaiso. Biochim. Biophys. Acta 1773, 59–68 [DOI] [PubMed] [Google Scholar]
- 50. Pieters T., van Roy F., van Hengel J. (2012) Functions of p120ctn isoforms in cell-cell adhesion and intracellular signaling. Front. Biosci. 17, 1669–1694 [DOI] [PubMed] [Google Scholar]
- 51. Zhang P. X., Wang Y., Liu Y., Jiang G. Y., Li Q. C., Wang E. H. (2011) p120-catenin isoform 3 regulates subcellular localization of Kaiso and promotes invasion in lung cancer cells via a phosphorylation-dependent mechanism. Int. J. Oncol. 38, 1625–1635 [DOI] [PubMed] [Google Scholar]
- 52. Paulson A. F., Mooney E., Fang X., Ji H., McCrea P. D. (2000) Xarvcf, Xenopus member of the p120 catenin subfamily associating with cadherin juxtamembrane region. J. Biol. Chem. 275, 30124–30131 [DOI] [PubMed] [Google Scholar]
- 53. Bessonov S., Anokhina M., Will C. L., Urlaub H., Lührmann R. (2008) Isolation of an active step I spliceosome and composition of its RNP core. Nature 452, 846–850 [DOI] [PubMed] [Google Scholar]
- 54. Behzadnia N., Golas M. M., Hartmuth K., Sander B., Kastner B., Deckert J., Dube P., Will C. L., Urlaub H., Stark H., Lührmann R. (2007) Composition and three-dimensional EM structure of double affinity-purified, human prespliceosomal A complexes. EMBO J. 26, 1737–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Krainer A. R., Conway G. C., Kozak D. (1990) Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 4, 1158–1171 [DOI] [PubMed] [Google Scholar]
- 56. Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P., Burge C. B. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grabowski P. (2011) Alternative splicing takes shape during neuronal development. Curr. Opin. Genet. Dev. 21, 388–394 [DOI] [PubMed] [Google Scholar]
- 58. Wang Y., Ma M., Xiao X., Wang Z. (2012) Intronic splicing enhancers, cognate splicing factors and context-dependent regulation rules. Nat. Struct. Mol. Biol. 19, 1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huelga S. C., Vu A. Q., Arnold J. D., Liang T. Y., Liu P. P., Yan B. Y., Donohue J. P., Shiue L., Hoon S., Brenner S., Ares M., Jr., Yeo G. W. (2012) Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 1, 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang Y., Xiao X., Zhang J., Choudhury R., Robertson A., Li K., Ma M., Burge C. B., Wang Z. (2013) A complex network of factors with overlapping affinities represses splicing through intronic elements. Nat. Struct. Mol. Biol. 20, 36–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Paul S., Dansithong W., Jog S. P., Holt I., Mittal S., Brook J. D., Morris G. E., Comai L., Reddy S. (2011) Expanded CUG repeats dysregulate RNA splicing by altering the stoichiometry of the muscleblind 1 complex. J. Biol. Chem. 286, 38427–38438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Park I., Han C., Jin S., Lee B., Choi H., Kwon J. T., Kim D., Kim J., Lifirsu E., Park W. J., Park Z. Y., Kim do H., Cho C. (2011) Myosin regulatory light chains are required to maintain the stability of myosin II and cellular integrity. Biochem. J. 434, 171–180 [DOI] [PubMed] [Google Scholar]
- 63. Wolf A., Krause-Gruszczynska M., Birkenmeier O., Ostareck-Lederer A., Hüttelmaier S., Hatzfeld M. (2010) Plakophilin 1 stimulates translation by promoting eIF4A1 activity. J. Cell Biol. 188, 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang C., Strobel P., Marx A., Hofmann I. (2013) Plakophilin-associated RNA-binding proteins in prostate cancer and their implications in tumor progression and metastasis. Virchows Arch. 463, 379–390 [DOI] [PubMed] [Google Scholar]
- 65. Licatalosi D. D., Darnell R. B. (2010) RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 11, 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Maynard T. M., Haskell G. T., Peters A. Z., Sikich L., Lieberman J. A., LaMantia A. S. (2003) A comprehensive analysis of 22q11 gene expression in the developing and adult brain. Proc. Natl. Acad. Sci. U.S.A. 100, 14433–14438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mas S., Bernardo M., Gasso P., Alvarez S., Garcia-Rizo C., Bioque M., Kirkpatrick B., Lafuente A. (2010) A functional variant provided further evidence for the association of ARVCF with schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 153, 1052–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mas S., Bernardo M., Parellada E., Garcia-Rizo C., Gassó P., Alvarez S., Lafuente A. (2009) ARVCF single marker and haplotypic association with schizophrenia. Prog. Neuropsychopharmacol Biol. Psychiatry 33, 1064–1069 [DOI] [PubMed] [Google Scholar]
- 69. Morikawa T., Manabe T. (2010) Aberrant regulation of alternative pre-mRNA splicing in schizophrenia. Neurochem. Int. 57, 691–704 [DOI] [PubMed] [Google Scholar]
- 70. Wu J. Q., Wang X., Beveridge N. J., Tooney P. A., Scott R. J., Carr V. J., Cairns M. J. (2012) Transcriptome sequencing revealed significant alteration of cortical promoter usage and splicing in schizophrenia. PLoS One 7, e36351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Laurent F. X., Sureau A., Klein A. F., Trouslard F., Gasnier E., Furling D., Marie J. (2012) New function for the RNA helicase p68/DDX5 as a modifier of MBNL1 activity on expanded CUG repeats. Nucleic Acids Res. 40, 3159–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. David C. J., Manley J. L. (2010) Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 24, 2343–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]