Abstract
Family-based genetic association studies of related individuals provide opportunities to detect genetic variants that complement studies of unrelated individuals. Most statistical methods for family association studies for common variants are single-marker-based, which test one SNP a time. In this paper, we consider testing the effect of a SNP set, e.g., SNPs in a gene, in family studies, for both continuous and discrete traits. Specifically, we propose a Generalized Estimating Equations (GEE)-based kernel association test, a variance component-based testing method, to test for the association between a phenotype and multiple variants in a SNP set jointly using family samples. The proposed approach allows for both continuous and discrete traits, where the correlation among family members is taken into account through the use of an empirical covariance estimator. We derive the theoretical distribution of the proposed statistic under the null and develop analytical methods to calculate the p-values. We also propose an efficient resampling method for correcting for small sample size bias in family studies. The proposed method allows for easily incorporating covariates and SNP-SNP interactions. Simulation studies show that the proposed method properly controls for type-I error rates under both random and ascertained sampling schemes in family studies. We demonstrate through simulation studies that our approach has superior performance for association mapping compared to the single marker based minimum p-value GEE test for a SNP set effect over a range of scenarios. We illustrate the application of the proposed method using data from the Cleveland Family GWAS Study.
Keywords: Family-based association, Generalized estimation equations, Kernel machine regression, Marginal models, Score test, Variance component
1. INTRODUCTION
Family-based design is commonly used in many genetic association studies. Current statistical methods for family data have mainly focused on individual-marker or single-SNP analysis [Chen and Yang 2010; Li, et al. 2011b; Namkung 2012]. These methods can be grouped into two major categories referred to as conditional methods and unconditional methods. The conditional family-based analysis is based on evaluating the association between a phenotype and the transmission of marker alleles within family members, such as the transmission disequilibrium test (TDT) method and its various extensions (QTDT, FBAT)[Laird and Lange 2006; Ott, et al. 2011]. These test statistics model the offspring genotypes conditional on parental genotypes (if informative) within each family/pedigree. Although inherently robust to population stratification, they can be less powerful than unconditional methods, which are adapted from population based analysis, where both within- and between-family variations can be incorporated. These methods directly model the associations between phenotypes and genotypes of all individuals. The correlation among family members is often taken into account in mixed models by including a random polygenic effect [Wang, et al. 2013] or in generalized estimating equations (GEE) [Chen and Yang 2010]. The unconditional methods also have gained increasing popularity recently because they are computationally efficient and easy to integrate data with both family and unrelated individuals.
As an important alternative to individual-marker-based tests, SNP-set association tests are believed to be advantageous in several ways. Examples of a SNP set include SNPs in a gene, pathway, network, or any region in the genome, such as a haplotype block. By incorporating linkage disequilibrium (LD) and haplotype information among the markers being tested, joint analysis of multiple markers can be more powerful in detecting associated variants with small effects, and offer the possibility of capturing underlying joint effects such as SNP-SNP interactions. In addition, the results obtained from SNP-set tests at the gene level can be more readily extended to and integrated with downstream functional and pathogenic investigation because a gene is the basic functional unit of inheritance[Li, et al. 2011a]. Several multi-marker methods have been proposed based on dimension reduction techniques, such as Fourier transformation [Wang and Elston 2007], principal component analysis [Wang and Abbott 2008] and partial least-squares regression [Chun, et al. 2011; Wang, et al. 2009]. Methods that are based on combining the p-values of single marker tests have also been proposed in view of their convenience in implementation and downstream analysis [Dudbridge and Koeleman 2003; Yu, et al. 2009; Zaykin, et al. 2002]. However, permutation procedures are often required for calculating p-values of these multi-marker tests, because one needs to consider correlations among individual-marker test p-values, which can be computationally expensive for large data sets. These SNP-set based methods are, however, limited to case-control samples. In addition, their extensions to incorporate family data may not be feasible. For example, permutation tests will be difficult to implement when there are different family sizes in family studies.
Recently, a new category of methods that are based on kernel machines regression has gained increasing popularity, such as the kernel machine (KM) test [Wu, et al. 2010; Wu, et al. 2011], pairwise similarity [Mukhopadhyay, et al. 2009; Tzeng, et al. 2011; Tzeng, et al. 2009; Wessel and Schork 2006] and the SSU test [Han and Pan 2010]. They provide a flexible and computationally efficient framework for testing the joint effect of SNPs in a SNP set, and have been shown as an attractive alternative to the standard multivariate test under a variety of settings. The KM test is a variance component score test that assumes a common distribution of regression coefficients of multiple SNPs and account for LD among SNPs, and can improve the power by borrowing information across multiple SNPs. The KM test has recently been extended to test for the effect of a SNP set in family based association studies using mixed models for continuous phenotypes [Chen, et al. 2012; Schifano, et al. 2012]. However these mixed model based methods are difficult to apply directly to discrete traits, such as binary traits, as logistic mixed models are more challenging to fit and their likelihood does not have a closed form. Furthermore, the mixed model based SNP set test requires the familial correlation to be correctly specified, which is difficult to ensure in practice due to the presence of unmeasured genetic or shared environmental factors.
To overcome these limitations, in this paper we propose to test for the effects of a SNP set in family based association studies for both quantitative and discrete phenotypes using the generalized estimation equation approach. Specifically, a KM-like estimating equation based statistic is constructed to test for the association between a phenotype and a SNP set. We assume that a continuous phenotype marginally follows a linear regression and a binary phenotype marginally follows a logistic regression. An advantage of the GEE based SNP set test is that it allows for the within-family correlation to be misspecified and uses the empirical covariance estimator to correct for possible misspecification of the within-family correlation.
We derive the asymptotic null distribution of the proposed test statistic and provide an analytic scheme to calculate the p-value of the test statistic. In order to correct for small sample sizes, an efficient re-sampling method is further proposed by matching the higher moments of the statistic with a chi-square statistic. We show through extensive simulations and analysis of actual data that the proposed methods control type I error rates well under both random and ascertainment sampling schemes. We also show that the suggested approach has higher power compared to the individual-marker based minimum p-value test for family studies.
The remainder of the paper is organized as follows. In Section 2, we describe the proposed model and the KM SNP set test in the GEE framework for family studies. In Section 3, we present simulation settings and results to evaluate the finite sample performance of the proposed method and compare the proposed approach to the single-SNP based minimum-p-value analysis. In Section 4, we apply the proposed method to the data from the Cleveland Family GWAS Study, followed by discussions.
2. METHODS
Assume there are n families, and family i has mi members (i=1,…,n). Suppose a SNP set, e.g., a gene or a genomic region, contains p variants. Let yij denote a continuous or discrete phenotype for the j th individual in the i th family; Xij = (1, xij1, xij2, …, xijq)T denote a (q + 1) × 1 vector of an intercept and covariates, such as sex, age and environmental factors; Zij = (zij1, zij2,…zijp)T denote a p × 1 genotype vector for the p SNPs or variants in the set, coded 0,1,2, reflecting the number of copies of minor allele (additive coding).
We model the mean of the phenotype of the ij th individual μij = E(yij | Xij, Zij) using the marginal generalized linear model
| (1) |
where α = [α0,α1,…,αq] is a q×1 vector of an intercept and regression coefficients for the covariates Xij, β is a p×1 vector of regression coefficients for the genotypes Zij, g(.) is a link function and g(μij) = μij for continuous phenotypes, and g(μij) = logit(μij) for dichotomous phenotypes. The Generalized Estimating Equations for the parameters θ = (αT,βT)T can be written as
where μi = (μi1,.., μimi)T, Di = ∂μi/∂θT, and is a working covariance matrix of yi, and Ai = diag{υ(μil),…,υ(μim)}, υ(μij) is a variance function, with υ(μij) = 1 for normally distributed phenotypes and υ (μij) = μij(1 − μij) for binary phenotypes. Here Ri (δ) is a working correlation matrix defined by a kinship matrix and a scale parameter δ, where for all j ≠ k, the (j,k)th element of Ri (δ) is 2ϕijkδ with ϕijk as the kinship coefficient between individuals j and k in ith family, e.g. 2ϕijk = 0.5 for sib–sib and parent–child pairs with ϕ and δ satisfying{(ϕ,δ): 0 ≤ ϕ ≤ 0.5; −1 ≤ 2ϕδ ≤1}. Further, Δi = diag{μ̇i1,…, μ̇im}, where μ̇ is the first derivative of g−1(.). We allow the working correlation matrix Ri(δ) to be misspecified.
Our primary interest is to test whether there is an overall genetic effect of a SNP set, i.e., the null hypothesis H0: β = 0. If a SNP set contains SNPs in a gene, this tests for the overall effects of the gene. Under H0 model (1) becomes . The estimator of α under H0 (denoted as α̃) is the solution to the GEE , which can be computed by iterating between a Fisher scoring algorithm for α̃ and the method of moments for estimating δ until convergence (Appendix).
To develop a GEE based score test for H0, we decompose the GEEs as , where Ux and Uz are of dimension p × 1 and q × 1, respectively and are the estimating functions for α and β respectively. The standard estimating equation based score statistic for testing H0: β = 0 is where Ũz is the value of Uz(θ) evaluated at θ̃ = (α̃,0). , where Ĩzz, Ĩzz, Ĩxx are the corresponding decomposed submatrices of Ĩ, where Equation. The standard GEE score statistic T asymptotically follows a central chi-square distribution with p degrees of freedom.
When p is large, this standard GEE score statistic has a large degree of freedom and loses power. To improve the power of the score test when the number of SNPs (p) is large and when some SNPs in a set are highly correlated, we assume the individual components of the regression coefficients βj (j=1, …, p) follow an arbitrary distribution with mean 0 and common variance τ. The null hypothesis H0: β = 0 is equivalent to testing H0:τ = 0. We propose the following GEE-based KM test as
where , and . When yi is a scalar, i.e., for population studies, TS reduces to the KM statistic given in Wu, et al. [2010].
Using the results in the Appendix, it can be shown that
where are independent random variables, and (λ1,λ2,…,λp) are eigenvalues defined in the Appendix estimated using the empirical covariance matrix.
Therefore, the asymptotic distribution of the score statistic TS under the null hypothesis is a mixture of chi-square distributions, which can be approximated by a scaled chi-square distribution through matching the first two moments using the Satterthwaite method [Satterthwaite 1946], or matching the third moments [Liu, et al. 2007], or using the exact methods such as the Davies method [Davies 1980; Duchesne and Lafaye De Micheaux 2010]. In our simulation studies below, we will use the Davies method to obtain the p-values of TS. As sample sizes in real family based studies are often relatively small, i.e., the number of families is often relatively small, e.g., in hundreds, the large sample based Davies method for calculating the p-value might not perform well in small samples. This is because the sample variance of TS can be considerably smaller than the asymptotic variance especially for binary traits.
To correct for small sample bias, the variance of the GEE-based score statistic needs to be adjusted using more accurate small sample variance calculations. Following Lee, et al [2012a], the p-value adjusted for small samples can be calculated as
| (2) |
where is the distribution function of and df = 12/γ̂ · μ̂T, ν̂T and γ̂ are the estimated small sample mean, variance and kurtosis of the statistic Ts under the null, respectively. As shown by Lee et al. [2012b], it is much more convenient to calculate these moments especially the kurtosis by re-sampling methods. When there are no covariates and all families have the same pedigree structure, a simple permutation method can be used. For more general settings in the presence of covariates and different pedigree structures among different families, a perturbation process can be applied as described in the Appendix, in which a realized statistic is calculated by , where Ũb is a perturbation of Ũz.
3. SIMULATION STUDIES
3.1 Simulation study using ASAH1 Gene
To evaluate the performance of the proposed method in terms of Type I error control and statistical power, we carried out simulations studies in a range of settings. We first present the simulation results based on ASAH1 gene, which is a region located on chromosome 8 with a length of around 28.6kb. Based on the LD structure of ASAH1, we generated genotypes of 100,000 samples (200,000 haplotypes) based on HapMap CEU samples using the software HAPGEN[Su, et al. 2011]. There are a total of 93 sites in the region, and 83 sites are left after removing non-variant sites. We selected 13 typed SNPs on Affy6 as the genotyped SNPs that can be used in the analysis.
In the first simulation setting, we generated a dataset containing 1,000 and 2,000 sib-pairs with random sampling, i.e., without ascertainment. The genotypes of each pedigree were generated using an allele dropping algorithm [Thornton and McPeek 2010]: we first simulated the genotype for each pedigree founder (parent) by randomly selecting two haplotypes (sampled with replacement from the previously obtained haplotype pool); the parental haplotypes are then transmitted to offspring with equal chance. The correlated binary phenotype were simulated using the method described in Park, et al. [1996], where the correlation between sibling outcomes was set at 2ϕijkδ = 2 × 0.25 × 0.6 = 0.3. The phenotype mean for each individual was generated conditional on genotypes and two continuous covariates under the logistic model: , where α=(α0, 0.01, 0.01)T and α0 was chosen to make the prevalence around 0.01. Xij includes two continuous covariates generated from standard normal distributions. The effect size of a causal SNP βcausal was set as 0 under the null model to study the type I error and 0.2 (a genetic OR of 1.22) under an alternative model to study power assuming the type I error rate is 0.05. Each of the 83 SNPs in the gene region was chosen in turn as the causal SNP.
In the second simulation setting, we used a rejection sampling to randomly ascertain n/2 (500 and 1000) affected sib-pairs (with at least one disease individual) and an equal number of unaffected sib-pairs. The genotypes and phenotypes were generated using the same procedure described above.
For type I error rate evaluation, we considered 1000 sib pairs and conducted simulations under the null logistic model in which βcausal = 0. To investigate whether the proposed statistic can preserve type I error for extremely small genome-wide threshold, each simulation was replicated 1,000,000 times.
Power evaluation was based on 400 replicates with sample sizes of 1000 and 2000 sib pairs, respectively assuming the type I error rate is 0.05 and the regression coefficient of the causal variant is 0.2. For a comparison purpose, each simulation replicate was also analyzed by the single-SNP-based minimum p-value GEE test to test for the effect of a gene, where the individual SNP p-value was calculated using the R package “gee” [Carey 2002] (a wrap-up function is also available in R package “GWAF”) and the minimum p-value of individual SNP p-values was calculated. We calculated the gene level p-value by correcting the minimum p-value using the modified Bonferroni correction based on an estimated effective number of independent tests [Gao, et al. 2010].
We repeated the simulation for smaller sample sizes (500 and 300 sib-pairs) under the random sampling scheme for sib-pairs. We also conducted an additional simulation for data with a larger family size (4 members per family).
3.2 Simulation Study Using Random Genes
We next evaluate the power of the proposed method under the third simulation setting by generating SNP sets based on randomly sampled genes where the LD block structure varies among different SNP sets. We generated 20,000 simulation scenarios based on 998 real genes on chromosome 6. In each scenario, one gene was randomly chosen to generate haplotype samples using HAPGEN and a HapMap SNP was chosen as the causal SNP. The genotype and the phenotype were simulated using the same ascertainment scheme described in the second simulation setting. We again selected the SNPs that are covered by Affy6 as genotyped SNPs in each SNP set and used them for SNP set analysis.
3.3 Simulation Results
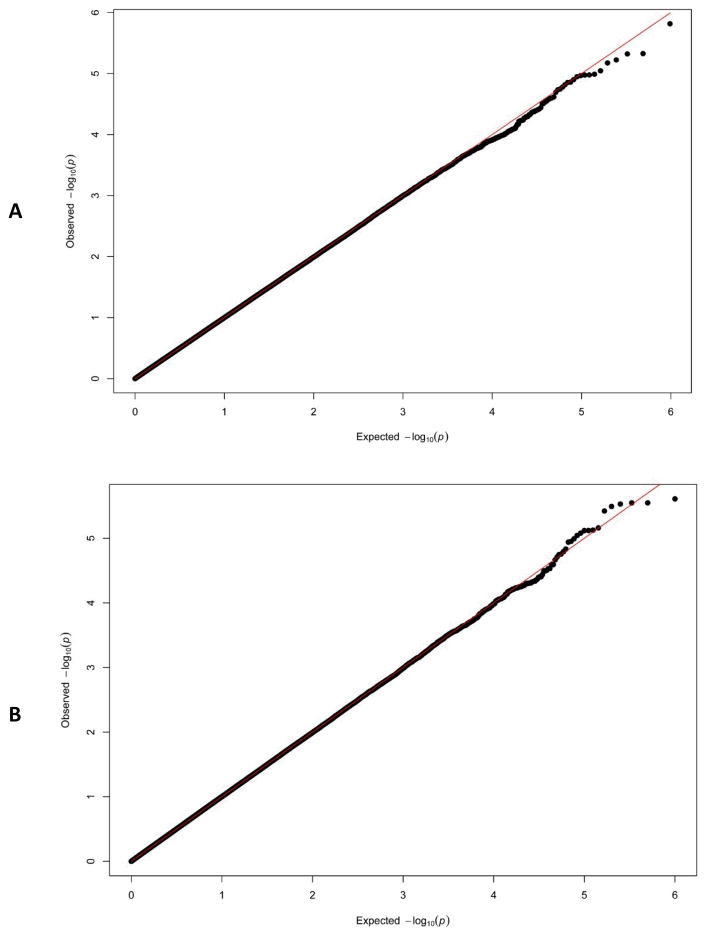
Figure 1 shows the quantile-quantile (Q-Q) plots of the observed p-values under the null to evaluate the performance of the proposed GEE-KM SNP set test in terms of type I error control (from the first two simulation settings). The Q-Q plot in Figure 1 plots the estimated p-values against what would be expected under the null. It suggests that Type I error rate remains well controlled for both random and ascertainment sampling schemes. When the sample size is small, as shown in Fig S1, the Davies based method tends to produce conservative results but works well with the proposed perturbation adjustment. Similar results are obtained for a larger family size (Fig. S2).
Fig. 1.
Quantile-quantile plot comparing empirical (−log10) p-values for testing the effects of a SNP set using the GEE KM test (based on 1,000,000 simulations under the null model) against those expected under the null from the first two simulation settings: A) randomly sampling scheme; B) ascertained sampling scheme. Each simulated data set has 1000 sibpairs. P-values were calculated using the perturbation based method.
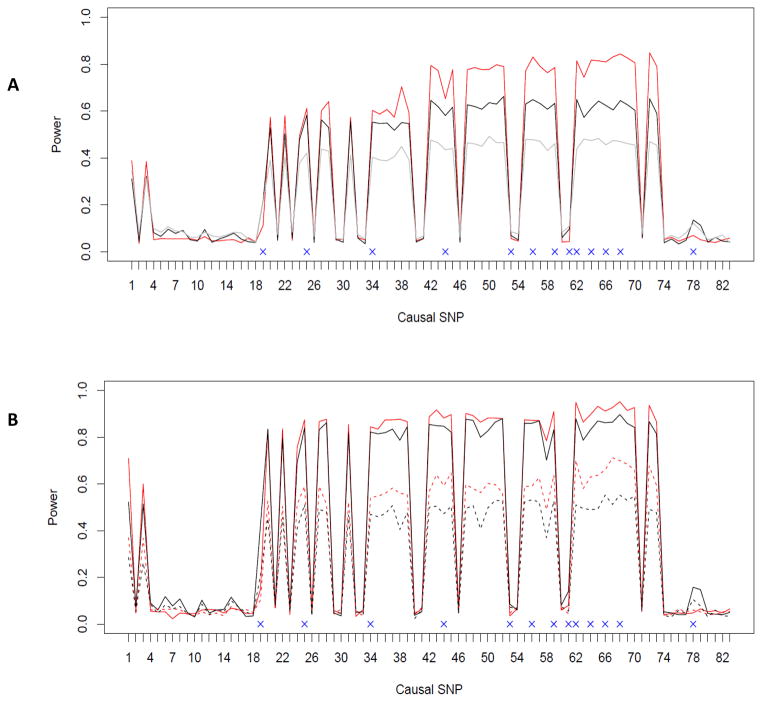
The results of empirical power based on gene ASAH1 are presented in Figure 2. The plots compare the powers of the GEE KM test and the minimum p-value method when each of the 83 sites was generated as the causal SNP. In the random sampling scheme (Fig 2A), both approaches have good power when the causal SNP is in high or moderate LD with the typed SNPs used in SNP set analysis, and have a power around the expected Type I error rate (0.05) when the causal SNP is not in LD with any of the typed ones (from 5 to 17 and 75 to 83). Generally, the proposed GEE KM test provides better performance than the minimum p-value approach. There is a significant increase in the detection power for both approaches when samples are ascertained (Fig 2B), but our approach remains superior compared to the individual SNP based minimum p-value test. The advantage becomes clearer when we lower the sample size (as indicated by the dashed lines in Fig 2B).
Fig. 2.
Empirical power for testing a SNP set using the ASAH1 gene: A) randomly sampling scheme; B) ascertained sampling scheme. Each of the 83 SNPs was generated as the causal variant in turn. The typed SNPs are denoted with a cross and are used in SNP set analysis. The red and black lines indicate the power curves for the proposed GEE KM test and the individual marker based minimum p-value method for testing the ASAH1 gene effects respectively when the sample size is n=2000. The gray line in Fig 2A indicates the power curves for the p degrees of freedom chi-square test (as implemented in the R package “geepack”) after removing 3 high LD SNPs. The solid and dashed lines (in Fig 2B) are observed powers for simulations with a sample size (number of sib pairs) of 2000 and 1000, respectively.
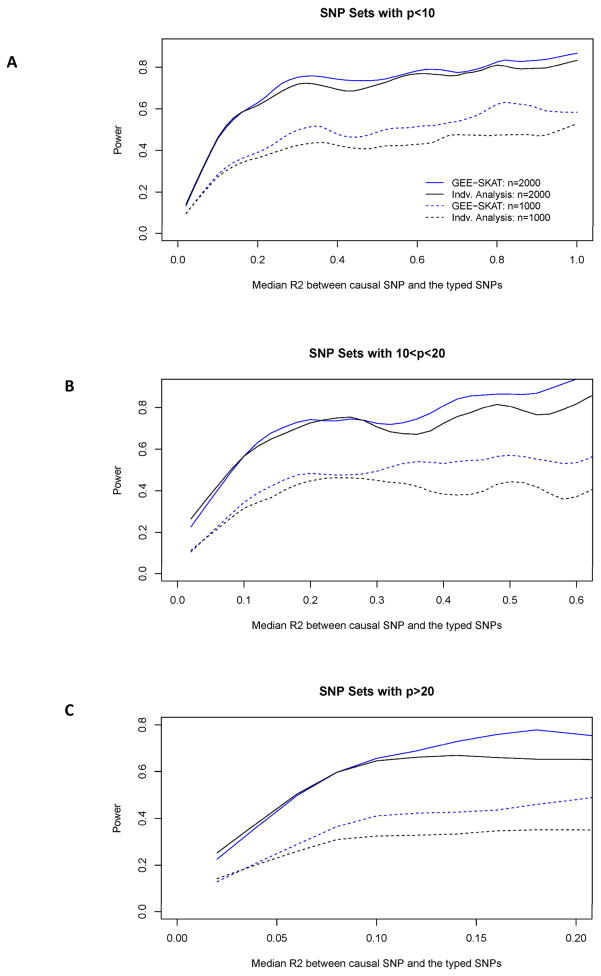
Figure 3 summarizes the results from the third simulation setting, i.e., the random gene simulation. Similar to Wu et al. [2010], we divided the simulation scenarios into three groups based on the number of typed SNPs within one gene. The empirical power was computed by first binning the simulations on the basis of the median R2 between the causal and the typed SNPs, where each group was evenly blocked into 50 subgroups. The power was then calculated as the proportion of p values less than 0.05. The smoothed curves of the power in Figure 3 show that, as expected, the power of the GEE KM test increases as the LD between the causal and typed SNPs increases. In all simulation scenarios, the GEE KM method tends to have higher power than the GEE individual-marker based minimum p-value analysis. The results from this simulation setting suggest that the proposed approach is robust in performance over a wide range of genes in real data.
Fig. 3.
Smoothed empirical power curve as a function of Median R2 between the causual SNP and the typed SNP for simulation scenarios based on randomly selected genes. Here n indicates the number of families consisting of sib-pairs.
4. Application to Cleveland Family Study
We applied the proposed methods to analyze the family samples collected in the Cleveland Family GWAS Study (CFS), which consists of first and second-degree relatives and spouses of a proband with either laboratory diagnosed obstructive sleep apnea or neighborhood control of an affected proband [Palmer, et al. 2003]. Blood pressure and hypertension related phenotypes were also collected. As part of the NHLBI’s Candidate-gene Association REsource (CARe) Study, a total of 630 African-American individuals from 143 families were genotyped on the Affymetrix 6.0 (Affy6.0) platform [Fox, et al. 2011; Zhu, et al. 2011]. Hypertension was analyzed as a binary trait which was defined as a systolic blood pressure higher than 140 mm Hg or diastolic blood pressure was higher than 90 mm Hg, or report of using antihypertensive medication. We performed a genome-wide association test on 16,406 gene regions. Each association test was adjusted for age, age2, gender and body mass index (BMI). We also adjusted for population stratification using principle component estimates derived from unrelated individuals selected from each family and projected to the rest of family members [Zhu, et al. 2008]. In addition to the proposed GEE KM approach, we also analyzed each gene region using the GEE individual marker based minimum p-value test by adjusting for multiple comparisons using the effective number of independent tests.
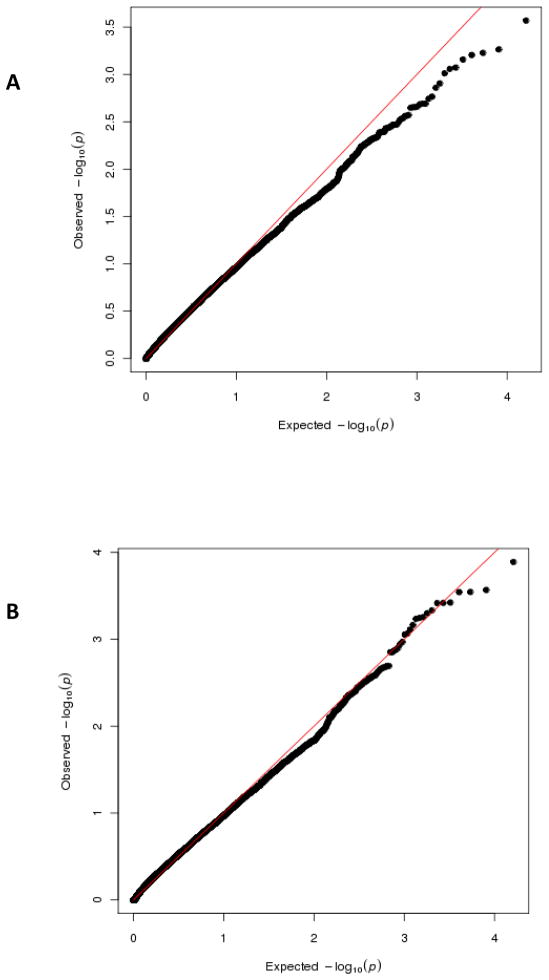
Figure 4 shows the Q-Q plots of −log10(p-value) from the genome-wide screen on gene-level analysis. The observed distribution of the score statistic shows no significant departure from the null. As expected, the score test tends to be conservative if small sample size adjustment is not applied. As the sample size is limited, none of the genes reached the genome-wide significance. Several genes have small p-values. We summarize the top list of genes that are associated with hypertension in Table 1. Interestingly, several genes among the list have been shown to be associated with hypertension related traits in previous studies with much larger sample sizes. For example, PLEKHG1 is the gene that has been identified in Continental Origins and Genetic Epidemiology Network (COGENT) meta-analysis with 30,000 African Ancestral individuals (to be published). Another gene in our list, MARCH5, is near the gene PLCE1, which was identified by the International Consortium for Blood Pressure (ICBP) which consists of ~200,000 European origin samples [Ehret, et al. 2011].
Fig. 4.
Genome wide quantile-quantile plot comparing (−log10) p-values of 16,406 gene regions against those expected under the null using the GEE KM method using the Cleveland Family Study data: A) without small sample size adjustment; B) perturbation-based correction method.
Table 1.
Top genes identified using the data from the Cleveland Family Study using the proposed GEE-KM method and the minimum p-value GEE method.
| GENE | Chr. No. | GEE-SKAT | MinP-GEE | |
|---|---|---|---|---|
| 1 | AP4S1 | 14 | 0.000129 | 0.0112 |
| 2 | TMEM98 | 17 | 0.000271 | 0.002938 |
| 3 | RNF144A | 2 | 0.000285 | 0.008314 |
| 4 | IFITM3 | 11 | 0.000287 | 0.0009285 |
| 5 | HNRNPA1L2 | 16 | 0.000378 | 0.004832 |
| 6 | MARCH5 | 10 | 0.000382 | 0.003501 |
| 7 | LAPTM5 | 1 | 0.000383 | 0.007125 |
| 8 | ACAA2 | 18 | 0.000465 | 0.003828 |
| 9 | CAPN10 | 2 | 0.000501 | 5.74E-06 |
| 10 | AK5 | 1 | 0.000555 | 0.004784 |
| 11 | C19orf45 | 19 | 0.000568 | 0.001163 |
| 12 | C5orf45 | 5 | 0.000582 | 0.0003733 |
| 13 | LOC338588 | 10 | 0.000685 | 0.01406 |
| 14 | MED29 | 19 | 0.000776 | 7.94E-05 |
| 15 | GORAB | 1 | 0.000862 | 0.01256 |
| 16 | B4GALT1 | 9 | 0.000884 | 0.0006125 |
| 17 | ANGPT4 | 20 | 0.001075 | 0.001693 |
| 18 | SLC37A4 | 11 | 0.001137 | 0.000565 |
| 19 | TTC30A | 2 | 0.001259 | 0.01363 |
| 20 | FBLIM1 | 1 | 0.001315 | 0.001056 |
| 21 | LY6K | 8 | 0.00135 | 0.001523 |
| 22 | MBOAT1 | 6 | 0.001407 | 0.01596 |
| 23 | SOCS5 | 2 | 0.001411 | 0.01689 |
| 24 | FAM129B | 9 | 0.002028 | 0.01949 |
| 25 | SLC35C2 | 20 | 0.002029 | 0.001766 |
| 26 | ZNF479 | 7 | 0.002072 | 0.008996 |
| 27 | LOC100128023 | 3 | 0.002103 | 0.05897 |
| 28 | KLRC1 | 12 | 0.002125 | 0.01421 |
| 29 | GNG8 | 19 | 0.002175 | 0.002144 |
| 30 | DLL4 | 15 | 0.002208 | 0.01759 |
| 31 | PLEKHG1 | 6 | 0.002319 | 0.1245 |
| 32 | LOC349114 | 7 | 0.002377 | 0.006306 |
| 33 | HIVEP3 | 1 | 0.002497 | 0.0005597 |
| 34 | DSG2 | 18 | 0.002616 | 0.03342 |
| 35 | ZZZ3 | 1 | 0.002619 | 9.72E-05 |
| 36 | CD63 | 12 | 0.002669 | 0.001587 |
| 37 | ZP1 | 11 | 0.00267 | 0.00567 |
| 38 | MRI1 | 19 | 0.0028 | 0.0006096 |
| 39 | LOC339788 | 2 | 0.002806 | 0.08376 |
| 40 | PRB1 | 12 | 0.00281 | 0.002012 |
5. DISCUSSION
A family-based design has several advantages compared to a population based design of unrelated subjects in genetic association studies. It offers better genotype quality control (such as Mendelian error checking), a better control for population stratification, and allows for a variety of genetic analyses to be performed, including the analysis of parent-of-origin effects, de novo variants, and combined linkage and association mapping [Ott, et al. 2011]. Under certain designs, family based association studies can be more powerful than population based studies using unrelated samples [Feng, et al. 2011; Laird and Lange 2006]. As an alternative to individual SNP analysis, we proposed the GEE-based KM test statistic to test the joint effects of multiple variants in a set on a phenotype in a family based association study. The correlations among family members are taken into account through the use of generalized estimating equations. The proposed methods can be conveniently applied to both continuous and binary traits while accounting for within-family correlation, and are robust to misspecification of within-family correlation. Further, by specifying an appropriate working correlation, the proposed method can be readily used to handle clustered data in population based studies, such as the data clustered by geographic regions, and longitudinal data with repeated measurements.
With the advent of next generation sequencing, it will be possible to extend the proposed method to study rare variant effects in family sequencing association studies. Family data can be more informative for identifying rare variants than unrelated samples because rare variants segregate within families [Zhu, et al. 2010]. When a child inherits a rare variant, he/she also inherits the haplotype segment surrounding the rare variant. Even when a region has multiple rare variants in different families, the inheritance patterns obtained from rare variants embedded in the same haplotype segments may still provide good information for the region to be detected. It is easy to construct a new statistic for studying rare variant effects using sequencing data by incorporating variants weight, i.e., , where W = diag(w1,…, wp) are variant weights that are based on external functional information or the minor allele frequency (MAF) of a variant. The null distribution of this new statistic can also be easily derived by plugging in the corresponding weight matrix. It will also be of interest in future studies to examine in detail the performance of the proposed method for testing rare variant effects in sequencing based family studies, and to compare with two recently developed kernel based methods that are based on conditional genotypes [Ionita-Laza, et al. 2013] and traditional score statistics [Schaid, et al. 2013], respectively.
We have demonstrated through simulations that the proposed test controls Type I error very well. Parallel to the findings in population based studies of unrelated samples, the proposed GEE-KM method is more powerful than the single-marker based minimum p-value test especially for testing a gene effect when SNPs are in moderate or high LDs. The proposed method is developed unconditional on parental genotypes, which increases use of information from all individuals. The unconditional method is not naturally robust to population structure, but population stratification can be easily adjusted in our model by incorporating principle components of population variation as covariates [Zhu, et al. 2008].
As a score-type test, only the null model needs to be fit when calculating the GEE-KM test. It is hence computationally efficient when scanning the genome especially for large sample sizes as the null model is the same for testing for the effects of different genes. The proposed method can be readily applied to data with different pedigree structures, while the current R packages such as ‘gee’ and ‘geepack’ can only define a working correlation matrix when all families have the same structure [Chen, et al. 2011].
We have also considered ascertained samples in the simulation study. The results show that given the same sample size, power using ascertained samples is higher compared to the random sampling scheme, while the type I error is well controlled. It suggests that our GEE-based approach has better robustness to ascertainment compared to the mixed model based ML and REML methods. Our approach provides a promising alternative to laborious conditional likelihood adjustment methods using the retrospective model approach [Pfeiffer, et al. 2008; Zheng, et al. 2010]. The robustness of our approach to ascertainment was also supported by analysis of actual data from related individuals in the CFS in which the genome wide Q-Q plot did not show any substantial departure.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health [R37 CA076404 and P01 CA134294 to XW and XL; K99 HL113264 (SL), HG003054 (XZ); R01 HL113338 (XW, XZ, SR and XL) and R01 HL46380 (SR)].
APPENDIX
The Fisher scoring algorithm and the method of moments for estimating α̃ and δ
At a given iteration, α̃ is updated iteratively by with and evaluated at the current parameter estimates. Define the Pearson residual , where μ̂ij is the estimate of μij from the current fit of the null model. The parameter δ at a given step is estimated by .
The asymptotic distribution of the score statistic TS
To derive the asymptotic distribution of the score statistic TS under the null hypothesis, denote , where θ0 = (α0,0)′ is the true value of θ.
Partition A as Axx, Axz, Azx, Azz according to the dimensions of α and β. From a Taylor series expansion, we get , where α̃ is the MLE of α under the null. A Taylor expansion of Uz (θ̃), where θ̃ = (α̃,0)′ about θ0 gives Let , then Uz≈CU(θ0).
Denote . As n → ∞, we have B−1/2U(θ0)→ N(0, I) in distribution. Hence,
where (λ1,λ2,…,λp) are the eigenvalues of B1/2CTCB1/2, and are independent random variables. Cov(yi) in B is estimated by {yi − μi(θ0)i} {yi − μi(θ0)i}T.
A perturbation process for small sample size adjustment
Analogous to the Rademacher bootstrap [Davidson and Flachaire 2008], the perturbed score Ũb equals to , where ri is a random variable generated from the Rademacher distribution (a discrete distribution where a random variate has a half chance of being either +1 or −1). Suppose a total of P samples of the perturbed score Tp are generated, the sample kurtosis γ̂ is calculated as , where and , and is the GEE KM test statistic from a perturbation sample. The perturbation p-value can then be calculated using equation (2).
References
- Carey VJ. R package version 4.13–10. 2002. gee: Generalized Estimation Equation Solver. Ported from S-PLUS to R by Thomas Lumley (versions 3.13 and 4.4) and Brian Ripley (version 4.13) [Google Scholar]
- Chen H, Meigs JB, Dupuis J. Sequence Kernel Association Test for Quantitative Traits in Family Samples. Genet Epidemiol. 2012;37:196–204. doi: 10.1002/gepi.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Liu X, Wei F, Larson MG, Fox CS, Vasan RS, Yang Q. A comparison of strategies for analyzing dichotomous outcomes in genome-wide association studies with general pedigrees. Genet Epidemiol. 2011;35:650–657. doi: 10.1002/gepi.20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun H, Ballard DH, Cho J, Zhao H. Identification of association between disease and multiple markers via sparse partial least squares regression. Genet Epidemiol. 2011;35:479–486. doi: 10.1002/gepi.20596. [DOI] [PubMed] [Google Scholar]
- Davidson R, Flachaire E. The wild bootstrap, tamed at last. J Econometrics. 2008;146:162–169. [Google Scholar]
- Davies R. The distribution of a linear combination of chi-square random variables. J R Stat Soc Ser C Appl Stat. 1980;29:323–333. [Google Scholar]
- Duchesne P, Lafaye De Micheaux P. Computing the distribution of quadratic forms: Further comparisons between the Liu-Tang-Zhang approximation and exact methods. Comput Stat Data Anal. 2010;54:858–862. [Google Scholar]
- Dudbridge F, Koeleman BPC. Rank truncated product of P values, with application to genomewide association scans. Genet Epidemiol. 2003;25:360–366. doi: 10.1002/gepi.10264. [DOI] [PubMed] [Google Scholar]
- Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng T, Elston RC, Zhu X. Detecting rare and common variants for complex traits: sibpair and odds ratio weighted sum statistics (SPWSS, ORWSS) Genet Epidemiol. 2011;35:398–409. doi: 10.1002/gepi.20588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox ER, Young JH, Li Y, Dreisbach AW, Keating BJ, Musani SK, Liu K, Morrison AC, Ganesh S, Kutlar A. Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Hum Mol Genet. 2011;20:2273–2284. doi: 10.1093/hmg/ddr092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Becker LC, Becker DM, Starmer JD, Province MA. Avoding the high Bonferroni penalty in genome-wide association studies. Genet Epidemiol. 2010;34:100–105. doi: 10.1002/gepi.20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Pan W. Powerful multi-marker association tests: unifying genomic distance-based regression and logistic regression. Genet Epidemiol. 2010;34:680–688. doi: 10.1002/gepi.20529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, Lin X. Family-based association tests for sequence data, and comparisons with population-based association tests. Eur J Hum Genet. 2013 doi: 10.1038/ejhg.2012.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird NM, Lange C. Family-based designs in the age of large-scale gene-association studies. Nat Rev Genet. 2006;7:385–394. doi: 10.1038/nrg1839. [DOI] [PubMed] [Google Scholar]
- Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, Christiani DC, Wurfel MM, Lin X. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet. 2012a doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Wu MC, Lin X. Optimal tests for rare variant effects in sequencing association studies. Biostatistics. 2012b;13:762–775. doi: 10.1093/biostatistics/kxs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M-X, Gui H-S, Kwan JS, Sham PC. GATES: a rapid and powerful gene-based association test using extended Simes procedure. Am J Hum Genet. 2011a;88:283–293. doi: 10.1016/j.ajhg.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Basu S, Miller MB, Iacono W, McGue M. A Rapid Generalized Least Squares Model for a Genome-Wide Quantitative Trait Association Analysis in Families. Hum Hered. 2011b;71:67–82. doi: 10.1159/000324839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Lin X, Ghosh D. Semiparametric Regression of Multidimensional Genetic Pathway Data: Least-Squares Kernel Machines and Linear Mixed Models. Biometrics. 2007;63:1079–1088. doi: 10.1111/j.1541-0420.2007.00799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay I, Feingold E, Weeks DE, Thalamuthu A. Association tests using kernel-based measures of multi-locus genotype similarity between individuals. Genet Epidemiol. 2009;34:213–221. doi: 10.1002/gepi.20451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namkung J. Single marker family-based association analysis not conditional on parental information. In: Elston RC, Satagopan JM, Sun S, editors. Statistical human genetics Methods and Protocols. Springer; 2012. p. 371. [DOI] [PubMed] [Google Scholar]
- Ott J, Kamatani Y, Lathrop M. Family-based designs for genome-wide association studies. Nat Rev Genet. 2011;12:465–474. doi: 10.1038/nrg2989. [DOI] [PubMed] [Google Scholar]
- Palmer LJ, Buxbaum SG, Larkin E, Patel SR, Elston RC, Tishler PV, Redline S. A whole-genome scan for obstructive sleep apnea and obesity. Am J Hum Genet. 2003;72:340–350. doi: 10.1086/346064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Park T, Shin D. A simple method fro generating correlated binary variates. Am Stat. 1996;50:306–310. [Google Scholar]
- Pfeiffer RM, Pee D, Landi MT. On combining family and case-control studies. Genet Epidemiol. 2008;32:638–646. doi: 10.1002/gepi.20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite FE. An approximate distribution of estimates of variance components. Biom Bull. 1946;2:110–114. [PubMed] [Google Scholar]
- Schaid DJ, McDonnell SK, Sinnwell JP, Thibodeau SN. Multiple Genetic Variant Association Testing by Collapsing and Kernel Methods With Pedigree or Population Structured Data. Genet Epidemiol. 2013;37:409–418. doi: 10.1002/gepi.21727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schifano ED, Epstein MP, Bielak LF, Jhun MA, Kardia SLR, Peyser PA, Lin X. SNP Set Association Analysis for Familial Data. Genet Epidemiol. 2012;36:797–810. doi: 10.1002/gepi.21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Marchini J, Donnelly P. HAPGEN2: simulation of multiple disease SNPs. Bioinformatics. 2011;27:2304–2305. doi: 10.1093/bioinformatics/btr341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton T, McPeek MS. ROADTRIPS: case-control association testing with partially or completely unknown population and pedigree structure. Am J Hum Genet. 2010;86:172–184. doi: 10.1016/j.ajhg.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng J-Y, Zhang D, Pongpanich M, Smith C, McCarthy MI, Sale MM, Worrall BB, Hsu F-C, Thomas DC, Sullivan PF. Studying gene and gene-environment effects of uncommon and common variants on continuous traits: a marker-set approach using gene-trait similarity regression. Am J Hum Genet. 2011;89:277–288. doi: 10.1016/j.ajhg.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng JY, Zhang D, Chang SM, Thomas DC, Davidian M. Gene-Trait Similarity Regression for Multimarker-Based Association Analysis. Biometrics. 2009;65:822–832. doi: 10.1111/j.1541-0420.2008.01176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Abbott D. A principal components regression approach to multilocus genetic association studies. Genet Epidemiol. 2008;32:108–118. doi: 10.1002/gepi.20266. [DOI] [PubMed] [Google Scholar]
- Wang T, Elston RC. Improved power by use of a weighted score test for linkage disequilibrium mapping. Am J Hum Genet. 2007;80:353–360. doi: 10.1086/511312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Ho G, Ye K, Strickler H, Elston RC. A partial least square approach for modeling gene gene and gene environment interactions when multiple markers are genotyped. Genet Epidemiol. 2009;33:6–15. doi: 10.1002/gepi.20351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Morris NJ, Zhu X, Elston RC. A variance component based multi-marker association test using family and unrelated data. BMC Genetics. 2013;14:17. doi: 10.1186/1471-2156-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel J, Schork NJ. Generalized genomic distance–based regression methodology for multilocus association analysis. Am J Hum Genet. 2006;79:792–806. doi: 10.1086/508346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, Hunter DJ, Lin X. Powerful SNP-set analysis for case-control genome-wide association studies. Am J Hum Genet. 2010;86:929–942. doi: 10.1016/j.ajhg.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Li Q, Bergen AW, Pfeiffer RM, Rosenberg PS, Caporaso N, Kraft P, Chatterjee N. Pathway analysis by adaptive combination of P values. Genet Epidemiol. 2009;33:700–709. doi: 10.1002/gepi.20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaykin DV, Zhivotovsky LA, Westfall PH, Weir BS. Truncated product method for combining P-values. Genet Epidemiol. 2002;22:170–185. doi: 10.1002/gepi.0042. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Heagerty PJ, Hsu L, Newcomb PA. On combining family-based and population-based case–control data in association studies. Biometrics. 2010;66:1024–1033. doi: 10.1111/j.1541-0420.2010.01393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Feng T, Li Y, Lu Q, Elston RC. Detecting rare variants for complex traits using family and unrelated data. Genet Epidemiol. 2010;34:171–187. doi: 10.1002/gepi.20449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Li S, Cooper RS, Elston RC. A unified association analysis approach for family and unrelated samples correcting for stratification. Am J Hum Genet. 2008;82:352–365. doi: 10.1016/j.ajhg.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Young J, Fox E, Keating BJ, Franceschini N, Kang S, Tayo B, Adeyemo A, Sun YV, Li Y. Combined admixture mapping and association analysis identifies a novel blood pressure genetic locus on 5p13: contributions from the CARe consortium. Hum Mol Genet. 2011;20:2285–2295. doi: 10.1093/hmg/ddr113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.