Background: Cell wall of Malassezia restricta is involved in interactions with human skin.
Results: Its core is composed of cross-linked polysaccharides such as chitin, chitosan, β-(1,3)-glucan and β-(1,6)-glucan.
Conclusion: The composition of cell wall polysaccharides of M. restricta is unique in the fungal kingdom.
Significance: The cell wall of M. restricta has evolved as a yeast that adapted to the skin microenvironment and host interactions.
Keywords: Carbohydrate Complex, Carbohydrate Structure, Cell Wall, Chitin, Yeast
Abstract
Malassezia species are ubiquitous residents of human skin and are associated with several diseases such as seborrheic dermatitis, tinea versicolor, folliculitis, atopic dermatitis, and scalp conditions such as dandruff. Host-Malassezia interactions and mechanisms to evade local immune responses remain largely unknown. Malassezia restricta is one of the most predominant yeasts of the healthy human skin, its cell wall has been investigated in this paper. Polysaccharides in the M. restricta cell wall are almost exclusively alkali-insoluble, showing that they play an essential role in the organization and rigidity of the M. restricta cell wall. Fractionation of cell wall polymers and carbohydrate analyses showed that the polysaccharide core of the cell wall of M. restricta contained an average of 5% chitin, 20% chitosan, 5% β-(1,3)-glucan, and 70% β-(1,6)-glucan. In contrast to other yeasts, chitin and chitosan are relatively abundant, and β-(1,3)-glucans constitute a minor cell wall component. The most abundant polymer is β-(1,6)-glucans, which are large molecules composed of a linear β-(1,6)-glucan chains with β-(1,3)-glucosyl side chain with an average of 1 branch point every 3.8 glucose unit. Both β-glucans are cross-linked, forming a huge alkali-insoluble complex with chitin and chitosan polymers. Data presented here show that M. restricta has a polysaccharide organization very different of all fungal species analyzed to date.
Introduction
Fungi of the Malassezia genus are members of the resident superficial cutaneous microbiota of healthy humans and warm-blooded animals. Malassezia species are also known as opportunistic pathogens, being found associated with diverse dermatological pathologies including pityriasis versicolor, seborrheic dermatitis, atopic dermatitis, and folliculitis (1–3). Among these species, Malassezia restricta was revealed to be the most abundant Malassezia species on the scalp, neck, face, and ears of healthy volunteers (4, 5). Recently, it has been shown that dandruff was associated with imbalance in the scalp microbiota population where M. restricta yeast significantly increases with dandruff severity (6).
Malassezia species are lipophilic with a cell morphology characterized by a thick and multilamellar cell wall. In electron microscopy ultrathin section, the inner surface of the cell wall appears as a serrated arrangement associated with invaginations of the plasma membrane (7, 8). The Malassezia cell wall appears as crossed electron-translucent bands with helicoidal ridges, making this cell wall morphology unique in the fungal kingdom (9). It is well known that the fungal cell wall is a highly complex and dynamic insoluble structure making a shield that protects the cell from changes in osmotic pressure and other environmental stresses while allowing the fungal cell to interact with its environment and mediate the adhesion to host cells or tissue (10). The stability and rigidity of fungal cell wall, essential to the integrity and viability of fungal cells, is based upon cross-linking between high molecular weight polysaccharides. In addition, these carbohydrate structures dominate immune recognition (11–13). Thus, gaining insight into cell wall composition and structure is an important part of understanding fungal behavior and devising strategies for controlling fungal growth. The composition of the Malassezia cell wall remains poorly studied. Water-extractable polysaccharides and glycoproteins have been described in Malassezia furfur, Malassezia pachydermatis, and Malassezia sympodialis, showing that Malassezia species were able to synthesize β-(1,3)-glucans and β-(1,6)-glucans, galactofuran, and mannan structures (14, 15). However, the water-extractable fraction represents only a small part of fungal cell wall, whose structural function is limited. The chemical composition and organization of the cell wall polysaccharide skeleton of Malassezia yeast has never been investigated.
In this study the cell wall of M. restricta has been isolated, and its polysaccharide organization has been investigated through specific chemical and enzymatic degradations. Chitin/chitosan, β-(1,3)-glucans, and β-(1,6)-glucans are essential components of the cell wall. The structures and cross-linking of these polymers inside the cell wall are unique to M. restricta.
MATERIALS AND METHODS
Strain and Cultivation Conditions
M. restricta strain CBS7877 was obtained from the fungal collection of the Institut Pasteur. Yeasts were grown in liquid or 1.2% agar modified Dixon's medium (3.6% malt extract (Oxoid, Dardilly, France), 0.6% mycological peptone (BD Biosciences), 2% ox bile (BD Biosciences), 1% Tween 40 (Sigma), 0.2% glycerol (Sigma), 0.2% oleic acid (Sigma)). Yeasts were inoculated at 107 cells/ml of liquid medium and incubated at 32 °C at 160 rpm for 4 days.
Cell Wall Preparation
Cells were collected from liquid culture by centrifugation (4500 × g for 10 min), washed 5 times with distilled water, and disrupted in 200 mm Tris-HCl, pH 8, buffer using a cell disruptor (B. Braun, Germany) with 0.3-mm diameter ceramic beads (W.A.B. Bale). The cell wall was collected by centrifugation (4500 × g for 10 min), washed 5 times with distilled water, and freeze-dried. Proteins and lipids were removed by the following treatment; 1 g of dried cell wall was treated twice in 50 mm Tris-HCl, pH 8, 5 mm EDTA, 2% SDS, 40 mm β-mercaptoethanol at 100 °C for 10 min (10 mg of cell wall/ml). Insoluble material was washed five times with distilled water and freeze dried. Lipids contained in the insoluble pellet were extracted overnight by a chloroform/methanol/water (10/10/3, v/v/v) mixture at room temperature under a magnetic bar stirring. Insoluble material was harvested and washed with water by centrifugation (2000 × g for 10 min) and finally freeze-dried.
The dried pellet was treated twice with 1 m sodium hydroxide (NaOH) at 65 °C for 1 h. Alkali-soluble (AS)3 and alkali-insoluble (AI) fractions were separated by centrifugation (4500 × g for 10 min). The AS supernatant was neutralized by adding acetic acid, dialyzed against water, and finally freeze-dried. The AI pellet was washed eight times with distilled water and freeze-dried. Cell wall extraction and fractionation were summarized in supplemental Fig. 1.
Carbohydrate Composition
Total neutral hexoses were quantified by the phenol-sulfuric acid procedure (16) using glucose as standard. Hexosamines were identified and quantified by high performance anion exchange chromatography (HPAEC) on a CarboPAC-PA1 column (Dionex) after acid hydrolysis (8 n HCl, 4 h at 100 °C) using glucosamine as a standard. Monosaccharides were analyzed by GLC as their alditol acetates obtained after hydrolysis (4 n trifluoroacetic acid or 8 n HCl, 100 °C, 4 h) followed by reduction with sodium borohydride (BH4Na) and peracetylation (17). Derivatized monosaccharides were identified and quantified by GLC.
Enzymatic Degradation of Cell Wall Polysaccharides
Polysaccharide fractions were subjected to several enzymatic treatments. Cellulase, amyloglucosidase, and amylase from Aspergillus niger were purchased from Sigma (stock solution at 1 mg/ml). Five recombinant enzymes were used: mutanase, an α-(1,3)-glucanase from Trichoderma harzianum, produced in Escherichia coli, a kind gift from C. Fuglsang (Novozymes A/S, Bagwaerd, Denmark) (18); laminarinase A, an endo-β-(1,3)-glucanase from Thermotoga neapolitana expressed in E. coli (19); an endo-β-(1,6)-glucanase (member of GH-5 family) from Schizosaccharomyces pombe expressed in Pichia pastoris (20, 21); a chitinase from Serratia marcescens produced in E. coli (22), and a chitosanase (Csn46A) from Streptomyces coelicolor expressed in E. coli (23). Specific activities and optimum pH values are indicated in supplemental Table 1.
Digestions were undertaken by treating 100 μg of the cell wall fraction with 25 μl of enzyme stock solution in 50 mm sodium acetate, 5 mm sodium azide at the optimum pH of each enzyme in a final volume of 250 μl at 37 °C for 24–96 h. Degradation products were analyzed by HPAEC and gel filtration chromatography as described below.
Chemical Treatments of Cell Wall Polysaccharides
The methylation procedure to analyze glycosidic linkages was performed by the lithium methyl sulfinyl carbanion/methyl iodide method modified by Fontaine et al. (24). Methyl ethers were analyzed by GLC-MS after acid hydrolysis (TFA 4 n, 4 h, 100 °C), reduction with NaBD4 and peracetylation.
The periodate oxidization was used to degrade β-(1,6)-glucans containing cell wall fractions. 50 mg of fraction were resuspended in 4 ml of 100 mm sodium meta-periodate, and the oxidization was performed for 2 days at room temperature in dark conditions under gentle stirring. Excess reagent was destroyed by adding 1 ml of glycerol. After dialysis against water (membrane cut-off, 1 kDa), the oxidized product was reduced by NaBH4 at 10 mg/ml in 0.1 m NaOH overnight. After neutralization by the addition of acetic acid, the sample was desalted by dialysis against 0.1% acetic acid (membrane cut-off 1 kDa) overnight. Smith degradation was then performed with 10% acetic acid at 100 °C for 1 h. Acetic acid was removed by freeze-drying. To analyze small oligosaccharides released during the periodate oxidization, the dialysis step was replaced by gel filtration chromatography on a G15-Sephadex column (1.5 × 90 cm, GE Healthcare) equilibrated with 0.25% acetic acid at a flow rate of 8 ml/h.
Chitin and chitosan in the AI fraction were degraded by nitrous deamination degradation. Before the degradation reactions, the AI fraction was fully de-N-acetylated by incubating 50 mg of AI fraction with 4 ml of 10 m NaOH at 100 °C for 4 h. After neutralization with acetic acid, salts were removed by extensive dialysis against water (membrane cut-off, 10 kDa), and the sample was freeze-dried. The material was then resuspended in 0.5 m sodium acetate pH 4 (5 mg/ml), and the nitrous deamination was performed by incubation with 2 n sodium nitrite (0.5 ml/mg fraction/h) at 50 °C for 3 h. Salts were removed by extensive dialysis against water (membrane cut-off, 10 kDa). After dialysis, the water soluble fraction (WSDN) was recovered by centrifugation (4500 × g, 10 min) and freeze-dried. The pellet was then submitted to an alkali extraction with 1 m NaOH as described above to obtain two other fractions, i.e. ASDN for alkali-soluble and AIDN for alkali-insoluble fractions (supplemental Fig. 1). 50 mg of the fully de-N-acetylated AI fraction were submitted to a chitosanase digestion as described above. Similarly to the nitrous deamination protocol, three fractions, WSCsn, ASCsn, and AICsn, were isolated for water-soluble, alkali-soluble, and alkali-insoluble fractions respectively (supplemental Fig. 1).
Gel Permeation of Cell Wall Fractions
WSDN, ASDN, and WSCsn fractions were dissolved in 8 m urea and analyzed by gel permeation chromatography on a Sephacryl S300 column (0.5 × 30 cm, GE Healthcare) equilibrated in 8 m urea at a flow rate of 0.33 ml/min. The column was calibrated with dextran molecular mass standards (10, 40, 70 and 500 kDa; GE Healthcare). Polysaccharide fractions were detected by the phenolsulfuric acid method (16). β-(1,3)-Glucan containing fractions was specifically quantified by Lam-A digestion followed by measuring the amount of released reducing sugars using the p-hydroxybenzoic acid hydrazide method (25).
HPAEC
Monosaccharides and oligosaccharides obtained after chemical or enzymatic treatments were analyzed by HPAEC using a CarboPAC PA-1 column (4 × 25 mm, Dionex ThermoScientific, Villebon sur Yvette, France) and a pulsed electrochemical detector. Monosaccharides were eluted at a flow rate of 1 ml/min with 18 mm NaOH for 15 min, then with a linear gradient to 300 mm AcONa in 100 mm NaOH for 20 min and finally under isocratic conditions with 300 mm AcONa in 100 mm NaOH for 5 min. The column was stabilized for 20 min at initial conditions before injection. Oligosaccharides obtained after enzymatic digestion were analyzed with the following gradient: isocratic step of 98% of eluent A (50 mm NaOH) and 2% of eluent B (500 mm AcONa in 50 mm NaOH) for 2 min, 2–15 min of a linear gradient (98% A/2% B to 80% A/20% B), 15–20 min of a linear gradient (80% A/20% B to 57% A/43% B), 20–22 min of a linear gradient (57% A/43% B to 0% A/100% B), 22–25 min under isocratic conditions with 100% B. The column was stabilized for 20 min at initial conditions before injection.
Gas Liquid Chromatography and Mass Spectrometry
GLC-MS analyses of monosaccharides and methyl ethers were recorded using a quadrupole mass spectrometer (model 5975C, Agilent Technologies, Les Ulis, France) interfaced with a gas chromatograph (model 7890A, Agilent Technologies). Electron ionization spectra were recorded using an ionization energy of 70 eV. The gas chromatograph was equipped with a HP5-MS capillary column (30 m × 0.25 mm, film 0.25 μm; Agilent Technologies). Helium was used as mobile phase at a flow rate of 1.2 ml/min. The column temperature was raised from 100 to 240 °C at a rate of 5 °C/min.
Matrix-assisted Desorption Ionization/Time of Flight (MALDI-TOF) Mass Spectrometry
MALDI-TOF mass spectra of oligosaccharides were acquired on a 4800 MALDI TOF/TOF Analyzer (Applied Biosystems/MDS SCIEX, Framingham, MA) equipped with a laser diode-pumped UV (335 nm-200Hz) and a gridless delayed extraction ion source. The spectrometer was operated in positive reflectron mode by delayed extraction with an accelerating voltage of 20 kV, a pulse delay time of 200 ns, and a grid voltage of 66%. Samples were prepared by mixing directly on the target 1 μl of oligosaccharide solution in water (10–50 pmol) with 1 μl of 2,5-dihydroxybenzoic acid matrix solution (5 mg/ml in 5 mm NaCl). The samples were dried for about 5 min at room temperature. 1000–5000 laser shots were averaged for every spectrum.
NMR Experiments
NMR spectra were acquired at 338 K on an Agilent spectrometer operating at a proton frequency of 500 MHz equipped with a triple resonance 1H{13C/15N} triax probe (Agilent Technologies). Samples were dissolved in 650 μl of D2O (99.97% 2H atoms, Euriso-top, Commissariat à l'Energie Atomique, Saclay, France) and transferred into a 5-mm NMR tube (Wilmad 535-PP, Interchim, Montluçon, France). 1H chemical shifts were referenced to external DSS (2,2-dimethyl-2-silapentane-5-sulfonate sodium salt; its methyl resonance was set to 0 ppm). 13C chemical shifts were then calculated from 1H chemical shifts and a γ ratio relative to 2,2-dimethyl-2-silapentane-5-sulfonate sodium salt. A 13C/1H γ ratio of 0.251449530 was used (26).
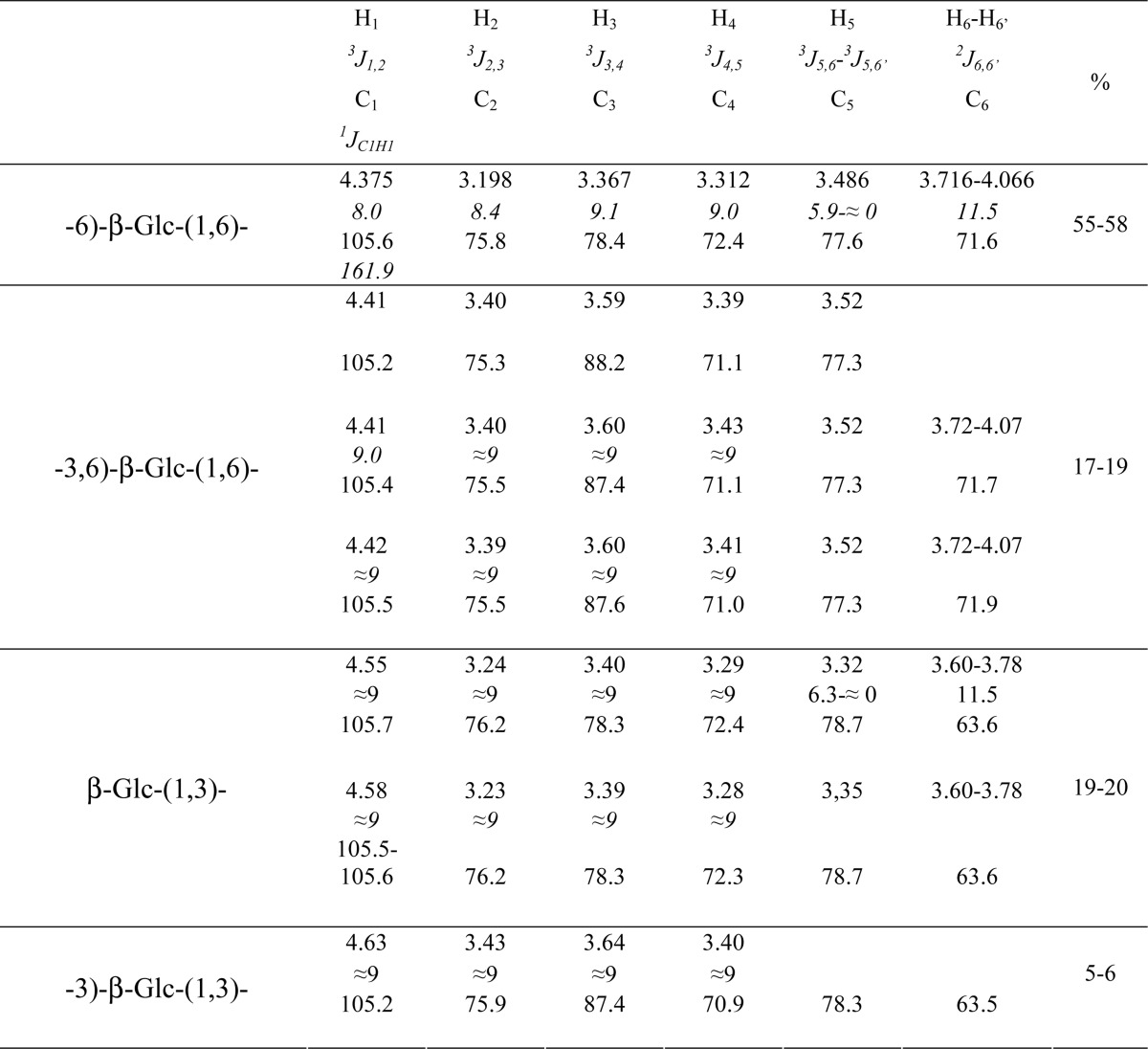
The following nuclei assignment strategy was adopted. First, proton resonances were assigned using two-dimensional COSY and RELAY (relayed COSY) experiments with one and two relays of 60 ms allowing connectivities from the anomeric proton to be followed (27, 28). Second, a 13C,1H-edited gHSQC experiment and a 13C,1H gHSQC-TOCSY experiment with a mixing time of 150 ms allowed 13C chemical shifts assignment from previously identified 1H resonances to be achieved (29, 30). Analysis of 1H,1H coupling constants from one-dimensional and/or COSY spectrum (1H resolution of 0.1 and 1.0 Hz, respectively) assessed the monosaccharide identity. Moreover, the anomeric configuration of the monosaccharide residues was established from the knowledge of 3J1,2 values and confirmed by the measurement of the 1JC1H1 heteronuclear coupling constants in the 1H dimension of the gHMBC spectrum (1H resolution of 0.8Hz) (29). Finally, glycosidic linkages were established via through-space dipolar interactions using an 1H,1H NOESY experiment (mixing time of 200 ms) (31) and/or via three-bond interglycosidic 13C,1H correlations using a 13C,1H gHMBC experiment (long range delay of 60 ms). The proportion of the different monosaccharide residues was estimated from a one-dimensional 1H spectrum acquired with a relaxation delay of at least 10 s and an acquisition time of 0.5 s.
RESULTS
Polysaccharides were purified from a crude cell wall extract by chloroform/methanol treatment and detergent extraction under reducing condition to eliminate contaminating lipids and proteins (supplemental Fig. 1). Alkali treatment of the resulting cell wall fraction solubilized only 8% of the total polysaccharides. Thus, in contrast to most fungi analyzed (10), the polysaccharide core of the M. restricta cell wall was extremely alkali-insoluble. HPLC and GLC analysis of sugar composition showed that both AS and AI fractions were composed of glucose and glucosamine residues only, at 94/6 and 75/25 ratios, respectively, suggesting the presence of glucans and chitin/chitosan in the cell wall. Global glycosidic linkages in the glucans in the AI fraction were analyzed by a methylation procedure, and the resulting methyl ethers are presented in Table 1. Four methyl ethers were identified: 2,3,4,6-tetra-O-methyl-glucitol, 2,4,6-tri-O-methyl-glucitol, 2,3,4-tri-O-methyl-glucitol, and 2,4-di-O-methyl-glucitol, suggesting the putative presence of α- or/and β-(1,3)-glucans and β-(1,6)-glucans. To identify cell wall polysaccharides, specific enzymatic digestions were performed on the AI fraction. Recombinant endo-β-(1,3)-glucanase, endo-β-(1,6)-glucanase, chitosanase, and chitinase partially degraded the AI fraction, indicating the presence of β-(1,3)-glucan, β-(1,6)-glucan, chitin, and chitosan (Table 2). Digestion with α-(1,3)glucanase, amylase, or amyloglucosidase did not release any degradation product, showing the absence of α-(1,3)-glucan and glycogen in the cell wall extract (data not shown). Chitosanase digestion solubilized 75% of the glucosamine content of the AI fraction, suggesting the presence of a high amount of chitosan. In agreement with the enzymatic degradation, nitrous deamination, which destroys the glucosamine residue but not the N-acetylated glucosamine residue (GlcNAc), degraded 80% of the glucosamine residues in the AI fraction (data not shown), indicating an average chitosan/chitin ratio of 80/20.
TABLE 1.
Percentage of methyl ethers obtained after permethylation, hydrolysis, reduction, and acetylation of the AI fraction of the cell wall of M. restricta
Percentages were based on the response of each methyl on GLC and flame ionization detection. As an example of nomenclature of methyl ethers, 2,3,4,6-Glc corresponded to 2,3,4,6-tetra-O-methyl-1,5-di-O-acetyl-glucitol.
| Methyl ethers | Sugar linkages | % |
|---|---|---|
| 2,3,4,6-Glc | Glc- | 15.7 |
| 2,4,6-Glc | -3Glc- | 16.8 |
| 2,3,4-Glc | -6Glc- | 50.8 |
| 2,4-Glc | -3,6Glc- | 16.8 |
TABLE 2.
Enzymatic digestions of cell wall fractions of M. restricta
Percentages were calculated from the amount of degraded products identified by HPAEC and sugar reducing assay using the p-hydroxybenzoic acid hydrazide reagent. AI is cell wall alkali-insoluble fraction, and WSDN and ASDN are water-soluble and alkali-soluble fractions obtained after nitrous deamination of the de-N-acetylated AI fraction. ND, no product detected.
| Enzymes | % of products released from cell wall fractions |
||
|---|---|---|---|
| AI | WSDN | ASDN | |
| Endo-β-(1,3)-glucanase (Lam-A) | 5 | 5 | 8 |
| Endo-β-(1,6)-glucanase | 18 | 79 | 67 |
| Chitinase | traces | ND | ND |
| Chitosanase | 16 | ND | ND |
Chitin and Chitosan Polymers Are Essential for the Alkali Insolubility of β-Glucans in M. restricta Cell Wall
After full de-N-acetylation by strong alkali treatment to convert chitin into chitosan and nitrous deamination, the AI fraction became soluble: 74% of the glucan moieties becoming water-soluble (WSDN fraction) and 26%, alkali-soluble (ASDN fraction). No glucan remained alkali-insoluble after the nitrous degradation of de-N-acetylated glucosamine polymers. When nitrous deamination was substituted by an enzymatic digestion with a chitosanase, 69 and 22% of glucan of the AI fraction became water-soluble (WSCsn fraction) and alkali-soluble (ASCsn), respectively. These data showed clearly that glucosamine/N-acetylglucosamine polymers were essential for the alkali insolubility of the β-glucans. Nitrous deamination of the native AI fraction (i.e. with the chitin still N-acetylated) resulted in the water and alkali solubilization of 54 and 39% of glucan moieties, respectively, leaving 7% of cell wall glucans alkali-insoluble. This confirms the notion that both chitin and chitosan polymers were involved in cross-linking of β-glucans.
β-(1,6)-Glucans Represent the Major Polysaccharide and β-(1,3)-Glucans Form Small Complexes of Branched β-Glucans in the Cell Wall of M. restricta
The WSDN fraction, obtained after de-N-acetylation-HNO2 treatment of the AI fraction, was submitted to specific enzymatic degradations, endo-β-(1,3)-glucanase (Lam-A) and endo-β-(1,6)-glucanase. The quantification of degraded product showed that β-(1,6)-glucans accounted for 84% of the total amount of glucan released by enzymatic degradations of WSDN (Table 2). This WSDN fraction was analyzed by 1H and 13C NMR experiments. The one-dimensional 1H spectrum exhibited two signals groups in the sugar anomeric region that are attributed to β-glucopyranose residues by 1H and 13C chemical shift analysis and 3JH,H and 1JC1,H1 values (Table 3, supplemental Fig. 2) (32). Two-dimensional 13C,1H-edited gHSQC and gHSQC-TOCSY experiments showed that the main signal corresponded to a 6-O-substituted glucose (supplemental Fig. 2). The H1/C6 correlation observed in the gHMBC experiment indicated the main following sequence motif: -6)-β-Glcp-(1,6)-β-Glcp. A second motif identified from the same anomeric protons group between 4.40 and 4.43 ppm correspond to -3,6)-β-Glcp-(1,6) (Table 3). In the second anomeric protons group, H1/H3 (4.56–4.58/3.59 ppm) and H1/C3 (4.55/88.2, 4.58/87.3, and 4.63/87.3 ppm) correlations, observed in NOESY and HMBC experiments, indicated that these residues were branched in the 3-O position on the next residue. Terminal β-Glcp-(1,3) residues were identified by the absence of shifted carbon signals and corresponded to the anomeric protons ranging from 4.55 to 4.58 ppm, whereas the observation of a downfield-shifted C3 signal (Table 3) for the glucose residue corresponding to the anomeric proton at 4.63 ppm indicated a -3)-β-Glcp-(1,3 sequence. These NMR data were similar to those from β-(1,6)-glucans isolated from Saccharomyces cerevisiae (33) showing their common chemical organization. The proportion of identified glucose residues was estimated from the ratios of the signal intensity of their corresponding anomeric protons (Table 3). The relative percentages clearly show that the β-(1,6)-glucans are composed of a linear chain of 6-O-linked β-glucose residues with side branches every 3.8 residues on average. The side branch is mainly composed of a single 3-O-linked β-glucose residue. Taken together, the NMR data show that β-(1,6)-glucans and β-(1,3)-glucans represent 95 and 5% of the WSDN fraction, respectively.
TABLE 3.
1H and 13C NMR chemical shifts (ppm) and coupling constants (JH,H and 1JC1H1, Hz) for the WSDN fraction
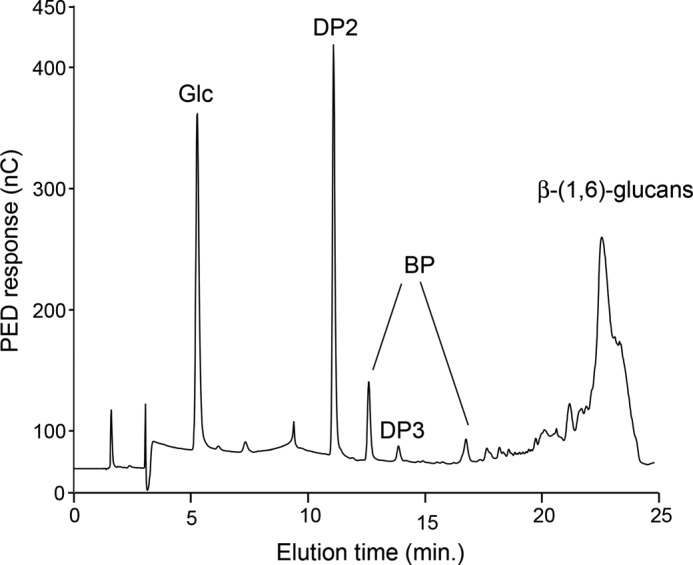
To complete its chemical characterization, the WSDN fraction was submitted to an endo-β-(1,6)-glucanase treatment followed by analysis using HPAEC. The chromatogram showed the release of glucose, linear gentio-oligosaccharides (DP 2–5), and branched gentio-oligosaccharides (Fig. 1). Linear gentio-oligosaccharides were characteristic of enzymatic digestion of pustulan, an unbranched β-(1,6)-glucan (20, 33), and the large number of species of branched oligosaccharides, eluted from 15 to 24 min, are resistant to endo-β-(1,6)-glucanase activity. These data indicated that β-glucosyl side chain are not homogeneously distributed along the β-(1,6)-glucan chain and suggested that β-(1,6)-glucan produced by M. restricta contained highly branched regions. To confirm these data, a periodate degradation followed by a reduction and Smith hydrolysis were performed on the WSDN fraction. Periodate treatment degraded terminal glucose and 6-O-linked glucose residues, whereas 3-O-linked glucose and branching point residues were resistant. The resistant oligosaccharidic fraction from periodate-treated WSDN was desalted by gel filtration chromatography on a Sephadex G15 column and then analyzed by MALDI-TOF mass spectrometry (Fig. 2). Mass spectra showed the presence of oligosaccharides of DP 4 up to 10 containing one glycerol residue. Methylation and GLC-MS analysis showed the presence of 2,3,4,6-tetra-O-methyl-glucitol and 2,3,4-tri-O-methyl-glucitol as the main products (data not shown). Glycerol residues came from periodate oxidization of 6-O-subsituted glucose residues. The presence of 6-O-substituted glucose residues after periodate treatment indicated that these residues were previously also 3-O-substituted by a glucosyl residue and corresponded to a branching point. These data showed that this oligosaccharidic fraction corresponded to sequences from native β-(1,6)-glucan containing a glucosyl β-(1,3)-linked on every β-(1,6)-glucose residue of the main chain, which is in agreement with the heterogeneous distribution of branch point of β-(1,6)-glucan chains observed by HPAEC analysis of β-(1,6)-glucanase-degraded products (Fig. 1).
FIGURE 1.
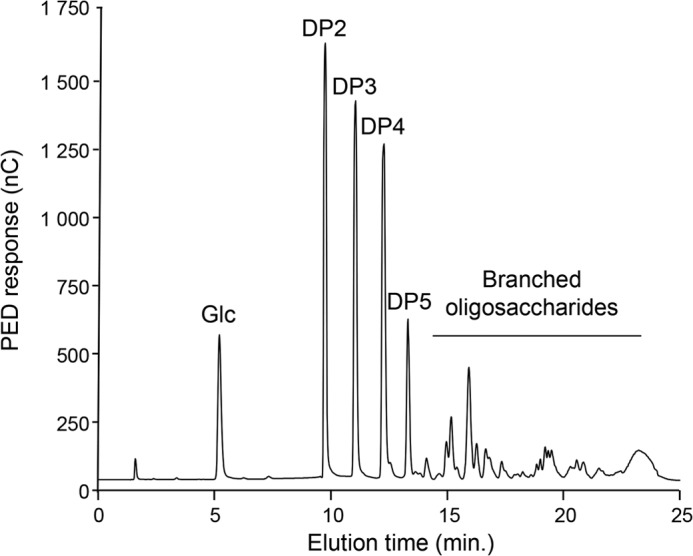
HPAEC analysis of oligosaccharides released by the endo-β-(1,6)-glucanase digestion of the WSDN fraction. Products were identified by their retention time as previously described (33). PED, pulsed electrochemical detector; nC, nanocoulomb.
FIGURE 2.
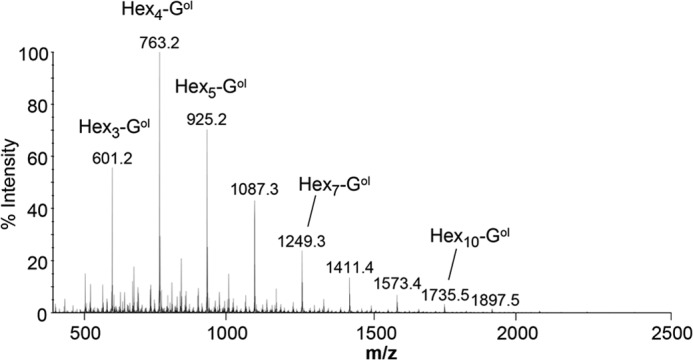
Analysis of the periodate-oxidized WSDN fraction by mass spectrometry. MALDI-TOF mass spectra of oligosaccharides obtained after periodate oxidization, reduction, and Smith hydrolysis of the WSDN fraction and purified by gel filtration chromatography on a Sephadex G15 column. Pseudomolecular mass, m/z = [M+Na]+ (Hex, hexose; Gol, glycerol).
Because of their small amount, the structure of β-(1,3)-glucans produced by M. restricta was analyzed after endo-β-(1,3)-glucanase digestion and periodate oxidization. HPAEC analysis of products obtained after laminarinase A digestion of the WSDN or AI fractions showed the release of glucose, laminaribiose, and two branching point oligosaccharides, which is characteristic of β-(1,6)-branched β-(1,3)-glucans (Fig. 3) (34). Based on the peak surface of these oligosaccharides, the branching level of cell wall β-(1,3)-glucan was estimated to be 14%. After the degradation of β-(1,6)-glucan by periodate oxidization of the WSDN fraction, resistant β-(1,3)-glucans were analyzed by gel filtration chromatography on a Sephacryl S300 column and were eluted as a polydisperse polymer of 3–30 kDa with an apparent average molecular mass of 10 kDa, corresponding to a DP of 60 glucose units (Fig. 4). The same chromatographic profile was obtained after periodate oxidization of the AI fraction (supplemental Fig. 3). This periodate-treated-resistant fraction was analyzed by HPLC after laminarinase A digestion. The HPLC profile revealed the same level of branch point, showing that β-(1,3)-glucan chains were not substituted by single β-(1,6)-glucosyl unit as described in scleroglucan (35).
FIGURE 3.
HPAEC analysis of oligosaccharides released by the endo-β-(1,3)-glucanase (Lam-A) digestion of the WSDN fraction. Products were identified by their retention time as previously described (34). PED, pulsed electrochemical detector; nC, nanocoulomb.
FIGURE 4.
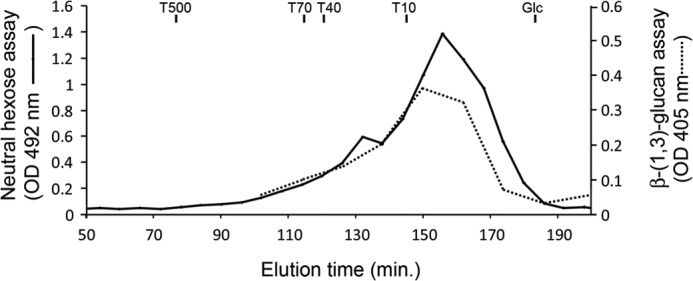
Analysis of β-(1,3)-glucans of M. restricta by gel filtration chromatography on a Sephacryl S300 column. β-(1,3)-Glucans were isolated after periodate oxidization, reduction, and Smith hydrolysis of WSDN fraction and desalted by dialysis (cut off of 1 kDa). Neutral sugars were detected by phenolsulfuric acid assay (16). β-(1,3)-Glucans were detected by Lam-A digestion and p-hydroxybenzoic acid hydrazide reagent (25). The Sephacryl S300 column was calibrated with dextrans (T500, T70, T40, and T10), and glucose.
β-(1,6)-Glucans and β-(1,3)-Glucans Are Covalently Linked
The HPAEC analysis of endo-β-(1,3)-glucanase-treated WSDN fraction showed the release of β-(1,6)-glucans eluted after 23 min (Fig. 3), suggesting an interconnection between β-(1,6)-glucans and β-(1,3)-glucans as previously described in S. cerevisiae (33). The WSDN fraction was also analyzed by gel permeation chromatography on a Sephacryl S300 column and was eluted as a single peak with an apparent average molecular mass of 500 kDa (Fig. 5A). The treatment of the samples with 1 m NaOH or strong sonication, known to dissociate polysaccharide aggregate, did not modify the elution profile of WSDN fraction, showing the absence of nonspecific interchain interaction. The endo-β-(1,3)-glucanase (Lam-A)-resistant WSDN fraction was eluted as a polydisperse polymer with an apparent molecular mass of 10–250 kDa with an average of 80 kDa (Fig. 5B), corresponding to an average DP of 500 glucose units/chain. Methylation analysis and endo-β-(1,6)-glucanase digestion showed that this fraction was composed of β-(1,6)-glucans (not shown). Similarly, specific degradation of β-(1,6)-glucans from the WSDN fraction by the endo-β-(1,6)-glucanase activity induced a delay of the elution time of the resistant β-glucan on the Sephacryl S300 column (Fig. 5C). The specific detection of β-(1,3)-glucans by the Lam-A/p-hydroxybenzoic acid hydrazide reagent showed that β-(1,6)-glucanase resistant β-(1,3)-glucan fraction was eluted mainly as a polymer with an apparent molecular mass of 15–120 kDa (Fig. 5C). These three gel filtration patterns show that the enzymatic degradation of each β-glucan induced a delay of gel filtration elution of the second one, indicating the presence of covalent linkages between both β-glucans in the WSDN fraction. After degradation of the β-(1,6)-glucan by periodate treatment, the remaining β-(1,3)-glucan was eluted after 140 min (Fig. 4), in agreement with the cross-linking between β-(1,6)-glucans and β-(1,3)-glucans. However, periodate-resistant β-(1,3)-glucans were eluted later than those after the treatment with β-(1,6)-glucanase (Fig. 5C), indicating incomplete enzymatic treatment, which was confirmed by methylation analysis (not shown). The ASDN fraction was also analyzed by gel filtration on a Sephacryl S300 column, and treatments similar to those described above for the WSDN fraction were carried out (supplemental Fig. 4). Gel filtration analyses of the degraded products after enzymatic digestion or periodate oxidization were similar to the WSDN data (Figs. 4 and 5). These data showed that ASDN and WSDN possess the same general chemical organization of β-glucans and that β-(1,6)-glucans and β-(1,3)-glucans are cross-linked in the M. restricta cell wall. Taking into account the knowledge on the cell wall polysaccharides reticulation in yeast, these information allowed us to propose a schematic representation of cell wall polysaccharides in M. restricta (Fig. 6).
FIGURE 5.
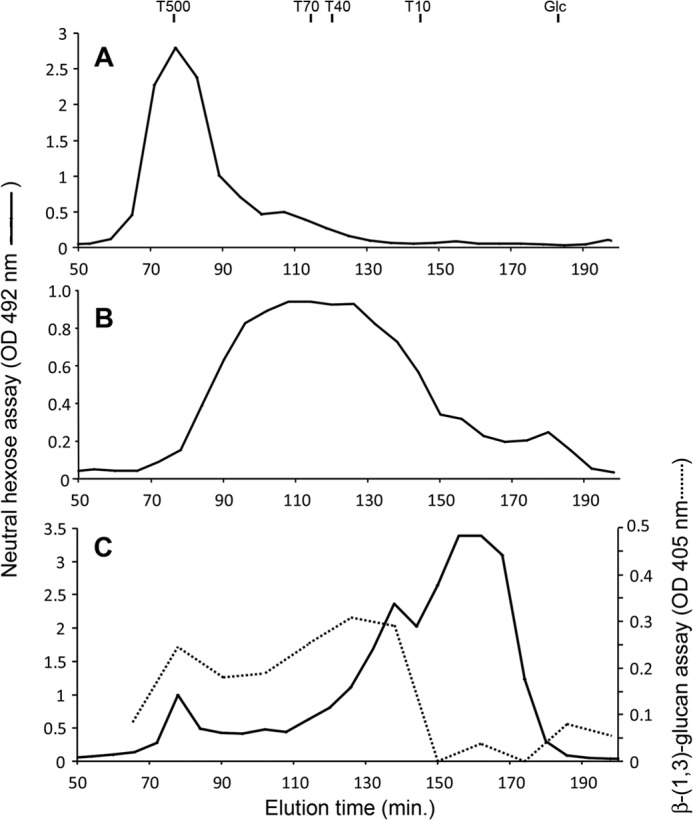
Gel filtration chromatography of the WSDN fraction on a Sephacryl S300 column. A, native fraction; B, endo-β-(1,3)-glucanase (Lam A)-treated fraction; C, endo-β-(1,6)-glucanase treated fraction. Neutral sugars were detected by phenolsulfuric acid assay (16). β-(1,3)-Glucans were detected by Lam-A digestion and p-hydroxybenzoic acid hydrazide reagent (25). The Sephacryl S300 column was calibrated with dextrans (T500, T70, T40, and T10) and glucose.
FIGURE 6.
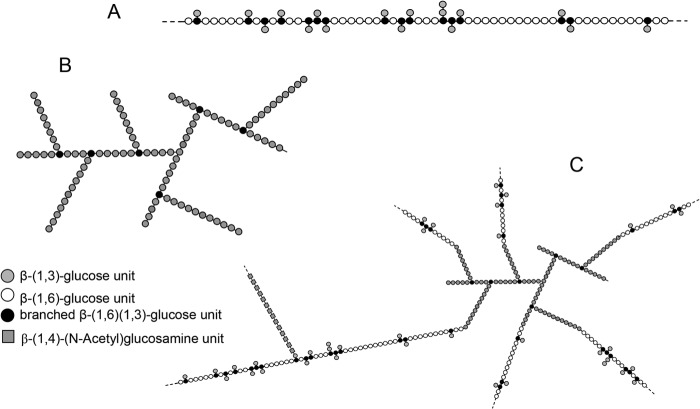
Proposed schematic representation of the polysaccharide organization of the alkali-insoluble fraction of the M. restricta cell wall. A, β-(1,6)-glucan chain with an average molecular mass of 80 kDa (around 500 glucose units/chain). β-(1,6)-Glucans are constituted by a main chain of β-(1,6)-linked glucose units with a side chain composed of β-(1,3)-glucosyl residue with an average of one branch point for 3.8 glucose units of the main β-(1,6)-glucan chain. B, branched β-(1,3)-glucan chains with an average of 10 kDa (around 60 glucose units). The branching level is 14%, and the length of the side chain of β-(1,3)-glucan remains unknown. C, scheme of the polysaccharide complex of the alkali-insoluble fraction. Based on their apparent molecular mass and their relative abundance, an average of six β-(1,6)-glucan chains were estimated to be linked to one β-(1,3)-glucan. In agreement with the cross-linking described in S. cerevisiae, the scheme represents β-(1,6)-glucan chains linked to the non-reducing end of β-(1,3)-glucan chain (37, 38). Chitin/chitosan polymers were cross-linked to the β-glucan complex, but the chemical bond remains unknown. These glucosamine polymers could be linked to β-(1,6)-glucan or β-(1,3)-glucan as previously described in S. cerevisiae (37, 38). Because β-(1,6)-glucan is the most abundant β-glucan, chitin/chitosan polymers were represented to be linked to β-(1,3)-glucosyl residue side chain of β-(1,6)-glucan as characterized in S. cerevisiae.
DISCUSSION
Fungal cell walls are organized around a polysaccharide core of glucans and chitin (10, 36). In this study fractionation and biochemical analyses showed that the polysaccharide core of the cell wall of M. restricta was quite exclusively alkali-insoluble and contained an average of 5% chitin, 20% chitosan, 5% β-(1,3)-glucan, and 70% β-(1,6)-glucan. These polymers are covalently interconnected together to form an alkali-insoluble complex. To date, polysaccharide reticulations in the fungal cell wall have been described in detail in S. cerevisiae and Aspergillus fumigatus (37–40), and polysaccharide branching and cross-linking are admitted to be essential in cell wall rigidity in many fungal species. The present data show that the cell wall of M. restricta is constructed with similar polymer interconnections, but in addition it possesses unique structural properties.
Chitin and its de-N-acetylated form account for 25% of the carbohydrate skeleton of the M. restricta cell wall, whereas chitin is a minor cell wall component in other yeasts (41). In yeasts, chitosan is usually only produced during sporulation. In the basidiomycete yeast Cryptococcus neoformans, chitosan is produced during vegetative growth with chitosan/chitin ratio similar to the one found in the cell wall of M. restricta, but the total chitosan/chitin amount did not exceed 3% of the cell wall-dried mass (42). Interestingly, the level of chitosan may be related to the high amount of lipid found in the cell wall of Malassezia as chitosan has been described to interact with lipids (43, 44).
The branching of β-(1,3)-glucan chains is essential for cell wall organization and stability, as it allows cross-linking to other polymers such as β-(1,6)-glucan, chitin, and galactomannan to occur (37–39). β-(1,3)-Glucans produced by M. restricta are relatively small molecules (∼30–100 glucose units/glucan chain; Fig. 3), whereas it has been estimated at 1500 glucose units/chain or more in S. cerevisiae (40, 45). M. restricta β-(1,3)-glucan possesses a higher branching level compared with other fungal species (14% instead of 3, 4, and 5% in S. cerevisiae, A. fumigatus, and S. pombe, respectively) (39, 45, 46). Moreover, in contrast to other fungi, β-(1,3)-glucan is a minor component of the M. restricta cell wall. In yeast, β-(1,6)-glucans are also cross-linked to chitin and glycoproteins (40, 47). β-(1,6)-Glucans of M. restricta are essential to the cross-linking to β-(1,3)-glucan, chitin, and chitosan chains, which are required to the alkali insolubility of β-glucans. In contrast to previously described yeast structures, the β-(1,6)-glucans from M. restricta are very large molecules (up to 1500 glucose units; Fig. 5) and are highly branched mainly with a β-glucosyl residue (average of 1 branch point for 3.8 glucose units of the main β-(1,6)-glucan chain) compared with S. cerevisiae (average of one branch point to five glucose units of the main chain) (33). The presence of some branching sequences on every glucose unit is similar to that previously observed in S. pombe (48). Based on data from gel filtration chromatography on Sephacryl S300, the apparent molecular mass of each β-glucan (i.e. average of 500 kDa for the β-glucan complex, 80 kDa for β-(1,6)-glucans, and 10 kDa for β-(1,3)-glucans) and their relative abundance in the cell wall allow us to conclude that an average of 6 β-(1,6)-glucan chains are cross-linked to one branched β-(1,3)-glucan polymer. However, the presence of linkage of β-(1,3)-glucan to β-(1,6)-glucan chain cannot be excluded. All these data allowed us to propose a chemical organization of the polysaccharide core of the cell wall of M. restricta (Fig. 6). Linkage responsible for cross-linking chitin/chitosan to β-glucans is currently investigated. In fungi, it has been shown that chitin polymers were connected to terminal glucose residue of β-(1,3)-glucan chain or to β-(1,3)-glucosyl side chain of β-(1,6)-glucan (37–39). Based on the relative abundance of each type of β-glucans in M. restricta cell wall, we propose that chitin and chitosan chains may be mainly connected to β-(1,6)-glucan (Fig. 6).
The genome of Malassezia species has been sequenced (49) and is available on the NCBI website (ncbi.nlm.nih.gov). Genes involved in cell wall biosynthesis have been identified by blast analysis (supplemental Table 2). The high amount of chitin and chitosan are in agreement with the presence of six chitin synthase and four chitin deacetylase genes in the M. restricta genome. Similarly, the absence of detection of α-(1,3)-glucan in the cell wall of M. restricta is related to the absence of α-glucan synthase gene. One FKS gene encoding the putative catalytic subunit (FKS) of the β-(1,3)-glucan synthase and only two KRE genes among the five involved in the β-(1,6)-glucan synthesis are present in the genomes of Malassezia species. The presence of Crh proteins as a putative transglycosidases involved in chitin-β-glucan cross-linking is in agreement with the structural data from the M. restricta cell wall (47). However, some fungal genes are involved in the remodeling of β-(1,3)-glucan were not identified in any Malassezia genome. Particularly, Gas/Gel proteins are β-(1,3)-glucanosyltransferases that are involved in the elongation of β-(1,3)-glucan chains (50, 51). Sun proteins have been recently described to bind and degrade cell wall β-(1,3)-glucan (52). Both families are involved in cell wall biogenesis (53–55). Their absence in Malassezia species may be related to the difference in size and branching of β-(1,3)-glucan isolated from its cell wall. In accordance to their environment that does not contain carbohydrate molecules, it was expected that the genomes of Malassezia species contain a low number of genes encoding for glycosylhydrolase (56). However, six genes belonging to GH5 family, homologous to ScEXG1, were identified in the M. restricta genome. EXG1 homologs have been characterized as exo-β-(1,3)-glucanases or endo-β-(1,6)-glucanases (20, 57), suggesting that these EXG1 orthologs may be also involved in cell wall β-glucan remodeling in Malassezia species.
M. restricta is a commensal yeast of the skin microbiota. Under steady state conditions, Malassezia yeasts manage to evade local immune responses and remain in balance with the human skin. Because fungal cell wall carbohydrates are widely conserved across the fungal kingdom, but absent in humans, these structural components of the cell wall are convenient targets for immune recognition and critical for host inflammation responses (12, 13). β-(1,3)-Glucan recognition has been extensively studied as receptors have been identified in a number of human cells including keratinocytes (e.g. langerrin, dectin-1, CR3) (11, 13, 58). In contrast, the effect of β-(1,6)-glucans on the immune system has been poorly studied (59). Chitin is recognized by the immune system and particularly the keratinocytes (60). Chitin and chitosan were also activators of the human immune system, but their recognition and signaling pathways are totally independent (13, 61). In addition, it has also been observed that large chitin fragments or the conversion of chitin to chitosan favors the escape from immune recognition (62, 63). The characterization of the cell wall polysaccharides paves the way for a thorough understanding of the role of M. restricta polysaccharides (specially β-(1,6)-glucan or chitosan) in local immune response or in masking minor polysaccharides (e.g. β-(1,3)-glucan and chitin) from host recognition.
Supplementary Material

This article contains supplemental Tables 1 and 2 and Figs. 1–4.
- AS
- alkali-soluble fraction of the cell wall
- AI
- alkali-insoluble fraction of the cell wall
- AICsn
- alkali-insoluble fraction obtained after chitosanase digestion of fully de-N-acetylated AI
- AIDN
- alkali-insoluble fraction obtained after nitrous deamination of fully de-N-acetylated AI
- ASCsn
- alkali-soluble fraction obtained after chitosanase digestion of fully de-N-acetylated AI
- ASDN
- alkali-soluble fraction obtained after nitrous deamination of fully de-N-acetylated AI
- Csn
- chitosanase from S. coelicolor
- GLC
- gas liquid chromatography
- gHMBC
- gradient selected heteronuclear multiple bond correlation
- gHSQC
- gradient selected heteronuclear single-quantum correlation
- HPAEC
- high performance anion exchange chromatography
- Lam-A
- laminarinase A from T. harzianum
- TOCSY
- total correlation spectroscopy
- WSCsn
- water-soluble fraction obtained after chitosanase digestion of fully de-N-acetylated AI
- WSDN
- water-soluble fraction obtained after nitrous deamination of fully de-N-acetylated AI
- DP
- degree of polymerization.
REFERENCES
- 1. Ashbee H. R., Evans E. G. (2002) Immunology of diseases associated with Malassezia species. Clin. Microbiol. Rev. 15, 21–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ashbee H. R. (2007) Update on the genus Malassezia. Med. Mycol. 45, 287–303 [DOI] [PubMed] [Google Scholar]
- 3. Gaitanis G., Magiatis P., Hantschke M., Bassukas I. D., Velegraki A. (2012) The Malassezia genus in skin and systemic diseases. Clin. Microbiol. Rev. 25, 106–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Paulino L. C., Tseng C. H., Blaser M. J. (2008) Analysis of Malassezia microbiota in healthy superficial human skin and in psoriatic lesions by multiplex real-time PCR. FEMS Yeast Res. 8, 460–471 [DOI] [PubMed] [Google Scholar]
- 5. Findley K., Oh J., Yang J., Conlan S., Deming C., Meyer J. A., Schoenfeld D., Nomicos E., Park M., NIH Intramural Sequencing Center Comparative Sequencing Program, Kong H. H., Segre J. A. (2013) Topographic diversity of fungal and bacterial communities in human skin. Nature 498, 367–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clavaud C., Jourdain R., Bar-Hen A., Tichit M., Bouchier C., Pouradier F., El Rawadi C., Guillot J., Ménard-Szczebara F., Breton L., Latgé J. P., Mouyna I. (2013) Dandruff is associated with disequilibrium in the proportion of the major bacterial and fungal populations colonizing the scalp. PLoS ONE 8, e58203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mittag H. (1995) Fine structural investigation of Malassezia furfur. II. The envelope of the yeast cells. Mycoses 38, 13–21 [DOI] [PubMed] [Google Scholar]
- 8. Guillot J., Guého E. (1995) The diversity of Malassezia yeasts confirmed by rRNA sequence and nuclear DNA comparisons. Antonie Van Leeuwenhoek 67, 297–314 [DOI] [PubMed] [Google Scholar]
- 9. David M., Gabriel M., Kopecka M. (2003) Unusual ultrastructural characteristics of the yeast Malassezia pachydermatis. Scripta Medica (BRNO) 76, 173–176 [Google Scholar]
- 10. Latgé J. P. (2007) The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 66, 279–290 [DOI] [PubMed] [Google Scholar]
- 11. Latgé J. P. (2010) Tasting the fungal cell wall. Cell Microbiol. 12, 863–872 [DOI] [PubMed] [Google Scholar]
- 12. Romani L. (2011) Immunity to fungal infections. Nat. Rev. Immunol. 11, 275–288 [DOI] [PubMed] [Google Scholar]
- 13. Gow N. A., Hube B. (2012) Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 15, 406–412 [DOI] [PubMed] [Google Scholar]
- 14. Kruppa M. D., Lowman D. W., Chen Y. H., Selander C., Scheynius A., Monteiro M. A., Williams D. L. (2009) Identification of (1→6)-β-d-glucan as the major carbohydrate component of the Malassezia sympodialis cell wall. Carbohydr. Res. 344, 2474–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shibata N., Saitoh T., Tadokoro Y., Okawa Y. (2009) The cell wall galactomannan antigen from Malassezia furfur and Malassezia pachydermatis contains β-1,6-linked linear galactofuranosyl residues, and its detection has diagnostic potential. Microbiology 155, 3420–3429 [DOI] [PubMed] [Google Scholar]
- 16. Dubois M., Gilles K. A., Hamilton J. K., Rebers P. A., Smith F. (1956) Colorimetric methods for determination of sugars and related substances. Anal. Chem. 28, 350–356 [Google Scholar]
- 17. Sawardeker J. S., Sloneker J. H., Jeanes A. (1965) Quantitative determination of monosaccharides as their alditol acetates by gas liquid chromatography. Anal. Chem. 37, 1602–1604 [Google Scholar]
- 18. Fuglsang C. C., Berka R. M., Wahleithner J. A., Kauppinen S., Shuster J. R., Rasmussen G., Halkier T., Dalboge H., Henrissat B. (2000) Biochemical analysis of recombinant fungal mutanases. A new family of α1,3-glucanases with novel carbohydrate-binding domains. J. Biol. Chem. 275, 2009–2018 [DOI] [PubMed] [Google Scholar]
- 19. Zverlov V. V., Volkov I. Y., Velikodvorskaya T. V., Schwarz W. H. (1997) Highly thermostable endo-1,3-β-glucanase (laminarinase) LamA from Thermotoga neapolitana: nucleotide sequence of the gene and characterization of the recombinant gene product. Microbiology 143, 1701–1708 [DOI] [PubMed] [Google Scholar]
- 20. Dueñas-Santero E., Martín-Cuadrado A. B., Fontaine T., Latgé J. P., del Rey F., Vázquez de Aldana C. (2010) Characterization of glycoside hydrolase family 5 proteins in Schizosaccharomyces pombe. Eukaryot. Cell 9, 1650–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brurberg M. B., Eijsink V. G., Nes I. F. (1994) Characterization of a chitinase gene (chiA) from Serratia marcescens BJL200 and one-step purification of the gene product. FEMS Microbiol. Lett. 124, 399–404 [DOI] [PubMed] [Google Scholar]
- 22. Vorgias C. E., Tews I., Perrrakis A., Wilson K. J., Oppenheim A. B. (1993) Purification and characterization of the recombinant chitin degrading enzymes, chitinase A, and chitobiase from Serratia marcescens in Chitin Enzymology (Muzzarelli R. A., ed) pp. 417–422, European Chitin Society, Antona, Italy [Google Scholar]
- 23. Heggset E. B., Dybvik A. I., Hoell I. A., Norberg A. L., Sørlie M., Eijsink V. G., Vårum K. M. (2010) Degradation of chitosans with a family 46 chitosanase from Streptomyces coelicolor A3(2). Biomacromolecules 11, 2487–2497 [DOI] [PubMed] [Google Scholar]
- 24. Fontaine T., Talmont F., Dutton G. G., Fournet B. (1991) Analysis of pyruvic acid acetal containing polysaccharides by methanolysis and reductive cleavage methods. Anal. Biochem. 199, 154–161 [DOI] [PubMed] [Google Scholar]
- 25. Fontaine T., Hartland R. P., Beauvais A., Diaquin M., Latge J. P. (1997) Purification and characterization of an endo-1,3-β-glucanase from Aspergillus fumigatus. Eur. J. Biochem. 243, 315–321 [DOI] [PubMed] [Google Scholar]
- 26. Wishart D. S., Bigam C. G., Yao J., Abildgaard F., Dyson H. J., Oldfield E., Markley J. L., Sykes B. D. (1995) 1H, 13C, and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6, 135–140 [DOI] [PubMed] [Google Scholar]
- 27. Wagner G. (1983) Two-dimensional relayed coherence transfer spectroscopy of a protein. J. Magn. Reson. 55, 151–156 [Google Scholar]
- 28. Rance M., Sørensen O. W., Bodenhausen G., Wagner G., Ernst R. R., Wüthrich K. (1983) Improved spectral resolution in cosy 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun. 117, 479–485 [DOI] [PubMed] [Google Scholar]
- 29. Willker W., Leibfritz D., Kerssebaum R., Bermel W. (1993) Gradient selection in inverse heteronuclear correlation spectroscopy. Magn. Reson. Chem. 31, 287–292 [Google Scholar]
- 30. Delay C., Gavin J. A., Aumelas A., Bonnet P. A., Roumestand C. (1997) Isolation and structure elucidation of a highly haemolytic saponin from the Merck saponin extract using high-field gradient-enhanced NMR techniques. Carbohydr. Res. 302, 67–78 [DOI] [PubMed] [Google Scholar]
- 31. Macura S., Huang Y., Suter D., Ernst R. R. (1981) Two-dimensional chemical exchange and cross-relaxation spectroscopy of coupled nuclear spins. J. Magn. Reson. 43, 259–281 [Google Scholar]
- 32. Bock K., Pedersen C., Heding H. (1974) A 13C NMR spectroscopic study of α- and β-streptomycin. J. Antibiot. 27, 139–140 [DOI] [PubMed] [Google Scholar]
- 33. Aimanianda V., Clavaud C., Simenel C., Fontaine T., Delepierre M., Latgé J. P. (2009) Cell wall β-(1,6)-glucan of Saccharomyces cerevisiae: structural characterization and in situ synthesis. J. Biol. Chem. 284, 13401–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gastebois A., Mouyna I., Simenel C., Clavaud C., Coddeville B., Delepierre M., Latgé J. P., Fontaine T. (2010) Characterization of a new β(1–3)-glucan branching activity of Aspergillus fumigatus. J. Biol. Chem. 285, 2386–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim Y. T., Kim E. H., Cheong C., Williams D. L., Kim C. W., Lim S. T. (2000) Structural characterization of β-D-(1→3,1→6)-linked glucans using NMR spectroscopy. Carbohydr. Res. 328, 331–341 [DOI] [PubMed] [Google Scholar]
- 36. Cabib E., Arroyo J. (2013) How carbohydrates sculpt cells: chemical control of morphogenesis in the yeast cell wall. Nat. Rev. Microbiol. 11, 648–655 [DOI] [PubMed] [Google Scholar]
- 37. Kollár R., Petráková E., Ashwell G., Robbins P. W., Cabib E. (1995) Architecture of the yeast cell wall. The linkage between chitin and β(1→3)-glucan. J. Biol. Chem. 270, 1170–1178 [DOI] [PubMed] [Google Scholar]
- 38. Kollár R., Reinhold B. B., Petráková E., Yeh H. J., Ashwell G., Drgonová J., Kapteyn J. C., Klis F. M., Cabib E. (1997) Architecture of the yeast cell wall. β(1→6)-glucan interconnects mannoprotein, β(1→)3-glucan, and chitin. J. Biol. Chem. 272, 17762–17775 [DOI] [PubMed] [Google Scholar]
- 39. Fontaine T., Simenel C., Dubreucq G., Adam O., Delepierre M., Lemoine J., Vorgias C. E., Diaquin M., Latgé J. P. (2000) Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem. 275, 27594–27607 [DOI] [PubMed] [Google Scholar]
- 40. Cabib E., Blanco N., Arroyo J. (2012) Presence of a large β(1–3)glucan linked to chitin at the Saccharomyces cerevisiae mother-bud neck suggests involvement in localized growth control. Eukaryot. Cell 11, 388–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Free S. J. (2013) Fungal cell wall organization and biosynthesis. Adv. Genet. 81, 33–82 [DOI] [PubMed] [Google Scholar]
- 42. Banks I. R., Specht C. A., Donlin M. J., Gerik K. J., Levitz S. M., Lodge J. K. (2005) A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 4, 1902–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dimzon I. K., Ebert J., Knepper T. P. (2013) The interaction of chitosan and olive oil: effects of degree of deacetylation and degree of polymerization. Carbohydr. Polym. 92, 564–570 [DOI] [PubMed] [Google Scholar]
- 44. Zhou K., Xia W., Zhang C., Yu L. (2006) In vitro binding of bile acids and triglycerides by selected chitosan preparations and their physico-chemical properties. Food Sci. Technol. 39, 1087–1092 [Google Scholar]
- 45. Manners D. J., Masson A. J., Patterson J. C. (1973) The structure of a β-(1–3)-d-glucan from yeast cell walls. Biochem. J. 135, 19–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. de Medina-Redondo M., Arnáiz-Pita Y., Clavaud C., Fontaine T., del Rey F., Latgé J. P., Vázquez de Aldana C. R. (2010) β(1,3)-glucanosyl-transferase activity is essential for cell wall integrity and viability of Schizosaccharomyces pombe. PLoS ONE 5, e14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cabib E., Farkas V., Kosík O., Blanco N., Arroyo J., McPhie P. (2008) Assembly of the yeast cell wall. Crh1p and Crh2p act as transglycosylases in vivo and in vitro. J. Biol. Chem. 283, 29859–29872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sugawara T., Takahashi S., Osumi M., Ohno N. (2004) Refinement of the structures of cell-wall glucans of Schizosaccharomyces pombe by chemical modification and NMR spectroscopy. Carbohydr. Res. 339, 2255–2265 [DOI] [PubMed] [Google Scholar]
- 49. Gioti A., Nystedt B., Li W., Xu J., Andersson A., Averette A. F., Münch K., Wang X., Kappauf C., Kingsbury J. M., Kraak B., Walker L. A., Johansson H. J., Holm T., Lehtiö J., Stajich J. E., Mieczkowski P., Kahmann R., Kennell J. C., Cardenas M. E., Lundeberg J., Saunders C. W., Boekhout T., Dawson T. L., Munro C. A., de Groot P. W., Butler G., Heitman J., Scheynius A. (2013) Genomic insights into the atopic eczema-associated skin commensal yeast Malassezia sympodialis. MBio. 4, e00572–00512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hartland R. P., Fontaine T., Debeaupuis J. P., Simenel C., Delepierre M., Latgé J. P. (1996) A novel β-(1–3)-glucanosyltransferase from the cell wall of Aspergillus fumigatus. J. Biol. Chem. 271, 26843–26849 [DOI] [PubMed] [Google Scholar]
- 51. Ragni E., Fontaine T., Gissi C., Latgè J. P., Popolo L. (2007) The Gas family of proteins of Saccharomyces cerevisiae: characterization and evolutionary analysis. Yeast 24, 297–308 [DOI] [PubMed] [Google Scholar]
- 52. Gastebois A., Aimanianda V., Bachellier-Bassi S., Nesseir A., Firon A., Beauvais A., Schmitt C., England P., Beau R., Prévost M. C., d'Enfert C., Latgé J. P., Mouyna I. (2013) SUN proteins belong to a novel family of β-(1,3)-glucan-modifying enzymes involved in fungal morphogenesis. J. Biol. Chem. 288, 13387–13396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gastebois A., Fontaine T., Latgé J. P., Mouyna I. (2010) β(1–3)Glucanosyltransferase Gel4p is essential for Aspergillus fumigatus. Eukaryot. Cell 9, 1294–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mouyna I., Fontaine T., Vai M., Monod M., Fonzi W. A., Diaquin M., Popolo L., Hartland R. P., Latgé J. P. (2000) Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 275, 14882–14889 [DOI] [PubMed] [Google Scholar]
- 55. Hiller E., Heine S., Brunner H., Rupp S. (2007) Candida albicans Sun41p, a putative glycosidase, is involved in morphogenesis, cell wall biogenesis, and biofilm formation. Eukaryot. Cell 6, 2056–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xu J., Saunders C. W., Hu P., Grant R. A., Boekhout T., Kuramae E. E., Kronstad J. W., Deangelis Y. M., Reeder N. L., Johnstone K. R., Leland M., Fieno A. M., Begley W. M., Sun Y., Lacey M. P., Chaudhary T., Keough T., Chu L., Sears R., Yuan B., Dawson T. L., Jr. (2007) Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc. Natl. Acad. Sci. U.S.A. 104, 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nebreda A. R., Vazquez C. R., Villa T. G., Villanueva J. R., del Rey F. (1987) Heterogeneous glycosylation of the EXG1 gene product accounts for the two extracellular exo-β-glucanases of Saccharomyces cerevisiae. FEBS Lett. 220, 27–30 [DOI] [PubMed] [Google Scholar]
- 58. Kobayashi M., Yoshiki R., Sakabe J., Kabashima K., Nakamura M., Tokura Y. (2009) Expression of toll-like receptor 2, NOD2 and dectin-1 and stimulatory effects of their ligands and histamine in normal human keratinocytes. Br J. Dermatol. 160, 297–304 [DOI] [PubMed] [Google Scholar]
- 59. Rubin-Bejerano I., Fraser I., Grisafi P., Fink G. R. (2003) Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 100, 11007–11012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Koller B., Müller-Wiefel A. S., Rupec R., Korting H. C., Ruzicka T. (2011) Chitin modulates innate immune responses of keratinocytes. PLoS ONE 6, e16594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bueter C. L., Lee C. K., Rathinam V. A., Healy G. J., Taron C. H., Specht C. A., Levitz S. M. (2011) Chitosan but not chitin activates the inflammasome by a mechanism-dependent upon phagocytosis. J. Biol. Chem. 286, 35447–35455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Da Silva C. A., Chalouni C., Williams A., Hartl D., Lee C. G., Elias J. A. (2009) Chitin is a size-dependent regulator of macrophage TNF and IL-10 production. J. Immunol. 182, 3573–3582 [DOI] [PubMed] [Google Scholar]
- 63. Goldman D. L., Vicencio A. G. (2012) The chitin connection. MBio 3, 00056–00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.