Background: An unknown third divinyl-chlorophyllide a reductase (DVR) was proposed among phototrophs.
Results: The double mutant lacking the conventional DVR and chlorophyllide a reductase (COR) of a purple bacterium uniquely accumulated divinyl-chlorophyllide a.
Conclusion: An additional activity of COR corresponds to the third DVR, which is conserved among other photosynthetic bacteria.
Significance: COR is suggested to be the evolutionarily oldest DVR among three DVRs.
Keywords: Evolution, Microbiology, Photosynthesis, Photosynthetic Pigments, Reductase, BciA, BciB, DVR, Photosynthetic Bacteria, Purple Bacteria
Abstract
Bacteriochlorophyll a is widely distributed among anoxygenic photosynthetic bacteria. In bacteriochlorophyll a biosynthesis, the reduction of the C8 vinyl group in 8-vinyl-chlorophyllide a is catalyzed to produce chlorophyllide a by an 8-vinyl reductase called divinyl reductase (DVR), which has been classified into two types, BciA and BciB. However, previous studies demonstrated that mutants lacking the DVR still synthesize normal bacteriochlorophyll a with the C8 ethyl group and suggested the existence of an unknown “third” DVR. Meanwhile, we recently observed that chlorophyllide a oxidoreductase (COR) of a purple bacterium happened to show the 8-vinyl reduction of 8-vinyl-chlorophyllide a in vitro. In this study, we made a double mutant lacking BciA and COR of the purple bacterium Rhodobacter sphaeroides in order to investigate whether the mutant still produces pigments with the C8 ethyl group or if COR actually works as the third DVR. The single mutant deleting BciA or COR showed production of the C8 ethyl group pigments, whereas the double mutant accumulated 8-vinyl-chlorophyllide, indicating that there was no enzyme other than BciA and COR functioning as the unknown third DVR in Rhodobacter sphaeroides (note that this bacterium has no bciB gene). Moreover, some COR genes derived from other groups of anoxygenic photosynthetic bacteria were introduced into the double mutant, and all of the complementary strains produced normal bacteriochlorophyll a. This observation indicated that COR of these bacteria performs two functions, reductions of the C8 vinyl group and the C7=C8 double bond, and that such an activity is probably conserved in the widely ranging groups.
Introduction
Chlorophyll (Chl)4 and bacteriochlorophyll (BChl) pigments are required by photosynthetic organisms to perform primary processes for capturing photons, migrating excited energy, and achieving charge separation. These pigment species possess distinctive electronic absorption bands that are dependent on their molecular structures and also influence their light-harvesting efficiencies in photosynthetic apparatuses. Oxygenic photosynthetic organisms (e.g. higher plants, algae, and cyanobacteria) contain Chls a, b, c, d, and f (1–3), whereas anoxygenic photosynthetic bacteria (e.g. purple bacteria, green sulfur bacteria, filamentous anoxygenic phototrophs, heliobacteria, and acidobacteria) possess BChls a, b, c, d, e, and g (4, 5). All of the Chl and BChl species are biosynthesized from 8-vinyl-protochlorophyllide a (8V-PChlide a) by a series of enzymes. Their structural variation is ascribable to the biosynthetic derivatization from the common pigment intermediate through several transformations, including reduction of C=C double bonds. Evolutionarily modified enzymes catalyze to produce the (B)Chl pigments, which are diverged in all of the phototrophs.
BChl a is found in light-harvesting antenna systems and reaction center complexes of anoxygenic photosynthetic bacteria, except for some purple bacteria giving BChl b and heliobacteria having BChl g. The biosynthetic pathways for BChl a have been well characterized using model organisms in purple bacteria that are genetically amendable (4), together with biochemical studies of some enzymes (6–8). In the genomes of purple bacteria, genes encoding enzymes for the BChl a biosynthesis are usually arranged in a large region called a photosynthesis gene cluster, which also contains genes for the reaction center, the light-harvesting complex, and carotenoid biosynthesis (9, 10). The photosynthetic gene cluster is conserved in many genome-sequenced purple bacteria and is thought to have been acquired by a horizontal gene transfer from a photosynthetically ancestral species.
In the biosynthetic pathways of BChl a, a vinyl group at the C8 position at the stage of 8V-PChlide a or 8-vinyl-chlorophyllide a (8V-Chlide a) is reduced to the C8 ethyl group by 8-vinyl reductase (also known as divinyl reductase (DVR)) (see Fig. 1). In an early study using the purple bacterium Rhodobacter capsulatus, the mutational analysis judged that a bchJ gene encodes one of the subunits for DVR (4, 11). However, recent investigations have indicated that BchJ is not DVR and would be a scaffold or chaperone protein for an unidentified route among several biosynthetic steps from 8V-PChlide a to BChl a (8, 12). In the case of Chl a synthesis, a AT5G18660 gene of the higher plant Arabidopsis thaliana was identified as a DVR gene because the gene mutant accumulated 8-vinyl-Chls (8V-Chls) a and b (13, 14); hereafter in this work, this DVR is called plant-type DVR. In the green sulfur bacterium Chlorobaculum tepidum, an ortholog of the plant-type DVR, denoted as BciA for BChl biosynthesis, was also identified; the bciA gene deletion mutant accumulated 8-vinylated derivatives (12). The recombinant enzymes of plant-type DVR and BciA showed reduction activities using NADPH as an electron donor (12, 13, 15). The in vitro enzymatic measurements of plant-type DVR showed that this enzyme had a higher activity with 8V-Chlide a than 8V-PChlide a. By contrast, in genomes of many cyanobacteria, no orthologs of plant-type DVR and BciA were found, but a nonhomologous slr1923 gene in the cyanobacterium Synechocystis sp. PCC6803 did code an enzyme showing DVR activity (16, 17), which is here called cyano-type DVR. Many orthologs of cyano-type DVR were also found in genomes of anoxygenic photosynthetic bacteria. One of those orthologs, the Ctha_1208 gene of the green sulfur bacterium Chloroherpeton thalassium, was observed (18); although a genetic manipulation of this bacterium is not available, the Ctha_1208 gene could complement the DVR ability in the bciA mutant of C. tepidum and was denoted as bciB for BChl synthetic pathways. Very recently, the recombinant BciB and cyano-type DVR were studied in vitro and showed the 8-vinyl reduction activity using ferredoxin as an electron donor (19, 20).
FIGURE 1.
Proposed BChl a biosynthetic pathway, including reduction process of C8 vinyl group.
As mentioned above, two classes of DVR, BciA and BciB, have been registered for synthesis of BChl species in most anoxygenic photosynthetic bacteria. The model purple bacterium R. sphaeroides has only BciA, as does C. tepidum, whereas another purple bacterium, Rhodopseudomonas palustris, has only BciB as its DVR, similar to C. thalassium (16, 18). Some green sulfur bacteria are known to have both the bciA and bciB genes. Interestingly, filamentous anoxygenic phototrophs Roseiflexus species do not possess either bciA or bciB in the genomes, although these bacteria produce normal 8-ethylated BChl a. Other examples of bacteria lacking DVR genes are BChl b-producing Blastochloris viridis and BChl g-producing Heliobacterium modesticaldum because the C8 vinyl group is directly converted to the C8 ethylidene group in the biosynthetic pathway of these pigments (21–23).
We reported that the bciA deletion mutant of C. tepidum still synthesized normal BChl a (24). Recently, Hunter and co-workers (25) also demonstrated that the bciA mutant of R. sphaeroides synthesized BChl a, and they claimed the presence of an unknown “third-type” DVR. On the other hand, we recently found that Chlide a oxidoreductase (COR) composed of BchX, -Y, and -Z subunits of R. capsulatus, the original function of which is the reduction of the C7=C8 double bond in a chlorin ring, could also reduce the C8 vinyl to ethyl group in vitro (22). To confirm the dual activities of the COR protein in vivo and to conclude the presence or absence of the third-type DVR, we constructed the R. sphaeroides mutant lacking both COR and BciA and investigated whether the pigment accumulated by the mutant is 8-ethylated or not. Additionally, the bchXYZ genes of various bacteria, including Roseiflexus castenholzii, were introduced into the double mutant to investigate whether the function of the 8-vinyl reduction is restored.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions
Bacterial strains used in this study are listed in Table 1. R. sphaeroides J001 and R. palustris J002 strains were rifampicin-resistant derivatives from R. sphaeroides 2.4.1 (26) and R. palustris CGA009 (27), respectively, and used as wild type strains in this study. These bacteria were grown in liquid or on solid PYS media (27) at 30 °C under aerobically and semiaerobically dark conditions for genetic manipulations and pigment analyses, respectively, or anaerobically illuminated conditions for phototrophic growth. The Escherichia coli strains, DH5α (28), JM109 λ-pir (29), and S17-1 λ-pir (30), were grown in liquid or on solid LB media at 37 °C.
TABLE 1.
Bacterial strains used in this study
| Bacterial strains | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | Host for cloning vectors | Ref. 28 |
| JM109 λ-pir | Host of pJSC plasmid for cloning | Ref. 29 |
| S17-1 λ-pir | Host for pJSC and pZJ102 derivatives to deliver them to J001 or J002 strain by conjugation | Ref. 30 |
| R. sphaeroides | ||
| J001 (wild type) | Plating rifampicin resistance derivative from R. sphaeroides 2.4.1 (Rfr) | This study |
| dA | bciA gene-deleted mutant (Rfr, Kmr) | This study |
| dXYZ | bchX, -Y, and -Z genes deleted mutant (Rfr, Smr) | This study |
| dA/XYZ | bciA, bchX, -Y, and -Z genes deleted mutant (Rfr, Kmr, Smr) | This study |
| dXYZ_102 | The dXYZ introduced the pZJ102 plasmid (Rfr, Kmr, Gmr) | This study |
| dXYZ_sphaXYZ | The dXYZ introduced the pZJ_sphaXYZ plasmid (Rfr, Kmr, Gmr) | This study |
| dXYZ_palXYZ | The dXYZ introduced the pZJ_palXYZ plasmid (Rfr, Kmr, Gmr) | This study |
| dXYZ_tepXYZ | The dXYZ introduced the pZJ_tepXYZ plasmid (Rfr, Kmr, Gmr) | This study |
| dXYZ_castXYZ | The dXYZ introduced the pZJ_castXYZ plasmid (Rfr, Kmr, Gmr) | This study |
| dA/XYZ_102 | The dA/XYZ introduced the pZJ102 plasmid (Rfr, Kmr, Smr, Gmr) | This study |
| dA/XYZ_sphaA | The dA/XYZ introduced the pZJ_sphaA plasmid (Rfr, Kmr, Smr, Gmr) | This study |
| dA/XYZ_palB | The dA/XYZ introduced the pZJ_palB plasmid (Rfr, Kmr, Smr, Gmr) | This study |
| dA/XYZ_sphaXYZ | The dA/XYZ introduced the pZJ_sphaXYZ plasmid (Rfr, Kmr, Smr, Gmr) | This study |
| dA/XYZ_palXYZ | The dA/XYZ introduced the pZJ_palXYZ plasmid (Rfr, Kmr, Smr, Gmr) | This study |
| dA/XYZ_tepXYZ | The dA/XYZ introduced the pZJ_tepXYZ plasmid (Rfr, Kmr, Smr, Gmr) | This study |
| dA/XYZ_castXYZ | The dA/XYZ introduced the pZJ_castXYZ plasmid (Rfr, Kmr, Smr, Gmr) | This study |
| R. palustris | ||
| J002 (wild type) | Plating rifampicin resistance derivative from Rps. palustris CGA009 (Rfr) | This study |
| dB | bciB gene deleted mutant (Rfr, Kmr) | This study |
a Gmr, gentamycin resistance; Kmr, kanamycin resistance; Rfr, rifampicin resistance; Smr, streptomycin resistance.
The following antibiotics with specified concentrations were added to the media: 25 μg/ml kanamycin, 100 μg/ml rifampicin, 25 μg/ml streptomycin, and 10 μg/ml gentamycin for R. sphaeroides; 100 μg/ml kanamycin and 100 μg/ml rifampicin for R. palustris; 100 μg/ml ampicillin, 25 μg/ml kanamycin, 20 μg/ml streptomycin, 10 μg/ml gentamycin, and 34 μg/ml chloramphenicol for E. coli.
Plasmid Constructions
All plasmids used in this study are listed in Table 2. The PCR primers used for the construction of plasmids and confirmation of mutants in this study are described in Table 3. To construct the R. sphaeroides mutant lacking BciA, the pJSC-sphaA-Km plasmid (Fig. 2A) was produced as follows. A 1.04-kbp DNA fragment containing the bciA gene was amplified from the genomic DNA of R. sphaeroides by PCR using sphaA-F and sphaA-R primers. The PCR was performed with KOD-plus-DNA polymerase (TOYOBO, Japan). The product was digested with EcoRI and cloned into the SmaI and EcoRI restriction sites of pUC118 vector (Takara), yielding pUC-sphaA plasmid. The neo gene, conferring resistance to kanamycin, was excised from pUCKM1 (31) with HindIII and SmaI and then blunted. The pUC-sphaA plasmid was digested with XhoI, blunted, and ligated with the blunt-ended neo gene fragment, creating pUC-sphaA-Km plasmid. The fragment containing neo and partial bciA genes was excised from pUC-sphaA-Km with SmaI and XbaI and subcloned into the same restriction sites of the pJSC vector (32), producing pJSC-sphaA-Km. The plasmid pJSC is a chloramphenicol-resistant suicide vector and has the sacB gene encoding the levansucrase; the expression of sacB in the presence of sucrose was lethal for most of the Gram-negative bacteria (33). To construct the bchX/Y/Z-deleted mutant of R. sphaeroides, pJSC-sphaXYZ-Sm plasmid (Fig. 2B) was created as follows. The aadA gene, conferring resistance to streptomycin, was amplified from the plasmid pHP45Ω (34) by PCR using the primer set aadA-F and aadA-R. It is noted that the bchXYZ genes are usually arranged next to each other in the genomes except for that of green sulfur bacteria (see Fig. 2, B, G, and H). A 4.08-kbp DNA fragment containing the bchX/Y/Z genes was amplified from the R. sphaeroides genome using sphaXYZ-F and -R primers. The PCR product and the SmaI-digested pUC118 were ligated using an In-Fusion® HD cloning kit (Clontech), yielding pUC-sphaXYZ. To amplify the DNA fragment from pUC-sphaXYZ without the inner portion of bchXYZ, the plasmid was used as the template for the inverse PCR with primers sphaXYZ-inf-F and -R (see Fig. 2B). The resulting PCR product and the PCR fragment containing the aadA gene were ligated with the In-Fusion cloning kit, producing pUC-sphaXYZ-Sm. The obtained pUC-sphaXYZ-Sm was used as the template for PCR with the sphaXYZ-F and -R primer set, and the PCR fragment was subcloned into the SmaI site of pJSC by In-Fusion cloning, creating the pJSC-sphaXYZ-Sm plasmid.
TABLE 2.
Plasmids used in this study
| Plasmids | Relevant characteristicsa | Reference or source |
|---|---|---|
| pUC118 | Cloning vector (Apr) | Takara |
| pUCKM1 | pUC plasmid with neo gene (Apr, Kmr) | Ref. 31 |
| pJSC | A derivative plasmid of suicide vector pJP5603 containing sacB and cat genes (Cmr) | Ref. 32 |
| pHP45Ω | pHP45 plasmid with aadA gene flanked by Ω-terminators (Apr, Smr) | Ref. 34 |
| pJZ102 | An expression vector in purple bacteria, possessing puc gene promoter (Gmr, Smr) | Ref. 35 |
| pUC-sphaA | 1.04-kbp DNA fragment containing bciA of Rba. sphaeroides cloned in pUC118 (Apr) | This study |
| pUC-sphaA-Km | (pUC118+bciA::neo). neo gene cloned in HindIII and SmaI sites within the bciA in the pUC-sphaA (Apr, Kmr) | This study |
| pJSC-sphaA-Km | (pJSC+ bciA::neo). DNA fragment containing neo and partial bciA genes from the pUC-sphaA-Km, subcloned into the pJSC (Apr, Kmr, Cmr) | This study |
| pUC-sphaXYZ | 4.08-kbp DNA fragment containing bchX, Y, and Z genes of Rba. sphaeroides cloned in pUC118 (Apr) | This study |
| pUC-sphaXYZ-Sm | (pUC118+bchXYZ::aadA). A part of bchXYZ genes in the pUC-sphaXYZ, replaced with the aadA gene (Apr, Smr) | This study |
| pJSC-sphaXYZ-Sm | (pJSC+bchXYZ::aadA). DNA fragment containing aadA and partial bchXZ genes from the pUC-sphaXYZ-Sm, subcloned into the pJSC (Apr, Kmr, Cmr) | This study |
| pUC-palB | 1.23-kbp DNA fragment containing bciB of Rps. palustris cloned in pUC118 (Apr) | This study |
| pUC-palB-Km | (pUC118+bciB::neo). neo gene cloned in SalI sites within the bciB in the pUC-sphaA (Apr, Kmr) | This study |
| pJSC-palB-Km | (pJSC+bchXYZ::aadA). DNA fragment containing aadA and partial bciB genes from the pUC-palB-Km, subcloned into the pJSC (Apr, Kmr, Cmr) | This study |
| pUC-tepXYZ | bchX, bchY, and bchZ genes of Cba. tepidum independently amplified each other, and then connected and cloned into the pUC118 (Apr) | This study |
| pUC-castXYZ | 7.06-kbp DNA fragment containing bchXYZ and Rcas3745 genes of Rfx. castenholzii, cloned into pUC118 (Apr) | This study |
| pUC-castXYZ2 | The Rcas3745 gene removed from pUC-castXYZ (Apr) | This study |
| pZJ-sphaA | DNA fragment containing bciA of Rba. sphaeroides from pUC-sphaA, subcloned into the pZJ102 (Gmr, Smr) | This study |
| pZJ-palB | DNA fragment containing bciB of Rps. palustris from pUC-sphaB, sub- cloned into the pZJ102 (Gmr, Smr) | This study |
| pZJ-sphaXYZ | DNA fragment containing bchXYZ of Rba. sphaeroides from pUC-spha- XYZ, subcloned into the pZJ102 (Gmr, Smr) | This study |
| pZJ-palXYZ | 4.06-kbp DNA fragment containing the bchXYZ genes of Rps. palustris amplified, and cloned into the pZJ102 (Gmr, Smr) | This study |
| pZJ-tepXYZ | DNA fragment containing bchXYZ of Cba. tepidum from pUC-tepXYZ, subcloned into the pZJ102 (Gmr, Smr) | This study |
| pZJ-castXYZ | DNA fragment containing bchXYZ of Rfx. castenholzii from pUC-cast- XYZ2, subcloned into the pZJ102 (Gmr, Smr) | This study |
a Apr, ampicillin resistance; Cmr, chloramphenicol resistance.
TABLE 3.
Sequences of primers for construction of the plasmids and confirmation of the mutants used in this study

FIGURE 2.
Constructions of plasmids and bacterial mutants used in this study. Schematic maps show bciA (A) or bchX/Y/Z (B) deletion mutants of R. sphaeroides J001 strain. The neo and aadA genes confer resistance to kanamycin and streptomycin, respectively. Arrows in A and B represent the primers sphaA-F (i), sphaA-R (ii), sphaA-comf-F (iii), sphaA-comf-R (iv), sphaXYZ-F (v), sphaXYZ-R (vi), sphaXYZ-inf-F (vii), sphaXYZ-inf-R (viii), sphaXYZ-comf-F (ix), and sphaXYZ-comf-R (x). C, schematic maps showing bciB deletion mutant of R. palustris J002 strain. Arrows in C represent the primers palB-F (xi), palB-R (xii), palB-comf-F (xiii), and palB-comf-R (xiv). D–F, PCR analyses using the genomic DNA extracted from J001 (lanes 1 and 5), dA (lanes 2 and 6), dXYZ (lanes 3 and 7), dA/XYZ (lanes 4 and 8), J002 (lane 9), and dB (lane 10) strains. D, agarose gel electrophoresis (AGE) shows amplified fragments around the bciA locus using the above-mentioned sphaA-comf-F and -R primers. The PCR products from J001 and dXYZ strains (lanes 1 and 3, respectively) were 1.17 kbp, whereas the fragments from dA and dA/XYZ strains (lanes 2 and 4, respectively) were 2.50 kbp. E, the image for agarose gel electrophoresis shows amplified fragments around bchX, -Y, and -Z loci using the above-mentioned sphaXYZ-comf-F and -R primers. The PCR products from J001 and dA (lanes 5 and 6, respectively) were 4.55 kbp, whereas the fragments from dXYZ and dA/XYZ strains (lanes 7 and 8, respectively) were 2.61 kbp. F, the agarose gel electrophoresis image shows amplified fragments around the bciB locus using the above-mentioned palB-comf-F and -R primers. The PCR products from J002 and dB strains were 1.38 and 2.21 kbp, respectively. Lane M, molecular size marker (the sizes of bands are indicated at the left). Cloning of bchX, -Y, and -Z genes from C. tepidum (G) and R. castenholzii (H) is shown. Arrows in G and H represent the primers tepX-F (xv), tepX-R (xvi), tepY-F (xvii), tepY-R (xviii), tepZ-F (xix), tepZ-R (xx), castXYZ-F (xxi), castXYZ-R (xxii), cast-remove-F (xxiii), and cast-remove-R (xxiv).
For construction of the BciB-lacking mutant of R. palustris, the plasmid pJSC-palB-Km (Fig. 2C) was created as follows. A 1.23-kbp DNA fragment containing bciB was amplified from the genome of R. palustris by PCR using palB-F and palB-R primers. The PCR product was digested with HindIII and cloned into the SmaI and HindIII sites of pUC118, yielding pUC-palB. The neo gene was excised from pUCKM1 by digestion with SmaI. The pUC-palB plasmid was digested with SalI, blunted, and ligated with the neo gene fragment, creating the plasmid pUC-palB-Km. The pUC-palB-Km and pJSC plasmids were digested with SmaI and XbaI and then ligated together to create the pJSC-palB-Km plasmid.
For complementation experiments, the plasmid pZJ-sphaA carrying bciA of R. sphaeroides and the plasmid pZJ-palB containing bciB of R. palustris were constructed as follows. The open reading frame regions of bciA and bciB in pUC-sphaA and pUC-palB, respectively, were digested by NdeI and XbaI, excised, and subcloned into the same sites of the pZJ102 vector (35), giving pZJ-sphaA and pZJ-palB, respectively. The pZJ102 was a derivative of pBBR1MCS5 (36), which contained the R. capsulatus puc promoter for a high level of expression in Rhodobacter species, and the ATG codon in the unique NdeI site of this vector was positioned as the start codon of the pucB gene. To perform the complementation experiments of the bchXYZ genes from R. sphaeroides, R. palustris, C. tepidum, and R. castenholzii, the expression plasmids pZJ-sphaXYZ, pZJ-palXYZ, pZJ-tepXYZ, and pZJ-castXYZ were created as follows. The fragment containing bchXYZ of R. sphaeroides was excised from pUC-sphaXYZ plasmid with NdeI and XbaI and then ligated with the NdeI- and XbaI-digested pZJ102, yielding pZJ-sphaXYZ plasmid. A 4.06-kbp DNA fragment containing the bchXYZ genes was amplified from the genomic DNA of R. palustris by PCR using palXYZ-F and -R primer sets. The PCR product was digested by NdeI and XbaI and cloned into the same sites of pJZ102, creating plasmid pZJ-palXYZ. To construct the pZJ-tepXYZ, the PCRs were performed to amplify the bchX, bchY, and bchZ genes of the green sulfur bacterium C. tepidum using primer sets tepX-F and tepX-R, tepY-F and tepY-R, and tepZ-F and tepZ-R, respectively (Table 3). The three PCR products and the SmaI-digested pUC118 were mixed and treated with the In-Fusion cloning kit, and then the bchX, -Y, and -Z genes were subcloned into pUC118 in this order, giving pUC-tepXYZ (Fig. 2G). The plasmids pUC-tepXYZ and pZJ102 were digested by NdeI and XbaI and then ligated together to construct the pZJ-tepXYZ plasmid. To create the pZJ-castXYZ plasmid, a 7.06-kbp DNA fragment containing the bchXYZ and adjacent Rcas3745 genes of R. castenholzii (see Fig. 2H) was amplified from its genomic DNA by PCR using castXYZ-F and castXYZ-R primers. The PCR product was ligated with the SmaI-digested pUC118 vector by In-Fusion cloning, giving the plasmid pUC-castXYZ. To amplify the bchXYZ-coding region without Rcas3745, the plasmid pUC-castXYZ was used as the template for PCR with primers cast-remove-F and -R (Fig. 2H and Table 3). The resulting PCR fragment was phosphorylated and self-ligated, producing pUC-castXYZ2 (Fig. 2H). The DNA fragment containing the R. castenholzii bchXYZ genes was excised from the pUC-castXYZ2 with NdeI and XbaI and ligated into the same sites of pZJ102, creating the plasmid pZJ-castXYZ.
Construction of the Mutant Strains of R. sphaeroides and R. palustris
The pJSC-based plasmids, pJSC-sphaA-Km, pJSC-sphaXYZ-Sm, and pJSC-palB-Km, were transformed into the mobilizing E. coli strain S17-1 λ-pir (30). To construct the bciA mutant of R. sphaeroides, the J001 was used as a parent strain. The plasmid pJSC-sphaA-Km was transferred into the J001 strain by a conjugation method with the E. coli S17-1 strain (32). The kanamycin-resistant colonies grown in the presence of 5% sucrose were selected as double-crossover candidates, and the chromosomal insertion into bciA by the neo gene was confirmed by PCR using sphaA-comf-F and -R primers (Fig. 2D). A DNA molecular weight marker, the λ-EcoT14I digest (Takara), was used for molecular mass estimation of the PCR products. Furthermore, DNA sequences of the PCR fragments were determined using an ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems). The mutant strain was designated the dA strain. To create the mutants of R. sphaeroides deleting bchXYZ and both regions of bchXYZ and bciA, the J001 and dA strains, respectively, were conjugated with S17-1 having the plasmid pJSC-sphaXYZ-Sm. In the presence of 5% sucrose, the streptomycin-resistant transconjugants were picked as candidate mutants, and the mutated alleles in the genome were confirmed by PCR in the manner mentioned above (Fig. 2E). The confirmed mutants were named dXYZ and dA/XYZ strains, respectively.
For construction of the R. palustris bciB mutant, the pJSC-palB-Km was transferred into the J002 strain, and transconjugants on a PYS plate containing kanamycin and 5% sucrose were selected. The PCR confirmation of the mutated allele using primer set palB-comf-F and -R is shown in Fig. 2F. The bciB deletion mutant of R. palustris was called the dB strain.
Complementation Experiments
The pZJ102-based plasmids, pZJ-sphaA, pZJ-palB, pZJ-sphaXYZ, pZJ-palXYZ, pZJ-tepXYZ, and pZJ-castXYZ, in the E. coli strain S17-1 were transformed into R. sphaeroides mutant strain dA/XYZ or dXYZ by conjugation (32, 35), and complemented strains carrying each plasmid were selected on PYS plates containing gentamycin and rifampicin. The resulting recovered strains are listed in Table 1.
Determination of Pigment Compositions of R. sphaeroides and R. palustris Mutant Strains
Two extraction and two HPLC methods were applied. One was tailored for lipophilic pigments (BChl a) and the other for hydrophilic pigments (Chlide a). For the analysis of lipophilic pigments, a mixture (2 ml) of acetone and methanol (1:1, v/v) was added to harvested cells (about 0.25 g of wet cells) and mixed using a vibrator. A mixture (2 ml) of diethyl ether and petroleum ether (1:1, v/v) and then distilled water (4 ml) were added to transfer the pigment components to the ether layer. The ether phase was collected and evaporated to dryness under a stream of N2 gas, and the residue was dissolved in a mixture (50 μl) of methanol and acetone (3:1, v/v). Twenty μl of the mixed solution was then used for the following HPLC analysis. For the analysis of hydrophilic pigments, the above funnel separation procedures with ether were omitted. The extracts of a mixture of acetone and methanol were immediately evaporated to dryness under a stream of N2 gas, the residue was dissolved in methanol (50 μl), and then 20 μl of the mixture was used for pigment analysis.
HPLC was performed using a Shimadzu Prominence liquid chromatograph system comprising a CBM communications bus module, an SPD-M20A photodiode array detector, an LC-20AD pump, a DGU-20A3 degasser, and a CTO-20AC column oven (Shimadzu, Japan). Reverse-phase HPLC was performed under the following conditions: for the analysis of lipophilic pigments, column, Cosmosil 5C18-AR-II (3.0 × 150 mm, 5 μm; Nacalai Tesque Inc.); eluent, methanol/acetone/H2O (75:15:10, v/v/v); flow rate, 0.75 ml/min; range of detection wavelength by photodiode array, 350–800 nm; and for the analysis of hydrophilic pigments, column, Inertsil ODS-EP (3.0 × 150 mm, 5 μm; GL Sciences Inc.); eluent, methanol/aqueous 50 mm ammonium acetate (pH 5.25) (70:30, v/v); flow rate, 0.50 ml/min; range of detection wavelength by photodiode array, 350–800 nm. Liquid chromatography mass spectrometry (LCMS) was performed under the above HPLC conditions, using a Shimadzu LCMS-2010EV system comprising a quadrupole mass spectrometer equipped with an atmospheric pressure chemical ionization (APCI) for lipophilic pigments or an electrospray ionization (ESI) probe for hydrophilic pigments as described (22, 37). All solvents used for the analytical HPLC were of HPLC grade quality and were purchased from Nacalai Tesque Inc. and Wako Pure Chemical Industries Ltd.
Authentic Chlide a and 8V-Chlide a were prepared from a commercially available cyanobacterium Spirulina containing Chl a (Spirulina powder S1K, DIC Lifetec Co., Ltd.) and the Δslr1923 mutant of Synechocystis sp. PCC6803 containing 8V-Chl a (16), respectively, as described previously (22).
RESULTS
Analysis of Pigment Compositions in bciA and/or bchXYZ Deletion Mutants of R. sphaeroides
As mentioned in the Introduction, there is a possibility that an unknown third type of DVR exists in Rhodobacter species and that COR has the potential to act as DVR for BChl a biosynthesis. To clarify these speculations, we constructed bciA- and/or bchXYZ-deleted mutants of R. sphaeroides and identified the resulting pigments from cultured cells. The PCR analyses of genomic DNAs confirmed that the targeted mutations in the genes accordingly occurred (Fig. 2, D and E). The mutant lacking sole BciA, the dA strain, could grow under anaerobic light conditions as well as aerobic dark conditions. On the other hand, the single bchXYZ mutant and the double bciA/bchXYZ mutant, the dXYZ and dA/XYZ strains, respectively, did not show phototrophic growth and grew under aerobic/semiaerobic conditions in the dark. For the identification of lipophilic pigments (BChl a) in the mutant strains, APCI mass spectrometry (positive ion mode) was applied. For the identification of hydrophilic pigments (Chlide a and 8V-Chlide a) in the mutant strains, ESI mass spectrometry (positive ion mode) as well as authentic standards were used.
Fig. 3 shows results of LC(MS) analysis of pigments extracted from semiaerobically cultured cells of constructed mutant strains. In the dA strain, BChl a possessing a phytyl group was detected (Fig. 3A, trace ii), and its intermediates were not detected within the detection limit of HPLC (Fig. 3B, trace ii). The presence of phytylated BChl a in the mutant cells was confirmed in terms of the identical retention time to BChl a in the J001 strain (Fig. 3A, trace i) as well as APCI mass spectrometry as follows. Fig. 3C shows the mass spectra of the BChl a components found in the J001 (i) and dA (ii) strains. Both of the BChl a components gave the same molecular ion peak at m/z = 911.3, which was consistent with the calculated mass number of 911.6 for phytylated BChl a as the protonated form. One of the fragment ion peaks was observed at m/z = 633.1 produced by the loss of the esterifying moiety in the 17-propionate residue, indicating that the pigment had a C20 phytyl group: 911.3 − 633.1 = 278.2 = C20H39–H. Therefore, peak 1 was confirmed to be phytylated BChl a.
FIGURE 3.
LCMS measurements of extracted pigments from the mutant cells of R. sphaeroides and R. palustris. Analyses for BChls a (A) and Chlide a derivatives (B) were monitored at 770 and 435 nm (solid lines), respectively. The dotted lines in A show mass chromatograms at m/z = 911.3 ([MH]+) for phytylated BChl a. i, R. sphaeroides J001 (wild type); ii, dA; iii, R. palustris J002 (wild type); iv, dB; v, dXYZ; vi, dA/XYZ; vii, standard applying Chlide a and 8V-Chlide a. Peak 1, BChl a; peak 2, geranylgeranylated BChl a; peak 3, dihydrogeranylgeranylated BChl a; peak 4, tetrahydrogeranylgeranylated BChl a; peak 5, Chlide a; peak 6, 8V-Chlide a. Peaks *5-1 and *5-2, allomers of Chlide a; peaks *6-1 and *6-2, allomers of 8V-Chlide a. C, APCI mass analysis of BChl a detected in R. sphaeroides J001 (i) and dA cells (ii). D, ESI mass spectra of Chlide a and 8V-Chlide a derivatives observed in B. Peaks *5-1, *5-2, 5, *6-1, *6-2, and 6 gave molecular ion peaks at m/z = 631.0, 631.2, 615.2, 629.2, 629.2, and 613.1, respectively, as their protonated forms ([MH]+). The calculated mass numbers of 631.24, 615.25, 629.23, and 613.23 are for allomer of Chlide a (and its epimer), Chlide a, allomer of 8V-Chlide a (and its epimer), and 8V-Chlide a. i, peak *5-1 found in dXYZ, allomer of Chlide a; ii, peak *5-2 found in dXYZ, allomer of Chlide a; iii, peak 5 found in dXYZ, Chlide a; iv, peak *6-1 found in dA/XYZ, allomer of 8V-Chlide a; v, peak *6-2 found in dA/XYZ, allomer of 8V-Chlide a; vi, peak 6 found in dA/XYZ, 8V-Chlide a.
On the other hand, the dXYZ strain accumulated Chlide a (Fig. 3B, trace v) and no BChl a (Fig. 3A, trace v). The presence of Chlide a was determined using ESI-LCMS as well as the authentic standard. The Chlide a component found in the dXYZ strain (peak 5) showed a retention time identical to that of the authentic standard (at 16.5 min in traces v and vii of Fig. 3B). Further, the component exhibited its molecular ion peak at m/z = 615.2 as shown in trace iii of Fig. 3D, which was consistent with that for protonated Chlide a (calculated mass number of 615.3); peak 5 was identified to be Chlide a.
A mutant deleting the bchX, -Y, or -Z gene of Rhodobacter species was reported to accumulate both Chlide a and 3-(1-hydroxyethyl)-Chlide a (4, 38). The latter pigment was not detected in the dXYZ strain, where all of the bchX, -Y, and -Z genes were absent, but the other components absorbing 435-nm light were observed as minor components, denoted as *5-1 and *5-2 in trace v of Fig. 3B. The *5-1 and *5-2 components exhibited the molecular ion peaks at m/z = 631.0 and 631.2, respectively, in their ESI mass spectra as shown in Fig. 3D, i and ii. The differences between mass numbers from peak *5-1/2 components and Chlide a (equal to peak 5) were 15.8/16.0 Da, indicating the addition of an oxygen atom to Chlide a for *5-1/2. Further, in on-line absorption spectra, the *5-1/2 components showed absorption spectra identical to that of Chlide a, suggesting the presence of the 3-vinyl group in *5-1/2 components (data not shown). In general, allomerization of (B)Chl pigments non-stereoselectively occurred and gave a mixture of epimers at the 132-position (39, 40). The pair of diastereomers was easily separated on reverse-phase HPLC. Thus, the *5-1 and *5-2 components were assigned to a pair of allomerized Chlide a, (132R/S)-hydroxy adducts.
The mutant lacking the BciA and COR, the dA/XYZ strain, had no Chlide a nor BChl a but showed the accumulation of a Chlide-like component (traces vi of Fig. 3, A and B). This pigment (peak 6 in trace vi of Fig. 3B) was detected at the same retention time with the standard of 8V-Chlide a (at 19.0 min in traces vi and vii of Fig. 3B) and exhibited a red-shifted Soret absorption at 440 nm and almost the same Qy absorption at 667 nm in the eluent as authentic Chlide a (Soret/Qy = 431/666 nm). In general, Chl and BChl species having a C8 vinyl group showed their red-shifted Soret absorption maxima and almost no shift of the Qy absorption peaks, compared with those of the corresponding C8-ethylated analogs (12, 13, 24). Furthermore, the component showed its molecular ion peak at m/z = 613.1 (Fig. 3D, vi), which corresponded with that for protonated 8V-Chlide a (calculated mass number of 613.2), assigning the peak 6 in the dA/XYZ strain as 8V-Chlide a. Similarly to the Chlide a in the dXYZ strain, (132R/S)-hydroxy allomers of 8V-Chlide a were detected (peaks *6-1 and *6-2) and showed their molecular ion peaks at 629.2 (Fig. 3D, iv and v, respectively). These results clearly indicated that both BciA and COR proteins catalyzed the reduction of the C8 vinyl group, and at least one of the two was requisite for the reduction in R. sphaeroides cells. Further, an unknown third-type DVR does not exist in this bacterium.
Analysis of Pigment Compositions in the Mutant Lacking BciB of R. palustris
In addition to the observation of BciA and COR in R. sphaeroides, we studied the bciB gene, an ortholog of the cyano-type DVR (16). Although direct mutation of bciB had not been observed among anoxygenic photosynthetic bacteria, complementation experiments were previously performed using green sulfur bacteria for identification of the gene function (18). The bciB gene is not found in the genome of R. sphaeroides but is found in the genome of R. palustris, which has no bciA homolog. Therefore, we constructed a bciB-deleted mutant of R. palustris, dB strain, and analyzed the pigment composition from semiaerobically cultured cells. LC(MS) analysis showed that the mutant synthesized BChl a with a phytyl group as the dominant pigment (peak 1 in the solid line; also see selective mass chromatograms at m/z = 911.3 in the dotted line of Fig. 3A, trace iv) and accumulated no Chlide a species (Fig. 3B, trace iv) as the wild type J002 strain did (Fig. 3, A and B, trace iii). In addition to the dominant phytylated BChl a, other pigments showing BChl a-like absorption were detected. These peaks were assigned to BChls a carrying a geranylgeranyl (peak 2), dihydrogeranylgeranyl (peak 3), or tetrahydrogeranylgeranyl group (peak 4) in the 17-propionate residues in the order of elution. The assignment of these three BChl a components was established in our previous studies (26, 27). The function of BciB could not be clarified using the dB strain, as in the situation of BciA using the dA strain.
Introduction of R. sphaeroides bciA and R. palustris bciB into dA/XYZ Mutant Cells
The mutational analysis of bciB in R. palustris indicates that the dB strain showed no phenotype, similar to the dA strain of R. sphaeroides. To address whether BciB of R. palustris has DVR activity or not, the bciB gene was introduced into the dA/XYZ mutant strain of R. sphaeroides (dA/XYZ_palB strain). The bciA gene of R. sphaeroides was also introduced into the dA/XYZ strain (dA/XYZ_sphaA), and pigment compositions of the complemented strains were similarly analyzed by ESI-LCMS as described. Fig. 4 summarizes the results. Briefly, the selective mass chromatograms at m/z = 615.1 (solid lines in Fig. 4B) appeared to show that these strains accumulated Chlide a, not 8V-Chlide a. The results indicate that the two DVRs work for the 8-vinyl reduction in BChl a biosynthesis in the R. sphaeroides cells. Moreover, it was suggested that the COR in R. palustris cells could also catalyze the reduction of the C8 vinyl group.
FIGURE 4.
HPLC profiles detected at 435-nm absorbance (A) and selective mass chromatograms at m/z = 615.1 ([MH]+) for Chlide a (solid lines) and at m/z = 613.1 ([MH]+) for 8V-Chlide a (dashed lines) (B) of extracted pigments from the complementary strains of dA/XYZ mutant with bciA of R. sphaeroides and bciB of R. palustris. i, dA/XYZ_102 (empty vector control); ii, dA/XYZ_sphaA; iii, dA/XYZ_palB; iv, standards for Chlide a and 8V-Chlide a. Peak 5, Chlide a; peak 6, 8V-Chlide a. Peaks *5-1 and *5-2, allomers of Chlide a; peaks *6-1 and *6-2, allomers of 8V-Chlide a. The detected peaks *5-1, *5-2, 5, *6-1, *6-2, and 6 in these strains showed the same mass spectra as i, ii, iii, iv, v, and vi in Fig. 3D, respectively.
Complementation of the dA/XYZ Mutant with bchXYZ of Various Bacteria
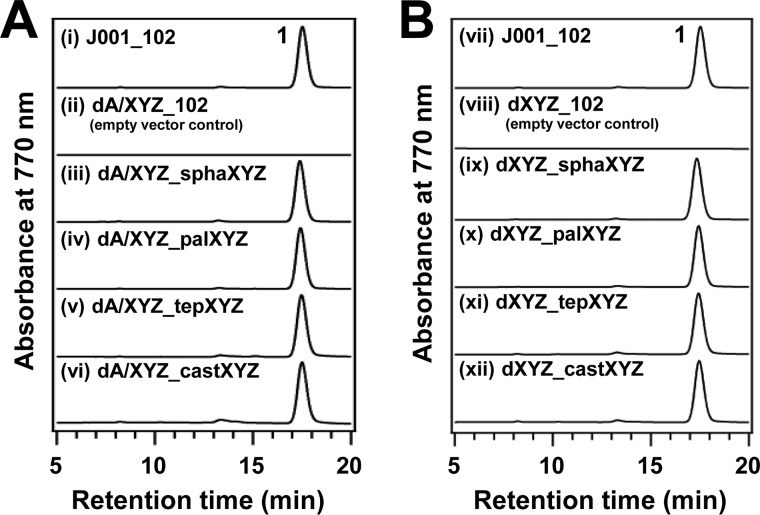
In BChl a biosynthesis of R. sphaeroides, COR plays a comparable role with BciA and then catalyzes the double reduction from 8V-Chlide a to 3-vinyl-bacteriochlorophyllide a (3V-BChlide a) via Chlide a in vivo (see Fig. 1): hydrogenation of C8-CH=CH2 to C8-CH2–CH3 and C7=C8 to C7H–C8H. Is this dual function of COR general in any anoxygenic photosynthetic bacteria producing BChl a? To answer the question, we performed complementation of the dA/XYZ strain with the bchXYZ genes from various phylogenetic groups of anoxygenic photosynthetic bacteria. We analyzed the pigment compositions extracted from the bchXYZ-introduced mutants, dA/XYZ_sphaXYZ, dA/XYZ_palXYZ, dA/XYZ_tepXYZ, and dA/XYZ_castXYZ, having bchXYZ derived from R. sphaeroides, R. palustris, C. tepidum, and R. castenholzii, respectively (Fig. 5A). BChl a with a phytyl group was detected in these strains as well as the wild type of R. sphaeroides (Fig. 5A, traces i and iii–vi). These strains were able to complement BChl a synthesis ability. No BChl a was detected in the control strain, dA/XYZ_102, grown under semiaerobic dark conditions (Fig. 5A, trace ii). These results indicated that all of the BchXYZ proteins examined here possessed 8-vinyl reduction activity together with COR activity and could convert from 8V-Chlide a to 3-vinyl-bacteriochlorophyllide a via Chlide a. Indeed, these strains recovered their photosynthetic ability and could grow under anaerobic light conditions. We also performed complementation experiments of the dXYZ strains introduced with bchXYZ of the various above-mentioned species and confirmed their original COR activities (Fig. 5B). The dual function of COR is general and conserved among purple bacteria, green sulfur bacteria, and filamentous anoxygenic phototrophs.
FIGURE 5.
HPLC elution profiles of extracted pigments from the complementary strains of dA/XYZ (A) and dXYZ (B) mutants with bchXYZ genes of various species. i and vii, R. sphaeroides J001 with pZJ102 plasmid; ii, dA/XYZ_102 (empty vector control); iii, dA/XYZ_sphaXYZ; iv, dA/XYZ_palXYZ; v, dA/XYZ_tepXYZ; vi, dA/XYZ_castXYZ; viii, dXYZ_102 (empty vector control); ix, dXYZ_sphaXYZ; x, dXYZ_palXYZ; xi, dXYZ_tepXYZ; xii, dXYZ_castXYZ. The elution of pigments was monitored at 770 nm. Peak 1, BChl a.
DISCUSSION
In this study, we observed the function of COR as DVR, using the mutants of R. sphaeroides. In BChl a biosynthesis, BciA and BciB are considered to be authentic DVRs using 8V-Chlide a or 8V-PChlide a as the substrate. Even if the BciA and BciB enzymes are absent in cells, COR acts as DVR and proceeds to further biosynthesis of BChl a. In the complementation experiments of dA/XYZ strain with bchXYZ genes of various species, the 8-vinyl reduction ability of COR was revealed to be conserved in R. sphaeroides, R. palustris, C. tepidum, and R. castenholzii. The results clearly indicated that this function of COR is general among purple bacteria, green sulfur bacteria, and filamentous anoxygenic phototrophs. Especially in R. castenholzii and Roseiflexus sp. RS-1, whose genomes carry neither bciA nor bciB gene (16), the DVR activity of their COR is inevitably requisite, and not a subfunction, to synthesize BChl a. From the results of the pigment analysis of the dA/XYZ strain of R. sphaeroides, we demonstrated that an “unknown” third-type DVR does not exist in this bacterium. We then conclude that COR is actually the third DVR, assigned following BciA and BciB.
In purple bacterial genomes, the bchXYZ genes are conserved within the photosynthetic gene cluster, which includes genes for reaction centers, light-harvesting complexes, and BChl and carotenoid biosynthesis. It is known that a set of genes in the photosynthetic gene cluster has been conserved from ancestral organisms of purple bacteria by horizontal transfer (9, 10). Interestingly, the bciA/bciB gene is not included in the gene cluster, which is the only case among genes for synthesizing photosynthetic apparatus in purple bacteria. The findings in this study indicate that the photosynthetic gene cluster without bciA and bciB is enough to produce BChl a. When the gene cluster is subjected to the horizontal gene transfer, it could give the ability of BChl a biosynthesis to a recipient organism. These pieces of evidence suggest that BciA and BciB were missing in ancestral anoxygenic photosynthetic bacteria and that COR could be the evolutionarily oldest type of DVR.
In that case, why are BciA and/or BciB present in almost all of the isolated phototrophs producing BChl a? The only exception lacking both the bciA and bciB genes is Roseiflexus species. We propose the following three possible reasons to answer this question. First, BciA/BciB is advantageous in terms of suppression of the energy usage for the 8-vinyl reduction. BciA and BciB require NADPH (E′0 = −0.32 V) and ferredoxin (E′0 = −0.43 V) as electron donors, respectively, for their enzymatic reactions (20). In contrast, COR needs ATP in addition to ferredoxin as a considerable electron donor in vivo for its enzymatic activity (7) and thus requires greater energy than BciA and BciB do. The second reason is the catalytic activity rates for the 8-vinyl reduction by COR and BciA/BciB. Although the efficiency of COR as DVR is still unknown, COR seems to have lower activities for the 8-vinyl reduction than BciA and BciB, because the dA and dB strains showed slightly slower growth than wild type strains under anaerobic low light conditions (data not shown). COR together with BciA and/or BciB would accelerate the whole BChl a biosynthesis. Third, COR and BciA/BciB have different roles in adapting to environmental changes, especially oxygen concentrations. COR carries out its enzymatic reduction under anaerobic conditions but shows no activity in the presence of dioxygen (7). In contrast, BciA and BciB are active even under aerobic conditions (12, 19). Most purple bacteria still synthesize BChl a under semiaerobic and dark conditions. BciA and BciB showed higher enzymatic activities than COR under these conditions.
Previously we demonstrated that CORs from B. viridis and H. modesticaldum utilize 8V-Chlide a (not Chlide a) as the substrate for biosyntheses of BChl b and BChl g, respectively (22, 23). In these cases, BciA and BciB are not required, and in fact these genes are absent in the genomes of the two bacteria (21).5 CORs from these species have probably been modified to synthesize bacteriochlorophyllide g (precursor for BChls b and g) from 8V-Chlide a in the course of evolution.
Green sulfur bacteria produce not only BChl a but also Δ2,6-phytadienylated Chl a and BChl c, d, or e (depending on species). Although BciA and BciB are not indispensable for BChl a biosynthesis, as shown in this study, the two enzymes are required for biosynthesis of Δ2,6-phytadienylated Chl a and BChl c/d/e. In our previous study, the bciA mutant of C. tepidum synthesized normal BChl a but accumulated 8-vinyl-Δ2,6-phytadienylated Chl a and 8V-BChl c (26). This suggests that COR would not release Chlide a after the 8-vinyl reduction of 8V-Chlide a and then would immediately proceed to the C7=C8 double bond reduction for further BChl a biosynthesis. Therefore, enzymes for biosynthesis of Chl a and BChl c could not interact with Chlide a and only react with 8V-Chlide a in the bciA mutant.
It was reported that the mutant of A. thaliana lacking plant-type (BciA-type) DVR accumulated 8V-Chls a and b and showed slow growth rates under high light conditions (13). The mutant of Synechocystis sp. PCC6803 lacking cyano-type (BciB-type) DVR accumulating 8V-Chl a was also reported to be sensitive to oxygen molecules and to grow slowly under high light conditions (16). Plant- and cyano-type DVRs are likely to be important for protection against oxygen stress even in oxygenic phototrophs. Because Prochlorococcus species of marine cyanobacteria utilize 8V-Chl a for the authentic photosynthetic pigment, their photosystems are useful to evaluate how to adapt to oxygen stress conditions with 8V-Chl a.
Before increasing oxygen concentrations on early earth, ancestral anoxygenic photosynthetic bacteria probably synthesized BChl a without bciA and bciB. Given that a loss-of-function mutation had occurred in any of bchX, -Y, or -Z, the resultant pigment accumulated in mutated bacteria is probably 8V-Chlide a, not Chlide a. Therefore, 8V-Chl a could be produced as an ancestral chlorophyllous pigment in the last universal ancestors of oxygenic phototrophs. Elevation of oxygen concentrations in early earth after the appearance of ancestral oxygenic phototrophs using 8V-Chl a might relate to the acquisition of oxygen-tolerant BciA/BciB as DVR for Chl a biosynthesis.
Acknowledgment
We thank Prof. Masato Noguchi (Kurume University School of Medicine) for useful discussions.
This work was supported in part by Grant-in-Aid for Scientific Research (A) 22245030 (to H. T.), Grant-in-Aid for Scientific Research (C) 24590366 (to T.M.), Grant-in-Aid for Scientific Research for Young Scientists (B) 24750169 (to J. H.), and Grant-in-Aid for Scientific Research on Innovative Areas “Artificial Photosynthesis (AnApple)” 24107002 (to H. T.) from the Japan Society for the Promotion of Science.
Y. Tsukatani, J. Harada, and H. Tamiaki, manuscript in preparation.
- Chl
- chlorophyll
- Chlide a
- chlorophyllide a
- APCI
- atmospheric pressure chemical ionization
- BChl
- bacteriochlorophyll
- COR
- chlorophyllide a oxidoreductase
- DVR
- divinyl reductase
- ESI
- electrospray ionization
- LCMS
- liquid chromatography mass spectrometry
- 8V-Chl
- 8-vinyl-chlorophyll
- 8V-Chlide a
- 8-vinyl-chlorophyllide a
- 8V-PChlide a
- 8-vinyl-protochlorophyllide a.
REFERENCES
- 1. Tanaka R., Tanaka A. (2007) Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346 [DOI] [PubMed] [Google Scholar]
- 2. Miyashita H., Ikemoto H., Kurano N., Adachi K., Chihara M., Miyachi S. (1996) Chlorophyll d as a major pigment. Nature 10.1038/383402a0 [DOI] [Google Scholar]
- 3. Chen M., Schliep M., Willows R. D., Cai Z.-L., Neilan B. A., Scheer H. (2010) A red-shifted chlorophyll. Science 329, 1318–1319 [DOI] [PubMed] [Google Scholar]
- 4. Bollivar D. W., Suzuki J. Y., Beatty J. T., Dobrowolski J. M., Bauer C. E. (1994) Directed mutational analysis of bacteriochlorophyll a biosynthesis in Rhodobacter capsulatus. J. Mol. Biol. 237, 622–640 [DOI] [PubMed] [Google Scholar]
- 5. Chew A. G., Bryant D. A. (2007) Chlorophyll biosynthesis in bacteria: the origins of structural and functional diversity. Annu. Rev. Microbiol. 61, 113–129 [DOI] [PubMed] [Google Scholar]
- 6. Nomata J., Swem L. R., Bauer C. E., Fujita Y. (2005) Overexpression and characterization of dark-operative protochlorophyllide reductase from Rhodobacter capsulatus. Biochim. Biophys. Acta 1708, 229–237 [DOI] [PubMed] [Google Scholar]
- 7. Nomata J., Mizoguchi T., Tamiaki H., Fujita Y. (2006) A second nitrogenase-like enzyme for bacteriochlorophyll biosynthesis: reconstitution of chlorophyllide a reductase with purified X-protein (BchX) and YZ-protein (BchY-BchZ) from Rhodobacter capsulatus. J. Biol. Chem. 281, 15021–15028 [DOI] [PubMed] [Google Scholar]
- 8. Sawicki A., Willows R. D. (2010) BchJ and BchM interact in a 1:1 ratio with the magnesium chelatase BchH subunit of Rhodobacter capsulatus. FEBS J. 277, 4709–4721 [DOI] [PubMed] [Google Scholar]
- 9. Swingley W. D., Blankenship R. E., Raymond J. (2009) in The purple photosynthetic bacteria (Hunter C. N., Daldal F., Thurnauer M. C., Beatty J. T., eds) pp. 17–29, Springer, Dordrecht, The Netherlands [Google Scholar]
- 10. Nagashima S., Nagashima K. V. (2013) in Advances in Botanical Research, Genome Evolution of Photosynthetic Bacteria (Beatty J. T., ed) pp. 152–174, Academic Press, Inc., Waltham, MA [Google Scholar]
- 11. Suzuki J. Y., Bauer C. E. (1995) Altered monovinyl and divinyl protochlorophyllide pools in bchJ mutants of Rhodobacter capsulatus: possible monovinyl substrate discrimination of light-independent protochlorophyllide reductase. J. Biol. Chem. 270, 3732–3740 [PubMed] [Google Scholar]
- 12. Chew A. G., Bryant D. A. (2007) Characterization of a plant-like protochlorophyllide a divinyl reductase in green sulfur bacteria. J. Biol. Chem. 282, 2967–2975 [DOI] [PubMed] [Google Scholar]
- 13. Nagata N., Tanaka R., Satoh S., Tanaka A. (2005) Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 17, 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakanishi H., Nozue H., Suzuki K., Kaneko Y., Taguchi G., Hayashida N. (2005) Characterization of the Arabidopsis thaliana mutant pcb2 which accumulates divinyl chlorophylls. Plant Cell Physiol. 46, 467–473 [DOI] [PubMed] [Google Scholar]
- 15. Nagata N., Tanaka R., Tanaka A. (2007) The major route for chlorophyll synthesis includes [3,8-divinyl]-chlorophyllide a reduction in Arabidopsis thaliana. Plant Cell Physiol. 48, 1803–1808 [DOI] [PubMed] [Google Scholar]
- 16. Ito H., Yokono M., Tanaka R., Tanaka A. (2008) Identification of a novel vinyl reductase gene essential for the biosynthesis of monovinyl chlorophyll in Synechocystis sp. PCC6803. J. Biol. Chem. 283, 9002–9011 [DOI] [PubMed] [Google Scholar]
- 17. Islam M. R., Aikawa S., Midorikawa T., Kashino Y., Satoh K., Koike H. (2008) slr1923 of Synechocystis sp. PCC6803 is essential for conversion of 3,8-divinyl(proto)chlorophyll(ide) to 3-monovinyl(proto)chlorophyll(ide). Plant Physiol. 148, 1068–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu Z., Bryant D. A. (2011) Multiple types of 8-vinyl reductases for (bacterio)chlorophyll biosynthesis occur in many green sulfur bacteria. J. Bacteriol. 193, 4996–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saunders A. H., Golbeck J. H., Bryant D. A. (2013) Characterization of BciB: a ferredoxin-dependent 8-vinyl-protochlorophyllide reductase from the green sulfur bacterium Chloroherpeton thalassium. Biochemistry 52, 8442–8451 [DOI] [PubMed] [Google Scholar]
- 20. Ito H., Tanaka A. (2014) Evolution of a new chlorophyll metabolic pathway driven by the dynamic changes in enzyme promiscuous activity. Plant Cell Physiol. 55, 593–603 [DOI] [PubMed] [Google Scholar]
- 21. Sattley W. M., Madigan M. T., Swingley W. D., Cheung P. C., Clocksin K. M., Conrad A. L., Dejesa L. C., Honchak B. M., Jung D. O., Karbach L. E., Kurdoglu A., Lahiri S., Mastrian S. D., Page L. E., Taylor H. L., Wang Z. T., Raymond J., Chen M., Blankenship R. E., Touchman J. W. (2008) The genome of Heliobacterium modesticaldum, a phototrophic representative of the Firmicutes containing the simplest photosynthetic apparatus. J. Bacteriol. 190, 4687–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsukatani Y., Yamamoto H., Harada J., Yoshitomi T., Nomata J., Kasahara M., Mizoguchi T., Fujita Y., Tamiaki H. (2013) An unexpectedly branched biosynthetic pathway for bacteriochlorophyll b capable of absorbing near-infrared light. Sci. Rep. 3, 1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsukatani Y., Yamamoto H., Mizoguchi T., Fujita Y., Tamiaki H. (2013) Completion of biosynthetic pathways for bacteriochlorophyll g in Heliobacterium modesticaldum: the C8-ethylidene group formation. Biochim. Biophys. Acta 1827, 1200–1204 [DOI] [PubMed] [Google Scholar]
- 24. Mizoguchi T., Harada J., Tamiaki H. (2012) Characterization of chlorophyll pigments in the mutant lacking 8-vinyl reductase of green photosynthetic bacterium Chlorobaculum tepidum. Bioorg. Med. Chem. 20, 6803–6810 [DOI] [PubMed] [Google Scholar]
- 25. Canniffe D. P., Jackson P. J., Hollingshead S., Dickman M. J., Hunter C. N. (2013) Identification of an 8-vinyl reductase involved in bacteriochlorophyll biosynthesis in Rhodobacter sphaeroides and evidence for the existence of a third distinct class of the enzyme. Biochem. J. 450, 397–405 [DOI] [PubMed] [Google Scholar]
- 26. Mizoguchi T., Harada J., Tamiaki H. (2006) Structural determination of dihydro- and tetrahydrogeranylgeranyl groups at the 17-propionate of bacteriochlorophylls-a. FEBS Lett. 580, 6644–6648 [DOI] [PubMed] [Google Scholar]
- 27. Harada J., Mizoguchi T., Yoshida S., Isaji M., Oh-Oka H., Tamiaki H. (2008) Composition and localization of bacteriochlorophyll a intermediates in the purple photosynthetic bacterium Rhodopseudomonas sp. Rits. Photosynth. Res. 95, 213–221 [DOI] [PubMed] [Google Scholar]
- 28. Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]
- 29. Penfold R. J., Pemberton J. M. (1992) An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118, 145–146 [DOI] [PubMed] [Google Scholar]
- 30. de Lorenzo V., Herrero M., Jakubzik U., Timmis K. N. (1990) Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172, 6568–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saeki K., Suetsugu Y., Tokuda K., Miyatake Y., Young D. A., Marrs B. L., Matsubara H. (1991) Genetic analysis of functional differences among distinct ferredoxins in Rhodobacter capsulatus. J. Biol. Chem. 266, 12889–12895 [PubMed] [Google Scholar]
- 32. Masuda S., Tsukatani Y., Kimura Y., Nagashima K. V., Shimada K., Matsuura K. (2002) Mutational analyses of the photosynthetic reaction center-bound triheme cytochrome subunit and cytochrome c2 in the purple bacterium Rhodovulum sulfidophilum. Biochemistry 41, 11211–11217 [DOI] [PubMed] [Google Scholar]
- 33. Gay P., Le Coq D., Steinmetz M., Berkelman T., Kado C. I. (1985) Positive selection procedure for entrapment of insertion sequence elements in Gram-negative bacteria. J. Bacteriol. 164, 918–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prentki P., Krisch H. M. (1984) In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29, 303–313 [DOI] [PubMed] [Google Scholar]
- 35. Harada J., Miyago S., Mizoguchi T., Azai C., Inoue K., Tamiaki H., Oh-oka H. (2008) Accumulation of chlorophyllous pigments esterified with the geranylgeranyl group and photosynthetic competence in the CT2256-deleted mutant of the green sulfur bacterium Chlorobium tepidum. Photochem. Photobiol. Sci. 7, 1179–1187 [DOI] [PubMed] [Google Scholar]
- 36. Kovach M. E., Elzer P. H., Hill D. S., Robertson G. T., Farris M. A., Roop R. M., 2nd, Peterson K. M. (1995) Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166, 175–176 [DOI] [PubMed] [Google Scholar]
- 37. Harada J., Mizoguchi T., Satoh S., Tsukatani Y., Yokono M., Noguchi M., Tanaka A., Tamiaki H. (2013) Specific gene bciD for C7-methyl oxidation in bacteriochlorophyll e biosynthesis of brown-colored green sulfur bacteria. PLoS One 8, e60026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hunter C. N., Coomber S. A. (1988) Cloning and oxygen-regulated expression of the bacteriochlorophyll biosynthesis genes bch E, B, A and C of Rhodobacter sphaeroides. J. Gen. Microbiol. 10.1099/00221287-134-6-1491 [DOI] [Google Scholar]
- 39. Walker J. S., Jie C., Keely B. J. (2003) Identification of diastereomeric chlorophyll allomers by atmospheric pressure chemical ionisation liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 17, 1125–1131 [DOI] [PubMed] [Google Scholar]
- 40. Müller A. H., Gough S. P., Bollivar D. W., Meldal M., Willows R. D., Hansson M. (2011) Methods for the preparation of chlorophyllide a: an intermediate of the chlorophyll biosynthetic pathway. Anal. Biochem. 419, 271–276 [DOI] [PubMed] [Google Scholar]