Abstract
Despite the significance of the relationships between embryophytes and their charophyte algal ancestors in deciphering the origin and evolutionary success of land plants, few chloroplast genomes of the charophyte algae have been reconstructed to date. Here, we present new data for three chloroplast genomes of the freshwater charophytes Klebsormidium flaccidum (Klebsormidiophyceae), Mesotaenium endlicherianum (Zygnematophyceae), and Roya anglica (Zygnematophyceae). The chloroplast genome of Klebsormidium has a quadripartite organization with exceptionally large inverted repeat (IR) regions and, uniquely among streptophytes, has lost the rrn5 and rrn4.5 genes from the ribosomal RNA (rRNA) gene cluster operon. The chloroplast genome of Roya differs from other zygnematophycean chloroplasts, including the newly sequenced Mesotaenium, by having a quadripartite structure that is typical of other streptophytes. On the basis of the improbability of the novel gain of IR regions, we infer that the quadripartite structure has likely been lost independently in at least three zygnematophycean lineages, although the absence of the usual rRNA operonic synteny in the IR regions of Roya may indicate their de novo origin. Significantly, all zygnematophycean chloroplast genomes have undergone substantial genomic rearrangement, which may be the result of ancient retroelement activity evidenced by the presence of integrase-like and reverse transcriptase-like elements in the Roya chloroplast genome. Our results corroborate the close phylogenetic relationship between Zygnematophyceae and land plants and identify 89 protein-coding genes and 22 introns present in the chloroplast genome at the time of the evolutionary transition of plants to land, all of which can be found in the chloroplast genomes of extant charophytes.
Keywords: charophytes, bryophytes, land plants, chloroplast genomics
Introduction
It is now established that land plants evolved from freshwater green algal ancestors of the charophyte algae (McCourt 1995; Karol et al. 2001; Wodniok et al. 2011). The transition of plants from an aquatic to the terrestrial environment is thought to have occurred about 425–490 Ma (Sanderson 2003; Wellman et al. 2003; Gensel 2008; Rubinstein et al. 2010) and was followed by a rapid diversification of plant lineages that resulted in dramatic changes to the Earth's biosphere (Kenrick and Crane 1997; Lenton et al. 2012). Given the great evolutionary significance of the colonization of land by plants and the fundamental role of plants in Earth's ecosystems, the characterization of the ancestor of embryophytes has long been of special interest to evolutionary biologists. From the cytological, physiological, and biochemical perspective, it is evident that some of the features typically associated with land plants have their molecular origins in the preterrestrial era. Such features include multicellularity and three-dimensional growth, cellulosic cell walls, phragmoplast formation during cell division, or intercellular communication mediated by plasmodesmata, and plant hormones (Leliaert et al. 2012). Although these features are indeed fundamental to land plants, some of them involve genes that appear to have orthologs in charophyte algae (Timme and Delwiche 2010; De Smet et al. 2011). A better understanding of the evolution of embryophyte design is therefore dependent upon an improved understanding of streptophyte relationships but is currently hindered by the paucity of charophyte nuclear and organellar genomic data available for study.
Phylogenetically, extant charophyte (Charophyta) lineages form a paraphyletic assemblage with the land plants (Embryophyta) and are together classified as the Streptophyta. However, elucidation of phylogenetic relationships among the charophyte groups, namely, Chlorokybophyceae, Mesostigmatophyceae, Klebsormidiophyceae, Zygnematophyceae, Charophyceae, and Coleochaetophyceae, with respect to the land plant clade, has been controversial. Early phylogenetic studies appeared to provide evidence for an intuitively elegant progression of increasing morphological complexity from single-cell organisms of the Chlorokybophyceae, Mesostigmatophyceae, and Klebsormidiophyceae, through the multicellular, filamentous, and thallose structured algae of the Zygnematophyceae (conjugating algae) and Coleochaetophyceae, with the most complex, and most land plant-like, species of the Charophyceae being most-closely related to the land plants (Karol et al. 2001; McCourt et al. 2004). This same tree topology where Charophyceae are the sister group to land plants was again obtained in a six-gene phylogenetic analysis by Qiu et al. (2006). However, in the same study, gene matrices derived from complete chloroplast genomes yielded a highly supported monophyletic Zygnematophyceae clade as the sister group to land plants. More recent analyses based on chloroplast (Turmel et al. 2006, 2007) and nuclear phylogenomic data (Wodniok et al. 2011; Laurin-Lemay et al. 2012; Timme et al. 2012) place Zygnematophyceae, or a clade uniting Zygnematophyceae and Coleochaetophyceae, as closest group to the land plants, whereas mitochondrial gene data sets remain inconclusive (Turmel et al. 2013). Currently, the best-supported hypothesis of charophyte branching order has a clade uniting Chlorokybus and Mesostigma at the base of the streptophyte tree with Klebsormidiophyceae, then Charophyceae, the next two diverging lineages, respectively, with the closest relatives of land plants, either the Zygnematophyceae alone or a clade consisting of both Zygnematophyceae and Coleochaetophyceae (Turmel et al. 2006; Wodniok et al. 2011; Laurin-Lemay et al. 2012; Timme et al. 2012).
Photosynthetic organelles have a clear functional continuity spanning the transition period between aquatic algal and terrestrial embryophytic lifestyles. With a typical genome size of between 115 and 170 kb and a gene complement of 100–120 unique genes (Green 2011; Wicke et al. 2011), the streptophyte plastid gene repertoire is relatively stable because retention of the core set of chloroplast genes is likely under strong selection, and gene gains are exceptional (Wicke et al. 2011). Although many of the genes necessary for chloroplast-specific functions have been transferred to the nucleus and have their products imported into chloroplasts from the cytoplasm, the genes encoding transmembrane polypeptides (subunits of atp, ndh, pet, psa, and psb complexes) tend to be retained by the chloroplast genome (cpDNA), presumably because importing the protein products of these genes would be difficult (Wicke et al. 2011). Other plastid genes exhibit high expression levels at early developmental stages (e.g., genes for structural RNAs, ribosomal proteins, and RNA polymerase), which likely favor their localization in the chloroplast rather than the nucleus (Wicke et al. 2011). A stable gene content of the chloroplast genome is accompanied by a conserved structural organization of its circular map whereby two inverted repeats (IRs) are separated by a large single-copy (LSC) region and a small single-copy (SSC) region. As this quadripartite architecture likely confers physical resistance to recombinational losses (Palmer and Thompson 1981), structural changes to chloroplast genomes are infrequent, and their identification and distribution can be used to supplement sequence data in the evaluation of phylogenetic hypotheses (Qiu et al. 2006; Turmel et al. 2006, 2007; Jansen et al. 2008; Grewe et al. 2013). Although gene losses are often homoplastic (Martin et al. 1998), other rarer genomic changes such as large inversions, insertion, and deletion events (indels), intron gain and loss, or gene order rearrangements may provide reliable phylogenetic information (Rokas and Holland 2000).
The gene complements of land plant chloroplasts do not differ substantially from those of charophyte algae (Turmel et al. 2006; Green 2011; Wicke et al. 2011). Moreover, most introns found in embryophyte chloroplast genes are also present in charophyte chloroplasts and had been acquired before the transition to land (Turmel et al. 2006). However, although the chloroplast gene order among land plant groups is fairly stable (fig. 2, Wicke et al. 2011), dozens of sequence inversions separate the known charophyte chloroplast genomes from one another and from the conserved gene order found in bryophytes (Turmel et al. 2005, 2006). Chloroplast genome rearrangements are especially abundant in Zygnematophyceae, and it has been suggested that their high occurrence is causally related to the loss of quadripartite structure in this class (Turmel et al. 2005). However, a satisfactory mechanistic explanation of such causality is lacking and a broader examination of the zygnematophycean cpDNA architecture has yet to be conducted.
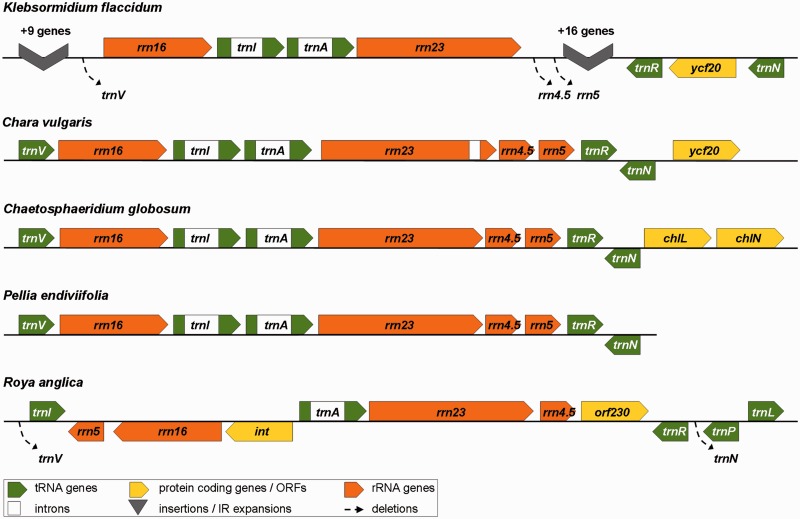
Fig. 2.—
IR regions of Klebsormidium and Roya, in comparison to Chaetosphaeridium and Chara (charophytes), and Pellia (a bryophyte).
Here, we report newly sequenced chloroplast genomes of three charophyte algae, namely, Klebsormidium flaccidum (Klebsormidiophyceae), Mesotaenium endlicheriannum (Zygnematophyceae), and Roya anglica (Zygnematophyceae). Klebsormidium flaccidum is a species from the last taxonomic class of charophyte algae to lack a completely sequenced chloroplast genome. The two zygnematophycean taxa are both saccoderm desmids of the previously unsampled family Mesotaeniaceae and thought to be early diverging or transitional forms of conjugating algae. The three genomes aid our understanding of the structural changes that occurred in chloroplasts during the evolution of early-diverging streptophyte clades. Moreover, comparisons of the genetic composition of chloroplast genomes ancestral to embryophytes with those of the Zygnematophyceae reveal several uniquely shared features that corroborate the close phylogenetic relationship of these plant groups.
Materials and Methods
Algal Cultures and Chloroplast Genome Sequencing
Cultures of K. flaccidum ([Kützing] P.C. Silva, K.R. Mattox, and W.H. Blackwell, 1972) and M. endlicherianum (Nägeli, 1849) were obtained from the SAG Culture Collection of Algae (accession numbers SAG121.80 and SAG12.97, respectively) and R. anglica (G.S. West in W.J. Hodgetts, 1920) (accession number ACOI799) from Algoteca de Coimbra (hereafter we refer to the samples as “Klebsormidium,” “Mesotaenium,” and “Roya,” for brevity). Klebsormidium and Mesotaenium cells were inoculated on Petri dishes with 1.5× Bold's basal medium (Andersen et al. 2005) supplemented with agar (1.5%, w/v) and cultivated for 10–14 days in a growth chamber under 14 h:10 h light:dark regime (100–120 µmol s−1 m−2 irradiation). Roya was grown in a liquid mixture of LC (Algoteca de Coimbra, Portugal) and Bold's basal medium (1:1) under the same light conditions as above. After approximately 1 month, the culture of Roya was passed through a 20–25 μm filter paper, the cells collected on the filter were rinsed with sterile 0.5× medium, and used for DNA extraction.
Approximately 1 g of cells was harvested for each taxon. The samples were briefly deep frozen in liquid nitrogen and used for DNA extraction without any further mechanical cell breaking. The frozen cells were resuspended in 5–10 ml of extraction buffer (0.1 M Tris; 20 mM Na2EDTA; 1.4 M NaCl; 2% CTAB [w/v]; 0.3% 2-ME; 0.1 mg ml−1 RNase A; pH ∼ 8.5) and incubated for 1 h at 65 °C with occasional vortexing. Subsequently, the tubes were chilled on ice, the DNA was extracted with equal volume of chloroform:isoamylalcohol (24:1) and precipitated with isopropanol for 1 h at −20 °C. The precipitate was collected and rinsed with wash buffer (70% ethanol; 0.12 M sodium acetate) and 70% ethanol. The pellet was dissolved in TE overnight, and the DNA was purified with High Pure PCR Product Purification Kit (Roche) according to the manufacturer's instructions. Quality of the DNA was checked on an agarose gel, and DNA quantity and purity were determined by nanodrop. Mesotaenium was sequenced on ½ picotiter plate with GS FLX Titanium (IGSP Genome Sequencing & Analysis Core Resource, Duke University), whereas Klebsormidium and Roya were sequenced on a single lane of Illumina HiSeq2000 (BGI Tech Solutions Co. Ltd, Hong Kong, China) along with four other samples not reported here. The library type for Illumina sequencing was 91 paired end, with approximately 500 bp fragment size.
Data Processing and Assembly
Roche 454 pyrosequencing and Illumina short-read data were imported into Geneious 5.6.3 (Biomatters, http://www.geneious.com, last accessed April 8, 2014) in sff and fastq formats, respectively. After the removal of oligonucleotide adapters, sequences were trimmed from both sides, discarding regions with >4% (sff) or >5% (fastq) chance of an error per base. As the data were from a whole-genome shotgun collection of sequences but only the chloroplast fraction was of interest, the assembly of the chloroplast genomes was undertaken in three stages. 1) For each taxon, a reference was chosen from the set of known chloroplast genomes of streptophytic algae. From each reference genome, protein-coding genes were extracted and used as templates for mapping of the sequence reads in Geneious. This reference-guided reconstruction typically yielded a set of short (0.1–1 kb), high-confidence chloroplast contigs representing <10% of the genome. 2) The full paired-read data sets were used for focused assembly by PRICE (Paired-Read Iterative Contig Extension, version 0.18; Ruby et al. 2013), utilizing the short chloroplast contigs as initial seeds. In PRICE assemblies, the minimal overlap was set to 30, and the minimal percent identity to 95 and 85 for Illumina and 454 data sets, respectively. For 454 sequence reads, the -spf argument was used to create false paired-end data file. Variable trimming and filtering options were applied. 3) Resulting contigs usually representing the whole reconstructed genome were imported back to Geneious, where the sequence reads were remapped onto the contigs. This third stage enabled the sequence coverage and base-assignment confidence to be evaluated, and the identification and adjustment of ambiguous sites and repeated regions.
Special attention was paid to the reconstruction of the IR regions of the chloroplast genomes. In a standard PRICE assembly of a quadripartite-structure chloroplast genome, one of the following problems may occur: two IRs are collapsed into a single contig; extension of the second IR stops due to reads mapping to an existing contig; and IRs and single-copy regions are joined incorrectly. To overcome these issues, a simple strategy was applied. After an IR was identified in preliminary runs of 2)–3) assembly steps, a “dead” IR contig was prepared and added to the initial seeds for another 2)–3) assembly run. The “dead” IR consisted of an IR region extended for approximately 500 cytosines on both ends, which effectively excludes this contig, as well as all the IR-mapping reads, from the PRICE assembly process. The remaining seeds are extended until the completion of SC regions, which contain short overlaps with the IRs, enabling the four regions to be joined correctly into a circle.
Annotation and Analyses of the Chloroplast Genome Content
The software DOGMA (Wyman et al. 2004) was used for initial gene annotations. Thereafter, a thorough examination of protein-coding gene content was performed as follows. Open reading frames (ORFs) in the assembled genomes were identified by getorf (part of the EMBOSS suite: minimal length 30 nucleotides, translations from start to stop codon retrieved), and BLASTp (Altschul et al. 1990) was used to detect similarities with a National Center for Biotechnology Information (NCBI) Reference Sequence (refseq) library of all known chloroplast proteins (downloaded in October 2012). After the annotation of known proteins, we further examined longer ORFs from conspicuously “empty” regions to determine whether these regions had unreported homologs. To facilitate these analyses, we built a custom BLAST database of plastid ORFs (determined by getorf: minimal length 100 nucleotides; translations from stop to stop codon retrieved) from all available Viridiplantae (chlorophytes and streptophytes) chloroplast genomes (downloaded from NCBI GenBank October 2012). This library consisted of 1.17 million ORFs and was used in BLASTp analyses to identify sequences with similarity (E value < 1e-4) to the ORFs identified from the “empty” regions of the newly assembled genomes. Introns were identified by comparison to gene alignments of other algae and representative bryophytes. Exon–intron borders were inferred with the aid of protein alignments and intron border consensus sequences (Sugita and Sugiura 1996). To determine the frequency of short repeats, one IR was removed from the quadripartite genomes, and direct and inverted repeats >20 bp were searched with a 1e-03 threshold using REPuter (Kurtz et al. 2001), and an average number of repeats per kb was calculated. The newly constructed genomes were visualized using circular genome maps created by OGDraw (Lohse et al. 2007).
Gene contents of the newly reconstructed chloroplast genomes were compared with the genomes of other streptophyte algae and a “hypothetical land plant ancestor” (HLPA). The gene content of the HLPA unit was inferred from a selection of taxa representing all major lineages of land plants (the same taxon set as used in the phylogenetic analyses below), assuming monophyly of land plants and only vertical transfer of genes. Genome rearrangements between charophytes and two land plants, namely, Pellia endiviifolia (a liverwort; NC_019628), and Isoetes flaccida (a lycophyte; NC_014675), were determined using multiple genome rearrangements (MGR: Bourque and Pevzner 2002) using analysis that ignored the transfer RNA (tRNA) genes and one of the IR in quadripartite genomes. Because the choice between the two IR copies is relevant for the gene order analyses, both alternative “single-IR” gene orders were considered for quadripartite genomes, and the arrangement leading to the most parsimonious result was chosen for pairwise genome comparisons in MGR.
Phylogenetic Analyses
Phylogenetic analyses of 83 protein-coding chloroplast genes from the newly assembled genomes of Roya, Mesotaenium, and Klebsormidium, plus 23 streptophytes and four chlorophyte outgroup taxa, were conducted. Maximum likelihood and Bayesian Markov chain Monte Carlo (MCMC) analyses were conducted using among-site (PhyloBayes CAT model; Lartillot and Philippe 2004) and among-lineage (P4 NDCH model; Foster 2004) composition models to determine the best-fitting models and the best-supported trees. Details of these analyses are presented elsewhere (Civáň et al. unpublished); the best-fitting PhyloBayes CAT-model using the gcpREV exchange rate model (Cox and Foster 2013) analysis of amino acid is presented here as a reference tree for the best-supported hypothesis of relationships based on these data.
Analyses of chloroplast genome structural features were based on 66 parsimony informative characters: The presence or absence of 30 monocistronic genes and 19 group II introns, plus the gene complement and gene order in 17 operons. tRNA genes and their introns were not considered, except for those tRNA genes located within polycistronic units. Introns were scored as Dollo characters with “absence” assumed to be the ancestral condition. Dollo character coding corresponds to a model in which each derived state is allowed to originate only once during evolution, and all homoplasy takes the form of reversals to the ancestral condition (Swofford and Begle 1993). The ancestral state of 28 monocistronic protein-coding genes was assumed to be “presence” with the characters treated as irreversible, therefore allowing multiple losses but no secondary gain of genes. The “presence” or “absence” of 77 additional genes within 17 operons was also evaluated (Sugita and Sugiura 1996; Wicke et al. 2013—information regarding the operonic organization is derived from model angiosperms but was adapted for the gene set observed here). Operons were coded as multistate characters defined by step matrices, with unspecified ancestral states. In the step matrices, every change in operon organization was of equal distance except irreversibility of genes lost from the genome (i.e., gene loss from an operon equals distance 1; gene gain in an operon from another cpDNA location equals distance 1; and gene gain in an operon from outside of cpDNA equals infinity). Structural characters were subjected to parsimony analysis in PAUP 4.0 (Swofford 2003), with optimal trees obtained using the branch-and-bound algorithm. Bootstrap analyses with 1,000 replicates were performed heuristically with default parameters. (A NEXUS formatted character matrix used for the structural data analyses is included in the supplementary material, Supplementary Material online.)
Results
Chloroplast Genome Assembly
For each of Klebsormidium, Mesotaenium, and Roya, assembly of the short-read data yielded a single large contiguous sequence for which it was possible to close into a circle. Because of high sequence read coverages, 153, 379, and 273 mean reads per site for Klebsormidium, Mesotaenium, and Roya, respectively, no gaps or ambiguous regions were present (supplementary fig. S1, Supplementary Material online). Summary statistics of the data and the genome assemblies are presented in table 1.
Table 1.
Summary Statistics of the Genome Assembly Data
| Platform | Number of Reads Obtained (Total) | Mean Read Length (After Trimming) (bp) | Proportion of Reads Mapping to the cp Genome (%) | Length of the cp Genome (bp) | Coverage (×) |
||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Min | Max | Standard Deviation | ||||||
| Klebsormidium flaccidum | Illumina HiSeq2000 | 46,124,918 | 86.5 | 0.66 | 176,832 | 152.7 | 6 | 235 | 23.6 |
| Mesotaenium endlicherianum | Roche 454 | 689,398 | 357.1 | 23.2 | 142,017 | 378.9 | 85 | 589 | 72.2 |
| Roya anglica | Illumina HiSeq2000 | 54,070,476 | 86.2 | 0.78 | 138,275 | 273.3 | 1a | 518 | 105.1 |
aThe 1× coverage was 10-bp long and located within an AT-rich intergenic region.
Klebsormidium flaccidum
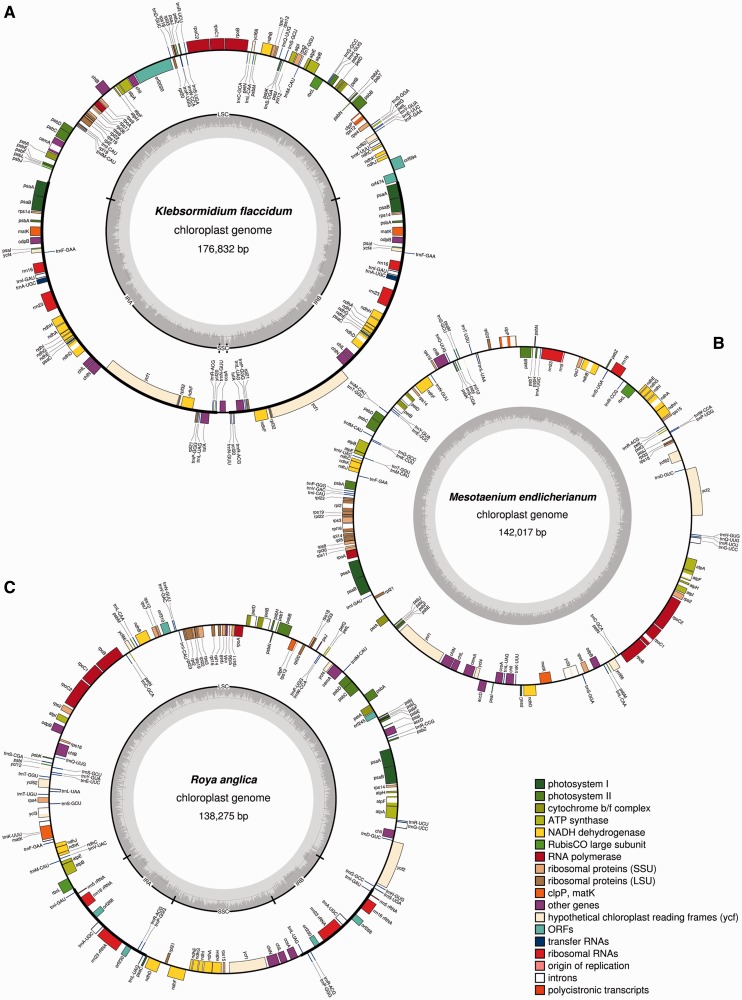
The chloroplast genome of Klebsormidium was assembled into a circular map of 176,832 bp (fig. 1A; NCBI GenBank accession number KJ461680); the third largest among currently sequenced streptophyte chloroplast genomes, smaller only than Pelargonium (Geraniaceae, Spermatophyta) and Chara (Charales, Charophyceae). The genome has a quadripartite organization, which differs from the typical embryophytic architecture by having exceptionally large IRs (51,118 bp each), a greatly reduced SSC region (1,817 bp), and a relatively shorter LSC region (72,779 bp). The expanded IR regions contain both small (rrn16) and large (rrn23) ribosomal RNA (rRNA) genes, seven tRNA genes typically found in streptophyte IRs, plus 23 additional protein-coding genes typically located in single-copy regions (fig. 2). Most remarkably, the rrn5 gene (5S rRNA) and the region homologous to the rrn4.5 gene in embryophytes (4.5S rRNA—in nonembryophyte streptophytes, the rrn4.5 gene-coding region forms an integral part of the 3′-end of the 23S ribosomal subunit) are absent from the genome (supplementary fig. S2, Supplementary Material online). The SSC region contains only a single gene (ccsA), whereas 59 protein-coding genes, and 21 tRNA genes, reside in the LSC region. Six ribosomal protein genes (rpl14, rpl16, rpl23, rps3, rps15, and rps16) usually present in streptophyte chloroplast genomes are missing, as are several other protein-coding genes (fig. 3). Two genes in the Klebsormidium genome, rps12 and psbA, require transsplicing for correct protein translation. In total, genes coding for two rRNA, 28 tRNA, and 82 proteins were identified in the Klebsormidium chloroplast genome. The GC content of the genome is relatively high (42%) compared among streptophytes (average 37%) but differs substantially between the IR and single-copy regions (46.0% and 36.5%, respectively). Mean intergenic spacer length was 358 bp (52,071 bp in total), with two conspicuous exceptions (6,340 and 4,231 bp). These two extended intergenic regions contain three unidentified ORFs (6,063, 1,785, and 1,425 bp), which had no strong matches (E value < 1e-4) among BLASTp searches of the refseq database or the custom ORF library. Group II introns were found in seven genes (table 2) and account for 3.7% of the total genome length. By comparison to the genome of Chara (Charophyceae) which has a larger overall size, the proportion of intergenic spacers and introns is several times lower, indicating that the large genome size of Klebsormidium can be attributed mainly to large IR regions.
Fig. 1.—
Chloroplast genome maps of Klebsormidium flaccidum (A), Mesotaenium endlicherianum (B), and Roya anglica (C).
Fig. 3.—
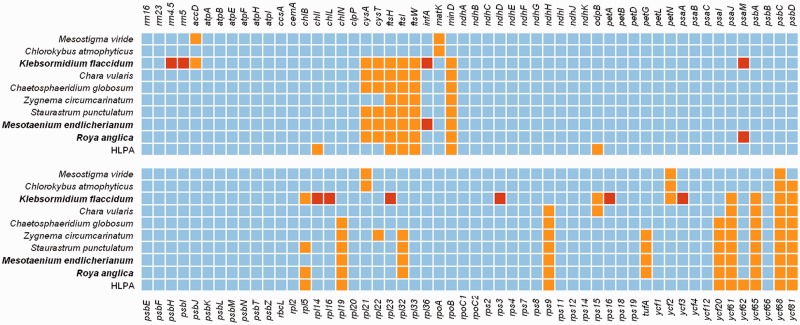
Chloroplast gene content among charophytes and an inferred HLPA. All rRNA and protein-coding genes found within the sample set of the phylogenetic analyses are included. Gene presence and absence are indicated by blue and orange shading, respectively. Novel absences of genes with respect to other charophyte genomes are highlighted in red. (Note that the disambiguation of ycf2/ftsH has been newly interpreted, see supplementary table S1, Supplementary Material online.)
Table 2.
Intron (Group II) Distribution among Charophytes and HLPA
| atpF i1 | cemA i1 | clpP i1 | clpP i2 | clpP i3 | ndhA i1 | ndhB i1 | ndhD i1 | petB i1 | petD i1 | psbA i1 | rpl2 i1 | rpl16 i1 | rpoC1 i1 | rps12 i1 | rps12 i2 | rps16 i1 | trnA(UGC) i1 | trnG(UCC) i1 | trnI(GAU) i1 | trnK(UUU) i1 | trnV(UAC) i1 | ycf3 i1 | ycf3 i2 | ycf66 i1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Klebsormidium | – | + | – | – | + | – | – | + | + | – | + | – | – | – | + | – | – | + | – | + | + | – | – | – | – |
| Mesotaenium | + | – | + | + | – | + | + | – | + | + | – | + | + | + | + | + | – | – | + | – | – | – | + | + | + |
| Roya | + | – | – | – | – | – | – | – | + | + | – | – | – | – | + | – | – | + | + | – | + | – | + | – | – |
| Zygnema | – | – | – | – | – | – | + | – | – | + | – | + | + | + | + | + | + | – | + | – | – | – | + | + | + |
| Staurastrum | + | – | – | – | – | – | – | – | – | – | – | – | + | – | + | – | – | + | + | – | + | – | + | – | – |
| Chara | – | – | + | – | – | + | + | – | + | – | – | + | + | – | + | – | + | + | + | + | + | + | + | + | + |
| Chaetosphaeridium | – | – | + | – | – | + | + | – | + | + | – | + | + | + | + | – | + | + | + | + | + | + | – | + | + |
| HLPA | + | – | + | + | – | + | + | – | + | + | – | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
Note.—Multiple introns occurring in the same gene are labeled numerically.
Mesotaenium endlicherianum
The chloroplast genome of Mesotaenium was assembled as a circular sequence comprising 142,017 bp (fig. 1B, NCBI GenBank accession number KJ461681) and lacks a quadripartite structure, as do the two previously published Zygnematophyceae chloroplast genomes (namely Zygnema and Staurastrum; Turmel et al. 2005). The Mesotaenium genome contains 88 protein-coding, 4 rRNA, and 34 tRNA genes, and although it is 23 and 15 kb shorter than Zygnema and Staurastrum, respectively, it does not contain fewer genes (fig. 3). Intergenic spacers occupy almost one-third of the genome length (46,765 bp), with a mean intergenic distance of 357 bp. Group II introns were found in 12 genes, with clpP and ycf3 having two introns each (table 2), and the group I intron typically found in the streptophyte trnL-UAA gene is present. With an average size of 669 bp, introns of Mesotaenium are similar in length to those of bryophytes (713 bp) rather than the longer introns in the other two zygnematophycean chloroplast genomes (966 bp). The overall genome GC content (42%) is notably higher than in the other chloroplast genomes of Zygnematophyceae (32%) or land plants (37%).
Roya anglica
The chloroplast genome of Roya was reconstructed as a circular sequence of 138,275 bp in length (fig. 1C, NCBI GenBank accession number KJ461682), making it the shortest of the four zygnematophycean chloroplast genomes sequenced so far (including Mesotaenium here). (One 10-bp region in the Roya genome had only 1X coverage: However, as this small stretch was in an AT-rich intergenic spacer and surrounded by well-supported paired reads, we did not verify the region via Sanger sequencing.) Unlike other zygnematophycean, the Roya genome has a quadripartite architecture. The genome sequence consists of SSC and LSC regions (20,213 bp and 92,926 bp, respectively) and a pair of IRs (12,568 bp each). The IRs of Roya bear some resemblance to a typical chloroplast IR in terms of gene content—all genes of the rRNA operon (rrn16–trnI-GAU–trnA-UGC–rrn23–rrn4.5–rrn5) are present—although, the integrity of this operon has been disrupted and the genes are merely neighboring units with jumbled order and orientation (fig. 2 and supplementary fig. S3, Supplementary Material online). At least two rearrangements would be necessary to restore the standard order of the rRNA operon. The IRs of Roya also contain three additional tRNA genes (trnR-ACG, trnP-GGG, and trnL-UAG) and two longer ORFs (orf268 and orf230). The protein translation of orf268 (807 bp) has high similarity (E value: 5e-14) to an IR-located int gene from the chloroplast genome of a chlorophycean algae Oedogonium cardiacum (Brouard et al. 2008). Because int encodes a protein belonging to the family of tyrosine recombinases (Brouard et al. 2008), the product of orf268 was labeled as a putative recombinase/integrase protein. In addition, orf268 has high similarity to ORF (46,439–46,717) in the Anthoceros (a hornwort) chloroplast genome (E value: 1e-13) although the latter is not located within the IR and is significantly shorter, suggesting that it may not be homologous by descent with orf268. The second unidentified reading frame orf230 (693 bp) shows high similarity to chloroplast ORFs present in two ferns of the Ophioglossaceae, namely Mankyua chejuensis and Ophioglossum californicum (E values: 3e-18 and 5e-06, respectively).
The single-copy regions of the Roya chloroplast genome contain 28 tRNA genes, 87 protein-coding genes, and two additional ORFs with high similarities to hypothetical proteins reported for other Zygnematophyceae. The first of these additional ORFs, orf245, has significant similarity to locus StPuCp039 of Staurastrum (E value: 1e-12) and locus ZyCiCp066 of Zygnema (E value: 2e-09), and the second, orf310, has similarity to a putative reverse transcriptase (RT) locus (StpuCp054) of Staurastrum (E value: 4e-16). RTs and integrases are not normally found in chloroplast genomes, consequently the presence of RT-like orf310 and int-like orf268 in Roya may suggest retroelement activity. The intergenic spacers occupy 30% of the genome (41,704 bp in total), with a mean intergenic distance of 297 bp: A similar gene density to the economically packed chloroplast genome of Mesotaenium. However, the overall GC content of the Roya genome (33%) more closely resembles the base composition in Zygnema (31%) and Staurastrum (33%) than Mesotaenium (42%).
Phylogenetic Analyses
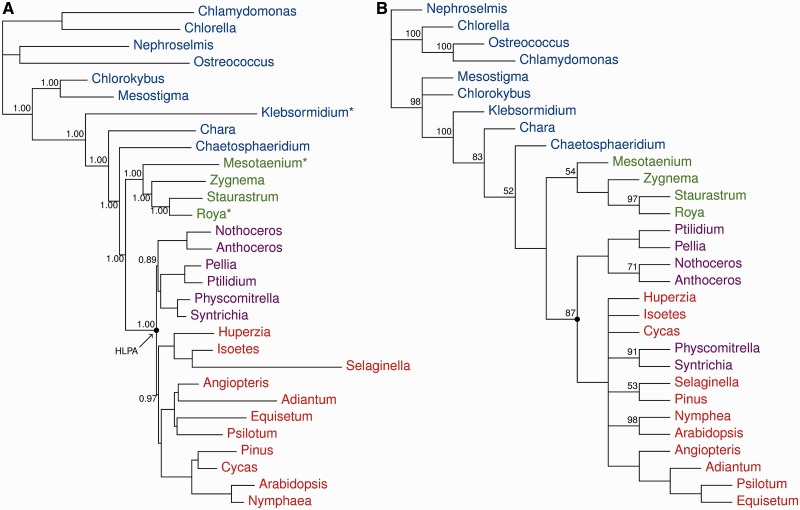
In figure 4A, a Bayesian MCMC analysis of the best-fitting model (PhyloBayes CAT + gcpREV + Γ4; −Lh = 244,645.3855) of amino acid data of the 83 proteins is presented. The tree shows strong support (>0.95 posterior probability) for the paraphyly of charophytes, with Klebsormidium branching early in the phylogenetic grade before Chara and Chaetosphaeridium, and with Zygnematophyceae as the sister group to land plants. Within the Zygnematophyceae, all relationships are strongly supported, with Mesotaenium forming the earliest-branching lineage, and Zygnema sister to a clade formed by Roya and Staurastrum. This finding is in conflict with the traditional placing of Roya in the family Mesotaeniaceae but is in agreement with other phylogenetic reconstructions of conjugating algae (Gontcharov et al. 2003; Gontcharov and Melkonian 2010). Parsimony analysis of the structural data (gene and intron content, and operon structure) identified six optimal trees (tree length 239 steps, consistency index 0.243, retention index 0.786): The strict consensus tree is presented in figure 4B. Nonparametric parsimony bootstrap analysis of the structural data poorly supports a monophyletic Zygnematophyceae (54% bootstrap proportion [BP]) with strong support for the sister-group relationship between Roya and Staurastrum (97% BP). The streptophytes as a whole are well supported (98% BP), with Mesostigma and Chlorokybus forming the earliest-branching lineage. The remaining streptophytes form a well supported (100% BP) clade within which Klebsormidium is the first diverging lineage (83%). Relationships among Chara, Chaetosphaeridium, Zygnematophyceae, and the land plant clade (itself 87% BP) are unsupported (or negligibly supported <70%), but the topology is nevertheless congruent with that of the protein tree.
Fig. 4.—
Phylogenetic analyses. (A) Bayesian MCMC phylogenetic analyses of 83 protein-coding chloroplast genes: PhyloBayes CAT + gcpREV + Γ, marginal likelihood: −Lh = 244,645.3855. (B) Strict consensus tree of six most parsimonious trees (length 239, consistency index = 0.243, retention index = 0.786) resulting from analysis of the structural data (gene and intron content, operon structure). Numbers at nodes are posterior probabilities and nonparametric bootstrap values for (A) and (B), respectively. The nodes representing the HLPA are highlighted.
Comparisons between Charophyte and Land Plant Chloroplast Genomes
The protein-coding gene complements of the Klebsormidium, Roya, and Mesotaenium chloroplast genomes are summarized in figure 3. The chloroplast genome of Klebsormidium, with a repertoire of only 82 unique protein-coding genes, has the lowest protein-coding gene content of any charophyte plastid genome reported to date. Hence, the Klebsormidium chloroplast gene set is more dissimilar to the estimated content of the HLPA (22 presence/absence differences) than are the genomes of Mesostigma or Chlorokybus (18 and 16 presence/absence differences, respectively). In contrast, the taxa with gene complements most closely resembling the HLPA are Roya and Chaetosphaeridium with eight presence/absence differences each: the other three Zygnematophyceae each have one additional difference. Comparisons of the presence and absence of group II chloroplast introns show that Chaetosphaeridium is the most similar to the HLPA with 17 introns at congruent positions (table 2). However, Mesotaenium is the next-most similar with 16 introns at common positions and also has the clpP-intron-2 that is common in land plant chloroplast genomes but has not previously been found in charophyte algae. When the operons (polycistronic units) of charophyte chloroplast genomes are compared with those of land plants, the operonic complements of Chara and Chaetosphaeridium show greater similarity to the HLPA (12 and 13 identical operons, respectively) than do Zygnematophyceae (11 or fewer identical operons). The operonic organization of Roya is the next-most similar to the HLPA (11 concordant operons), whereas the other three Zygnematophyceae bear as few operons of early land plants as do more distantly related streptophyte algae, such as Klebsormidium. This lack of maintenance of operonic integrity among Zygnematophyceae (excepting Roya) is consistent with the high number of implied genome rearrangements identified by MGR analysis (supplementary fig. S4, Supplementary Material online). The syntenic structure of the Staurastrum chloroplast genome implies 20 and 23 rearrangements to match the gene order in Pellia (liverwort) and Isoetes (lycopod), respectively. Roya appears to have the least number of rearrangements among the known zygnematophycean chloroplast genomes with a minimum of 18 and 21 changes implied by comparison to the Pellia and Isoetes genomes, respectively. However, Roya and Staurastrum are also highly rearranged with respect to each other, with 18 implied rearrangements. An even greater number of rearrangements separate the chloroplast genome of Pellia from Mesotaenium and Zygnema (at least 25 and 32 changes, respectively). By comparison, the gene order of Chaetosphaeridium is more similar to land plants than those of other charophytes and requires as few as 10 changes to match the operonic organization of Pellia and Isoetes. Although the abundance of short sequence repeats has previously been implicated as a possible mediator of genome arrangements, numbers of short repeats are not exceptionally high in the two zygnematophycean genomes reported here. In Klebsormidium and Roya, short sequence repeats were relatively rare (0.24 and 0.38 repeats/kb, respectively) and similar to the numbers found in Chaetosphaeridium, Staurastrum, Mesostigma, and Chlorokybus (all <1 repeat/kb). A greater number (1.68 repeats/kb) were recorded in Mesotaenium; however, the amount is still fewer than in Chara and Zygnema (3.16 and 25.73 repeats/kb, respectively).
Discussion
New Insights into the Chloroplast Genomics of Charophytes
The absence of the 5S rRNA gene from the Klebsormidium chloroplast genome, and the region homologous to the 4.5S rRNA gene of embryophytes, from the 3′-end of the 23S rRNA gene, is the first report of an incomplete set of rRNA genes in either chloroplast or mitochondrial genomes; even within the greatly reduced chloroplast genomes of parasitic plants, the rRNA operon remains intact (Krause 2008). Because the usual complement of rRNA subunit genes is assumed vital to the assembly and function of ribosomes, it seems likely that the 4.5S homologous region and 5S rRNA genes have been translocated to the nuclear genome of Klebsormidium and that their products are imported into the chloroplast stroma. Nevertheless, multiple losses among eukaryotes of the 5S gene in the mitochondrion (Adams and Palmer 2003) suggest that complete loss of these ribosomal subunits cannot be entirely ruled out. If the assumption of rRNA translocation from the nucleus is correct, chloroplast-directed rRNA import renders plastid protein synthesis in Klebsormidium ultimately dependent on the nucleus and raises questions concerning the mechanisms of inter-compartmental RNA trafficking. Additionally, if the 4.5S rRNA is being imported into the chloroplast, then it is also acting as a separate 4.5S rRNA species as in the embryophytes and is not an integral part of the 23S rRNA as is implied by its annotation in nonembryophyte streptophyte chloroplast genomes. The transport of nuclear mRNA into the chloroplast is known to occur (Nicolaï et al. 2007), and indirect evidence suggests that tRNAs are imported from outside the plastid in some parasitic plants (Bungard 2004). However, to date, the import of rRNA into the chloroplast has not been demonstrated. Although, the mechanism(s) of chloroplast-directed RNA import remain uncharacterized, two candidate pathways are currently considered plausible. First, the import of rRNA into the chloroplast could be facilitated by a protein precursor utilizing the protein import pathway, as is the case of tRNA transport into mitochondria (Schneider 2011). Alternatively, short noncoding RNA sequences may be responsible for chloroplast localization of nuclear transcripts (Gómez and Pallás 2010). In either case, the chloroplast genome of Klebsormidium is unusual in lacking the 5S rRNA gene, 4.5S-homologous region, and six ribosomal protein genes typically present in streptophyte chloroplast genomes and displays a unique dependency on the nucleus for chloroplast protein synthesis.
The distribution of group II introns among land plants is generally conserved and considered phylogenetically informative (Kelchner 2002; Qiu et al. 2006; Turmel et al. 2006, 2007; Brouard et al. 2008). Positional homologs of land plant group II introns have yet to be identified in the chloroplast genomes of nongreen algae or chlorophytes (Turmel et al. 2007), and they are also missing from the early-branching streptophyte chloroplast genomes of Mesostigma and Chlorokybus. The presence of five shared group II intron positional homologs identifies Klebsormidium as the earliest-branching streptophyte with land plant group II introns. Previous reconstructions have inferred that all embryophyte chloroplast group II introns were present in charophyte ancestors of land plants, with the exception of the clpP-2-intron, which was inferred to have been gained during the colonization of land (Turmel et al. 2006). Of the 21 group II introns of land plants, 16 and 17 were identified in the chloroplast genomes of Chara and Chaetosphaeridium (Turmel et al. 2006), respectively, whereas a minimum of 15 were inferred to be present in the common ancestor of Zygnematophyceae (Turmel et al. 2005). However, the chloroplast genome of the zygnematophycean alga Mesotaenium reported here has 16 group II introns positionally homologous to those of land plants, and, clpP-2-intron, the only group II intron thought to be unique to land plants, is also present. Comparisons among the intron sets of the four known zygnematophycean chloroplast genomes indicate that at least 19 of 21 embryophyte group II introns were present in the last common ancestor of conjugating algae; the two absent introns being trnIGAU-intron and trnVUAC-intron (table 2). Intron losses are unusually common in the Zygnematophyceae, because a minimum of 20 losses (most parsimoniously) are necessary to explain the intron distribution in the class given the tree topology in figure 4A. This is in sharp contrast with other streptophyte clades—for example, in bryophytes, a single intron loss is observed among the six representatives analyzed here. Retroposition (reverse transcription of a spliced RNA copy, followed by recombination-dependent insertion into the genome) is considered a likely cause of frequent intron losses in the chloroplast genomes (Turmel et al. 2005; Chumley et al. 2006; see later).
Based on the reconstructed chloroplast genomes of Zygnema and Staurastrum (Turmel et al. 2005) and the cpDNA map of Spirogyra maxima (Manhart et al. 1990), it has been hypothesized that one of the IRs was lost early in the evolution of the conjugating algae, and hence, members of Zygnematophyceae are expected to lack the typical quadripartite structure (Turmel et al. 2005). Moreover, it has been observed that loss of the IR regions and quadripartite structure are correlated with increased structural rearrangements (Palmer et al. 1987; Strauss et al. 1988; Turmel et al. 2005; Bélanger et al. 2006). Consequently, the structure and gene complement of the zygnematophycean chloroplast genomes might be assumed to have little importance with respect to reconstructing the ancestral chloroplast genome of land plants and the changes occurred during the transition to land. However, our discovery of a quadripartite chloroplast genome in Roya is a challenge to this interpretation. Phylogenetic analyses strongly support the sister-group relationship of Roya and Staurastrum, which together form a derived group with respect to the earlier-branching Zygnema and Mesotaenium (fig. 4A). Consequently, the hypothesis of an IR loss by a single event in early Zygnematophyceae has to be questioned. If we assume that IR gain is rare and effectively irreversible once lost (there are no reports of secondary IR gain in plant chloroplasts) and therefore that the IRs of streptophytes and Roya are homologous, then the phylogeny suggest that the IR has been lost independently at least three times in the Zygnematophyceae, specifically, in the lineages leading to Mesotaenium, Zygnema, and Staurastrum. Given the extraordinary stability of the quadripartite structure across Viridiplantae, the hypothesis of three independent losses within a single taxonomic class might seem difficult to defend, but it is possible that the last common ancestor of Zygnematophyceae possessed a quadripartite chloroplast genome predisposed to structural instability (see later). Alternatively, and most parsimoniously, if we consider gain and loss of the IRs to be equally probable, then the IR regions of Roya are inferred to have been acquired de novo, perhaps as a result of increased selection for genome stability that was lost along with the quadripartite structure early in the evolution of the Zygnematophyceae. An important observation is that the gene order and gene composition of the IRs in Roya are different to the consensus of the IRs of land plants and other green algae. Although the span of IRs is variable in streptophytes, rearrangements within IRs are uncommon, and the six gene (rrn16, trnIGAU, trnAUGC, rrn23, rrn4.5, and rrn5) rRNA operon is always present (excepting Klebsormidium reported here). In Roya, all six rRNA genes are present in the IR, but they do not constitute a single operon. Moreover, the IRs of Roya also contain two ORFs (orf268 and orf230) not previously found in the IRs of other streptophyte chloroplast genomes. Both of these observations could support the interpretation that the IR regions of Roya have been reconstituted de novo from an ancestor lacking an IR. At present, it is not strictly clear which of these two hypotheses is correct, and therefore although the streptophyte and Roya IR regions are perhaps most likely homologous, some doubt remains.
The quadripartite architecture of the Roya chloroplast genome also highlights those factors thought to be necessary for structural stability. It has long been recognized that the chloroplast genomes without IRs tend to have highly rearranged gene orders with respect to close relatives that possess IRs, suggesting that the quadripartite structure aids genome stability (Palmer et al. 1987; Strauss et al. 1988; Turmel et al. 2005; Bélanger et al. 2006). However, there is no satisfactory mechanistic explanation for the inferred causality, and many observations do not support a correlation between the IR-loss and the degree of genome rearrangement. Although there are examples of highly rearranged chloroplast genomes that have lost the IR (e.g., Fabaceae: Palmer et al. 1987; Chlorophyta: Bélanger et al. 2006), many IR-containing, but highly rearranged, genomes are also known (e.g., Campanulaceae: Haberle et al. 2008; Geraniaceae: Chumley et al. 2006). Moreover, loss of the quadripartite structure does not necessarily lead to changes in gene order, as is the case for the angiosperms Medicago and Cicer (Jansen et al. 2008) or the gymnosperm Pinus species (our observation). These observations cast doubt on the assumption of causality between the IR-loss and structural instability and imply a role of other, and possibly multiple, triggers of chloroplast genome rearrangements. Some authors have suggested that rearrangements are due to recombinations mediated by dispersed repeats (Milligan et al. 1989; Kawata et al. 1997); the number of DNA repeats (>20 bp and >60 bp) is significantly correlated with the degree of genome rearrangements in angiosperm Geraniaceae family (Weng et al. 2014). However, our results failed to identify a relationship between >20 bp repeat abundance and the number of genome rearrangements in charophyte chloroplast genomes. All four zygnematophycean chloroplast genomes are highly rearranged with respect to each other yet extremely variable in the repeat content (0.24 Roya–25.73 Zygnema) with low repeat content values being similar to those of charophyte taxa with IRs and more conserved genomic structure. It is important to note that the factors affecting chloroplast genome stability may not necessarily be found in the chloroplasts alone and could involve mediation by nuclear genes as well. Some authors have suggested that elevated mutability of chloroplast genomes could be a consequence of faulty DNA repair mechanisms (Guisinger et al. 2011; Wicke, Schäferhoff, et al. 2013). Evidence supporting such a hypothesis includes the demonstration that a mutation in a nuclear-encoded, chloroplast-targeted recA homolog results in altered structural forms of cpDNA in Arabidopsis (Rowan et al. 2010), and mutations of other recA homologs lead also to large-scale genome rearrangements of mtDNA in Arabidopsis and Physcomitrella (reviewed in Maréchal and Brisson 2010).
The chloroplast genome of Roya contains a RT-like ORF (orf310) that is a putative homolog of a RT-like ORF found previously in the chloroplast genome of Staurastrum (Turmel et al. 2005). Because RTs are recognized as the key factors of intron removal (Cohen et al. 2012), it is possible that the frequent intron losses observed in the Zygnematophyceae clade are indicative of this enzyme's activity in ancestral chloroplasts of conjugating algae. The Roya chloroplast genome also has an ORF (orf268), which contains a catalytic domain typical of integrase/recombinases (INT_REC_C, cd01182, NCBI Conserved Domain Database, Marchler-Bauer et al. 2013). It is likely this ORF originates from outside of the chloroplast, as it is most similar to ORFs found in the mitochondrial genomes of the lycopod Huperzia squarrosa and charophyte Chaetosphaeridium globosum (E values: 5e-28 and 9e-10, respectively) and the chloroplast genome of a distant chlorophyte Oedogonium cardiacum (E value: 2e-12) (Brouard et al. 2008). As in Roya, the O. cardiacum chloroplast genome has both int- and RT-like ORFs and also appears to have undergone considerable genome rearrangement (Brouard et al. 2008). Importantly, RTs and integrases are both essential components of the replicative machinery of retroelements (i.e., retroviruses and autonomous retrotransposons) (Wilhelm M and Wilhelm F-X 2001) that are widely recognized as the causative agents of genome rearrangements (Mieczkowski et al. 2006; Gogvadze and Buzdin 2009; Yu et al. 2011). Consequently, we suggest that the RT-like orf310 and int-like orf268 found in the chloroplast genome of Roya are remnant traces of retroelements whose activity shaped ancestral zygnematophycean chloroplast genomes leading to the loss of the IRs and quadripartite structure, reduced numbers of group-II introns, and loss of ancestral gene order and synteny. It is also possible that the hypothesized retroelements were present in the chloroplasts of the last common ancestor of Zygnematophyceae and embryophytes and were responsible for the structural differences that separate the chloroplast genomes of early land plants from their charophyte counterparts.
Zygnematophyceae Are the Closest Relatives of Land Plants
Our results indicate that the ancestral zygnematophycean chloroplast genome was most likely quadripartite but predisposed to structural instability. It contained at least 87 out of 89 protein-coding genes estimated to be present in the HLPA, five additional genes not found in the land plants (fig. 3), an almost complete land plant intron set (including clpP-intron-2; table 1), and at least 12 operons found in land plants. Other cpDNA features that may corroborate the genetic proximity of Zygnematophyceae and land plants are the two ORFs of Roya (orf268 and orf230), which have a high sequence similarity to some land plants but not to other charophytes.
Phylogenetic analyses of the structural character data (fig. 4B) and the chloroplast genes (fig. 4A) show that Zygnematophyceae and land plants share a more recent common ancestor than do land plants and any other group of streptophyte algae. Previous phylogenetic inferences based on chloroplast genes have suggested the Charales (Karol et al. 2001), Chaetosphaeridiales (Turmel et al. 2008 [amino acids]), or the Zygnematophyceae (Qiu et al. 2006; Turmel et al. 2008 [nucleotides]; Gao et al. 2010; Karol et al. 2010; Chang and Graham 2011) as the sister group to land plants. However, the analyses presented here (fig. 4B) are the first large-scale (83 gene) analyses to include all major charophyte lineages (Klebsormidium is included here for the first time) and are further strengthened by the inclusion of two additional Zygnematophyceae from previously unsampled family Mesotaeniaceae. Moreover, our phylogenetic analyses of structural genomic characters are congruent with respect to relationships among charophyte lineages and are similar to recent analyses based on nuclear genes (Wodniok et al. 2011; Laurin-Lemay et al. 2012; Timme et al. 2012). Hence, the congruence of results among data from multiple genomes and their high statistical probability (in this study and elsewhere) suggests the sister-group relationship between Zygnematophyceae and land plants is not an analytical artifact.
Despite the strength of the phylogenetic results, the sister-group relationship of Zygnematophyceae and land plants is somewhat surprising considering the morphology of charophyte algae. Early phylogenies, although based on few genes (e.g., Karol et al. 2001; Qiu et al. 2006), tended to place the Charales as the sister group to land plants. Because the Charales possess, at least superficially, similar morphological traits to the land plants such as multicellular, branching thalli, with archegonia remaining attached to, and protected by, the gametophyte, there appeared an intuitively elegant evolutionary progression from simple unicellular streptophyte algae such as Klebsormidium and Zygnematophyceae to more complex forms which eventually gave rise to land plants. In contrast, the Zygnematophyceae are morphologically simple, being either unicellular (e.g., desmids) or forming filaments (e.g., Spirogyra) and do not have a sexual cycle with motile male gametes (hence they entirely lack flagella) and a sessile archegonium but instead reproduce by cellular conjugation in a manner analogous to bacteria. Given the phylogenetic position of Zygnematophyceae, the lack of flagellate gametes and reproduction by conjugation are identified as derived states (apomorphies) unique to the lineage. In short, although Zygnematophyceae are the most diverse of the charophyte groups (∼3,000 species), they possess few macroscopic morphological similarities with land plants, hence they are a highly derived lineage of freshwater algae that appear to have retained few macromorphological characters that were present in their immediate ancestor with land plants. Their identification as the freshwater alga most closely to land plants will hopefully spur genomic research of this morphologically diverse and evolutionarily important group of organisms.
Supplementary Material
Supplementary material, figures S1–S4, and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Fundação para a Ciência e a Technologia (FCT), Portugal (grant number PTDC/BIA-EVF/113129/2009 to C.J.C.), by the European Research Council Advanced Investigator Programme to T.M.E., and in part by the European Regional Development Fund (ERDF) through the COMPETE—Operational Programme Competitiveness and national funds through FCT—under the project Pest-C/MAR/LA0015/2011 to C.J.C.
Literature Cited
- Adams KL, Palmer JD. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol. 2003;29:380–395. doi: 10.1016/s1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andersen RA, Berges JA, Harrison PJ, Watanabe MM. Recipes for freshwater and seawater media. In: Andersen RA, editor. Algal culturing techniques. Burlington (MA): Elsevier Academic Press; 2005. . p. 429–538. [Google Scholar]
- Bélanger A-S, et al. Distinctive architecture of the chloroplast genome in the chlorophycean green alga Stigeoclonium helveticum. Mol Gen Genomics. 2006;276:464–477. doi: 10.1007/s00438-006-0156-2. [DOI] [PubMed] [Google Scholar]
- Bourque G, Pevzner P. Genome-scale evolution: reconstructing gene orders in the ancestral species. Genome Res. 2002;12:26–36. [PMC free article] [PubMed] [Google Scholar]
- Brouard J-S, Otis C, Lemieux C, Turmel M. Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer. BMC Genomics. 2008;9:290. doi: 10.1186/1471-2164-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungard RA. Photosynthetic evolution in parasitic plants: insight from the chloroplast genome. BioEssays. 2004;26:235–247. doi: 10.1002/bies.10405. [DOI] [PubMed] [Google Scholar]
- Chang Y, Graham SW. Inferring the higer-order phylogeny of mosses (Bryophyta) and relatives using a large, multigene plastid data set. Am J Bot. 2011;95:839–849. doi: 10.3732/ajb.0900384. [DOI] [PubMed] [Google Scholar]
- Chumley TW, et al. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- Cohen NE, Shen R, Carmel L. The role of reverse transcriptase in intron gain and loss mechanisms. Mol Biol Evol. 2012;29:179–186. doi: 10.1093/molbev/msr192. [DOI] [PubMed] [Google Scholar]
- Cox CJ, Foster PG. A 20-state empirical amino-acid substitution model for green plant chloroplasts. Mol Phylogenet Evol. 2013;68:218–220. doi: 10.1016/j.ympev.2013.03.030. [DOI] [PubMed] [Google Scholar]
- De Smet I, et al. Unraveling the evolution of auxin signaling. Plant Physiol. 2011;155:209–221. doi: 10.1104/pp.110.168161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PG. Modeling compositional heterogeneity. Syst Biol. 2004;53:485–495. doi: 10.1080/10635150490445779. [DOI] [PubMed] [Google Scholar]
- Gao L, Su Y-J, Wang T. Plastid genome sequencing, comparative genomics, and phylogenomics: current status and prospects. J Syst Evol. 2010;48:77–93. [Google Scholar]
- Gensel PG. The earliest land plants. Annu Rev Ecol Evol Syst. 2008;39:459–477. [Google Scholar]
- Gogvadze E, Buzdin A. Retroelements and their impact on genome evolution and functioning. Cell Mol Life Sci. 2009;66:3727–3742. doi: 10.1007/s00018-009-0107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez G, Pallás V. Noncoding RNA mediated traffic of foreign mRNA into chloroplasts reveals a novel signaling mechanism in plants. PLoS One. 2010;5:e12269. doi: 10.1371/journal.pone.0012269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontcharov AA, Marin B, Melkonian M. Molecular phylogeny of conjugating green algae (Zygnematophyceae, Streptophyta) inferred from SSU rDNA sequence comparisons. J Mol Evol. 2003;56:89–104. doi: 10.1007/s00239-002-2383-4. [DOI] [PubMed] [Google Scholar]
- Gontcharov AA, Melkonian M. Molecular phylogeny and revision of the genus Netrium (Zygnematophyceae, Streptophyta): Nucleotaenium gen. nov. J Phycol. 2010;46:346–362. [Google Scholar]
- Green BR. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011;66:34–44. doi: 10.1111/j.1365-313X.2011.04541.x. [DOI] [PubMed] [Google Scholar]
- Grewe F, et al. Complete plastid genomes from Ophioglossum californicum, Psilotum nudum, and Equisetum hyemale reveal an ancestral land plant genome structure and resolve the position of Equisetales among monilophytes. BMC Evol Biol. 2013;13:8. doi: 10.1186/1471-2148-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 2011;28:583–600. doi: 10.1093/molbev/msq229. [DOI] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol. 2008;66:350–361. doi: 10.1007/s00239-008-9086-4. [DOI] [PubMed] [Google Scholar]
- Hodgetts WJ. Roya anglica G.S. West a new desmid; with an emended description of the genus Roya. J Bot. 1920;58:65–69. [Google Scholar]
- Jansen RK, et al. Complete plastid genome sequence of the chickpea (Cicer arientinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae) Mol Phylogenet Evol. 2008;48:1204–1217. doi: 10.1016/j.ympev.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karol KG, McCourt RM, Cimino MT, Delwiche CF. The closest living relatives of land plants. Science. 2001;294:2351–2353. doi: 10.1126/science.1065156. [DOI] [PubMed] [Google Scholar]
- Karol KG, et al. Complete plastome sequences of Equisetum arvense and Isoetes flaccida: implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evol Biol. 2010;10:321. doi: 10.1186/1471-2148-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata M, et al. Short inverted repeats function as hotspots of intermolecular recombination giving rise to oligomers of deleted plastid DNAs (ptDNAs) Curr Genet. 1997;31:179–184. doi: 10.1007/s002940050193. [DOI] [PubMed] [Google Scholar]
- Kelchner SA. Group II introns as phylogenetic tools: structure, function, and evolutionary constraints. Am J Bot. 2002;89:1651–1669. doi: 10.3732/ajb.89.10.1651. [DOI] [PubMed] [Google Scholar]
- Kenrick P, Crane PR. The origin and early evolution of plants on land. Nature. 1997;389:33–39. [Google Scholar]
- Krause K. From chloroplasts to “cryptic” plastids: evolution of plastid genomes in parasitic plants. Curr Genet. 2008;54:111–121. doi: 10.1007/s00294-008-0208-8. [DOI] [PubMed] [Google Scholar]
- Kurtz S, et al. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurin-Lemay S, Brinkmann H, Philippe H. Origin of land plants revisited in the light of sequence contamination and missing data. Curr Biol. 2012;22:R594. doi: 10.1016/j.cub.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Leliaert F, et al. Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci. 2012;31:1–46. [Google Scholar]
- Lenton TM, et al. First plants cooled the Ordovician. Nat Geosci. 2012;5:86–89. [Google Scholar]
- Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW)—a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- Manhart JR, Hoshaw RW, Palmer JD. Unique chloroplast genome in Spirogyra maxima (Chlorophyta) revealed by physical and gene mapping. J Phycol. 1990;26:490–494. [Google Scholar]
- Marchler-Bauer A, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;4(D1):D384–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Brisson N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010;186:299–317. doi: 10.1111/j.1469-8137.2010.03195.x. [DOI] [PubMed] [Google Scholar]
- Martin W, et al. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- McCourt RM. Green algal phylogeny. Trends Ecol Evol. 1995;10:159–163. doi: 10.1016/s0169-5347(00)89027-8. [DOI] [PubMed] [Google Scholar]
- McCourt RM, Delwiche CF, Karol KG. Charophyte algae and land plant origins. Trends Ecol Evol. 2004;19:661–666. doi: 10.1016/j.tree.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Mieczkowski PA, Lemoine FJ, Petes TD. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair. 2006;5:1010–1020. doi: 10.1016/j.dnarep.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Milligan BG, Hampton JN, Palmer JD. Dispersed repeats and structural reorganization in subclover chloroplast DNA. Mol Biol Evol. 1989;6:355–368. doi: 10.1093/oxfordjournals.molbev.a040558. [DOI] [PubMed] [Google Scholar]
- Nägeli C, et al. Gattungen einzelliger Algen, physiologisch und systematisch bearbeitet. Neue Denkschriften der Allg. Schweizerischen Gesellschaft für die Gesammten Naturwissenschaften. 1849;10:1–139. [Google Scholar]
- Nicolaï M, et al. Higher plant chloroplasts import the mRNA coding for the eucaryotic translation initiation factor 4E. FEBS Lett. 2007;581:3921–3926. doi: 10.1016/j.febslet.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Osorio B, Aldrich J, Thompson WF. Chloroplast DNA evolution among legumes: loss of a large inverted repeat occurred prior to other sequence rearrangements. Curr Genet. 1987;11:275–286. [Google Scholar]
- Palmer JD, Thompson WF. Rearrangements in the chloroplast genomes of mung bean and pea. Proc Natl Acad Sci U S A. 1981;78:5533–5537. doi: 10.1073/pnas.78.9.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y-L, et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc Natl Acad Sci U S A. 2006;103:15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Holland PWH. Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 2000;15:454–459. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- Rowan BA, Oldenburg DJ, Bendich AJ. RecA maintains the integrity of chloroplast DNA molecules in Arabidopsis. J Exp Bot. 2010;61:2575–2588. doi: 10.1093/jxb/erq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein CV, et al. Early middle ordovician evidence for land plants in Argentina (eastern Gondwana) New Phytol. 2010;188:365–369. doi: 10.1111/j.1469-8137.2010.03433.x. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Bellare P, DeRisi JL. PRICE: software for the targeted assembly of components of (meta)genomic sequence data. G3. 2013;3:865–880. doi: 10.1534/g3.113.005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson MJ. Molecular data from 27 proteins do not support a Precambrian origin of land plants. Am J Bot. 2003;90:954–956. doi: 10.3732/ajb.90.6.954. [DOI] [PubMed] [Google Scholar]
- Schneider A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu Rev Biochem. 2011;80:1033–1053. doi: 10.1146/annurev-biochem-060109-092838. [DOI] [PubMed] [Google Scholar]
- Silva PC, Mattox KR, Blackwell WH. The generic name Hormidium as applied to green algae. Taxon. 1972;2:639–645. [Google Scholar]
- Strauss HS, Palmer JD, Howe GT, Doerksen AH. Chloroplast genomes of two conifers lack a large inverted repeat and are extensively rearranged. Proc Natl Acad Sci U S A. 1988;85:3898–3902. doi: 10.1073/pnas.85.11.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita M, Sugiura M. Regulation of gene expression in chloroplasts of higher plants. Plant Mol Biol. 1996;32:315–326. doi: 10.1007/BF00039388. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods) 2003. Version 4. Sunderland (MA): Sinauer Associates. [Google Scholar]
- Swofford DL, Begle DP. PAUP phylogenetic analysis using parsimony. 1993. Version 3.1. User's manual. Washington (DC): Laboratory of Molecular Systematics, Smithsonian Institution. [Google Scholar]
- Timme RE, Bachvaroff TR, Delwiche CF. Broad phylogenomic sampling and the sister lineage of land plants. PLoS One. 2012;7:e29696. doi: 10.1371/journal.pone.0029696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Delwiche CF. Uncovering the evolutionary origin of plant molecular processes: comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biol. 2010;10:96. doi: 10.1186/1471-2229-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The complete chloroplast DNA sequences of the charophycean green algae Staurastrum and Zygnema reveal that the chloroplast genome underwent extensive changes during the evolution of the Zygnematales. BMC Biol. 2005;3:22. doi: 10.1186/1741-7007-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The chloroplast genome sequence of Chara vulgaris sheds new light into the closest green algal relatives of land plants. Mol Biol Evol. 2006;23:1324–1338. doi: 10.1093/molbev/msk018. [DOI] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. Tracing the evolution of streptophyte algae and their mitochondrial genome. Genome Biol Evol. 2013;5:1817–1835. doi: 10.1093/gbe/evt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Pombert J-F, Charlebois P, Otis C, Lemieux C. The green algal ancestry of land plants as revealed by the chloroplast genome. Int J Plant Sci. 2007;168:679–689. [Google Scholar]
- Turmel M, et al. Deep division in the Chlorophyceae (Chlorophyta) revealed by chloroplast phylogenomic analyses. J Phycol. 2008;44: 739–750. doi: 10.1111/j.1529-8817.2008.00510.x. [DOI] [PubMed] [Google Scholar]
- Wellman CH, Osterloff PL, Mohiuddin U. Fragments of the earliest land plants. Nature. 2003;425:282–285. doi: 10.1038/nature01884. [DOI] [PubMed] [Google Scholar]
- Weng M-L, Blazier JC, Govindu M, Jansen RK. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol Biol Evol. 2014;31:645–659. doi: 10.1093/molbev/mst257. [DOI] [PubMed] [Google Scholar]
- Wicke S, Schäferhoff B, dePamphilis CW, Müller KF. Disproportional plastome-wide increases of substitution rates and relaxed purifying selection in genes of carnivorous Lentibulariaceae. Mol Biol Evol. 2013;31:529–545. doi: 10.1093/molbev/mst261. [DOI] [PubMed] [Google Scholar]
- Wicke S, et al. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, et al. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 2013;25:3711–3725. doi: 10.1105/tpc.113.113373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Wilhelm F-X. Reverse transcription of retroviruses and LTR retrotransposons. Cell Mol Life Sci. 2001;58:1246–1262. doi: 10.1007/PL00000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodniok S, et al. Origin of land plants: do conjugating green algae hold the key? BMC Evol Biol. 11. 2011:104. doi: 10.1186/1471-2148-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Yu C, Zhang J, Peterson T. Genome rearrangements in maize induced by alternative transpositions of reversed Ac/Ds termini. Genetics. 2011;188:59–67. doi: 10.1534/genetics.111.126847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.