Abstract
Scavenger receptors represent an important class of pattern recognition receptors shown to mediate both beneficial and detrimental roles in host defense against microbial pathogens. The role of the major macrophage scavenger receptor, scavenger receptor A (SRA), in the immune response against the pathogenic fungus, Cryptococcus neoformans, is unknown. To evaluate the role of SRA in anticryptococcal host defenses, SRA+/+ mice and SRA−/− mice were infected intratracheally with C. neoformans. Results show that infection of SRA−/− mice resulted in a reduction in the pulmonary fungal burden at the efferent phase (3 wk) compared with SRA+/+ mice. Improved fungal clearance in SRA−/− mice was associated with decreased accumulation of eosinophils and greater accumulation of CD4+ T cells and CD11b+ dendritic cells. Additional parameters were consistent with enhanced anti-cryptococcal immunity in the infected SRA−/− mice: 1) increased expression of the costimulatory molecules CD80 and CD86 by lung APCs, 2) decreased expression of Th2 cytokines (IL-4 and IL-13) and IL-10 in lung leukocytes and in cryptococcal Ag-pulsed splenocytes, 3) diminished IgE production in sera, and 4) increased hallmarks of classical pulmonary macrophage activation. These effects were preceded by increased expression of early pro-Th1 genes in pulmonary lymph nodes at the afferent phase (1 wk). Collectively, our data show that SRA can be exploited by C. neoformans to interfere with the early events of the afferent responses that support Th1 immune polarization. This results in amplification of Th2 arm of the immune response and subsequently impaired adaptive control of C. neoformans in the infected lungs.
Cryptococcus neoformans is a fungal pathogen that commonly causes infection in HIV-positive individuals and in other immunosuppressed patients, such as those receiving cancer chemotherapy or transplant-related immunoconditioning and patients with hematologic malignancies (1, 2). Globally, C. neoformans causes an estimated 1 million cases of cryptococcosis per year in AIDS patients, leading to >600,000 deaths (3). Recent reports show that C. neoformans increasingly infects the immunocompetent individuals, including evidence that C. neoformans causes long-term latent infections in noncompromised individuals (4). These data suggest that C. neoformans can evade and/ or modulate the host defenses to promote its persistence even in immunocompetent hosts.
Studies in mouse models show that altering Th-polarization bias and macrophage activation status are strategies by which C. neoformans promotes its persistence. Strong Th1 polarization and the development of Th1 cell–mediated immunity correlates with optimal clearance rate, whereas shifting of the response characteristics toward Th2 and away from Th1 response components is associated with a worsening of clearance outcomes (5–9). Less is known about the role of Th17 cells in anticryptococcal defenses, but IL-17 appears to contribute to immune protection against C. neoformans, at least in some stages of the clearance process (9–12). Together, multiple studies demonstrate that the effective elimination of C. neoformans relies on the development and sustenance of a strong Th1 or mixed Th1/Th17 response.
The development of the protective adaptive immune response also requires an early and robust innate immune response. The balance between Th-polarizing cytokine networks induced by innate mechanisms directs the development of the adaptive immune response toward either Th1, Th2, or Th17 (13, 14). This early cytokine environment is triggered by sensing of cryptococcal molecular patterns by pathogen recognition receptors such as TLRs, mannose receptors, and collectin receptors (15, 16). A recent study documented that scavenger receptors like SCARF1 and CD36 play a role in the innate sensing of C. neoformans (17). This study showed that CD36 mediated macrophage activation was linked to the development of the protective immune response against cryptococcal infection in mice (17). However, the role of another major macrophage scavenger receptor, scavenger receptor A (SRA), in anticryptococcal host defense has not been studied in vivo, and the effect of scavenger receptors on the adaptive immune response is unknown.
SRA, also designated as CD204, is a pattern recognition receptor capable of binding to a broad range of ligands, including modified low-density lipoprotein, bacterial surface components, and heat shock proteins (18). SRA can play a beneficial or detrimental role in clearance of various pathogens. In infections with Listeria monocytogenes, Neisseria meningitides, Streptococcus pneumoniae, and HSV, SRA promotes resistance to the infection (19–22). In contrast, SRA was reported to be detrimental in Mycobacterium tuberculosis and Pneumocystis carinii infection models (23, 24), demonstrating that some pathogens can exploit SRA to promote their survival in the host. Early in vitro experiments have not detected SRA-mediated direct activation of macrophages with C. neoformans infection (17); however, these studies did not evaluate the effects of SRA on C. neoformans clearance in vivo. C. neoformans expresses numerous heat shock proteins, many as immunodominant proteins, which are known to bind to SRA. Thus, we sought to evaluate the role of SRA during the cryptococcal infection in vivo.
Our results show that SRA signaling impairs cryptococcal clearance during the efferent, rather than afferent, phase of the immune response. Specifically, SRA signaling interferes with the development of the protective Th1 immune response and the classical activation of effector macrophages. SRA-mediated skewing of the adaptive immune response in the lungs away from protective Th1 immune response and toward nonprotective and deleterious Th2 response was preceded by early change in the polarization patterns within pulmonary lymph nodes. Our findings show that C. neoformans can exploit SRA to induce nonprotective immune deviation, which promotes its persistence.
Materials and Methods
Mice
Female wild type 129/SVJ mice were obtained from Jackson Laboratories (Bar Harbor, ME). SRA−/− mice on a 129/SVJ background, originally obtained as the generous gift of Dr. Willem J. S. de Villiers (University of Kentucky Medical Center, Lexington, KY), were bred at the University of Michigan/Ann Arbor Veterans Administration Medical Center using microisolator cages covered with a filter top, with food/water provided ad libitum. Mice were aged to 8–10 wk at the time of infection. At the time of data collection, mice were humanely euthanized by CO2 inhalation. All experiments were approved by the University Committee on the Use and Care of Animals and the Veterans Administration Institutional Animal Care and Use Committee.
C. neoformans
C. neoformans strain H99 (ATCC 208821) and lac1Δ mutant (H99 with a targeted LAC1 gene deletion, as described by Dr. J. Andrew Alspaugh from Duke University, Durham, NC) (25) was recovered from 10% glycerol frozen stocks stored at −80°C and grown to stationary phase at 37°C in Sabouraud dextrose broth (1% Neopeptone, 2% dextrose; Difco, Detroit, MI) on a shaker. The cultures were then washed in nonpyrogenic saline (Travenol, Deerfield, IL), counted on a hemocytometer, and diluted to 3.3 × 105 yeast cells/ml in sterile nonpyrogenic saline.
Intratracheal inoculation of C. neoformans
Mice were anesthetized via i.p. injection of ketamine/xylazine (100/6.8 mg/kg body weight) and were restrained on a foam plate. A small incision was made through the skin covering the trachea. The underlying salivary glands and muscles were separated. Infection was performed by intratracheal injection of 30 μl (104 CFU) via 30-gauge needle actuated from a 1-ml tuberculin syringe with C. neoformans suspension (3.3 × 105/ml). After inoculation, the skin was closed with cyanoacrylate adhesive, and the mice were monitored during recovery from the anesthesia.
Lung CFU assay
For determination of microbial burden in the lungs, small aliquots of dispersed lungs were collected following the digestion procedure. Series of 10-fold dilutions of the lung samples were plated on Sabouraud dextrose agar plates in duplicates in 10-μl aliquots and incubated at room temperature. C. neoformans colonies were counted 2 d later, and the number of CFUs was calculated on a per-organ basis.
Bronchoalveolar lavage
Euthanized mice were lavaged after cannulation of the trachea with poly-ethylene tubing (PE50), which was attached to a 23-gauge needle on a tuberculin syringe. The lungs were lavaged twice with 1 ml PBS containing 5 × 10−5 M 2-ME (Sigma, St. Louis, MO) and protein inhibitor mixture (Roche Molecular Biochemicals, Indianapolis, IN). The recovered fluid (1.8–1.9 ml total) was spun at 500 × g for 10 min, and the supernatant was removed and analyzed for cytokines by ELISA using DuoSet kits (R&D Systems, Minneapolis, MN) following the manufacturer’s specifications. All plates were read on a Versamax plate reader (Molecular Devices, Sunnyvale, CA).
Lung leukocyte isolation
The lungs from each mouse were excised, washed in RPMI 1640, minced with scissors, and digested enzymatically at 37°C for 30 min in 5 ml/mouse digestion buffer (RPMI 1640, 5% FBS, penicillin and streptomycin [Invitrogen, Grand Island, NY]; 1 mg/ml collagenase A [Roche Diagnostics, Indianapolis, IN]; and 30 μg/ml DNase [Sigma]) and processed as previously described (26, 27). The cell suspension and tissue fragments were further dispersed by repeated aspiration through the bore of a 10-ml syringe and were centrifuged. Erythrocytes in the cell pellets were lysed by addition of 3 ml NH4Cl buffer (0.829% NH4Cl, 0.1% KHCO3, and 0.0372% Na2EDTA, pH 7.4) for 3 min followed by a 10-fold excess of RPMI 1640. Cells were resuspended and a second cycle of syringe dispersion and filtration through a sterile 100-μm nylon screen (Nitex, Kansas City, MO) was performed. The filtrate was centrifuged for 25 min at 1500 × g in the presence of 20% Percoll (Sigma) to separate leukocytes from cell debris and epithelial cells. Leukocyte pellets were resuspended in 5 ml complete RPMI 1640 media and enumerated on a hemocytometer after dilution in trypan blue (Sigma).
Lung-associated lymph node leukocyte isolation
Individual lung-associated lymph nodes (LALNs) were excised. To collect LALN leukocytes, we dispersed nodes using a 3-ml sterile syringe plunger and flushed them through a 70-μm cell strainer (BD Falcon, Bedford, MA) with complete media into a sterile tube, as described previously. After being centrifuged at 5000 rpm/min for 10 min, the supernatant was removed and the cell pellets were saved at −70°C for gene expression analysis by quantitative real-time RT-PCR (qRT-PCR).
Pulmonary cytokine production
Isolated lung leukocytes were diluted to 5 × 106 cells/ml and were cultured in 24-well plates with 2 ml complete RPMI 1640 medium at 37°C and 5% CO2 for 24 h. Supernatants were separated from cells by centrifugation, collected, and frozen until tested. The cytokines were quantified by ELISA using DuoSet kits (R&D Systems, Minneapolis, MN) following the manufacturer’s specifications. All plates were read on a Versamax plate reader (Molecular Devices, Sunnyvale, CA).
Ag-specific cytokine production by splenocytes
Spleens were excised and dispersed using a 3-ml sterile syringe plunger and flushed through a 70-μm cell strainer (BD Falcon) with complete media. Isolated spleen cells were diluted to 5 × 106 cells/ml and were cultured in media alone or with heat-killed C. neoformans (lac1Δ) in a ratio of 1:2 in 24-well plates with 2 ml complete RPMI 1640 medium at 37°C and 5% CO2 for 48 h. Supernatants were stored and analyzed for cytokine levels as described earlier. The Ag-specific cytokine production was calculated as a net gain of cytokine level compared with unstimulated controls of the same sample.
Total serum IgE
Serum was obtained from the blood samples collected by severing the vena cava of the mice before lung excision. Blood samples were then allowed to clot and were spun to separate serum. Serum samples were diluted 100-fold and assayed for total IgE levels using a mouse IgE sandwich ELISA kit (BioLegend, San Diego, CA) following the manufacturer’s specifications. All plates were read on a Versamax plate reader (Molecular Devices, Sunnyvale, CA).
qRT-PCR
Total RNA was prepared using RNeasy Plus Mini Kit (Qiagen, Valencia, CA), and first-strand cDNA was synthesized using SuperScriptIII (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Cytokine or other mRNA was quantified with SYBR green–based detection using an MX 3000P system (Stratagene, La Jolla, CA) according to the manufacturer’s protocols. Forty cycles of PCR (94°C for 15 s followed by 60°C for 30 s and 72°C for 30 s) were performed on a cDNA template. The data were normalized to GAPDH mRNA levels, compared with baseline expression level in corresponding samples from the uninfected mice, and expressed as fold induction.
Abs and flow cytometric analysis
For flow cytometry experiments, Abs were purchased from BioLegend (San Diego, CA), including rat anti-murine CD16/CD32 (Fc block), rat anti-murine CD45 conjugated to allophycocyanin, hamster anti-murine CD11c or CD8 conjugated to Pacific blue, rat anti-murine CD11b or CD4 conjugated to allophycocyanin-Cy7, rat anti-murine Ly6G conjugated to PE-Cy7, rat anti-murine CD3 or CD19 conjugated to PerCP-Cy5.5, rat anti-murine CD19 or Ly6C conjugated to FITC, and rat anti-mouse MHC class II (IA), CD80, or CD86 conjugated to PE.
Ab cell staining was performed as previously described (28). Data were collected on a FACS LSR2 flow cytometer using FACSDiva software (Becton Dickinson Immunocytometry Systems, Mountain View, CA). A minimum of 20,000 cells were evaluated from a predominantly leukocytic population identified by CD45+-stained cells per sample. The flow data are analyzed by FlowJo software (Tree Star, San Carlos, CA). An established gating strategy was used to identify subsets of lung myeloid cells (29). In brief, the total CD45+ lung leukocytes were identified; then a series of selective gates were used to remove lymphocytes (CD3+/CD19+), eosinophils (SSChigh /CD11cint), and neutrophils (Ly6G+/CD11b+). Consecutive gates were next used to identify CD11c+ cells: autofluorescent (AF+) macrophages were distinguished from non-AF (AF−) dendritic cells (DCs). Thereafter, the relative expression of CD11b was used to separate CD11blow alveolar macrophages (AMs) from CD11bhigh exudate macrophages (ExMs; within the autofluorescent population) and to identify CD11bhigh DCs (within the nonautofluorescent population). Last, CD11b+ Ly6C+ monocytes were identified within the CD11c−/FSClow population. Total numbers of each cell population were calculated by multiplying the frequency of the population by the total number of leukocytes (the percentage of CD45+ cells multiplied by the original hemocytometer count of total cells).
Histology
Histology was performed as previously described (9). In brief, lungs were fixed by inflation with 1 ml of 10% neutral buffered formalin, excised en bloc, and immersed in neutral buffered formalin. After paraffin embedding, 5-mm sections were cut and stained using H&E with mucicarmine. Sections were analyzed with light microscopy and microphotographs taken using Digital Microphotography system DFX1200 with ACT-1 software (Nikon, Tokyo, Japan).
Bone marrow–derived macrophage culture
Bone marrow was isolated as previously described (30). To obtain BMDMs, we cultured bone marrow cells from uninfected SRA+/+ or SRA−/− mice in RPMI 1640, 10% FCS, GlutaMAX, MEM nonessential amino acids, sodium pyruvate, and 20 ng/ml GM-CSF (PeproTech, Rocky Hill, NJ) at 37°C and 5% CO2. The medium was replaced every 3 d. Cells were harvested after 7 d of culture.
Calculations and statistics
Statistical significance was calculated using Student t test for individual paired comparisons. All values are reported as mean ± SEM. Means with p values <0.05 were considered significantly different. All statistical calculations were performed using Primer of Biostatistics (McGraw-Hill, New York, NY).
Results
SRA-deficient mice exhibit better control of cryptococcal lung infection during the efferent, but not afferent, phase of the immune response
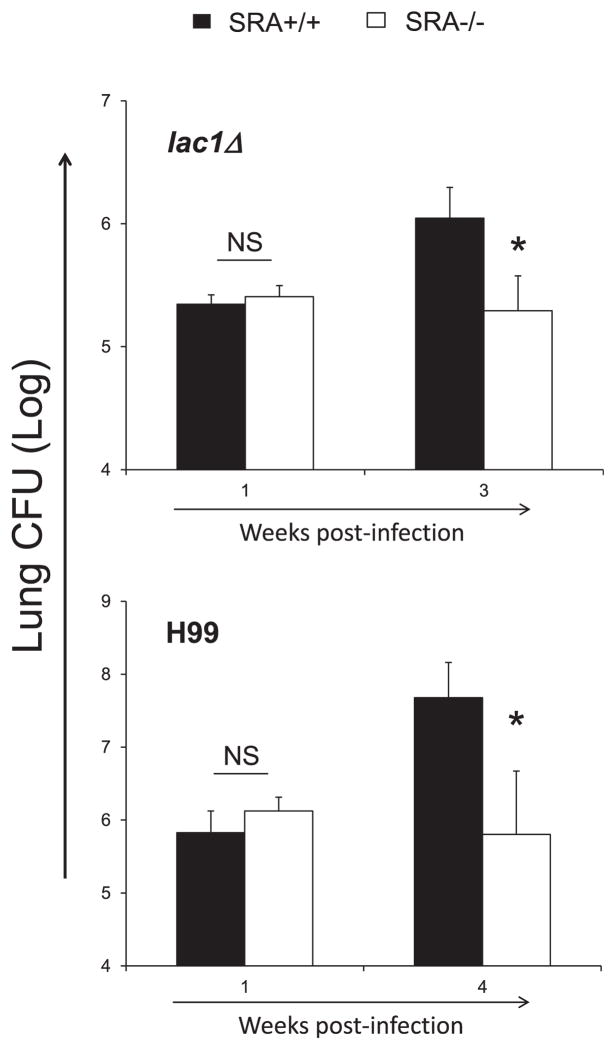
To evaluate the role of SRA in pulmonary cryptococcal infection, we first determined the effect of SRA on the pulmonary growth of C. neoformans. SRA+/+ and SRA−/− mice were infected with C. neoformans lac1Δ, and the fungal lung burdens were evaluated at 1 and 3 wk postinfection (wpi). We chose the lac1Δ mutant strain because it can grow progressively in the lungs without disseminating into the CNS, which would complicate interpretation by causing early death (6, 9, 10). Equivalent growth of yeast in the lungs was noted in both SRA+/+ and SRA−/− mice at 1 wk, indicating that SRA expression did not affect the initial afferent response to C. neoformans (Fig. 1). However, significantly improved cryptococcal clearance was found in infected SRA−/− mice during the efferent phase (3 wk) compared with their wild-type counterparts (Fig. 1). The improved containment of C. neoformans lac1Δ in SRA−/− mice corresponded to delayed onset of mice mortality at day 67: 60% of SRA+/+ mice died of crypto-coccal infection, whereas 100% of SRA−/− mice survived at this time. These findings suggest that SRA expression interferes with control of C. neoformans growth in the infected lungs during the efferent phase of the immune response.
FIGURE 1.
Effect of SRA on fungal control in the lungs. SRA+/+ and SRA−/− mice were inoculated intratracheally with 104 C. neoformans strain H99 or lac1Δ. Fungal lung burden was analyzed postinfection. Note the best improvement in fungal control is achieved in the lungs of SRA−/− mice infected with lac1Δ at 3 wpi, compared with SRA+/+ mice infected with lac1Δ. Also, a similar result was observed in H99-infected mice at 4 wpi. Data represent mean ± SEM pooled from two to three separate matched experiments; n ≥ 6 for each of the analyzed parameters. *p <0.05 in comparison between SRA+/+ and SRA−/− mice.
These findings were further confirmed with the wild-type strain H99. Consistent with Δlac1 infection, no differences in fungal lung burdens were observed at 1 wpi during WT strain H99 infection. Although differences in fungal load were less pronounced at 3 wpi (data not shown), a significant increase in pulmonary fungal load was observed at 4 wpi (Fig. 1). This increase corresponded to rapid onset of mortality in H99-infected SRA+/+ mice, which was delayed in SRA−/− mice. By day 36, 60% of WT-infected SRA+/+ mice died, whereas 100% of SRA−/− mice still survived. Collectively, SRA plays a detrimental role in the control of cryptococcal growth in the lungs during the efferent phase of the immune response to cryptococcal infection. Furthermore, these data reveal that the effect of SRA on cryptococcal growth in the lungs occurred independently of laccase expression.
SRA-deficient mice develop different microenvironmental features of cryptococcal lung infection during the efferent phase
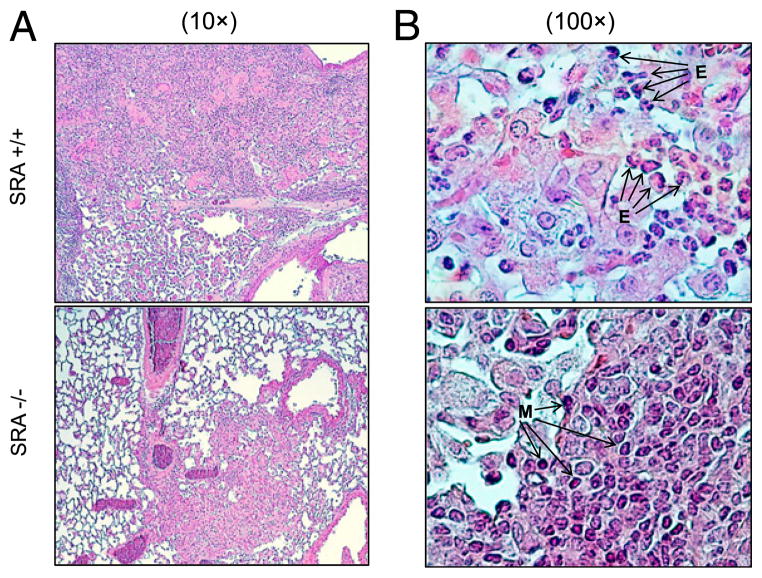
Because SRA promotes cryptococcal growth in the lung independently of laccase expression and lac1Δ grows in the lungs progressively without disseminating into the CNS, we chose C. neoformans lac1Δ for further experiments as a model to dissect the role of SRA in the pulmonary response. We next determined the role of SRA in lung histopathology during cryptococcal infection. Microscopic examination of uninfected lungs of SRA+/+ and SRA−/− mice showed typical murine lung morphology, indicating that SRA gene deletion did not cause any baseline alterations in lung tissue morphologic appearance (data not shown). Histologic analysis of infected lungs with C. neoformans at 3 wpi revealed robust leukocyte infiltration indicating that both strains of mice had induced a substantial inflammatory response. However, there were differences in the distribution of inflammatory responses between infected SRA+/+ and SRA−/− lungs. The leukocyte infiltrates in SRA−/− mice were dense, localized to bronchovascular structures, and were clearly demarcated from the uninfected lung portions (Fig. 2A). In contrast, leukocyte infiltrates in the lungs of SRA+/+ mice were more diffuse with less well-defined boundaries (Fig. 2A). High-power examination revealed that the leukocyte infiltrate composition was different. Although leukocyte infiltrates in both infected SRA+/+ and SRA−/− mice were composed of multiple cell types, numerous eosinophils were easily identified within the loose infiltrates observed in SRA+/+ mice, whereas the more tightly packed infiltrates seen in SRA−/− mice appeared enriched with smaller mononuclear cells (Fig. 2B). Thus, differences in the microenvironmental features of inflammation were observed in SRA+/+ and SRA−/− mice with cryptococcal lung infection at the efferent phase.
FIGURE 2.
Effect of SRA on microanatomic features of the host response against cryptococcal lung infection. Lungs from infected SRA+/+ and SRA−/− mice were perfused with buffered formalin, fixed, and processed for histology at 3 wpi. Representative photomicrographs of H&E mucicarmine-stained slides taken at ×10 (A) and ×100 (B) objective power. Note that the observed pattern of inflammation in SRA+/+ mice consisted of loose alveolar infiltrates enriched for eosinophils (E). In contrast, infiltrates in the lungs of infected SRA−/− mice appeared more discrete, were localized adjacent to bronchovascular structures, and were enriched for mononuclear cells (M).
SRA-deficient mice accumulate fewer eosinophils and more lymphocytes and pulmonary DCs in response to cryptococcal lung infection at efferent phase
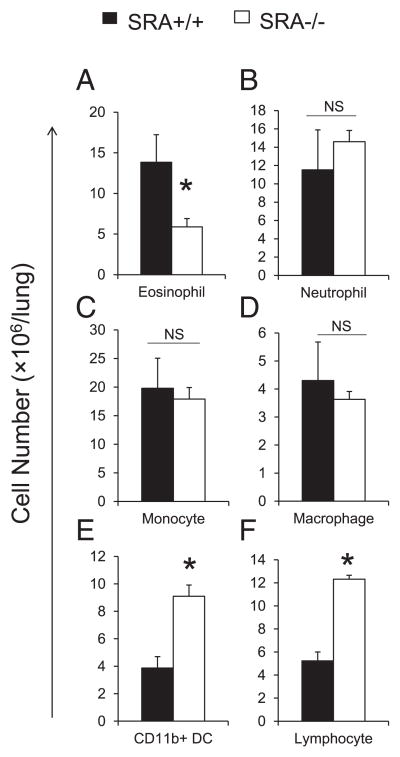
To further explore the differences in the microenvironmental features of the inflammatory response in SRA+/+ and SRA−/− mice, we quantified lung leukocyte populations using flow cytometric analysis and leukocyte subset-specific gating strategies (as described in Materials and Methods). Cryptococcal infection triggered a significant accumulation of CD45+ leukocytes in both SRA+/+ and SRA−/− mice at 3 wpi (72.18 ± 8.53 × 106/lung versus 83.68 ± 5.05 ×106/lung, respectively), compared with uninfected SRA+/+ and SRA−/− mice (27.37 ± 2.56 ×106/lung versus 27.64 ± 2.84 ×106/lung, respectively), consistent with the development of a robust immune response in both strains of mice. However, note that no significant difference was observed in the total numbers of CD45+ leukocytes within the infected lungs of SRA+/+ and SRA−/− mice at this time point during the efferent phase of the host response. Next, individual subsets of CD45+ leukocytes were identified by selective gating strategies using flow cytometry at 3 wpi. Accumulation of eosinophils, markers of a Th2 response in the lungs, was significantly reduced in SRA−/− mice compared with SRA+/+ mice (Fig. 3A), whereas no difference in the accumulation of neutrophils was observed (Fig. 3B). Total numbers of lung monocytes (Fig. 3C) and macrophages (Fig. 3D) did not differ between infected SRA+/+ and SRA−/− mice. In contrast, increased accumulation of CD11b+ pulmonary DCs and lymphocytes was observed in infected SRA−/− mice compared with infected SRA+/+ mice (Fig. 3E, 3F). These data suggest that increased accumulation of pulmonary DCs and lymphocytes contributed to the dense mononuclear cell infiltrates observed in infected SRA−/− mice (refer to Fig. 2).
FIGURE 3.
Effect of SRA on the accumulation of pulmonary leukocytes. Lung leukocytes were isolated from infected SRA+/+ and SRA−/− mice at 3 wpi, Ab stained, and analyzed using flow cytometry as per Materials and Methods. In brief, leukocytes were identified as CD45+ cells. Next, additional gates were set up to identify lymphocytes (CD3+ and CD19+), neutrophils (Ly6G+/CD11b+), eosinophils (SSChigh/CD11cint), Ly6C+ monocytes (CD11c−/Ly6C+/CD11b+), macrophages (autofluorescence+/CD11c+), and CD11b+ DCs (autofluorescence−/CD11c+/CD11b+). The total number of CD45+ leukocytes was determined by multiplying the frequency of CD45+ by the total number cells. Total numbers of eosinophils (A), neutrophils (B), monocyte (C), macrophages (D), CD11b+ DCs (E), and lymphocytes (F) were determined by multiplying the frequency of each subset by the total number of CD45+ leukocytes at 3 wpi. Data represent mean ± SEM pooled from one separate experiments; n ≥ 6 for each of the analyzed parameters; *p <0.05 in comparison between SRA+/+ and SRA−/− mice. NS, No significant difference between SRA+/+ and SRA−/− mice.
SRA-deficient mice enhance activation of pulmonary DCs and precursor Ly6C+ monocytes and increase total pulmonary CD4+ T cells, but diminish Th2 cytokine production during cryptococcal lung infection at the efferent phase
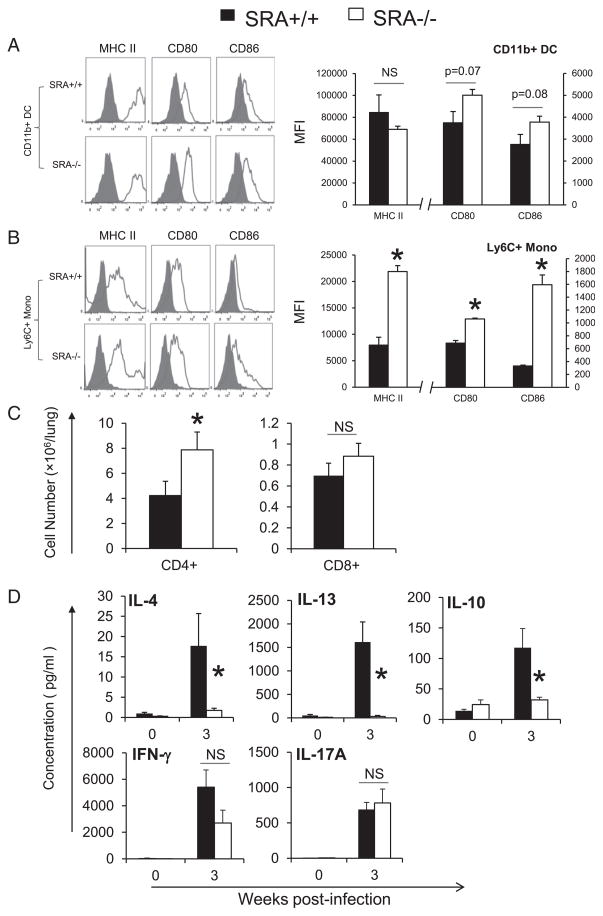
T cells, including CD4+ and CD8+ subsets, are required for the clearance of C. neoformans lung infection (31–36). The interaction between these T cells and APCs in the infected lungs, especially pulmonary DCs, is responsible for Ag-specific cytokine secretion and optimal immune polarization (28, 37). We have previously shown that increased numbers of DCs and lymphocytes, and the presence of dense bronchovascular infiltrates are associated with activated DCs and protective Th1 immune responses in mice with cryptococcal lung infection (28). Thus, given our observation of alterations in patterns of lung inflammation and total numbers of pulmonary DCs and lymphocytes in infected SRA+/+ and SRA−/− mice, we next examined the role of SRA in modulating the adaptive immune response to crypto-coccal lung infection. First, we analyzed the expression of MHC class II and two costimulatory molecules (CD80 and CD86) on CD11b+ DCs and their Ly6C+ precursors using flow cytometry analysis following established protocols (38). We found that SRA deletion slightly, yet nonsignificantly, increased the expression of CD80 and CD86 on CD11b+ DCs (Fig. 4A) and significantly increased the expression of all three molecules on Ly6C+ monocytes (Fig. 4B).
FIGURE 4.
Effect of SRA on pulmonary immune polarization. Leukocytes were isolated from the lungs of infected SRA+/+ and SRA−/− mice at 3 wpi. CD11b+ DCs and precursor Ly6C+ monocytes were identified by flow cytometry (as described in Materials and Methods and Fig. 3). The activation phenotype of CD11b+ DCs (A) and Ly6C+ monocytes (B) was evaluated by the surface expression of MHC class II and costimulatory molecules (CD80 and CD86). Stained samples are shown as solid lines and isotype controls as shaded histograms. The bar graph presents mean frequencies of positive cells derived from these histograms; the populations of CD4+ and CD8+ T lymphocytes were identified and enumerated using flow cytometry (C); the cytokine production by leukocytes isolated from infected lung at 3 wpi was evaluated by ELISA as described in Materials and Methods (D). Data were pooled from two to three separate matched experiments; n ≥ 6 for each of the analyzed parameters. *p <0.05 in comparison between SRA+/+ and SRA−/− mice. NS, No significant difference between SRA+/+ and SRA−/− mice.
Next, the CD4+ and CD8+ T cells numbers were evaluated in infected SRA+/+ and SRA−/− mice at 3 wpi by flow cytometry as previously described (39). Numbers of CD4+ T cells were increased almost 2-fold in SRA−/− mice relative to SRA+/+ mice (Fig. 4C). In contrast, no difference in the accumulation of CD8+ T cells in the lung was observed between infected SRA+/+ and SRA−/− mice (Fig. 4C). Note that no difference was observed in the uninfected lungs between SRA+/+ and SRA−/− mice in the total numbers of CD4+ and CD8+ T cells (data not shown).
To further determine whether the improved activation of CD11b+ DCs and accumulation of CD4+ T cells in infected SRA-deficient mice observed at 3 wpi altered the Th-immune polarization profile at this time point, we measured lung leukocyte cytokine profiles by ELISA as described in Materials and Methods. We observed that SRA deletion significantly diminished expression of Th2-type cytokines (IL-13 and IL-4) and an immunoregulatory cytokine (IL-10) compared with their counterparts in SRA+/+ mice (Fig. 4D). Interestingly, no difference was found in the expression of Th1 (IFN-γ) and Th17 cytokines (IL-17A; Fig. 4D). However, the changes in IL-4 secretion were sufficient to significantly improve IFN-γ/IL-4 ratio in SRA−/− mice to the level consistent with the improved clearance (data not shown). To further verify these observed changes in ex vivo cytokine production, we also performed qPCR for cytokine mRNA expression on freshly isolated lung leukocytes (at 3 wpi) and obtained similar results (data not shown).
Collectively, these results demonstrate that the improved fungal clearance observed in SRA−/− mice was strongly associated with increased activation of lung APCs and decreased hallmarks of nonprotective Th2 responses.
SRA-deficient mice improve classical activation of pulmonary macrophages during cryptococcal lung infection at the efferent phase
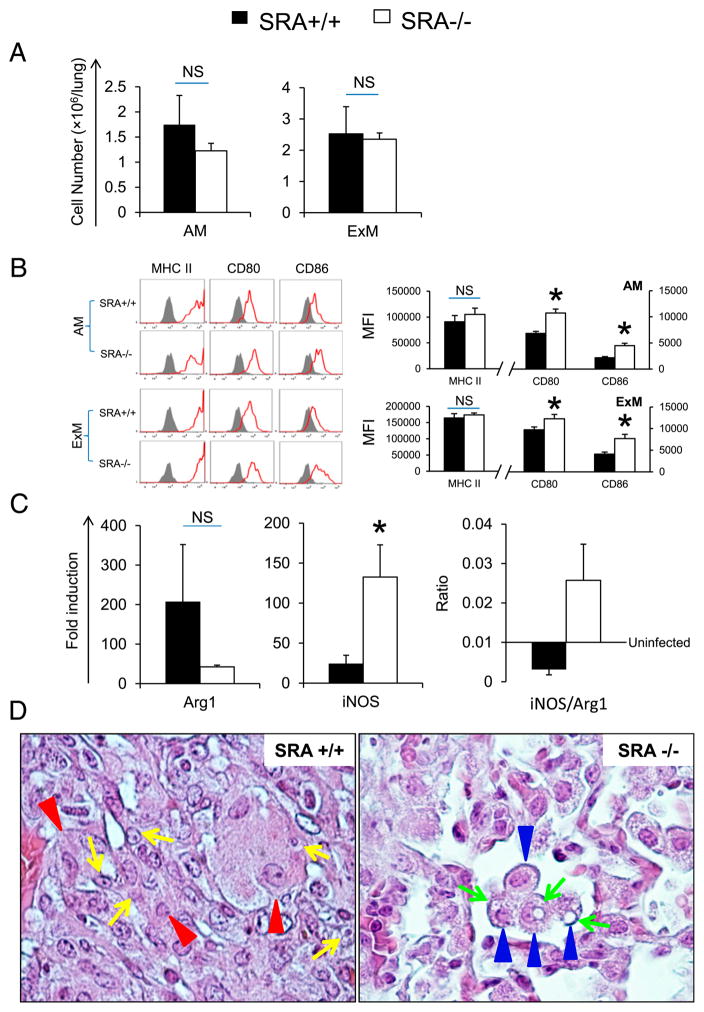
The balance between Th1/Th2 cytokine influences macrophage activation status, which, in turn, affects the microbicidal potential of these cells (40, 41). We next sought to determine whether SRA expression affects the total numbers or activation status of specific pulmonary macrophage populations during the cryptococcal lung infection. First, we examined the total numbers of AMs and ExMs in the lungs of SRA+/+ and SRA−/− mice at 3 wpi and their expression of MHC class II and the costimulatory molecules (CD80 and CD86) by flow cytometry using an established gating strategy (29). SRA deletion has no effects on the total accumulation of AM and ExM in the lung (Fig. 5A). However, SRA deletion was associated with increased expression of costimulatory molecules CD80 and CD86 on AMs and ExMs (Fig. 5B). Interestingly, no difference in the expression of MHC class II was found between infected SRA+/+ mice and SRA−/− mice at 3 wpi (Fig. 5B).
FIGURE 5.
Effect of SRA on the activation of pulmonary macrophages. Pulmonary macrophages including AMs and ExMs were isolated from infected SRA+/+ and SRA−/− mice at 3 wpi using flow cytometry as per Materials and Methods. In brief, total macrophages (autofluorescence+/CD11c+) were gated as described in Fig. 3. Next, AMs (CD11c+/CD11blow) and ExMs (CD11c+/CD11bhigh) were identified based on the expression of CD11c versus CD11b. Total numbers of AMs and ExMs were determined by multiplying the frequency of each subset by the total number of CD45+ leukocytes (A). Thereafter, the activation phenotype of AM and ExM was evaluated by the surface expression of MHC class II and costimulatory molecules (CD80 and CD86). Stained samples are shown as solid lines and isotype controls as shaded histograms. The bar graph presents mean frequencies of positive cells derived from these histograms (B). Gene expression of alternative macrophage activation (Arg1) and classical macrophage activation (iNOS) by total adherent pulmonary macrophages was evaluated by qPCR (as described in Materials and Methods) (C). Data were pooled from two to three separate matched experiments; n ≥ 6 for each of the analyzed parameters. *p <0.05 in comparison between SRA+/+ mice and SRA−/− mice. NS, No significant difference between SRA+/+ mice and SRA−/− mice. The morphologic appearance of pulmonary macrophages was evaluated (at 3 wpi by histology as described in Fig. 2). (D) Representative photomicrographs of H&E and mucicarmine-stained slides taken at 100× objective power. Note that the macrophages identified within infected lungs of SRA+/+ mice (red arrowheads) were large, sometimes multinu-cleated, and frequently contained intact intracellular cryptococci (yellow arrows). In contrast, macrophages (blue arrows) in the lungs of SRA−/− mice appeared smaller and often harbored degraded cryptococci (green arrows).
Next, to further assess the effect of SRA on macrophage activation and polarization of pulmonary macrophages from SRA+/+ and SRA−/− mice (at 3 wpi), we evaluated the expression of mRNA for M1 and M2 hallmarks (inducibe NO synthase [iNOS] and arginase 1 [Arg1], respectively) by qRT-PCR (Fig. 5C). Freshly isolated macrophages from infected SRA−/− mice showed an increased iNOS expression compared with SRA+/+ mice and a strong trend (however, not significant) toward diminished Arg1 expression. This leads to an “inversion” of iNOS/ Arg1 ratio (Fig. 5C) in favor of iNOS, indicating a shift from alternative activation to classical activation of macrophages in the absence of SRA. Moreover, a histopathologic examination of the lungs from infected SRA+/+ and SRA−/− mice (at 3 wpi) was performed to determine how SRA gene expression influences the morphologic features of pulmonary macrophages. Macrophages in the lungs of infected wild type SRA+/+ mice were large, sometimes multinucleated, and often contained one or more intracellular viable cryptococci (Fig. 5D, left panel). In sharp contrast, macrophages in the lungs of SRA−/− mice were smaller and often contained evidence of degraded intracellular cryptococci (Fig. 5D, right panel). Collectively, these data indicate that SRA gene expression promotes the alternative activation of pulmonary macrophages that likely contributes to the impairment in fungal clearance observed in SRA+/+ mice (relative to SRA−/− mice).
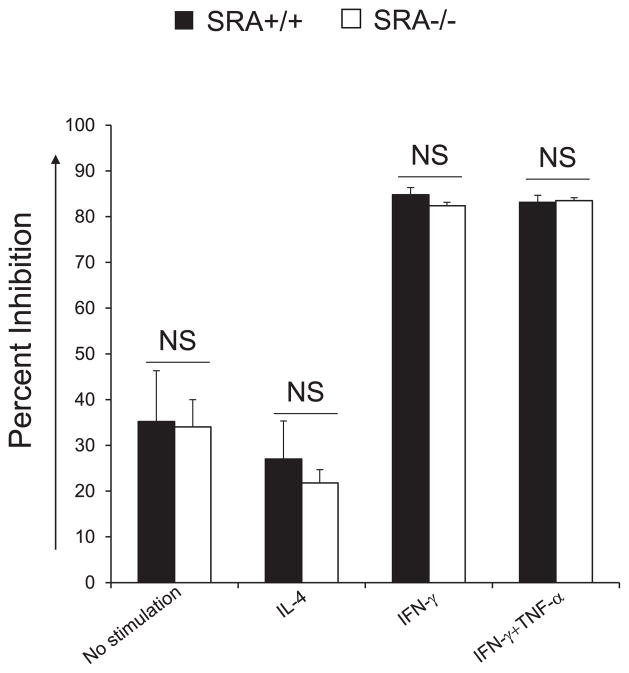
The cytokine milieu rather than SRA expression affects macrophage fungicidal function in vitro
To evaluate whether SRA expression has a direct role in macrophage–C. neoformans interactions, we examined the microbial growth inhibition and the NO production by macrophages stimulated with or without cytokines. BMDMs from SRA+/+ and SRA−/− mice were infected with opsonized C. neoformans and incubated at 37°C with or without cytokine stimulation. After 24 h, NO production and fungal growth were evaluated. The microbial growth inhibition by macrophages was measured relative to yeast growth in media alone. Macrophages under resting and IL-4–stimulated conditions showed minimal (30%) fungal inhibition (Fig. 6), whereas macrophages stimulated with IFN-γ alone and a combination of IFN-γ and TNF-α showed >80% of fungal inhibition. In all cases, there were no differences between SRA+/+ and SRA−/− macrophages in the levels of fungal inhibition (Fig. 6). The level of NO production by macrophages was assessed by Griess assay in supernatants from the cultures studied earlier. Minimal amounts of NO were detected in non-treated SRA+/+ or SRA−/− macrophages (0.76 ± 0.19 versus 0.32 ± 11 μM) or IL-4–treated (1.02 ± 0.29 versus 0.22 ± 0.01 μM) SRA+/+ or SRA−/− macrophages. In comparison, both SRA+/+ and SRA−/− macrophages showed robust NO production in response to either IFN-γ alone (87.49 ± 2.75 versus 67.30 ± 2.46 μM) or a combination of IFN-γ and TNF-α (93.60 ± 1.99 versus 76.55 ± 1.97 μM). Collectively, these data indicate that cytokine milieu rather than SRA expression defines macrophage fungicidal function in vitro.
FIGURE 6.
Effect of SRA on fungicidal activity of macrophages. BMDMs were generated from uninfected SRA+/+ or SRA−/− mice as per Materials and Methods. After harvest, BMDMs were seeded on 24-well plate with 1 × 106 cells/well, then infected with 1 × 105 Abopsonized C. neoformans lac1Δ, and incubated for 24 h at 37°C and 5% CO2 in media alone or with recombinant mouse cytokines: IL-4 (20 ng/ml), IFN-γ (100 ng/ml), or a combination of IFN-γ (20 ng/ml) and TNF-α (20 ng/ml). After the incubation, the yeast were collected by lysing the macrophages and enumerated by plating 10-fold serial dilutions for each sample onto Sabouraud dextrose agar plates. Colonies were counted after 48 h. CFU values in individual treatment groups were compared with culture wells without macrophages and expressed as percent of C. neoformans–growth inhibition. Data represent mean ± SEM pooled from several separate experiments; n ≥ 3 for each of the analyzed parameters. NS, No significant difference between SRA+/+ and SRA−/− mice.
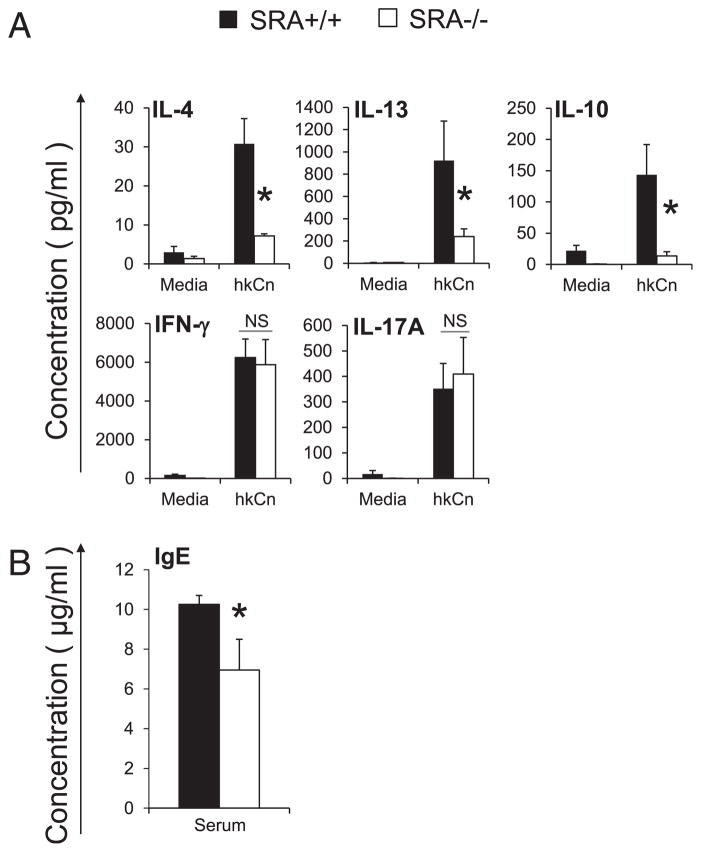
SRA-deficient mice decrease Th2 cytokine production in splenocytes and IgE production in the sera during cryptococcal lung infection at the efferent phase
Our data demonstrated that SRA expression influenced the local immune response in the lungs, which, in turn, defined the cytokine environment and macrophage fungicidal activity. Next, we sought to determine whether SRA also altered systemic immunity against cryptococcal lung infection. To accomplish this, we first isolated splenocytes from SRA+/+ and SRA−/− mice at 3 wpi. Splenocytes were pulsed with cryptococcal Ags (heat-killed Cryptococcus cells), and cytokine production was analyzed in cell culture supernatants in comparison with cytokine levels in the unstimulated cultures. Consistent with the results found in the lungs, SRA deletion resulted in diminished Ag-specific production of IL-13, IL-4, and IL-10 (Fig. 7A). Expression of IFN-γ and IL-17A was unaffected by SRA gene deletion (Fig. 7A). Next, we measured the IgE production in serum by ELISA (at 3 wpi). Consistent with diminished expression of Th2/regulatory cytokines by splenocytes, infected SRA−/− mice displayed a significant decrease in IgE compared with infected SRA+/+ mice (Fig. 7B). Thus, these results indicate that SRA promotes Th2 responses to cryptococcal infection at the local (lung) and systemic level.
FIGURE 7.
Effect of SRA on systemic immune polarization. (A) Effect of SRA on cytokine production by splenocytes from C. neoformans–infected mice. Splenocytes were isolated from infected SRA+/+ and SRA−/− mice at 3 wpi and cultured for 48 h at 5 × 106 cells/ml with or without heat-killed C. neoformans (hkCn) in a ratio of 1:2 in 24-well plates. Secreted cytokines levels were evaluated by ELISA in cell culture supernatants. Note a significant increase in production of Th2 cytokines and IL-10 after Ag stimuli in splenocytes from infected SRA+/+ mice, but no difference in the expression of IFN-γ and IL-17A. (B) Effect of SRA on production of serum IgE from C. neoformans–infected mice, SRA+/+ mice, and SRA−/− mice at 3 wpi (measured by ELISA). Data were pooled from two to three separate matched experiments; n ≥ 6 for each of the analyzed parameters. *p <0.05 in comparison between SRA+/+ and SRA−/− mice. NS, No significant difference between SRA+/+ and SRA−/− mice.
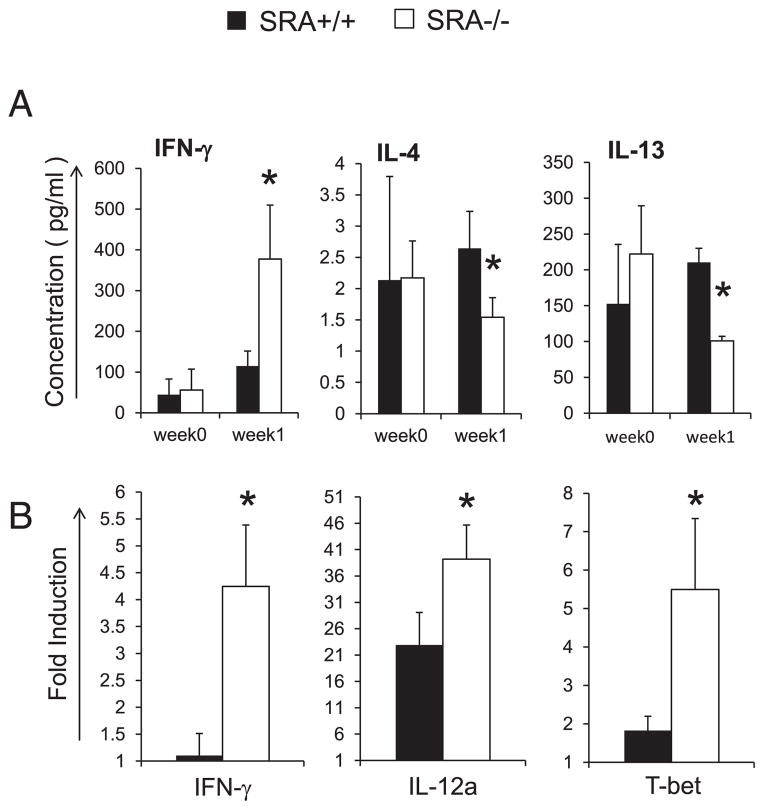
SRA-deficient mice promote induction of IFN-γ in bronchoalveolar lavage and Th1 skewing in LALN during cryptococcal lung infection at the afferent phase
Development of the adaptive immune response against C. neoformans infection is orchestrated by early innate responses and the interplay of cytokine signals at the site of primary infection in the lung (16, 42). Thus, our next goal was to determine whether SRA influenced the cytokine production profile during the early phase of the infection. Therefore, we obtained bronchoalveolar lavage (BAL) fluid from uninfected and infected SRA+/+ and SRA−/− mice at 1 wpi and assessed cytokine production by ELISA (Fig. 8A). Note that the fungal burden is equivalent in both strains of mice at this time point (refer to Fig. 1). Results show that compared with SRA+/+ mice, infection in SRA−/− mice was associated with a marked increase in IFN-γ, whereas levels of IL-4 and IL-13 were significantly decreased. These findings demonstrate that SRA signaling affected the lung cytokine balance during the very early stage of cryptococcal infection.
FIGURE 8.
Effect of SRA on the immune response in the lung and LALNs at 1 wpi. The BAL fluid and LALNs were collected from uninfected and infected SRA+/+ and SRA−/− mice. The BAL fluid was analyzed by ELISA to assess cytokines expression (A). Total RNA extracted from the cells of LALN was converted to cDNA and analyzed by real-time RT-PCR for the production of cytokine and transcript regulators of CD4+ T cells (B). Note a robust increase of IFN-γ and a significant deficiency in the expression of IL-4 and IL-13, in the BAL of infected SRA−/− mice, compared with infected SRA+/+ mice. And also note a significant induction of IFN-γ, IL-12a, and Th1 transcript regulator T-bet in the LALNs of infected SRA−/− mice. Data were pooled from two to three separate matched experiments; n ≥ 6 for each of the analyzed parameters. *p <0.05 in comparison between SRA+/+ and SRA−/− mice.
Lastly, we sought to further understand how SRA influenced early cytokine signals that translated to more profound alterations in Th immune polarization. The draining lymph nodes are another site in which early T cell polarization profiles are influenced by Ag uptake and presentation. Therefore, we evaluated expression of genes associated with Th polarization in mRNA obtained from pulmonary lymph nodes of SRA+/+ and SRA−/− mice at 1 wpi. Results demonstrate that SRA gene deletion was associated with a significant upregulation of IFN-γ, more robust induction of IL-12p35, and increased expression of Th1 master transcription factor T-bet (Fig. 8B). In contrast, SRA deletion did not significantly affect the expression of GATA-3, Foxp3, and IL-13 compared with infected SRA+/+ mice (data not shown). Collectively, these results suggest that the SRA signaling pathway impairs cryptococcal clearance by inhibiting early local and regional immunologic events, resulting in a skewing of the adaptive immune response toward Th2 polarization, which is nonprotective.
Discussion
This study provides novel information regarding the role of SRA signaling in pulmonary C. neoformans infection. Our data demonstrate that SRA expression interferes with clearance of C. neoformans by contributing to nonprotective Th2 bias evidenced by: 1) changes in pulmonary and systemic cytokine levels; 2) accumulation of eosinophils and alternatively activated macrophages that replace protective cellular players: activated CD4+ T cells, CD11b+ DCs, and classically activated macrophages; 3) diffuse inflammatory pathology in the lungs; and 4) uncontrolled expansion of C. neoformans in the infected lungs. These detrimental effects of SRA signaling were attributed to inhibition of afferent signals, such as innate IFN-γ, which are needed for the development of a protective Th1 immune response and prevention of detrimental Th2 in the infected lungs. Collectively, these data provide novel demonstration that C. neoformans can exploit the innate host-defense receptor to promote nonprotective immunity and its persistence in the infected host.
SRA is an innate receptor of myeloid leukocytes (macrophages and DC) known to play an important role in various infections and inflammation (18). Interestingly, the initial in vitro study documented that SRA had no role in activation of macrophages by C. neoformans (17). In this study, in a mouse model of cryptococcal pneumonia, we demonstrate improved cryptococcal clearance at the efferent phase of the immune response (3 wpi) in the lungs of SRA-deficient mice. These data demonstrate that SRA signaling, instead of benefiting the host, contributes to the loss of protection during the efferent phase of the immune response. Although our observation could be considered counterintuitive, it is not completely unexpected. Pathogens can exploit loopholes in the immune system to gain an advantage over the immune host defenses. SRA is not an exception; although beneficial in clearance of L. monocytogenes and S. pneumoniae, it was found to be exploited by M. tuberculosis and P. carinii (20, 22–24). Our study reveals that the opportunistic yeast C. neoformans can exploit SRA to increase its virulence in the infected host.
The timing of observed changes in cryptococcal clearance provided a clue that SRA signaling affected the efferent phase of the immune response, whereas differential characteristics of cellular response indicated that SRA signaling contributed to an immune dysregulation. Although the overall magnitude of inflammatory response in the C. neoformans–infected lungs was not different between SRA+/+ and SRA−/− mice (as shown in recruitment of total lung CD45+ cells), SRA signaling contributed to the accumulation of eosinophils, and diminished accumulation of lymphocytes and CD11b+ DCs (Fig. 3A, 3E, 3F). The suboptimal composition of cellular infiltrate in the presence of SRA and only a moderate expression of costimulatory molecules (CD80 and CD86) by mononuclear phagocytes were associated with more diffused appearance of cellular infiltrates in the lungs for SRA+/+ mice compared with SRA−/− mice at 3 wpi (Fig. 4A, 4B, 5B). In contrast, SRA−/− mice showed an improved pulmonary accumulation of CD4+ T cells and CD11b+ DCs, improved expression of costimulatory molecules, and formation of tight mononuclear cell infiltrates, in which direct interaction between CD4+ T cells, CD11b+ DCs, and macrophages are known to occur during protective anticryptococcal responses (28, 37). Together, these data provide evidence that SRA interfered with execution of optimal effector immune response in the infected lungs.
Although studies have suggested that other pulmonary pathogens such as M. tuberculosis and P. carinii can exploit SRA signaling and subsequently interfere with the development of T cell–mediated responses (23, 24), these studies have not identified modulation of the immune polarization as a potential mechanism. Our data clearly point out that the interference of SRA with microbial clearance is tightly linked with changes in the immune polarization. SRA deletion significantly reduced production of cytokines observed in the nonprotective Th2 response (IL-4, IL-13, and IL-10) both within the lungs and in cryptococcal Ag-induced splenocyte cytokine response assay. These data, along with a more robust accumulation of serum IgE in SRA+/+ mice, provide strong evidence that SRA signaling in the context of cryptococcal infections contributed to an expansion of the Th2 arm of the immune response. To our knowledge, this is a first example of SRA being exploited by a pathogen to mediate this immunomodulatory effect by enhancing the Th2 component of the immune response.
The effect of C. neoformans interaction with SRA had significant consequences on macrophage activation profiles. As expected, more pronounced Th2 response in SRA+/+ mice was associated with more pronounced expression of alternative activation markers by pulmonary macrophages. The improved C. neoformans clearance in SRA−/− mice was associated with improved expression of markers of classically activated macrophages and histologic evidence of improved intracellular killing of C. neoformans by macrophages. This finding is not only consistent with previous reports that linked Th1 response, classical activation of macrophages to containment of C. neoformans (15, 43–45), but is also evidence that SRA expression in the context of crypto-coccal infection could promote alternative activation of macrophages. Interestingly, however, the direct interaction between C. neoformans and SRA on macrophages is not responsible for the shift in macrophage activation status. First, we do not observe any effect of SRA deletion on macrophage polarization during the afferent phase of infection at 1 wpi (data not shown), a time point that precedes recruitment of polarized T cell into C. neoformans–infected mice. Second, SRA expression had no effect on macrophage responses to C. neoformans in vitro (17), including the expression of alternative versus classical activation genes (data not shown). This observation is in contrast with the profound differences in macrophage activation pattern at 3 wpi, which occurs in parallel with the changes of macrophage-polarizing cytokine profiles in the infected lungs. Lastly, our in vitro data indicate SRA deletion had no direct effect on C. neoformans uptake by macrophages (data not shown), or on their fungicidal capacity under the same cytokine environments, indicating that the SRA-induced effects on fungal control are induced indirectly through the changes of Th-immune bias, rather than by interfering with intrinsic fungicidal functions of macrophages. These outcomes support the conclusion that the mechanism(s) by which SRA alters macrophage polarization is/are indirect, that is, induced via modulation of T cell polarization and the subsequent changes in cytokine profiles in the infected lungs (46, 47).
The final group of data seeks to identify the upstream mechanisms linked to the effects of SRA on the immune polarization. The induction of proinflammatory cytokine signals, including IFN-γ, at the afferent phase is important to the development of the protective immune response against cryptococcal infection (35, 37, 45, 48, 49). In this article, we demonstrate that, albeit without effect on early growth of C. neoformans in the lungs, SRA signaling results in diminished secretion of IFN-γ in the lung at 1 wpi. This afferent effect on afferent-phase IFN-γ production translates into a reduced induction of IFN-γ, IL-12p35, and Th1 master regulator T-bet in the LALN at the afferent phase. These data provide evidence that SRA signaling interferes with the afferent mechanisms that orchestrate the subsequent immune polarization. SRA acts as an immune repressor of crucial cytokines such as innate IFN-γ during the cryptococcal lung infection.
Modulation of the immune response is an important strategy used by C. neoformans to evade host defenses in immunocompromised or immunocompetent individuals (16). C. neoformans developed multiple strategies to modulate the innate and adaptive immune response via expression of different virulence factors. As previously described by our group, the highly virulent strain H99 promotes Th2 polarization by expression of different virulence factors including laccase (6, 8, 10, 50). Interestingly, SRA interferes with the pulmonary fungal growth during the efferent phase both in the presence and absence of cryptococcal laccase expression, indicating that laccase expression is not required for C. neoformans to interfere with SRA-induced effects. At present, it is not clear which cryptococcal factor(s) is/are involved in binding to SRA and which intracellular signaling pathway(s) is/are involved in immune modulation through SRA during cryptococcal lung infection. Thus, future studies are needed to address these points. In this article, we provide an example of SRA exploitation by C. neoformans to promote a nonprotective Th2 immune response. These findings provide important evidence that SRA can act as an immune repressor to promote cryptococcal virulence and persistence.
Acknowledgments
This work was supported by Merit Review funding (Grant 1I01BX000656 to M.A.O.) and a Career Development Award (to J.J.O.) from the Biomedical Laboratory Research and Development Service, Department of Veterans Affairs, and Multidisciplinary Training Program in Pulmonary Diseases Grant T32-HL07749-19 (to M.J.D.) from the U.S. Public Health Service.
We acknowledge the contributions of Drs. Fuyuan Wang, Yanmei Zhang, Xiumiao He, and Benjamin Murdock, and the technical assistance of Joudeh Freij, Mengmeng Jie, Zach Hadd, Daniel Lyons, Daniel Meister, Joanne Sonstein, and Priya Vedula. We thank the Undergraduate Research Opportunity Program at the University of Michigan for supporting the undergraduate students working in the laboratory.
Abbreviations used in this article
- AM
alveolar macrophage
- Arg1
arginase 1
- BAL
bronchoalveolar lavage
- BMDM
bone marrow–derived macrophage
- ExM
exudate macrophage
- iNOS
inducibe NO synthase
- LALN
lung-associated lymph node
- qRT-PCR
quantitative real-time RT-PCR
- SRA
scavenger receptor A
- wpi
weeks postinfection
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Chuck SL, Sande MA. Infections with Cryptococcus neoformans in the acquired immunodeficiency syndrome. N Engl J Med. 1989;321:794–799. doi: 10.1056/NEJM198909213211205. [DOI] [PubMed] [Google Scholar]
- 2.Pappas PG, Perfect JR, Cloud GA, Larsen RA, Pankey GA, Lancaster DJ, Henderson H, Kauffman CA, Haas DW, Saccente M, et al. Cryptococcosis in human immunodeficiency virus-negative patients in the era of effective azole therapy. Clin Infect Dis. 2001;33:690–699. doi: 10.1086/322597. [DOI] [PubMed] [Google Scholar]
- 3.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 4.Kronstad JW, Attarian R, Cadieux B, Choi J, D’Souza CA, Griffiths EJ, Geddes JM, Hu G, Jung WH, Kretschmer M, et al. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat Rev Microbiol. 2011;9:193–203. doi: 10.1038/nrmicro2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackstock R, Murphy JW. Role of interleukin-4 in resistance to Cryptococcus neoformans infection. Am J Respir Cell Mol Biol. 2004;30:109–117. doi: 10.1165/rcmb.2003-0156OC. [DOI] [PubMed] [Google Scholar]
- 6.He X, Lyons DM, Toffaletti DL, Wang F, Qiu Y, Davis MJ, Meister DL, Dayrit JK, Lee A, Osterholzer JJ, et al. Virulence factors identified by Cryptococcus neoformans mutant screen differentially modulate lung immune responses and brain dissemination. Am J Pathol. 2012;181:1356–1366. doi: 10.1016/j.ajpath.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoag KA, Street NE, Huffnagle GB, Lipscomb MF. Early cytokine production in pulmonary Cryptococcus neoformans infections distinguishes susceptible and resistant mice. Am J Respir Cell Mol Biol. 1995;13:487–495. doi: 10.1165/ajrcmb.13.4.7546779. [DOI] [PubMed] [Google Scholar]
- 8.Osterholzer JJ, Surana R, Milam JE, Montano GT, Chen GH, Sonstein J, Curtis JL, Huffnagle GB, Toews GB, Olszewski MA. Cryptococcal urease promotes the accumulation of immature dendritic cells and a non-protective T2 immune response within the lung. Am J Pathol. 2009;174:932–943. doi: 10.2353/ajpath.2009.080673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Wang F, Tompkins KC, McNamara A, Jain AV, Moore BB, Toews GB, Huffnagle GB, Olszewski MA. Robust Th1 and Th17 immunity supports pulmonary clearance but cannot prevent systemic dissemination of highly virulent Cryptococcus neoformans H99. Am J Pathol. 2009;175:2489–2500. doi: 10.2353/ajpath.2009.090530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu Y, Davis MJ, Dayrit JK, Hadd Z, Meister DL, Osterholzer JJ, Williamson PR, Olszewski MA. Immune modulation mediated by cryptococcal laccase promotes pulmonary growth and brain dissemination of virulent Cryptococcus neoformans in mice. PLoS ONE. 2012;7:e47853. doi: 10.1371/journal.pone.0047853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valdez PA, Vithayathil PJ, Janelsins BM, Shaffer AL, Williamson PR, Datta SK. Prostaglandin E2 suppresses antifungal immunity by inhibiting interferon regulatory factor 4 function and interleukin-17 expression in T cells. Immunity. 2012;36:668–679. doi: 10.1016/j.immuni.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wozniak KL, Hardison SE, Kolls JK, Wormley FL. Role of IL-17A on resolution of pulmonary C. neoformans infection. PLoS ONE. 2011;6:e17204. doi: 10.1371/journal.pone.0017204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu J, Paul WE. Peripheral CD4+ T-cell differentiation regulated by networks of cytokines and transcription factors. Immunol Rev. 2010;238:247–262. doi: 10.1111/j.1600-065X.2010.00951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torchinsky MB, Garaude J, Martin AP, Blander JM. Innate immune recognition of infected apoptotic cells directs T(H)17 cell differentiation. Nature. 2009;458:78–82. doi: 10.1038/nature07781. [DOI] [PubMed] [Google Scholar]
- 15.Voelz K, May RC. Cryptococcal interactions with the host immune system. Eukaryot Cell. 2010;9:835–846. doi: 10.1128/EC.00039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olszewski MA, Zhang Y, Huffnagle GB. Mechanisms of cryptococcal virulence and persistence. Future Microbiol. 2010;5:1269–1288. doi: 10.2217/fmb.10.93. [DOI] [PubMed] [Google Scholar]
- 17.Means TK, Mylonakis E, Tampakakis E, Colvin RA, Seung E, Puckett L, Tai MF, Stewart CR, Pukkila-Worley R, Hickman SE, et al. Evolutionarily conserved recognition and innate immunity to fungal pathogens by the scavenger receptors SCARF1 and CD36. J Exp Med. 2009;206:637–653. doi: 10.1084/jem.20082109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Areschoug T, Gordon S. Scavenger receptors: role in innate immunity and microbial pathogenesis. Cell Microbiol. 2009;11:1160–1169. doi: 10.1111/j.1462-5822.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki H, Kurihara Y, Takeya M, Kamada N, Kataoka M, Jishage K, Ueda O, Sakaguchi H, Higashi T, Suzuki T, et al. A role for macrophage scavenger receptors in atherosclerosis and susceptibility to infection. Nature. 1997;386:292–296. doi: 10.1038/386292a0. [DOI] [PubMed] [Google Scholar]
- 20.Arredouani MS, Yang Z, Imrich A, Ning Y, Qin G, Kobzik L. The macrophage scavenger receptor SR-AI/II and lung defense against pneumococci and particles. Am J Respir Cell Mol Biol. 2006;35:474–478. doi: 10.1165/rcmb.2006-0128OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plüddemann A, Hoe JC, Makepeace K, Moxon ER, Gordon S. The macrophage scavenger receptor A is host-protective in experimental meningococcal septicaemia. PLoS Pathog. 2009;5:e1000297. doi: 10.1371/journal.ppat.1000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishiguro T, Naito M, Yamamoto T, Hasegawa G, Gejyo F, Mitsuyama M, Suzuki H, Kodama T. Role of macrophage scavenger receptors in response to Listeria monocytogenes infection in mice. Am J Pathol. 2001;158:179–188. doi: 10.1016/S0002-9440(10)63956-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollifield M, Bou Ghanem E, de Villiers WJ, Garvy BA. Scavenger receptor A dampens induction of inflammation in response to the fungal pathogenPneumocystis carinii. Infect Immun. 2007;75:3999–4005. doi: 10.1128/IAI.00393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sever-Chroneos Z, Tvinnereim A, Hunter RL, Chroneos ZC. Prolonged survival of scavenger receptor class A-deficient mice from pulmonary Mycobacterium tuberculosis infection. Tuberculosis (Edinb) 2011;91(Suppl 1):S69–S74. doi: 10.1016/j.tube.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pukkila-Worley R, Gerrald QD, Kraus PR, Boily MJ, Davis MJ, Giles SS, Cox GM, Heitman J, Alspaugh JA. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot Cell. 2005;4:190–201. doi: 10.1128/EC.4.1.190-201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olszewski MA, Huffnagle GB, McDonald RA, Lindell DM, Moore BB, Cook DN, Toews GB. The role of macrophage inflammatory protein-1 alpha/CCL3 in regulation of T cell-mediated immunity to Cryptococcus neoformans infection. J Immunol. 2000;165:6429–6436. doi: 10.4049/jimmunol.165.11.6429. [DOI] [PubMed] [Google Scholar]
- 27.Olszewski MA, Huffnagle GB, Traynor TR, McDonald RA, Cook DN, Toews GB. Regulatory effects of macrophage inflammatory protein 1alpha/CCL3 on the development of immunity to Cryptococcus neoformans depend on expression of early inflammatory cytokines. Infect Immun. 2001;69:6256–6263. doi: 10.1128/IAI.69.10.6256-6263.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osterholzer JJ, Curtis JL, Polak T, Ames T, Chen GH, McDonald R, Huffnagle GB, Toews GB. CCR2 mediates conventional dendritic cell recruitment and the formation of bronchovascular mononuclear cell infiltrates in the lungs of mice infected withCryptococcus neoformans. J Immunol. 2008;181:610–620. doi: 10.4049/jimmunol.181.1.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osterholzer JJ, Chen GH, Olszewski MA, Zhang YM, Curtis JL, Huffnagle GB, Toews GB. Chemokine receptor 2-mediated accumulation of fungicidal exudate macrophages in mice that clear cryptococcal lung infection. Am J Pathol. 2011;178:198–211. doi: 10.1016/j.ajpath.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arora S, Olszewski MA, Tsang TM, McDonald RA, Toews GB, Huffnagle GB. Effect of cytokine interplay on macrophage polarization during chronic pulmonary infection withCryptococcus neoformans. Infect Immun. 2011;79:1915–1926. doi: 10.1128/IAI.01270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchanan KL, Doyle HA. Requirement for CD4(+) T lymphocytes in host resistance against Cryptococcus neoformans in the central nervous system of immunized mice. Infect Immun. 2000;68:456–462. doi: 10.1128/iai.68.2.456-462.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huffnagle GB, Lipscomb MF, Lovchik JA, Hoag KA, Street NE. The role of CD4+ and CD8+ T cells in the protective inflammatory response to a pulmonary cryptococcal infection. J Leukoc Biol. 1994;55:35–42. doi: 10.1002/jlb.55.1.35. [DOI] [PubMed] [Google Scholar]
- 33.Huffnagle GB, Yates JL, Lipscomb MF. T cell-mediated immunity in the lung: a Cryptococcus neoformans pulmonary infection model using SCID and athymic nude mice. Infect Immun. 1991;59:1423–1433. doi: 10.1128/iai.59.4.1423-1433.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huffnagle GB, Yates JL, Lipscomb MF. Immunity to a pulmonary Cryptococcus neoformans infection requires both CD4+ and CD8+ T cells. J Exp Med. 1991;173:793–800. doi: 10.1084/jem.173.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawakami K, Tohyama M, Teruya K, Kudeken N, Xie Q, Saito A. Contribution of interferon-gamma in protecting mice during pulmonary and disseminated infection withCryptococcus neoformans. FEMS Immunol Med Microbiol. 1996;13:123–130. doi: 10.1016/0928-8244(95)00093-3. [DOI] [PubMed] [Google Scholar]
- 36.Mody CH, Chen GH, Jackson C, Curtis JL, Toews GB. Depletion of murine CD8+ T cells in vivo decreases pulmonary clearance of a moderately virulent strain ofCryptococcus neoformans. J Lab Clin Med. 1993;121:765–773. [PubMed] [Google Scholar]
- 37.Qiu Y, Zeltzer S, Zhang Y, Wang F, Chen GH, Dayrit J, Murdock BJ, Bhan U, Toews GB, Osterholzer JJ, et al. Early induction of CCL7 downstream of TLR9 signaling promotes the development of robust immunity to cryptococcal infection. J Immunol. 2012;188:3940–3948. doi: 10.4049/jimmunol.1103053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osterholzer JJ, Chen GH, Olszewski MA, Curtis JL, Huffnagle GB, Toews GB. Accumulation of CD11b+ lung dendritic cells in response to fungal infection results from the CCR2-mediated recruitment and differentiation of Ly-6Chigh monocytes. J Immunol. 2009;183:8044–8053. doi: 10.4049/jimmunol.0902823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Wang F, Bhan U, Huffnagle GB, Toews GB, Standiford TJ, Olszewski MA. TLR9 signaling is required for generation of the adaptive immune protection in Cryptococcus neoformans-infected lungs. Am J Pathol. 2010;177:754–765. doi: 10.2353/ajpath.2010.091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Classen A, Lloberas J, Celada A. Macrophage activation: classical versus alternative. Methods Mol Biol. 2009;531:29–43. doi: 10.1007/978-1-59745-396-7_3. [DOI] [PubMed] [Google Scholar]
- 41.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Herring AC, Hernández Y, Huffnagle GB, Toews GB. Role and development of TH1/TH2 immune responses in the lungs. Semin Respir Crit Care Med. 2004;25:3–10. doi: 10.1055/s-2004-822300. [DOI] [PubMed] [Google Scholar]
- 43.Jain AV, Zhang Y, Fields WB, McNamara DA, Choe MY, Chen GH, Erb-Downward J, Osterholzer JJ, Toews GB, Huffnagle GB, Olszewski MA. Th2 but not Th1 immune bias results in altered lung functions in a murine model of pulmonary Cryptococcus neoformans infection. Infect Immun. 2009;77:5389–5399. doi: 10.1128/IAI.00809-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hardison SE, Ravi S, Wozniak KL, Young ML, Olszewski MA, Wormley FL., Jr Pulmonary infection with an interferon-gamma-producing Cryptococcus neoformans strain results in classical macrophage activation and protection. Am J Pathol. 2010;176:774–785. doi: 10.2353/ajpath.2010.090634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arora S, Hernandez Y, Erb-Downward JR, McDonald RA, Toews GB, Huffnagle GB. Role of IFN-gamma in regulating T2 immunity and the development of alternatively activated macrophages during allergic bronchopulmonary mycosis. J Immunol. 2005;174:6346–6356. doi: 10.4049/jimmunol.174.10.6346. [DOI] [PubMed] [Google Scholar]
- 46.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schenten D, Medzhitov R. The control of adaptive immune responses by the innate immune system. Adv Immunol. 2011;109:87–124. doi: 10.1016/B978-0-12-387664-5.00003-0. [DOI] [PubMed] [Google Scholar]
- 48.Bava AJ, Afeltra J, Negroni R, Diez RA. Interferon gamma increases survival in murine experimental cryptococcosis. Rev Inst Med Trop Sao Paulo. 1995;37:391–396. doi: 10.1590/s0036-46651995000500003. [DOI] [PubMed] [Google Scholar]
- 49.Lovchik JA, Lyons CR, Lipscomb MF. A role for gamma interferon-induced nitric oxide in pulmonary clearance ofCryptococcus neoformans. Am J Respir Cell Mol Biol. 1995;13:116–124. doi: 10.1165/ajrcmb.13.1.7598935. [DOI] [PubMed] [Google Scholar]
- 50.Noverr MC, Cox GM, Perfect JR, Huffnagle GB. Role of PLB1 in pulmonary inflammation and cryptococcal eicosanoid production. Infect Immun. 2003;71:1538–1547. doi: 10.1128/IAI.71.3.1538-1547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]