Background: The C-terminal Src kinase (Csk) is known as a tumor suppressor, but Src-independent function is unclear.
Results: eEF2 is a new protein substrate of Csk.
Conclusion: eEF2 phosphorylation and SUMOylation promote its proteolytic cleavage and nuclear localization.
Significance: Our findings suggest that a tyrosine kinase can be both a tumor suppressor and a promoter through regulation of different substrate proteins.
Keywords: Cancer Biology, Nuclear Translocation, Phosphorylation, Sumoylation, Translation Elongation Factors, Csk, Nuclear Pleomorphism, Proteolytic Cleavage
Abstract
Protein-tyrosine kinase C-terminal Src kinase (Csk) was originally purified as a kinase for phosphorylating Src and other Src family kinases. The phosphorylation of a C-terminal tyrosine residue of Src family kinases suppresses their kinase activity. Therefore, most physiological studies regarding Csk function have been focused on Csk as a negative regulator of Src family tyrosine kinases and as a potential tumor suppressor. Paradoxically, the protein levels of Csk were elevated in some human carcinomas. In this report, we show that eukaryotic elongation factor 2 (eEF2) is a new protein substrate of Csk and could locate in the nucleus. We demonstrate that Csk-mediated phosphorylation of eEF2 has no effect on its cytoplasmic function in regulating protein translation. However, phosphorylation of eEF2 enhances its proteolytic cleavage and the nuclear translocation of the cleaved eEF2 through a SUMOylation-regulated process. Furthermore, we show that cleaved fragments of eEF2 can induce nuclear morphological changes and aneuploidy similar to those in cancer cells, suggesting that there is an additional mechanism for Csk in tumorigenesis through regulation of eEF2 subcellular localization.
Introduction
Protein-tyrosine kinase Csk4 phosphorylates Src and other Src family kinases at their C-terminal tyrosine residues (1, 2). This phosphorylation suppresses the kinase activity of Src family tyrosine kinases (3, 4). Csk is ubiquitously expressed in mammalian cells and is evolutionarily conserved from the early diverged metazoan Hydra to humans (5). The Csk−/− mouse embryos exhibited defects in neurulation, an inability to complete the turning process, and a failure of the allantois to connect with the chorion, preventing the formation of the umbilical cord and placenta (6, 7). The Csk-deficient mouse embryos died around day 10 post-gestation. Mouse genetic studies revealed that Src−/−Csk−/− mouse embryos showed partial rescue of Csk−/− phenotypes (8). However, Src−/−Csk−/− mouse embryos still died around E10 to E11, implying Src-dependent and -independent functions for Csk (8). Indeed, cellular and biochemical studies suggested that Csk also interacts with other proteins, such as protein-tyrosine phosphatase PTP-PEST (9). Given the essential role of Csk for mouse development, it is essential to systematically identify the protein targets of Csk and the functional effects of Csk-mediated phosphorylation on these protein targets.
Deregulation of the activity of Src family tyrosine kinases can lead to oncogenic properties (10). Src family members are found overexpressed or deregulated in human melanoma and carcinoma (10). Human oncogenic alleles of Src kinases have been identified (11). All of these data indicate that Csk could potentially function as a tumor suppressor (12, 13). However, the role of Csk in cancer is more complex. Increased expression of Csk proteins has been observed in 5–20% of carcinoma patients (14). In tumor cells from these patients, the Src activity was not affected (14, 15). The role and mechanism linking high levels of expression of Csk to tumorigenesis are not known.
Eukaryotic elongation factor 2 (eEF2) is one of the three protein factors involved in polypeptide chain elongation during protein translation (16). The activity of eEF2 in translation is modulated by protein phosphorylation. Phosphorylation by kinases such as protein kinase R and Ca2+/calmodulin-dependent kinase III (also called eEF2 kinase) inhibits the activity of eEF2 (17). In addition, eEF2 has been observed to be proteolytically cleaved in many cell types and different stages of cell growth (18). The generation of small fragments of eEF2 could be induced by oxidative stress, aging, and irradiation (19–21). The function of these cleaved fragments of eEF2 is not clear. Recently, eEF2 has been found to be overexpressed in lung adenocarcinoma but not in the neighboring nontumor lung tissue (22). Overexpression of eEF2 has also been observed in gastrointestinal cancers (23). Patients with high levels of eEF2 have a higher incidence of early tumor recurrence and a worse prognosis (22).
In this report, we identified eEF2 as a new Csk substrate using a systematic approach. This phosphorylation event did not change the function of eEF2 in protein translation. Rather, this phosphorylation, together with SUMOylation, led to the cleavage of eEF2. Furthermore, we showed that the cleaved small fragments of eEF2 caused the morphological changes of the nuclei and led to aneuploidy. These changes in the architecture of cell nuclei are similar to those observed in cancer cells (24). These data suggest that Csk also can act as a tumor promoter by changing the morphology of nuclei through regulation of eEF2 cleavage and nuclear translocation.
EXPERIMENTAL PROCEDURES
Cells, Plasmids, and siRNAs
HEK293T and HeLa cells were purchased from ATCC. MEF and Csk−/− cells were described previously (25). Cells were transfected in 6- or 12-well plates using FuGENE® HD (Roche Applied Science) for plasmids and Lipofectamine 2000 for siRNAs and using calcium phosphate for plasmids in 10-cm plates. Cells were harvested 24 or 48 h after transfection for immunoblotting, immunofluorescence, immunoprecipitation, and flow cytometry analysis. HA-tagged SUMO1, SUMO2, and FLAG-tagged Ubc9 plasmids were obtained from Dr. C. Lima at the Sloan-Kettering Cancer Institute. Full-length human eEF2 plasmid was purchased from ATCC. FLAG-tagged Csk was generated by inserting Csk cDNA sequence into pCMV-tag2B vector with BamHI and XhoI. EGFP and Myc-tagged eEF2 fragments were created by PCR subcloning of appropriated PCR products into EGFP-C1 and pcDNA3.0 vectors. eEF2 mutants were generated by one-step PCR-based site-directed mutagenesis. All mutations were confirmed by DNA sequencing. For SUMO1, SUMO2/3, Csk, and eEF2 RNA interference, siRNAs were used. The sequences used were as follows: SUMO1, 5′-CACATCTCAAGAAACTCAA-3′; SUMO2/3, 5′-GTCAATGAGGCAGATCAGA-3′; Csk, 5′-CCGGUGUCUCCUCAAGUUCUCGCUATT-3′.
Kinase Assay and Mass Spectrometry
Recombinant Csk was purified from Escherichia coli as described previously (25–27). eEF2 was purified from rabbit reticulocytes as described previously (28). Csk in vitro kinase assay was performed as described previously (25). For substrate identification, purified Csk kinase was added to Csk−/− MEF whole cell lysates for a 5-min reaction in 30 mm HEPES, pH 7.5, 10 mm MgCl2, 5 mm MnCl2, and 100 nm sodium orthovanadate along with 5 μCi of [γ-32P]ATP (3000 Ci/mmol) (PerkinElmer Life Sciences) for 5 min at 30 °C. The reaction was stopped by adding SDS-PAGE sample buffer and incubated at 90 °C for 5 min. Proteins were then separated by two-dimensional gel electrophoresis. The gels were dried and autoradiographed. Mass spectrometry was used to identify the potential protein substrates with a mass spectrometer operated as described previously (29).
Cytosol and Nuclear Fractionation
Cell pellets were collected and washed with PBS three times. After resuspending in Buffer A (10 mm Tris-HCl, 10 mm NaCl, 5 mm MgCl2, 1 mm DTT, pH 7.4) for 15 min on ice, Buffer B (10 mm Tris-HCl, 10 mm NaCl, 5 mm MgCl2, 1 mm DTT, 10% IgePal CA-630, pH 7.4) was added to make the IgePal CA-630 final concentration to 0.5%. Supernatants were collected as cytosol fraction, and the pellets were lysed with Buffer C (20 mm HEPES-KOH, 1.5 mm MgCl2, 0.5 m NaCl, 1 mm DTT, 0.2 mm EDTA, 20% (V/V) glycerol, pH 7.9) as the nuclear fraction.
Immunoblotting, Immunofluorescence, and Immunoprecipitation
Antibodies against N-terminal eEF2 (catalog no. 2332, Cell Signaling Technology, Inc.) and C-terminal eEF2 (sc-13004, Santa Cruz Biotechnology), Csk (601179, BD Biosciences), SUMO1 and SUMO2/3 (Neweast Biosciences), phosphotyrosine mouse mAb (Tyr(P)-100, Cell Signaling Technology, Inc.), phosphotyrosine (clone 4G10, Millipore), HA monoclonal antibody (Roche Applied Science), FLAG monoclonal antibody (M2008M, Abmart), lamin A/C (catalog no. 2921, Epitomics), and GAPDH (M20006, Abmart) were used for immunoblotting. Antibodies against eEF2 (catalog no. 2332, Cell Signaling Technology, Inc.) and lamin A/C (sc-6215, Santa Cruz Biotechnology) were used for immunofluorescence. Antibodies against eEF2 and Myc tag were used for immunoprecipitation coupled with protein G-Sepharose 4 Fast Flow from GE Healthcare.
Whole cell extracts were prepared as follows: confluent cells were harvested from 10-cm plates, washed twice with cold phosphate-buffered saline, and pellets resuspended in 0.8 ml of lysis buffer (150 mm NaCl, 20 mm Tris, pH 7.4, l mm EDTA, l mm EGTA, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 0.2 mm sodium orthovanadate, 0.03 mg/ml leupeptin). Resuspended pellets were sonicated and centrifuged at 5000 rpm for 5 min at 4 °C to remove insoluble material, and the supernatant was saved as the whole cell extract. Protein concentrations were measured by the Bradford assay, and equal amounts of protein were loaded onto a gel. After SDS-PAGE, protein samples were transferred to nitrocellulose membrane filters. Membrane filters were incubated in 1× Tris-buffered saline and 5% milk for 1 h and then incubated in primary antibody for 2 h at room temperature. Blots were washed three times with Tris-buffered saline/Tween 20 and one time with Tris-buffered saline and then incubated with secondary antibody for 1 h at room temperature. Blots were washed again, and signal was detected with ECL (Millipore).
Cells were seeded onto coverslips coated with poly-lysine and allowed to grow overnight. After 24 h of transfection, cells were fixed with 3.7% formaldehyde in PBS at room temperature and washed with PBS three times. Then the cells were permeabilized and blocked with PBS containing 0.3% Triton X-100, 5% FBS for 1 h. Following a 4-h primary antibody incubation at room temperature (anti-N-terminal eEF2 antibody from Cell Signaling Technology, Inc. or anti-lamin A/C antibody from Santa Cruz Biotechnology at 1:200 dilution), cells were washed with PBS and incubated with fluorescence-conjugated secondary antibody for 2 h. Actin cytoskeleton was stained with phalloidin (P1951, Sigma), and nuclei were stained with DAPI. The coverslips were mounted onto glass slides and imaged with a confocal microscope (Olympus FV1000).
Flow Cytometry
Cells were harvest and fixed for propidium iodide staining as described previously (29). Cells were washed two times with ice-cold PBS, fixed with 70% ethanol at −20 °C, collected by centrifugation, digested with RNase A, and stained with propidium iodide. Stained cells were then analyzed by flow cytometer (FACS AriaTM III, BC Biosciences). Two color channels were used as follows: FITC channel for selection of EGFP-positive cells and propidium iodide for DNA content analysis. Using FlowJo software, only EGFP-positive cells (control vector-only expression cells) were analyzed for their DNA content.
RESULTS
Identification of eEF2 as a New Substrate for Csk
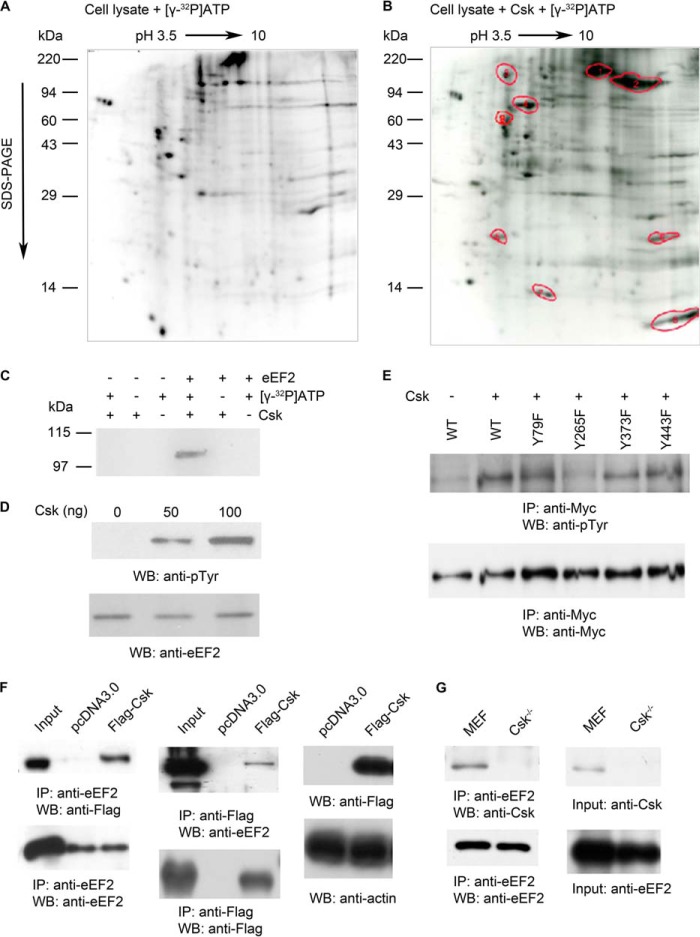
To fully understand the physiological functions of Csk, it is essential to identify the protein substrates of Csk in addition to Src family tyrosine kinases. In general, identification of physiologically relevant protein substrates for protein kinases is difficult. However, Csk has a unique property; it recognizes the three-dimensional structure of its substrates, rather than a linear amino acid sequence (30). For example, Csk phosphorylates Tyr-527 on full-length Src but would not efficiently phosphorylate this Tyr residue when it is included in a short peptide (31, 32). Taking this advantage, we used purified full-length Csk and cell lysates prepared from Csk−/− mouse embryonic fibroblast cells (MEF cells) in the presence of [γ-32P]ATP, in combination with two-dimensional electrophoresis and mass spectrometry, to identify potential Csk protein substrates. The reason to use the full-length Csk, rather than the kinase domain of Csk, is that the isolated kinase domain of Csk displayed a catalytic activity ∼100-fold lower than that of the full-length Csk (26). To avoid the potential problem of pre-phosphorylation of protein substrates by endogenous Csk, we used cell lysates prepared from Csk−/− cells (25). To minimize the potential cascade effect (Csk acts on other kinases that phosphorylate substrate proteins), the phosphorylation reaction was carried out for a short time (5 min). For control, a similar reaction was carried out in the absence of Csk but with [γ-32P]ATP (Fig. 1, A and B)
FIGURE 1.
Identification of new protein substrates for tyrosine kinase Csk. A and B, two-dimensional gel electrophoresis analysis of Csk−/− MEF cell lysates in the presence of [γ-32P]ATP alone (A) or in the presence of purified Csk and [γ-32P]ATP (B). C, phosphorylation of purified eEF2 by purified Csk in vitro. D, different concentrations of purified Csk (0, 50, and 100 ng) phosphorylate purified eEF2 in vitro, and anti-phosphotyrosine and anti-N-terminal eEF2 antibodies were used for blotting. E, identification of phosphorylated tyrosine residues on eEF2 by Csk. Mutant and wild-type eEF2 plasmids were tagged with Myc and transfected into HEK293T cells with or without Csk plasmid. Anti-Myc antibody was used for immunoprecipitation (IP). The blot was probed with either anti-phosphotyrosine monoclonal antibody (top panel) or with anti-Myc antibody (bottom panel). F, co-immunoprecipitation of Csk and eEF2. Plasmids of pcDNA3.0 and FLAG-Csk were transfected into HEK293T cells. Left panel, blots showed that anti-C-terminal eEF2 antibody co-immunoprecipitated FLAG-tagged Csk; middle panel, anti-FLAG antibody (for FLAG-tagged Csk) also co-immunoprecipitated eEF2. Input, whole cell lysates from cells transfected with FLAG-tagged Csk. Right panel, top, Western blot (WB) with anti-FLAG antibody shows the expression of FLAG-tagged Csk. Bottom, Western blot with anti-actin antibody shows similar amounts of total cell lysates were used in all immunoprecipitation experiments. G, interaction of endogenous eEF2 and Csk. Left panel, anti-C-terminal eEF2 antibody that targets endogenous eEF2 in MEF cells could co-immunoprecipitate endogenous Csk. Right panel, Western blot with anti-Csk antibody shows the absence of Csk expression in Csk−/− MEF cells (top), and anti-C-terminal eEF2 antibody shows similar levels of eEF2. Data are representative of three independent experiments.
Reaction samples were separated by two-dimensional gels with the first dimension of the pH gradient from pH 3.5 to 10 and the second dimension of regular SDS-PAGE. After comparing these two-dimensional gels, nine spots showed increased 32P labeling by Csk (Fig. 1, A and B). The proteins contained in these nine spots were identified by LC-MS-MS. In addition to c-Src, the major phosphorylated protein is eEF2 (spots 1 and 2) (Fig. 1B). To confirm the direct phosphorylation of eEF2 by Csk, we used purified Csk and purified eEF2 in an in vitro kinase assay. Purified eEF2 was incubated with or without Csk and with or without [γ-32P]ATP. Only when eEF2, Csk, and [γ-32P]ATP were present, eEF2 was phosphorylated (Fig. 1C). Phosphorylation of eEF2 was increased with increasing concentrations of Csk (Fig. 1D). Therefore, Csk could directly phosphorylate eEF2.
To identify the phosphorylation sites on eEF2, we mutated tyrosine residues in eEF2 that have been identified to be phosphorylated in large scale global phospho-proteomic studies (33, 34). The phosphorylation of these mutated eEF2 proteins by Csk in transfected cells was examined. Representative data were shown in Fig. 1E. From these studies, Tyr-265 and Tyr-373 are two phosphorylation sites on eEF2 by Csk (also see Fig. 5B). In addition to phosphorylating its substrates, Csk could directly bind its substrates (30–32). Therefore, we investigated whether Csk could bind eEF2 in cells. We transfected FLAG-tagged Csk into cells and performed co-immunoprecipitation experiments. As shown in Fig. 1F, Csk was co-immunoprecipitated with eEF2, and eEF2 was co-immunoprecipitated with Csk. Endogenous interaction of eEF2 and Csk was also examined. Anti-C-terminal eEF2 antibody, which targets endogenous eEF2 in MEF cells, could co-immunoprecipitate endogenous Csk (Fig. 1G). Therefore, Csk and eEF2 could interact with each other in cells. Altogether, these results indicate that Csk can interact with eEF2 and phosphorylate eEF2 on specific tyrosine residues.
FIGURE 5.
Cross-talk between eEF2 phosphorylation and SUMOylation. A, siRNA against Csk was introduced into HEK293T cells, fractionated after 24 h, immunoprecipitated (IP) with anti-C-terminal eEF2 antibody, and then blotted with the indicated antibodies. Knockdown Csk level led to decreased SUMOylation level of eEF2. Input: whole cell lysates. CE, cytosolic fraction; NE, nuclear fraction; WB, Western blot. B, HEK293T cells were transfected with WT eEF2 and the indicated eEF2 mutants, fractionated after 24 h, immunoprecipitated with anti-Myc antibody, and then blotted with the indicated antibodies. All of the phosphorylation and SUMOylation mutants showed decreased phosphorylation and SUMOylation levels of eEF2. Data are representative of three independent experiments.
Nuclear Localization of Cleaved eEF2 C-terminal Fragment
eEF2 is an essential factor in protein synthesis, and it promotes the translocation of the nascent peptide chain from the A-site to the P-site on the ribosome in a GTP-dependent manner (16). The activity of eEF2 in translation is modulated by protein phosphorylation. Serine phosphorylation by kinases such as protein kinase R and Ca2+/calmodulin-dependent kinase III (also called eEF2 kinase) inhibits the activity of eEF2 (17, 35, 36). To investigate the biological effect of Csk tyrosine phosphorylation on eEF2, we first studied the possible effect on protein translation. From studies with various in vitro protein translation systems, we did not observe any effect of Csk on protein translation (data not shown). These suggest that eEF2 may have other functions.
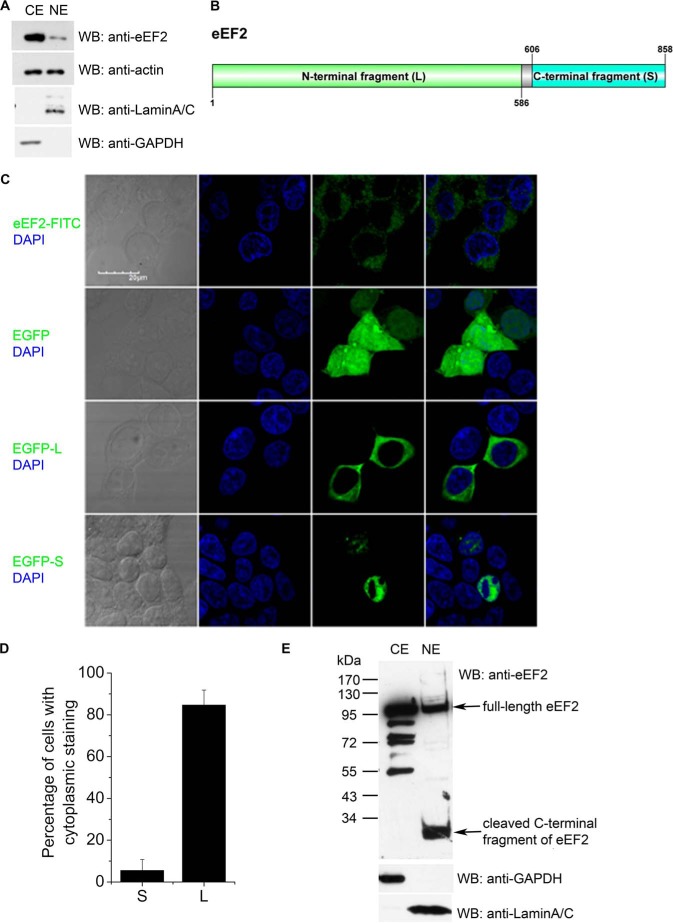
Eukaryotic translation elongation factors were considered to locate in the cytoplasm due to their major function in protein synthesis. However, it has been reported that ∼10% elongation factor 1A subunit (EF1A) is present in the nucleus. This implies additional functions for EF1A. For example, it was reported that EF1A could export several proteins, including the von Hippel-Lindau tumor suppressor and the poly(A)-binding protein (PABP1) (37). Yet it has never been tested whether eEF2 was present in the nucleus. We found that a small proportion (<10%) of eEF2 could locate in nucleus (Fig. 2A). Using the cNLS Mapper program to search for nuclear localization sequences (NLSs) in eEF2, we found that there were 2 monopartite NLSs and 31 bipartite NLSs in eEF2. One monopartite NLS with the highest score is located at the C terminus of eEF2 (amino acid residues 837–846). A recent proteomics study also identified eEF2 as a potential cargo of importin-β (38), implying a new eEF2 function in nucleus. It has been previously reported that eEF2 could be proteolytically cleaved at Glu-586/Ser-587, Lys-605/Ala-606, or Gly-612/Leu-613 to generate small fragments of ∼32/33 kDa (Fig. 2B). The function of these small fragments of eEF2 is not clear (39). To visualize the subcellular localization of these fragments, the larger N-terminal fragment (residues 1–586) and the smaller C-terminal fragment (residues 606–858) were tagged with GFP and expressed in HEK293T cells (Fig. 2C). The large N-terminal fragment (contains the functional guanine nucleotide-binding domain for protein translation regulation) was in the cytosol, whereas the smaller C-terminal fragment was in the nucleus (Fig. 2C). These results were quantified and shown in Fig. 2D. Although, in ∼84% of the transfected cells, the large N-terminal fragment was in the cytosol, the small C-terminal fragment was mainly in the nucleus. This nucleus-specific location of the C-terminal fragment of eEF2 was further confirmed with Western blot analyses of endogenous eEF2 (Fig. 2E).
FIGURE 2.
Nuclear localization of full-length eEF2 and its small cleaved fragment. A, eEF2 could translocate into nucleus. HEK293T cells were fractionated and blotted with indicated antibodies. CE, cytosolic fraction; NE, nuclear fraction. Anti-C-terminal eEF2 showed the different distribution of eEF2 in cytosolic and nuclear fractions. Anti-actin antibody showed similar amounts of cytosolic and nuclear proteins. Lamin A/C and GAPDH were used as nuclear and cytosolic markers, respectively. WB, Western blot. B, diagram of the eEF2 plasmid constructs. Large and small fragments of eEF2 were constructed as indicated. This diagram was drawn by Group-based prediction System (DOG 2.0). C, HEK293T cells were stained with anti-N-terminal eEF2 antibody (top row, green color) or transfected with EGFP control plasmid, EGFP-L, and EGFP-S plasmids. The images were taken with confocal microscopy (GFP, green color). Nuclei were stained with DAPI (blue color). D, quantification of data in C. Results are mean ± S.D. of at least 50 transfected cells. E, cleaved small fragment of eEF2 accumulated in nucleus. HEK293T cells were fractionated. Anti-C-terminal eEF2 antibody showed the cleaved eEF2 in nucleus. GAPDH was used as a cytosolic marker, and lamin A/C was used as nuclear marker. Data are representatives of three independent experiments.
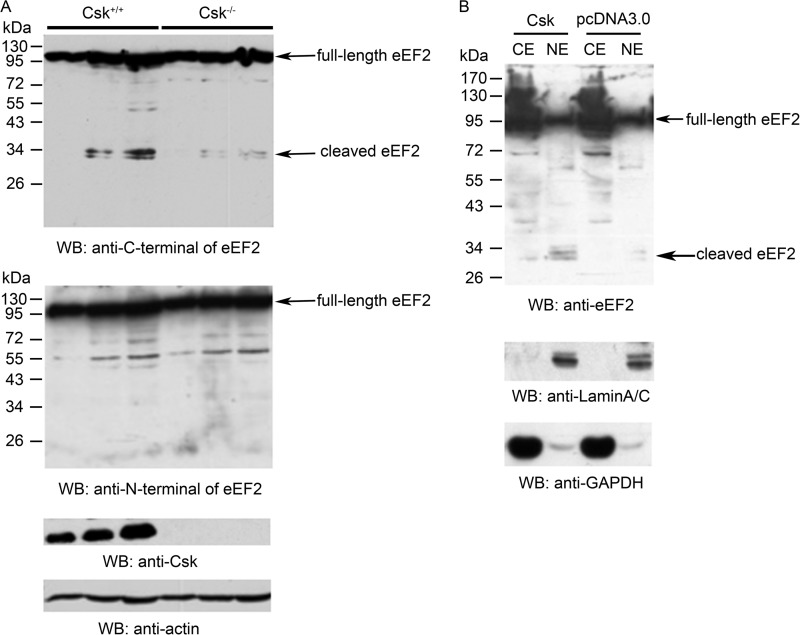
We next investigated the possible effect of Csk on the cleavage of eEF2. In Csk−/− MEF cells, there were lower levels of cleaved eEF2 fragments (Fig. 3A). When Csk was overexpressed in HEK293T cells, the levels of the cleaved eEF2 fragments were increased (Fig. 3B). Together, these data demonstrate the accumulation of the cleaved C-terminal small fragment of eEF2 in the nucleus, and Csk could enhance the proteolytic cleavage of eEF2.
FIGURE 3.
Csk could increase the cleavage of eEF2. A, eEF2 cleavage was decreased in Csk−/− MEF cells. Western blot (WB) analyses of whole cell lysates from wild-type MEF and Csk−/− MEF cells loaded at three different concentrations with the indicated antibodies. Anti-C and anti-N termini of eEF2 antibodies were used to show the full-length and cleaved eEF2 change. Actin was used as loading control. B, whole cell lysates from Csk or control vector transfected HEK293T cells were analyzed by Western blotting. Csk transfection positively increased eEF2 cleavage as detected by the anti-C-terminal eEF2 antibody. Lamin A/C and GAPDH were used as nuclear and cytosolic markers. Data are representative of three independent experiments. CE, cytosolic fraction; NE, nuclear fraction.
SUMOylation of eEF2
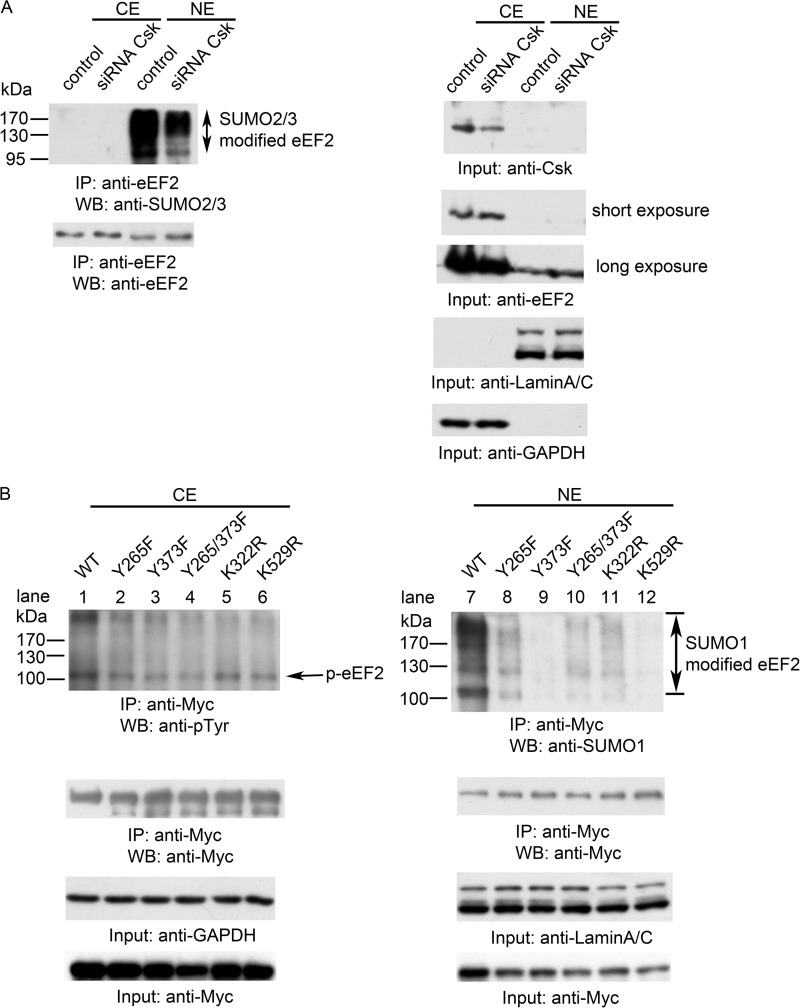
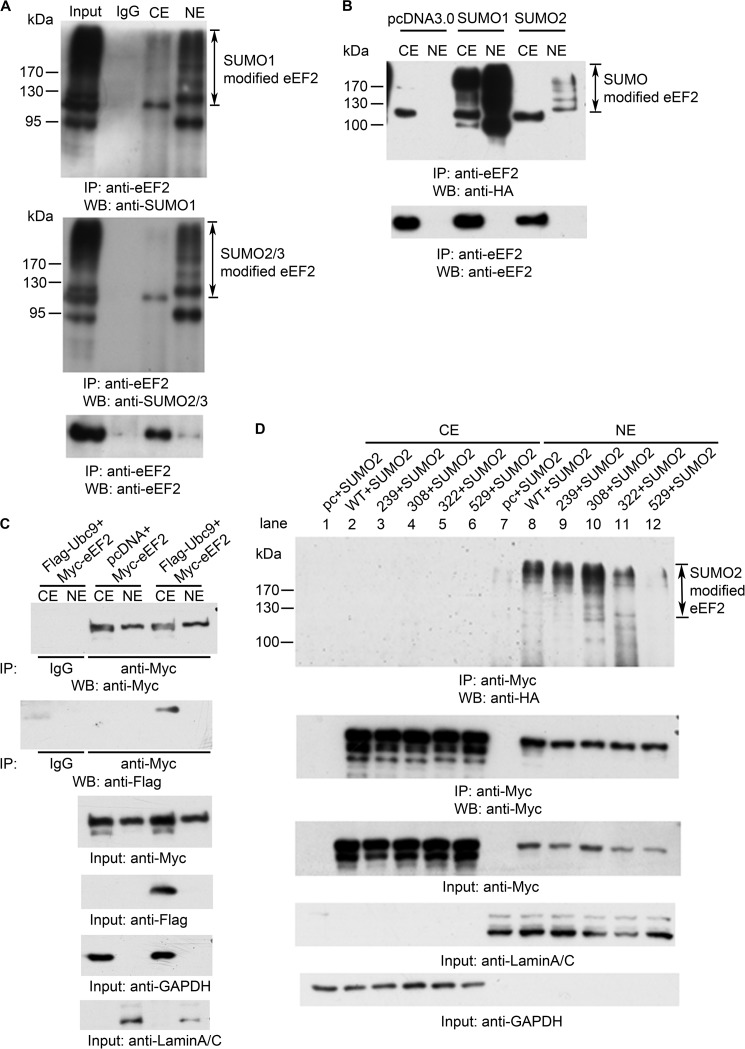
We next explored the mechanism by which Csk modulates the proteolytic cleavage of eEF2 and the nuclear translocation of eEF2. Protein SUMOylation has been shown to regulate nuclear trafficking of several proteins such as RanGAP, PML, and p53 (40–42). SUMO1, -2, and -3 are members of the ubiquitin-like protein family (43). The covalent attachment of SUMO1, -2, or -3 to target proteins is analogous to ubiquitination. We first examined whether eEF2 is SUMOylated. After cell fractionation, endogenous eEF2 proteins were immunoprecipitated and analyzed by Western blotting with anti-SUMO1 or with an antibody for both SUMO2 and SUMO3 (Fig. 4A). Strikingly, SUMOylated eEF2 was mainly in the nucleus as indicated by the presence of multiple bands recognized by anti-SUMO antibodies (Fig. 4A). The multiple bands represent different SUMOylation states (multiple sites and polySUMOylated) of eEF2. We also expressed SUMO1 and SUMO2 in cells, and immunoprecipitated endogenous eEF2. As shown in Fig. 4B, HA-tagged SUMO co-immunoprecipitated with eEF2 and mainly accumulated in nucleus. All three SUMO proteins use the same Ubc9 E2 conjugation enzyme. To investigate whether Ubc9 could bind to eEF2, we co-transfected Myc-tagged eEF2 and FLAG-tagged Ubc9 into HEK293T cells, following immunoprecipitation, and Western blot analyses showed the interaction of Ubc9 and eEF2, which occurred in the cytosol (Fig. 4C).
FIGURE 4.
SUMOylation of eEF2. A, whole cell lysates from HEK293T cells were fractionated, immunoprecipitated (IP) with an antibody against the C terminus of eEF2, and analyzed by Western blotting (WB) using anti-SUMO antibodies as indicated. CE, cytosolic fraction; NE, nuclear fraction. B, direct detection of SUMOylated eEF2. HEK293T cells were transfected with pcDNA3.0, HA-SUMO1, and HA-SUMO2, fractionated after 24 h, immunoprecipitated with anti-C-terminal eEF2 antibody, and blotted with anti-HA and anti-C-terminal eEF2 antibodies. C, anti-Myc antibody (for Myc-tagged eEF2) could co-immunoprecipitate FLAG-tagged Ubc9 in cytosolic fraction. HEK293T cells were co-transfected with either Myc-eEF2 and pcDNA3.0 or Myc-eEF2 and FLAG-ubc9. Cells were fractionated after 24 h, and the cytoplasmic extract (CE) and nuclear extract (NE) were immunoprecipitated with mouse IgG or anti-Myc antibody. Input, total cell lysates. D, pcDNA3.0, WT eEF2, and the indicated eEF2 mutants were co-transfected with SUMO2 into HEK293T cells, fractionated after 24 h, immunoprecipitated with anti-Myc antibody, and then blotted with indicated antibodies. Data are representative of three independent experiments.
To identify the SUMOylation sites on eEF2, we mutated several lysine residues based on the consensus sequences of SUMOylation sites and examined their SUMOylation. As shown in Fig. 4D, after co-transfecting eEF2 mutants with HA-tagged SUMO2, compared with WT (lane 8), mutation of Lys-529 to Arg decreased the SUMOylation of eEF2 (lane 12), and the K322R mutant also had a slightly decreased SUMOylation level (lane 11), whereas mutations of Lys-239 and Lys-308 to Arg had no effect on eEF2 SUMOylation (lanes 9 and 10). Therefore, Lys-322 and Lys-529 are the SUMOylation sites on eEF2. Altogether, these data suggest that full-length eEF2 could be SUMOylated by SUMO1 and SUMO2/3. SUMOylated eEF2 was then translocated into the nucleus.
Cross-talk between Csk-mediated Phosphorylation and SUMOylation of eEF2
We investigated the connection between Csk-mediated phosphorylation and SUMOylation of eEF2. First, we examined the effect of the Csk expression level on eEF2 SUMOylation. We used the siRNA technique to knock down the Csk protein level. As shown in Fig. 5A, after transfection with Csk siRNA, SUMOylation level of eEF2 was decreased. Second, we examined the effect of various eEF2 mutants, either on the phosphorylation sites or SUMOylation sites, on eEF2 phosphorylation and SUMOylation. When mutants of eEF2 with Tyr-265, Tyr-373, and the double mutant Y265F/Y373F (the phosphorylated sites by Csk) were expressed in cells, the level of SUMOylation of eEF2 was decreased (Fig. 5B, right panel, compare lane 7 with lanes 8–10). Conversely, mutation of the SUMOylation sites (K322R and K529R) decreased the tyrosine phosphorylation of eEF2 (Fig. 5B, compare lane 1 with lanes 5 and 6).
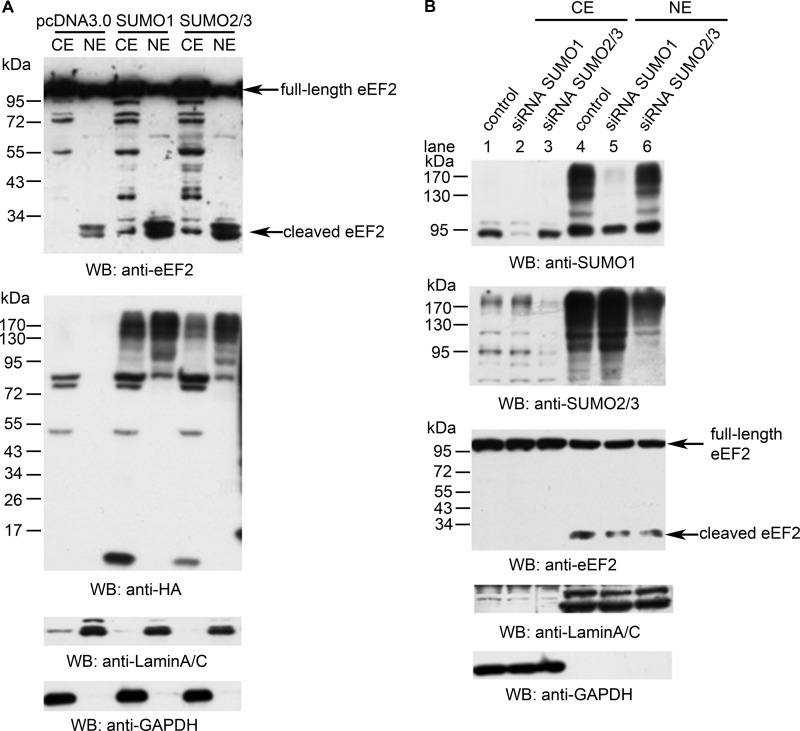
We also found that SUMOylation level can influence eEF2 cleavage. Overexpression of SUMO1 or SUMO2 increased the levels of the cleaved eEF2 fragments in the nucleus (Fig. 6A). Knockdown of SUMO1 or SUMO2/3 by RNA interference decreased the levels of cleaved eEF2 in the nucleus (Fig. 6B, compare lane 4 with lane 5 or 6). These data demonstrate an interplay between tyrosine phosphorylation and SUMOylation of eEF2, which in turn leads to the proteolytic cleavage of eEF2.
FIGURE 6.
Effect of SUMOylation on the cleavage of eEF2. A, HeLa cells were transfected with SUMO1, SUMO2, or control vector and blotted with anti-C-terminal eEF2 antibody. Increased level of cleaved eEF2 was observed in the nuclear extracts of SUMO-transfected cells. HA blotting indicated the transfect efficiency of SUMO plasmids. Lamin A/C and GAPDH were used as nuclear and cytosolic markers, respectively. B, HeLa cells were transfected with siRNAs against SUMO1 and SUMO2/3, then fractionated after 48 h, blotted with indicated antibodies that showed consistency of decreased amounts of cleaved eEF2, and decreased the SUMOylation level. Data are representatives of three independent experiments. CE, cytosolic fraction; NE, nuclear fraction; WB, Western blot.
Nuclear Deformation and Aneuploidy Induced by the Small Fragment of eEF2
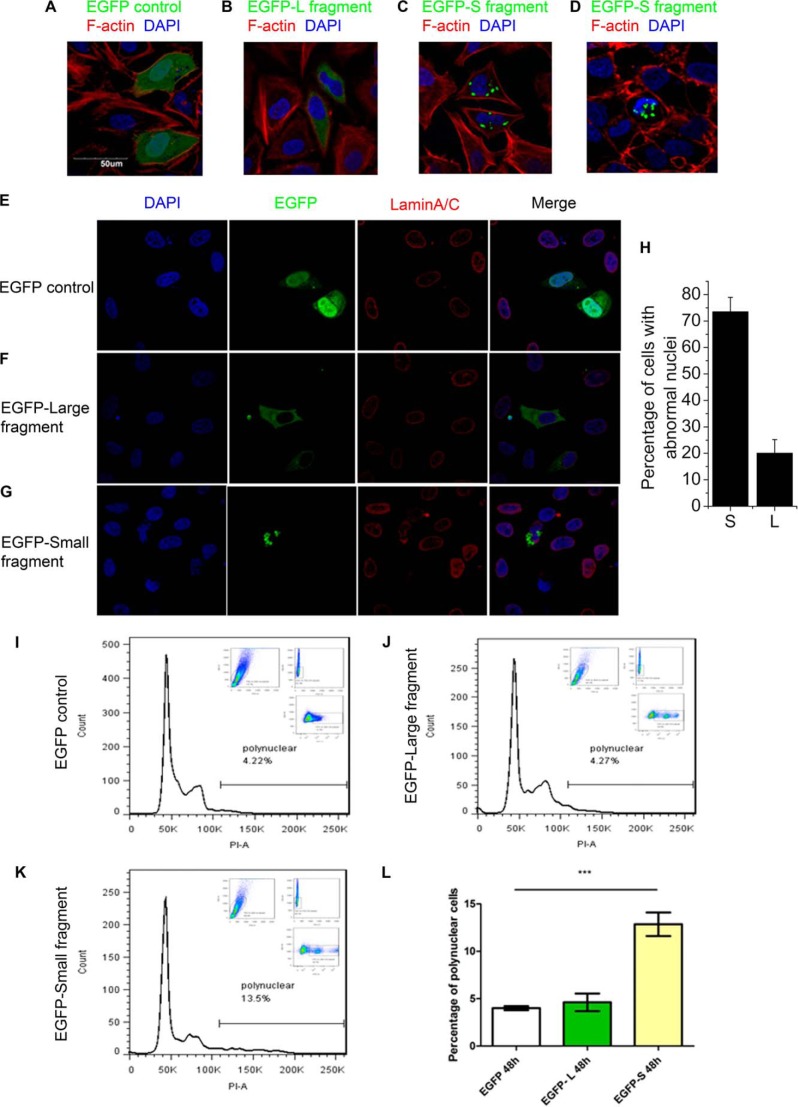
Finally, we studied the biological implication of the eEF2 cleavage and nuclear localization. Expression of eEF2 N-terminal large fragment (amino acids 1–686) or control GFP did not cause obvious changes on the nuclear morphology (Fig. 7, A and B). However, the C-terminal small fragment of eEF2 (amino acids 606–858) displayed punctate staining in the nucleus (Fig. 7, C and D). In some cells, this cleaved eEF2 caused an enlarged nucleus, consistent with aneuploidy. In other cells, it led to deformed nucleus. The effect of the C-terminal small fragment of eEF2 on the nuclear morphology was further confirmed with the lamin A/C staining (Fig. 7, E–G). Lamin A/C nuclear lamins are intermediate filament proteins that constitute the lattice-like matrix at the inner face of the nuclear membrane. In control cells transfected with GFP, lamin A/C staining indicated that these cells were with a smooth and oval- or round-shaped nuclear membrane (Fig. 7E). Similar results were observed in cells transfected with the N-terminal large fragment of eEF2 (fused with GFP) (Fig. 7F). However, cells transfected with the C-terminal small fragment of eEF2 (fused with GFP) displaced abnormal nuclear morphology with enlarged and deformed nuclei (Fig. 7G). Cells transfected with the C-terminal small fragment had a higher percentage of abnormal nuclei (∼74%) than cells transfected with the N-terminal large fragment (∼20%) (Fig. 7H). The cleaved eEF2 accumulated as specks between the lamin A/C matrix and the nuclear membrane and distorted the smooth appearance of lamin matrix. Enlarged sizes of the nuclei are usually caused by an increase in DNA content as a result of aneuploidy (24). Therefore, we investigated whether the C-terminal small fragment indeed led to aneuploidy. Flow cytometric analysis was performed after the cells were stained with propidium iodide as a DNA content indicator (Fig. 7, I–L). In GFP-transfected cells or the N-terminal large fragment of eEF2-transfected cells, the majority of cells was in the 2n (diploid) stages (only ∼4% in >4n stages) (Fig. 7, I, J, and L). In contrast, cells transfected with the C-terminal small fragment of eEF2 had a much higher (∼14%) percentage of cells in >4n stages, indicating aneuploidy (Fig. 7, K and L). Thus, the C-terminal small fragment of eEF2 leads to nuclear pleomorphism.
FIGURE 7.
Overexpression of eEF2 small fragment causes nuclear deformation and aneuploidy. A–D, HeLa cells were transfected with the indicated expression plasmids, fixed, and stained with phalloidin for F-actin polymers and DAPI for nuclei after 24 h. E–G, HeLa cells were transfected with the indicated expression plasmids, fixed, and stained with lamin A/C for nuclear membrane and DAPI for nuclei after 24 h. H, quantification of data in E–G. Results are a mean ± S.D. of at least 50 transfected cells. I–K, HEK293T cells were transfected with indicated expression plasmids, and analyzed by flow cytometry for DNA contents after 48 h. One representative of three similar experiments is shown. L, summary of the data from I–K. Data are presented as mean ± S.D. (n = 3).
DISCUSSION
In this report, we have identified a new protein substrate for Csk and a surprising link between eEF2, a cytoplasmic protein known for its function in protein translation, and nuclear deformation and aneuploidy. Csk phosphorylation of eEF2 did not affect eEF2 cytoplasmic function in protein translation. Rather, this phosphorylation is correlated with the SUMOylation of eEF2 and cleavage of eEF2. The molecular mechanisms by which Csk phosphorylation affects eEF2 SUMOylation and by which the C-terminal small fragment causes nuclear pleomorphism need further investigation.
The well known substrates of Csk are Src family tyrosine kinases, which are negatively regulated by Csk. Src family tyrosine kinases were once thought of as the only substrates of Csk. However, there are several reports suggesting that Csk has Src-independent functions. Here, we have identified and investigated a new substrate for Csk. Our results indicate that, in addition to acting as a tumor suppressor by inhibiting the activity of Src family tyrosine kinases, Csk may also participate in cell transformation through promoting the accumulation of the small fragments of eEF2 leading to nuclear morphological changes. Our data are in line with several other reports showing elevated expression of Csk in cancer cells without decreasing Src activity as well as Csk-independent inhibition of Src (15, 44, 45).
eEF2 was found highly expressed in tumor samples and cancer cells (22). However, the explanation for this overexpression of eEF2 in cancer cells is not clear. It was proposed that eEF2 could render a cancer cell growth advantage by inhibiting apoptosis, and eEF2 is involved in cell cycle progression in cancer cells (22, 23). Our data suggest another possible mechanism by which eEF2 influences cell transformation. eEF2 is cleaved into small functional fragments. The accumulation of these fragments in the nucleus causes nuclear deformation and aneuploidy. Nuclear instability and aneuploidy are directly linked to malignant transformation of cells (46). Although the exact molecular mechanism by which the small fragments induce nuclear instability needs further investigation, one possibility is that the small fragments interfere with the function of nuclear lamins, as we observed irregular shapes of nuclear membrane in cells expressing the small fragments. It is known that nuclear lamins are important in maintaining genome stability (47–49).
The production of the cleaved small fragments of eEF2 is tightly regulated by post-translational modifications on eEF2. SUMOylation of eEF2 is positively correlated with the amounts of the small fragments. Overexpression of SUMO proteins led to increased levels of small fragments, whereas knockdown of SUMO proteins decreased the level of small fragments. The levels of small fragments were also modulated by Csk-mediated phosphorylation on eEF2, as overexpression of Csk positively correlated with the amounts of the small fragments. In Csk−/− MEF cells, lower levels of the small fragments were observed compared with wild-type MEF cells. It is also possible that Csk regulates other processes that lead to destabilization of eEF2. Taken together, these data indicate a positive correlation between SUMOylation and tyrosine phosphorylation of eEF2 with the cleavage of eEF2. The relationship between SUMOylation and Csk-mediated tyrosine phosphorylation on eEF2 is cross-regulated. Knockdown Csk protein expression could decrease SUMOylation level of eEF2, and mutations in one type of modification (such as phosphorylation) led to the decrease of another type of modification (such as SUMOylation). How SUMOylation and phosphorylation modulate the cleavage of eEF2 needs further investigation. Considering the role of SUMOylation in modulating protein-protein interaction, it is possible that proteases that cleave eEF2 only recognize SUMOylated and phosphorylated eEF2, so that the cleavage is under surveillance to avoid excessive cleaved eEF2 fragments (50). Thus, SUMOylation could control protein shuttling between the cytoplasm and nucleus (51). It is also possible that the cleavage of eEF2 could only take place in the nucleus, and SUMOylation controls nuclear translocation of full-length eEF2. We observed that both the nuclear full-length eEF2 and its cleaved form can be positively influenced by SUMOylation (data not shown).
Protein post-translational modifications, such as phosphorylation, ubiquitination, SUMOylation, play important roles in regulating protein functions. Most studies have focused on individual modifications on proteins. However, it is now clear that interplays among multiple modifications are important in regulating protein functions. Large scale proteomic analyses have revealed global cross-talks among different types of modifications in regulating protein functions (52). We recently reported a global cross-talk between SUMOylation and phosphorylation (29). Others have reported cross-talks between glycosylation and phosphorylation (53). Furthermore, a phosphorylation-dependent SUMOylation motif has been identified (54). Acetylation within the SUMO interaction motifs controls SUMO-mediated interactions (55). Hence, multiple protein modifications can be integrated into modulating the functions of proteins and signaling networks.
Protein cleavage, an irreversible protein modification, also plays important regulatory roles in regulating protein functions. In addition to the well known functions of caspase-mediated cleavage cascades in apoptosis, several recent studies indicate that cleaved protein fragments could have rather different functions compared with its intact form. Full-length c-Myc protein, an oncogenic protein, is a transcription factor regulating a variety of genes involved in cell cycle and other cellular processes. However, c-Myc could be cleaved into a functional fragment that translocates to the cytoplasm, where it promotes acetylation of tubulin and cell differentiation (56). The TGFβ receptor 1, as an intact protein, receives a signal from the ligand TGFβs and transduces the signal. Its cleaved form translocates to the nucleus and directly regulates gene expression involved in tumor invasiveness (57). All these studies point to broader regulatory functions for protein cleavage.
Acknowledgments
We thank Yan Wang for assistance in flow cytometry analysis, Ying Hu for assistance in confocal microscopy analysis, and C. Lima for the SUMO1, SUMO2, and Ubc9 plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grant HL091525 (to X.-Y. H.). This work was also supported by National Science Foundation of China Grants 31028015 and 31221061, National Basic Research Program of China Grant 2013CB911102, and the 111 Project of China Grant B06018 (to L. G.).
- Csk
- C-terminal Src kinase
- MEF
- mouse embryonic fibroblast cell
- NLS
- nuclear localization sequence
- EGFP
- enhanced GFP
- SUMO
- small ubiquitin-related modifier.
REFERENCES
- 1. Okada M., Nada S., Yamanashi Y., Yamamoto T., Nakagawa H. (1991) CSK: a protein-tyrosine kinase involved in regulation of Src family kinases. J. Biol. Chem. 266, 24249–24252 [PubMed] [Google Scholar]
- 2. Nada S., Okada M., MacAuley A., Cooper J. A., Nakagawa H. (1991) Cloning of a complementary DNA for a protein-tyrosine kinase that specifically phosphorylates a negative regulatory site of p60c-src. Nature 351, 69–72 [DOI] [PubMed] [Google Scholar]
- 3. Cooper J. A., Gould K. L., Cartwright C. A., Hunter T. (1986) Tyr527 is phosphorylated in pp60c-src: implications for regulation. Science 231, 1431–1434 [DOI] [PubMed] [Google Scholar]
- 4. Cole P. A., Shen K., Qiao Y., Wang D. (2003) Protein-tyrosine kinases Src and Csk: a tail's tale. Curr. Opin. Chem. Biol. 7, 580–585 [DOI] [PubMed] [Google Scholar]
- 5. Miller M. A., Malik I. A., Shenk M. A., Steele R. E. (2000) The Src/Csk regulatory circuit arose early in metazoan evolution. Oncogene 19, 3925–3930 [DOI] [PubMed] [Google Scholar]
- 6. Imamoto A., Soriano P. (1993) Disruption of the csk gene, encoding a negative regulator of Src family tyrosine kinases, leads to neural tube defects and embryonic lethality in mice. Cell 73, 1117–1124 [DOI] [PubMed] [Google Scholar]
- 7. Nada S., Yagi T., Takeda H., Tokunaga T., Nakagawa H., Ikawa Y., Okada M., Aizawa S. (1993) Constitutive activation of Src family kinases in mouse embryos that lack Csk. Cell 73, 1125–1135 [DOI] [PubMed] [Google Scholar]
- 8. Thomas S. M., Soriano P., Imamoto A. (1995) Specific and redundant roles of Src and Fyn in organizing the cytoskeleton. Nature 376, 267–271 [DOI] [PubMed] [Google Scholar]
- 9. Davidson D., Cloutier J. F., Gregorieff A., Veillette A. (1997) Inhibitory tyrosine protein kinase p50csk is associated with protein-tyrosine phosphatase PTP-PEST in hemopoietic and non-hemopoietic cells. J. Biol. Chem. 272, 23455–23462 [DOI] [PubMed] [Google Scholar]
- 10. Bolen J. B. (1993) Nonreceptor tyrosine protein kinases. Oncogene 8, 2025–2031 [PubMed] [Google Scholar]
- 11. Irby R. B., Mao W., Coppola D., Kang J., Loubeau J. M., Trudeau W., Karl R., Fujita D. J., Jove R., Yeatman T. J. (1999) Activating SRC mutation in a subset of advanced human colon cancers. Nat. Genet. 21, 187–190 [DOI] [PubMed] [Google Scholar]
- 12. Masaki T., Okada M., Tokuda M., Shiratori Y., Hatase O., Shirai M., Nishioka M., Omata M. (1999) Reduced C-terminal Src kinase (Csk) activities in hepatocellular carcinoma. Hepatology 29, 379–384 [DOI] [PubMed] [Google Scholar]
- 13. Nakagawa T., Tanaka S., Suzuki H., Takayanagi H., Miyazaki T., Nakamura K., Tsuruo T. (2000) Overexpression of the csk gene suppresses tumor metastasis in vivo. Int. J. Cancer 88, 384–391 [PubMed] [Google Scholar]
- 14. Bénistant C., Bourgaux J. F., Chapuis H., Mottet N., Roche S., Bali J. P. (2001) The COOH-terminal Src kinase Csk is a tumor antigen in human carcinoma. Cancer Res. 61, 1415–1420 [PubMed] [Google Scholar]
- 15. Watanabe N., Matsuda S., Kuramochi S., Tsuzuku J., Yamamoto T., Endo K. (1995) Expression of C-terminal src kinase in human colorectal cancer cell lines. Jpn. J. Clin. Oncol. 25, 5–9 [PubMed] [Google Scholar]
- 16. Bermek E. (1978) Mechanisms in polypeptide chain elongation on ribosomes. Prog. Nucleic Acid Res. Mol. Biol. 21, 63–100 [DOI] [PubMed] [Google Scholar]
- 17. Ryazanov A. G., Shestakova E. A., Natapov P. G. (1988) Phosphorylation of elongation factor 2 by EF-2 kinase affects rate of translation. Nature 334, 170–173 [DOI] [PubMed] [Google Scholar]
- 18. Giovane A., Servillo L., Quagliuolo L., Balestrieri C. (1987) Purification of elongation factor 2 from human placenta and evidence of its fragmentation patterns in various eukaryotic sources. Biochem. J. 244, 337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ayala A., Parrado J., Bougria M., Machado A. (1996) Effect of oxidative stress, produced by cumene hydroperoxide, on the various steps of protein synthesis. Modifications of elongation factor-2. J. Biol. Chem. 271, 23105–23110 [DOI] [PubMed] [Google Scholar]
- 20. Parrado J., Bougria M., Ayala A., Castaño A., Machado A. (1999) Effects of aging on the various steps of protein synthesis: fragmentation of elongation factor 2. Free Radic. Biol. Med. 26, 362–370 [DOI] [PubMed] [Google Scholar]
- 21. Miura Y., Kano M., Yamada M., Nishine T., Urano S., Suzuki S., Endo T., Toda T. (2007) Proteomic study on x-irradiation-responsive proteins and ageing: search for responsible proteins for radiation adaptive response. J. Biochem. 142, 145–155 [DOI] [PubMed] [Google Scholar]
- 22. Chen C. Y., Fang H. Y., Chiou S. H., Yi S. E., Huang C. Y., Chiang S. F., Chang H. W., Lin T. Y., Chiang I. P., Chow K. C. (2011) Sumoylation of eukaryotic elongation factor 2 is vital for protein stability and anti-apoptotic activity in lung adenocarcinoma cells. Cancer Sci. 102, 1582–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamura J., Aoyagi S., Nanchi I., Nakatsuka S., Hirata E., Shibata S., Fukuda M., Yamamoto Y., Fukuda I., Tatsumi N., Ueda T., Fujiki F., Nomura M., Nishida S., Shirakata T., Hosen N., Tsuboi A., Oka Y., Nezu R., Mori M., Doki Y., Aozasa K., Sugiyama H., Oji Y. (2009) Overexpression of eukaryotic elongation factor eEF2 in gastrointestinal cancers and its involvement in G2/M progression in the cell cycle. Int. J. Oncol. 34, 1181–1189 [PubMed] [Google Scholar]
- 24. Zink D., Fischer A. H., Nickerson J. A. (2004) Nuclear structure in cancer cells. Nat. Rev. Cancer 4, 677–687 [DOI] [PubMed] [Google Scholar]
- 25. Lowry W. E., Huang J., Ma Y. C., Ali S., Wang D., Williams D. M., Okada M., Cole P. A., Huang X. Y. (2002) Csk, a critical link of G protein signals to actin cytoskeletal reorganization. Dev. Cell 2, 733–744 [DOI] [PubMed] [Google Scholar]
- 26. Sondhi D., Cole P. A. (1999) Domain interactions in protein-tyrosine kinase Csk. Biochemistry 38, 11147–11155 [DOI] [PubMed] [Google Scholar]
- 27. Ma Y. C., Huang J., Ali S., Lowry W., Huang X. Y. (2000) Src tyrosine kinase is a novel direct effector of G proteins. Cell 102, 635–646 [DOI] [PubMed] [Google Scholar]
- 28. Price N. T., Redpath N. T., Severinov K. V., Campbell D. G., Russell J. M., Proud C. G. (1991) Identification of the phosphorylation sites in elongation factor-2 from rabbit reticulocytes. FEBS Lett. 282, 253–258 [DOI] [PubMed] [Google Scholar]
- 29. Yao Q., Li H., Liu B. Q., Huang X. Y., Guo L. (2011) SUMOylation-regulated protein phosphorylation, evidence from quantitative phosphoproteomics analyses. J. Biol. Chem. 286, 27342–27349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Levinson N. M., Seeliger M. A., Cole P. A., Kuriyan J. (2008) Structural basis for the recognition of c-Src by its inactivator Csk. Cell 134, 124–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sondhi D., Xu W., Songyang Z., Eck M. J., Cole P. A. (1998) Peptide and protein phosphorylation by protein-tyrosine kinase Csk: insights into specificity and mechanism. Biochemistry 37, 165–172 [DOI] [PubMed] [Google Scholar]
- 32. Lee S., Ayrapetov M. K., Kemble D. J., Parang K., Sun G. (2006) Docking-based substrate recognition by the catalytic domain of a protein-tyrosine kinase, C-terminal Src kinase (Csk). J. Biol. Chem. 281, 8183–8189 [DOI] [PubMed] [Google Scholar]
- 33. Gevaert K., Staes A., Van Damme J., De Groot S., Hugelier K., Demol H., Martens L., Goethals M., Vandekerckhove J. (2005) Global phosphoproteome analysis on human HepG2 hepatocytes using reversed-phase diagonal LC. Proteomics 5, 3589–3599 [DOI] [PubMed] [Google Scholar]
- 34. Iliuk A. B., Martin V. A., Alicie B. M., Geahlen R. L., Tao W. A. (2010) In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion-functionalized soluble nanopolymers. Mol. Cell. Proteomics 9, 2162–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hincke M. T., Nairn A. C. (1992) Phosphorylation of elongation factor 2 during Ca(2+)-mediated secretion from rat parotid acini. Biochem. J. 282, 877–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mitsui K., Brady M., Palfrey H. C., Nairn A. C. (1993) Purification and characterization of calmodulin-dependent protein kinase III from rabbit reticulocytes and rat pancreas. J. Biol. Chem. 268, 13422–13433 [PubMed] [Google Scholar]
- 37. Khacho M., Mekhail K., Pilon-Larose K., Pause A., Côté J., Lee S. (2008) eEF1A is a novel component of the mammalian nuclear protein export machinery. Mol. Biol. Cell 19, 5296–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kimura M., Okumura N., Kose S., Takao T., Imamoto N. (2013) Identification of cargo proteins specific for importin-β with importin-α applying a stable isotope labeling by amino acids in cell culture (SILAC)-based in vitro transport system. J. Biol. Chem. 288, 24540–24549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kleifeld O., Doucet A., auf dem Keller U., Prudova A., Schilling O., Kainthan R. K., Starr A. E., Foster L. J., Kizhakkedathu J. N., Overall C. M. (2010) Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 28, 281–288 [DOI] [PubMed] [Google Scholar]
- 40. Matunis M. J., Coutavas E., Blobel G. (1996) A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135, 1457–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Duprez E., Saurin A. J., Desterro J. M., Lallemand-Breitenbach V., Howe K., Boddy M. N., Solomon E., de Thé H., Hay R. T., Freemont P. S. (1999) SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci. 112, 381–393 [DOI] [PubMed] [Google Scholar]
- 42. Gostissa M., Hengstermann A., Fogal V., Sandy P., Schwarz S. E., Scheffner M., Del Sal G. (1999) Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 18, 6462–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schwartz D. C., Hochstrasser M. (2003) A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem. Sci. 28, 321–328 [DOI] [PubMed] [Google Scholar]
- 44. Oneyama C., Hikita T., Enya K., Dobenecker M. W., Saito K., Nada S., Tarakhovsky A., Okada M. (2008) The lipid raft-anchored adaptor protein Cbp controls the oncogenic potential of c-Src. Mol. Cell 30, 426–436 [DOI] [PubMed] [Google Scholar]
- 45. Lutz M. P., Esser I. B., Flossmann-Kast B. B., Vogelmann R., Lührs H., Friess H., Büchler M. W., Adler G. (1998) Overexpression and activation of the tyrosine kinase Src in human pancreatic carcinoma. Biochem. Biophys. Res. Commun. 243, 503–508 [DOI] [PubMed] [Google Scholar]
- 46. Duesberg P., Fabarius A., Hehlmann R. (2004) Aneuploidy, the primary cause of the multilateral genomic instability of neoplastic and preneoplastic cells. IUBMB Life 56, 65–81 [DOI] [PubMed] [Google Scholar]
- 47. Gonzalez-Suarez I., Redwood A. B., Perkins S. M., Vermolen B., Lichtensztejin D., Grotsky D. A., Morgado-Palacin L., Gapud E. J., Sleckman B. P., Sullivan T., Sage J., Stewart C. L., Mai S., Gonzalo S. (2009) Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J. 28, 2414–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Capo-chichi C. D., Cai K. Q., Simpkins F., Ganjei-Azar P., Godwin A. K., Xu X. X. (2011) Nuclear envelope structural defects cause chromosomal numerical instability and aneuploidy in ovarian cancer. BMC Med. 9, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Capo-chichi C. D., Cai K. Q., Smedberg J., Ganjei-Azar P., Godwin A. K., Xu X. X. (2011) Loss of A-type lamin expression compromises nuclear envelope integrity in breast cancer. Chin. J. Cancer 30, 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y. (2004) Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sedek M., Strous G. J. (2013) SUMOylation is a regulator of the translocation of Jak2 between nucleus and cytosol. Biochem. J. 453, 231–239 [DOI] [PubMed] [Google Scholar]
- 52. Guo Z., Kanjanapangka J., Liu N., Liu S., Liu C., Wu Z., Wang Y., Loh T., Kowolik C., Jamsen J., Zhou M., Truong K., Chen Y., Zheng L., Shen B. (2012) Sequential posttranslational modifications program FEN1 degradation during cell-cycle progression. Mol. Cell 47, 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang Z., Gucek M., Hart G. W. (2008) Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proc. Natl. Acad. Sci. U.S.A. 105, 13793–13798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hietakangas V., Anckar J., Blomster H. A., Fujimoto M., Palvimo J. J., Nakai A., Sistonen L. (2006) PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. U.S.A. 103, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ullmann R., Chien C. D., Avantaggiati M. L., Muller S. (2012) An acetylation switch regulates SUMO-dependent protein interaction networks. Mol. Cell 46, 759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Conacci-Sorrell M., Ngouenet C., Eisenman R. N. (2010) Myc-nick: a cytoplasmic cleavage product of Myc that promotes α-tubulin acetylation and cell differentiation. Cell 142, 480–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mu Y., Sundar R., Thakur N., Ekman M., Gudey S. K., Yakymovych M., Hermansson A., Dimitriou H., Bengoechea-Alonso M. T., Ericsson J., Heldin C. H., Landström M. (2011) TRAF6 ubiquitinates TGFβ type I receptor to promote its cleavage and nuclear translocation in cancer. Nat. Commun. 2, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]