Abstract
Deinococcus deserti is a desiccation- and radiation-tolerant desert bacterium. Differential RNA sequencing (RNA-seq) was performed to explore the specificities of its transcriptome. Strikingly, for 1,174 (60%) mRNAs, the transcription start site was found exactly at (916 cases, 47%) or very close to the translation initiation codon AUG or GUG. Such proportion of leaderless mRNAs, which may resemble ancestral mRNAs, is unprecedented for a bacterial species. Proteomics showed that leaderless mRNAs are efficiently translated in D. deserti. Interestingly, we also found 173 additional transcripts with a 5′-AUG or 5′-GUG that would make them competent for ribosome binding and translation into novel small polypeptides. Fourteen of these are predicted to be leader peptides involved in transcription attenuation. Another 30 correlated with new gene predictions and/or showed conservation with annotated and nonannotated genes in other Deinococcus species, and five of these novel polypeptides were indeed detected by mass spectrometry. The data also allowed reannotation of the start codon position of 257 genes, including several DNA repair genes. Moreover, several novel highly radiation-induced genes were found, and their potential roles are discussed. On the basis of our RNA-seq and proteogenomics data, we propose that translation of many of the novel leaderless transcripts, which may have resulted from single-nucleotide changes and maintained by selective pressure, provides a new explanation for the generation of a cellular pool of small peptides important for protection of proteins against oxidation and thus for radiation/desiccation tolerance and adaptation to harsh environmental conditions.
Keywords: protein translation initiation, genome evolution, small peptides, desiccation tolerance, protein protection, transcription start sites
Introduction
Exposure of cells to radiation and increased oxidative stress results in damage of cellular macromolecules, including DNA, proteins, and lipids. However, different organisms are not equally sensitive to radiation. Although high doses of ionizing radiation (e.g., >2 kGy) are lethal for most organisms, a few known species show various levels of radiation tolerance. Deinococcus bacteria, which belong to an ancient and distinct lineage on the phylogenetic tree (Makarova et al. 2001), are well known for their extraordinary tolerance to gamma and UV radiation as well as to prolonged desiccation, which is related to their ability to repair massive DNA damage including hundreds of radiation- or desiccation-generated DNA double-strand breaks (Mattimore and Battista 1996; Battista 1997).
More than 40 Deinococcus species have been described to date. Of these, Deinococcus radiodurans has been studied most extensively. Analysis of its genome sequence revealed only a classical set of prokaryotic DNA repair proteins (Makarova et al. 2001). The use of microarrays uncovered various hypothetical genes highly induced following irradiation or desiccation, and the contribution to DNA repair and/or radiation resistance was demonstrated for five of these genes, designated pprA and ddrA to ddrD (Tanaka et al. 2004). Another gene, the constitutively expressed and Deinococcus-specific irrE, was also shown to be required for radiation tolerance (Earl et al. 2002; Hua et al. 2003). IrrE-dependent upregulation of 210 genes and 31 proteins was highlighted after irradiation of D. radiodurans by means of microarray-based transcriptomics and two-dimensional protein gel electrophoresis, respectively (Lu et al. 2009, 2012).
Microscopy images of several Deinococcus species revealed a highly condensed nucleoid structure, which may facilitate DNA repair by limiting diffusion of DNA fragments generated by radiation or desiccation (Levin-Zaidman et al. 2003; Zimmerman and Battista 2005). To be able to repair massive DNA damage and survive, at least some of the cell components should maintain their integrity and activity following irradiation. Deinococcus and other radiotolerant bacteria have a high intracellular Mn/Fe concentration ratio, which has been correlated with protection of proteins from oxidative damage during irradiation and desiccation (Daly et al. 2004, 2007; Fredrickson et al. 2008). With relatively low doses of ionizing radiation (i.e., ≈1.5 kGy), massive and lethal protein damage occurred in radiation-sensitive Escherichia coli, but protein oxidation was prevented in D. radiodurans. For the latter, only extremely high doses of radiation (i.e., >10 kGy) resulted in protein oxidation levels that caused cell death (Krisko and Radman 2010). Antioxidant protection of proteins was also correlated to the radiation tolerance of the rotifer Adineta vaga, a freshwater invertebrate (Krisko et al. 2012). In vitro experiments have shown that protein-free filtrated cell extract of D. radiodurans was extremely protective against radiation-induced protein oxidation (Daly et al. 2010). Compared with nonprotective cell extracts of radiation sensitive bacteria (e.g., E. coli), cell extracts of D. radiodurans are enriched in manganese (Mn2+), phosphate, and especially small peptides of 7–22 residues (Daly et al. 2010). In vitro, a synthetic decapeptide interacted synergistically with Mn2+ and phosphate and preserved activity of enzymes exposed to radiation (Daly et al. 2010). Taken all together, these different studies indicate that radiation tolerance of Deinococcus results from a combination of different molecular mechanisms and physiological determinants (Cox and Battista 2005; Blasius et al. 2008; Slade and Radman 2011; Daly 2012).
We have isolated Deinococcus deserti VCD115 from upper sand layers of the Sahara after exposure of the sand samples to 15 kGy of gamma irradiation (de Groot et al. 2005). Its genome was sequenced and annotated with the help of experimental data and proteogenomic approaches (de Groot et al. 2009; Baudet et al. 2010). Deinococcus deserti has in common with other Deinococci a highly condensed nucleoid, a high cellular Mn/Fe ratio, and several of the Deinococcus-specific radiation tolerance-associated genes, for example, ddrA to ddrD, pprA, and irrE (de Groot et al. 2009). Comparative genomics showed some interesting differences between D. deserti and other sequenced Deinococcus species. For example, D. deserti possesses supplementary DNA repair genes that code for mutagenic translesion DNA polymerases and two functionally different RecA proteins (Dulermo et al. 2009), whereas it lacks homologs of several radiation-induced genes in D. radiodurans (e.g., ddrP encoding a putative DNA ligase).
Proteomics allowed correction of various prediction errors (initiation codon, gene orientation, and unpredicted genes) in D. deserti. Our work on D. deserti also highlighted many annotation errors in D. radiodurans and Deinococcus geothermalis, for example, for radiation-induced genes ddrB, ddrC, and ddrH (de Groot et al. 2009; Baudet et al. 2010). Obviously, correction of prediction errors and an accurate genome annotation are crucial for the identification of radiation tolerance associated-genes by global approaches and their subsequent characterization by genetic, biochemical, and structural studies (e.g., see the expression attempts of DdrB of D. radiodurans in E. coli, which was only successful with the corrected 11-residue shorter DdrB [Norais et al. 2009]). However, gene prediction and proteomics have their limitations, especially with respect to small genes/proteins, which are difficult to predict and detect by mass spectrometry.
Here, we further characterized D. deserti using RNA sequencing (RNA-seq), a powerful method to study transcriptomes (Croucher and Thomson 2010; Sorek and Cossart 2010; van Vliet 2010) and complemented these data with proteomics specifically targeted on low molecular weight proteins. RNA-seq allows detection of transcription units without a priori genome annotation information and has a large dynamic range because the number of sequencing reads that map to unique regions of the genome does not have an upper limit. Strand-specific RNA-seq was performed with D. deserti bacteria grown in standard condition, as well as with cells recovering from exposure to gamma radiation. In addition, differential RNA-seq was applied, that is, part of the RNA samples from each condition was enriched for primary transcripts, allowing genome-wide identification of transcription start sites (TSSs), and hence, of promoter regions and 5′-untranslated regions (5′-UTRs) of mRNAs (Sharma et al. 2010). In prokaryotes, the 5′-UTR is present in the majority of known mRNAs and generally contains the Shine–Dalgarno (SD) sequence important for ribosome binding and selection of the correct translation initiation codon (Shine and Dalgarno 1974). The 5′-UTR may also form secondary structures, for example, riboswitches, implicated in regulation of transcription or translation. Strikingly, in D. deserti, we found a very high number of mRNAs that lack a 5′-UTR. These leaderless mRNAs include not only hundreds of previously annotated genes but also a high number of additional transcripts for novel peptides and proteins. Data analysis resulted in numerous start codon reannotations and in the identification of many new genes in D. deserti, of which several have unannotated homologs in other Deinococci. Novel radiation-induced genes were also found. The possible implications of these new results for radiation and desiccation tolerance are discussed.
Materials and Methods
Bacterial Strain, Growth Conditions, and Irradiation
Deinococcus deserti strain RD19 is a spontaneous streptomycin-resistant derivative of the wild-type strain VCD115 (Vujicic-Zagar et al. 2009). It is routinely grown at 30 °C with aeration in 10-fold diluted tryptic soy broth supplemented with trace elements (Vujicic-Zagar et al. 2009). For RNA-seq, a 100-ml culture of RD19 was grown to exponential phase (OD600 0.5) and then divided in two. One part (45 ml) was exposed to 1 kGy gamma irradiation at room temperature (23 Gy/min, 60Co source; CEA/Cadarache, France) and then recovered for 30 min. The other part (45 ml) was not irradiated but otherwise treated in the same manner. After recovery, 15 ml of each culture was added to RNAprotect Bacteria Reagent (Qiagen) to stabilize RNA, following the instructions of the manufacturer. RNAprotect-treated cells were centrifuged and cell pellets stored at −80 °C.
RNA Isolation, cDNA Library Construction, and Illumina Sequencing
Total RNA isolation and construction and sequencing of cDNA libraries were performed by Vertis Biotechnologie AG (Germany). Briefly, cell pellets were pretreated with lysozyme and proteinase K. Total RNAs were isolated using the mirVana RNA isolation kit (Ambion) including DNase treatment. From each RNA preparation, two cDNA syntheses were carried out, one with and one without Terminator exonuclease (TEX, Epicentre) treatment. For the +TEX protocol, RNA samples were incubated with TEX, which specifically degrades RNA species that carry a 5′-monophosphate (5′P). The exonuclease-resistant RNA (primary transcripts with 5′PPP) was poly(A)-tailed using poly(A) polymerase and treated with tobacco acid pyrophosphatase (TAP), which degrades 5′PPP to 5′P. Then an RNA adapter was ligated to the 5′P of the “de-capped” RNA. First-strand cDNA synthesis was performed using an oligo(dT)-adapter primer and M-MLV reverse transcriptase. The resulting cDNAs were polymerase chain reaction (PCR) amplified to about 30 ng/μl using a high-fidelity DNA polymerase. For the -TEX protocol, the RNA samples were directly poly(A) tailed using poly(A) polymerase, followed by TAP treatment. An RNA adapter was then ligated to the 5′P of the total RNA samples. First-strand cDNA synthesis was performed using an oligo(dT)-adapter primer and M-MLV H-reverse transcriptase. The resulting cDNAs were PCR amplified to about 60–90 ng/μl using a high-fidelity DNA polymerase. For both protocols, the cDNAs were purified using the Agencourt AMPure XP kit (Beckman Coulter Genomics) and analyzed by capillary electrophoresis. The primers used for PCR amplification were designed for TruSeq sequencing according to the instructions of Illumina, with the 3′-sequencing adapter containing a barcode specific for each library. For Illumina sequencing, the -TEX cDNA samples were pooled at approximately equimolar amounts and size fractionated in the range between 250 and 500 bp on Agarose Gel. Also the +TEX cDNA samples were pooled at approximately equimolar amounts, and the cDNA pool was used for sequencing without further treatment. The + and –TEX cDNA pool were sequenced on a Illumina HiSeq 2000 machine (read length: 100 bp).
RNA-Seq Analysis
Transcriptomic high-throughput sequencing data were analyzed using a bioinformatic pipeline implemented in the MicroScope platform (http://www.genoscope.cns.fr/agc/microscope/, last accessed April 12, 2014) (Vallenet et al. 2013). The current pipeline is a “Master” shell script that launches the various parts of the analysis (i.e., a collection of Shell/Perl/R scripts) and controls for all tasks having been completed without errors. In a first step, the RNA-seq data quality was assessed by including option-like reads trimming or merging/split paired-end reads. In a second step, reads were mapped onto the D. deserti VCD115 genome sequence (GenBank accession numbers CP001114, CP001115, CP001116, and CP001117 for the chromosome and plasmids P1, P2, and P3, respectively) using the SSAHA2 package (Ning et al. 2001) that combines the SSAHA searching algorithm (sequence information is encoded in a perfect hash function) aiming at identifying regions of high similarity and the cross-match sequence alignment program (Ewing et al. 1998), which aligns these regions using a banded Smith–Waterman–Gotoh algorithm (Smith and Waterman 1981). An alignment score equal to at least half of the read is required for a hit to be retained. To lower false-positive discovery rate, the SAMtools (v.0.1.8) (Li et al. 2009) were then used to extract reliable alignments from SAM formatted files. The number of reads matching each genomic object harbored by the reference genome was subsequently computed with the Bioconductor-GenomicFeatures package (Carlson et al. 2011). If reads matched several genomic objects, the count number was weighted to keep the same total number of reads. The Bioconductor-DESeq package (Anders and Huber 2010) with default parameters was used to analyze raw counts data and test for differential expression between conditions. The complete data set from this study has been deposited in National Center for Biotechnology Information’s Gene Expression Omnibus and is accessible through GEO Series accession number GSE56058. TSSs were annotated manually as described (Kröger et al. 2012), with enrichment in the +TEX libraries as the first criterion. In the case of no enrichment, a TSS was assigned if the four libraries agreed on the nucleotide position and if its location was plausible in relation to an adjacent open reading frame (ORF). Rapid amplification of cDNA ends (5′-RACE) was performed as described (Tillett et al. 2000). Sequences of the primers used for 5′-RACE are available on request.
Gene Prediction and Other Bioinformatic Analyses
New gene prediction was carried out using AMIGene (Bocs et al. 2003). Similarity searches were performed using various BLAST programs (Altschul et al. 1997) at NCBI or ExPASy. Multiple alignments were made with ClustalW at ExPASy. Structure-based homology search was performed using Phyre2 (Kelley and Sternberg 2009). Motifs were searched using MEME (Bailey and Elkan 1994).
Enrichment of Low Molecular Weight Proteins
Deinococcus deserti cells were grown in two fermentors and harvested during either exponential phase or stationary phase as described previously (de Groot et al. 2009). For each condition, a total of 2.5 g of wet material were resuspended in 25 ml of lysis buffer consisting in 50 mM TRIS/HCl buffered at pH 8.0 at 20 °C and containing 0.1 M NaCl and Complete Mini protease inhibitor mixture (Roche Applied Science, one tablet/7 ml). Both samples were then disrupted by means of a BasicZ cell disrupter (Constant Systems Ltd.) operated at 1,000 bars. After disruption, each cell extract was centrifuged at 20,000 × g and 4 °C for 30 min, and the soluble proteins were withdrawn. A volume of 10 ml of soluble proteins was treated with 2 µl of Benzonase (500 units) from Sigma for 30 min at 4 °C under gentle agitation. The proteins were then subjected to ammonium sulphate precipitation using 20 ml of a saturated solution of (NH4)2SO4 and further incubated for 1.5 h at 4 °C under agitation. The precipitated proteins were then pelleted by centrifugation at 18,000 g and 4 °C for 30 min. The resulting pellet was resuspended in 8 ml of 50 mM TRIS/HCl buffered at pH 8.0 and containing 2.5 mM EDTA, 1.5 M (NH4)2SO4 (Buffer A). A volume of 8 ml of solubilized proteins was applied at a flow rate of 1 ml per min onto a 5 ml Phenyl HP column (GE Healthcare) previously equilibrated with Buffer A and operated with an Äkta Purifier FPLC system (Amersham Biosciences). After column wash with Buffer A, proteins were eluted over a 60 ml linear gradient comprising 1,500–0 mM (NH4)2SO4. Proteins eluted over 16 fractions were precipitated as follows: A volume of 500 µl of each fraction was supplemented with a 50% aqueous trichloroacetic acid solution (10% final), vortexed, and then centrifuged for 30 min at 10 °C. Each resulting protein pellet was dissolved in 50 µl of Tricine SDS solution (Invitrogen) and 10 µl of reductor buffer (Invitrogen) upon sonication with a UP50H compact lab homogenizer (Hielscher) operated at 40% amplitude for 30–60 s. Each fraction was analyzed by SDS-PAGE on 16% Novex Tricine gels (Invitrogen) carried out as recommended. After migration, gels were stained with SimplyBlue SafeStain (Invitrogen). The low molecular weight proteome from each lane was then excised into two 4 × 5 × 2 mm thick pieces from bottom to top and further subdivided into two duplicates for trypsin and chymotrypsin proteolysis, respectively. In-gel proteolysis with trypsin (de Groot et al. 2009) and chymotrypsin (Baudet et al. 2010) was carried out as described. The 64 resulting peptide mixtures were analyzed by nano-liquid chromatography coupled to tandem mass spectrometry (nano-LC-MS/MS).
Nano-LC-MS/MS Analysis and Proteomic Data Processing
Peptide samples were analyzed by nano-LC-MS/MS using an LTQ-Orbitrap XL hybrid mass spectrometer (ThermoFisher) coupled to an UltiMate 3000 LC system (Dionex-LC Packings) in similar conditions as described previously (Rubiano-Labrador et al. 2014). The recorded MS/MS spectra were processed as described for peptide assignment (Christie-Oleza et al. 2013). Briefly, peak lists were generated with the Mascot Daemon software (version 2.3.2; Matrix Science) using the extract_msn.exe data import filter (Thermo) from the Xcalibur FT package (version 2.0.7; Thermo). Data import filter options were set to 400 (minimum mass), 5,000 (maximum mass), 0 (grouping tolerance), 0 (intermediate scans), and 1,000 (threshold). The search was performed using the following criteria: Tryptic peptides with a maximum of two miscleavages, mass tolerances of 5 ppm on the parent ion and 0.5 Da on the MS/MS, fixed modification for carbamidomethylated cysteine, and variable modification for methionine oxidation. The home-made polypeptide sequence database used here was described previously (de Groot et al. 2009). This database comprises 65,801 polypeptide sequences, totaling 6,040,642 amino acids, derived from a six-frame translation of the D. deserti genome sequence but restricted to ORFs (defined from STOP to STOP) with at least 33 amino acids. Mascot results were parsed using the IRMa 1.28.0 software (Dupierris et al. 2009) with a P-value threshold below 0.05 for peptide identification.
Results
Global Results of RNA-Seq Reads Mapped to D. deserti’s Genome
RNA was isolated from D. deserti strain RD19 grown in standard condition (nonirradiated, NI) and from cells recovering from exposure to gamma radiation (irradiated, IR). Part of the RNA samples from each condition was enriched for primary transcripts by incubation with Terminator exonuclease (TEX), which only degrades processed RNA molecules that carry a 5′-monophosphate but not primary transcripts that have a 5′-triphosphate. The 5′-end of a primary transcript corresponds to the TSS. Thus, RNA-seq reads were obtained from four samples, called RD19 NI, RD19 NI + TEX, RD19 IR, and RD19 IR + TEX.
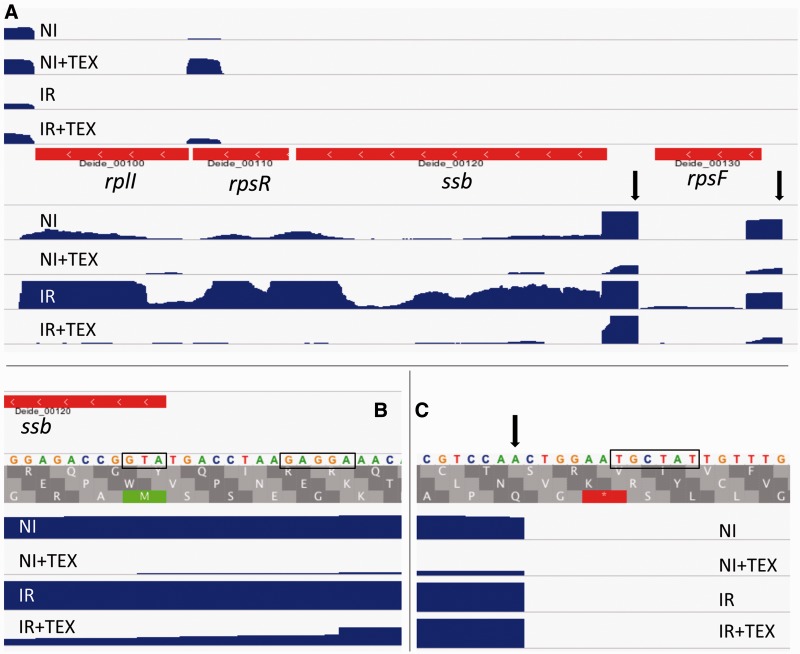
The RNA-seq reads were mapped to the genome of D. deserti, which consists of four replicons: The main chromosome (2.82 Mb) and three large plasmids called P1 (325 kb), P2 (314 kb), and P3 (396 kb) (de Groot et al. 2009). Before the present work was started, 3,459 coding sequences (CDSs) were annotated on the genome, as well as 12 rRNAs and 48 tRNAs. The read coverage for the entire genome can be visualized using the Integrative Genomics Viewer (Thorvaldsdottir et al. 2013) via the MicroScope platform (Vallenet et al. 2013). An example of such displayed read coverage for a 2.5 kb region is shown in figure 1A, where the data indicate a monocistronic transcript for rpsF and a polycistronic transcript for ssb-rpsR-rplI. An overview of the obtained read numbers mapped to D. deserti’s genome is presented in supplementary table S1, Supplementary Material online. To compare the relative transcription levels of the four replicons, the mapped read numbers (without reads for rRNA and tRNA genes) were normalized for the size of each replicon. When the normalized expression level was set at 100 for the chromosome, these levels were 53, 79, and 30 for plasmids P1, P2, and P3, respectively (average of the four RNA-seq samples). These normalized expression levels were similar for the nonirradiated and irradiated cultures. Highest global expression was thus found for the chromosome and plasmid P2, and the lowest for plasmid P3. This correlates well with previous proteome data where the highest percentage of the theoretical proteome was detected for proteins encoded on the chromosome and plasmid P2 (de Groot et al. 2009).
Fig. 1.—
Read coverage at the ssb region. Read coverage is indicated in blue (image taken from Integrative Genomics Viewer genome browser). Coverage of reads that map to the forward and reverse DNA strand are shown above and below the genes (in red), respectively. The four RNA samples are indicated: NI, nonirradiated cells; IR, irradiated cells; +TEX, RNA treated with terminator exonuclease. A TSS (arrows) is evident upstream of the operon containing ssb, rpsR, and rplI and upstream of rpsF. Panels (B) and (C) are images zoomed at the translation and transcription start of ssb, respectively. Start codon, SD sequence, and -10 motif are boxed.
The number of reads mapping sense and antisense to each annotated gene was determined (excluding rRNA genes) (supplementary table S2, Supplementary Material online). As expected, more sense than antisense reads were found for the majority of the genes, with an average of 14% antisense reads in the four RNA-seq samples. For several of the annotated genes, however, the amount of antisense reads is clearly higher than that of sense reads. The antisense reads may derive from adjacent gene expression or from antisense RNAs. For some cases, the antisense reads revealed gene prediction errors, which were subsequently corrected (see later). High levels of antisense transcription have also been observed in other bacteria, and roles of antisense RNAs in gene regulation have been reported (Georg and Hess 2011).
High Abundance of Leaderless mRNAs in D. deserti
TEX treatment of RNA samples resulted in enrichment of primary transcripts and thus of RNA-seq reads at TSSs (e.g., fig. 1). A TSS at a position between 0 and 300 nucleotides (nt) upstream of an annotated gene was classified as a gTSS for that gene. Supplementary table S3, Supplementary Material online, contains a list of all annotated protein-coding genes, and the gTSSs are indicated with their position relative to the first nucleotide of the start codon. Potential TSSs internal (iTSS) or antisense (aTSS) to each gene are also indicated. Supplementary table S4, Supplementary Material online, contains a fourth group of TSSs, that is, orphan TSSs in intergenic regions.
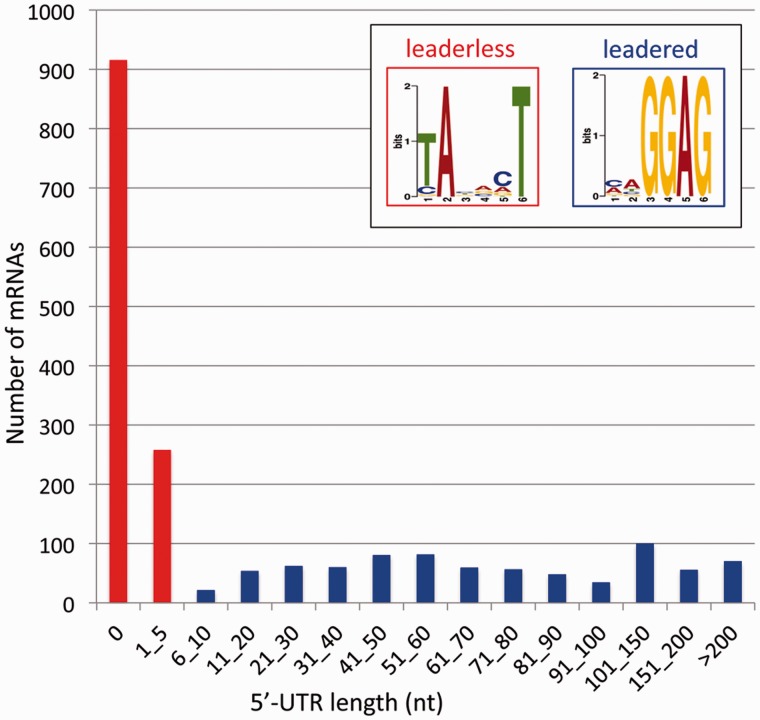
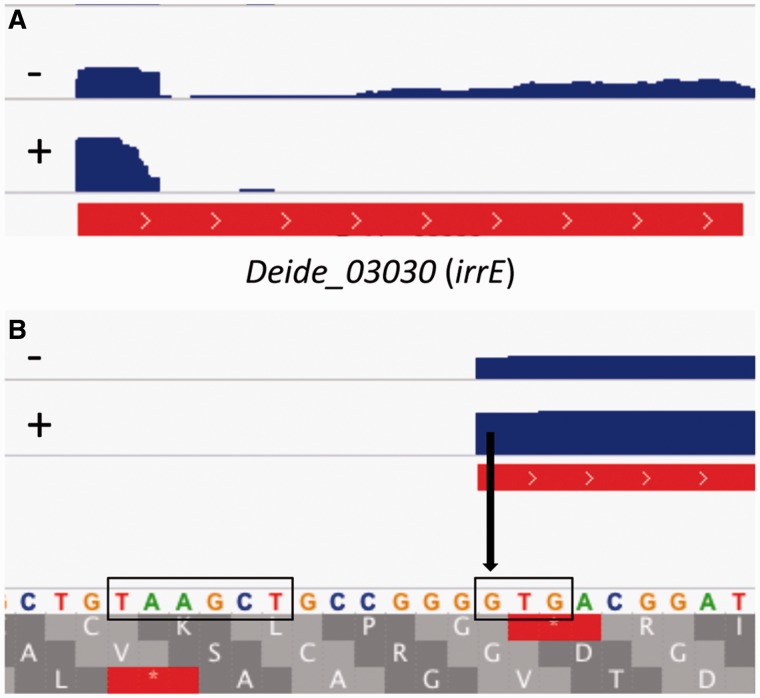
Unexpectedly, for numerous genes, the gTSS was found at exactly the first nucleotide of the translation initiation codon ATG or GTG, or within a few nt upstream of the start codon (fig. 2; supplementary table S3, Supplementary Material online). Figure 3 shows an example of such a leaderless gene, with the TSS at the GTG start codon of irrE (Deide_03030), a gene essential for radiation tolerance (Vujicic-Zagar et al. 2009). Leaderless mRNAs lack an SD sequence or other regulatory structures that are generally present in the 5′-untranslated region (5′-UTR) of leadered mRNAs. Previous studies have indicated that the upper limit for a 5′-UTR to allow usage of the leaderless translation pathway is around 5 nt, with most efficient translation when the start codon is directly at the 5′-terminus (Hering et al. 2009; Krishnan et al. 2010). Therefore, mRNAs with a 5′-UTR of less than 6 nt were classified as leaderless. For the total genome, 1,174 (60%) of the 1,958 identified gTSSs correspond to leaderless mRNA (5′-UTR < 6 nt), with 916 (47%) of these gTSSs located exactly at the first nucleotide of the translation initiation codon (5′-UTR = 0 nt) (table 1). An even higher percentage of leaderless mRNA was found for the main chromosome (table 1). Using MEME (Bailey and Elkan 1994), a conserved motif resembling the -10 box TATAAT was found directly upstream of 94% of the TSSs (fig. 2; supplementary table S5, Supplementary Material online; see also figs. 1, 3, 5, and 6). A widely conserved -35 motif was not detected using the same approach. While the -10 motif was thus found directly upstream of the start codon of leaderless genes, the SD motif shown in figure 2 was found upstream of the start codon in 62% of the leadered mRNAs (supplementary table S6, Supplementary Material online; see also figures 1 and 6), which supports the identification of two types of mRNAs, that is, leaderless and leadered. The remaining 38% of the leadered mRNAs may contain a less conserved SD motif or might be translated in an SD-independent manner.
Fig. 2.—
Frequency of 5′-UTR lengths. Data for leaderless and leadered genes are indicated in red and blue, respectively. The inset shows the -10 and SD motifs found upstream of the start codon of, respectively, leaderless and leadered genes.
Fig. 3.—
Deide_03030 (irrE), a leaderless gene with a GTG start codon. Panel (B) is a zoom of the TSS in panel (A). GTG start codon and -10 motif are boxed. Treatment (+) or not (–) of RNA with TEX is indicated. Only read coverage for RD19-IR is shown. The identified TSS (arrow) was also found by independent 5′-RACE.
Table 1.
TSSs of mRNAs
| gTSSa | Chromosome | Plasmid P1 | Plasmid P2 | Plasmid P3 | All |
|---|---|---|---|---|---|
| Total | 1,586 | 131 | 107 | 134 | 1,958 |
| 5′-UTR = 0 nt | 806 (51%) | 36 (27%) | 36 (34%) | 38 (28%) | 916 (47%) |
| 5′-UTR 1–5 nt | 226 (14%) | 14 (11%) | 8 (7%) | 10 (7%) | 258 (13%) |
| 5′-UTR > 5 nt | 554 (35%) | 81 (62%) | 63 (59%) | 86 (64%) | 784 (40%) |
aThe data show the number of identified mRNA transcription start sites (in total and for the indicated 5′-UTR length).
Fig. 5.—
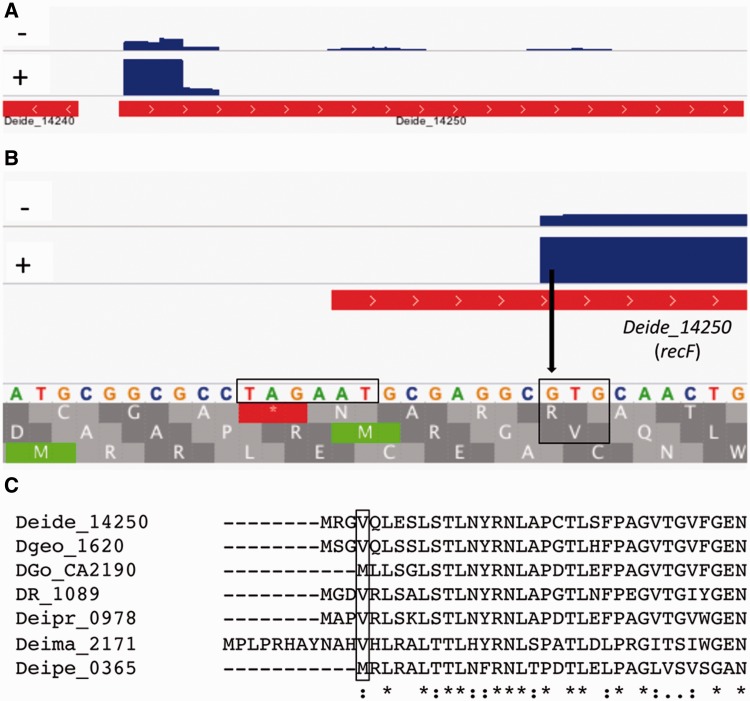
TSS reveals new start codon for DNA repair protein RecF (Deide_14250). (A) Read coverage for recF in the RD19-IR sample. Treatment (+) or not (–) of RNA with TEX is indicated. The first part of Deide_14240, on the reverse strand, is also visible at the left of recF. (B) Zoom at the TSS of recF. New start codon and -10 motif are boxed. The identified TSS (arrow) was also found by independent 5′-RACE. (C) Multiple alignment indicates similar RecF start correction in four other deinococcal homologs (only the N-terminus of the proteins is shown). The annotated or proposed start (boxed) for each of these 7 RecF proteins corresponds to a GTG codon.
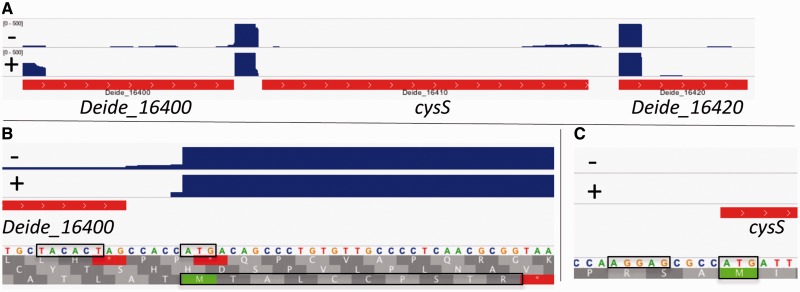
Fig. 6.—
The 5′-end of cysS mRNA encodes a ten-amino-acid-long leader peptide with two cysteine residues. cysS codes for cysteinyl-tRNA synthetase. Start codons, -10 motif (upstream of TSS), SD sequence (upstream of cysS start codon), and 10 aa peptide are boxed. Panel (B) is a zoom at the TSS for cysS, which is also the translation start of the predicted leader peptide. Panel (C) is a zoom at the translation start of cysS. Treatment (+) or not (–) of RNA with TEX is indicated.
Table 2 shows that leaderless genes in D. deserti almost exclusively contain either an ATG (83%) or GTG (17%) start codon. Only three leaderless genes (0.3%) with a predicted TTG start codon were found. Also for leadered genes, the majority of start codons are ATG (81%) and GTG (13%), but TTG (6%) and the rare start codons CTG and ATC are also used. For the entire CDSs (all codons), the overall relative synonymous codon usage of the leaderless genes is similar to that of the leadered genes (supplementary fig. S1, Supplementary Material online). The average amino acid composition of the leaderless gene products is also similar to that of the leadered genes (supplementary fig. S2, Supplementary Material online).
Table 2.
Start Codons and Proteome Data of Leaderless and Leadered Genes
| Start Codona | Leaderless 5′-UTR = 0 nt | Leaderless 5′-UTR 1–5 nt | Leadered 5′-UTR > 5 nt |
|---|---|---|---|
| 916 | 258 | 784 | |
| ATG | 740 | 237 | 632 |
| proteome; Nter peptide | 472; 120 | 143; 30 | 377; 86 |
| GTG | 176 | 18 | 103 |
| proteome; Nter peptide | 102; 16 | 6; 1 | 42; 4 |
| TTG | — | 3 | 47 |
| proteome; Nter peptide | 1; 0 | 22; 3 | |
| CTG | — | — | 1 |
| proteome; Nter peptide | 1; 1 | ||
| ATC | — | — | 1 |
| proteome; Nter peptide | 1; 1 |
aThe data show the number of mRNAs with the indicated start codon and the number of mRNAs for which the protein product and N-terminal peptide have been detected by proteomics.
To determine whether the identified leaderless mRNAs are well translated, the TSS data were compared with results of proteomics obtained in this work (see later) or in previous studies (de Groot et al. 2009; Dulermo et al. 2009; Baudet et al. 2010; Toueille et al. 2012; Bouthier de la Tour et al. 2013; Dedieu et al. 2013) (supplementary table S3, Supplementary Material online). For 167 leaderless genes, including genes with an ATG or GTG start codon and with or without a short 5′-UTR, the corresponding N-terminal peptide has been experimentally identified (table 2), confirming translation initiation at or very near the 5′-mRNA end. Of the 1,167 proteins that were detected and for which a gTSS was identified, 724 are translated from a leaderless mRNA (5′-UTR < 6 nt) (table 2). Moreover, of the 100 proteins most highly detected by shotgun proteomics (de Groot et al. 2009), 27 are produced from a leaderless mRNA and 37 from a leadered mRNA (supplementary table S7, Supplementary Material online). Of these 27 leaderless mRNAs, 23 have the TSS at the first nucleotide of the ATG (19 cases) or GTG (4 cases) start codon, and 4 contain a short 5′-UTR of 1–3 nt upstream of the AUG. Among their translation products are nucleoid-associated proteins, cell envelope proteins, response regulators, enzymes involved in posttranslational modification and in various metabolic functions, and uncharacterized proteins. Two of these were also identified among the 19 most abundant proteins on the basis of protein spot intensity after proteome fractionation on 2D gels (Dedieu et al. 2013). Together, these data show that leaderless mRNAs are efficiently translated in D. deserti, and that products from leaderless transcripts are present among the most abundant proteins.
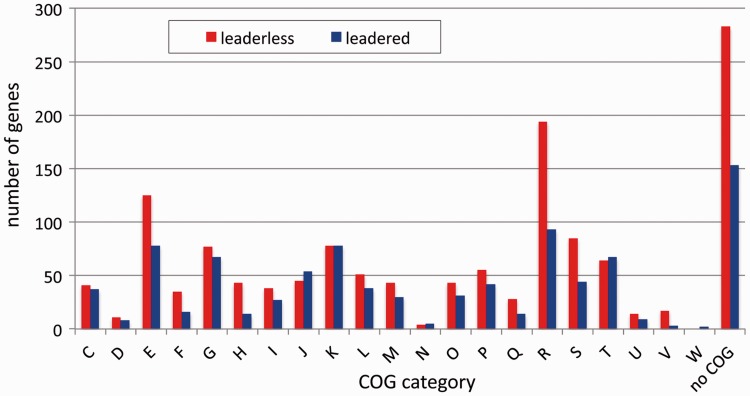
To see whether leaderless genes are over- or underrepresented in certain functions, the distribution of leaderless and leadered genes in clusters of orthologous groups (COGs) was analyzed. The result shows that both leaderless and leadered genes are found for the different COG categories, with the percentage of leaderless varying from 45% (translation, ribosomal structure, and biogenesis) to 75% (coenzyme transport and metabolism) (fig. 4). A high percentage (85%) of leaderless genes was also found for group V (defense mechanisms), but this group contains only 20 genes with a TSS. As an example, clpP (Deide_19570) and the closely located lon (Deide_19590) encode proteins from the same category (protein turnover), but ClpP is translated from a leaderless mRNA and Lon from a leadered mRNA (supplementary fig. S3, Supplementary Material online). Concerning proteins and processes that have been studied in Deinococcus, among the leaderless genes are those encoding IrrE (Deide_03030) (fig. 3), PolA (Deide_15130), RarA (Deide_04980), RecF (Deide_14250), RecN (Deide_12310), RuvA (Deide_09360), RuvB (Deide_18350), RuvC (Deide_20630), SbcD (Deide_16180), UvrA2 (Deide_2p02060), UvrD (Deide_12100), MutS (Deide_15540), HU1 (Deide_2p01940), HU2 (Deide_3p00060), Dps (Deide_21200), DdrA (Deide_09150), DdrC (Deide_23280), and DdrD (Deide_01160). For comparison, examples of proteins produced from leadered mRNAs are Ssb (Deide_00120) (fig. 1), UvrA (Deide_12760), UvrB (Deide_03120), GyrA (Deide_12520), GyrB (Deide_15490), TopA (Deide_07410), RecAP1 (Deide_1p01260), HU3 (Deide_00200), DdrB (Deide_02990), PprA (Deide_2p01380), SodA (Deide_07760), catalase (Deide_2p00330), Lon protease (Deide_05670 and Deide_19590), and ClpC (Deide_12680).
Fig. 4.—
Distribution of leaderless and leadered genes in COG functional categories. C, energy production and conversion; D, cell cycle control, cell division, chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination and repair; M, cell wall/membrane/envelope biogenesis; N, cell motility; O, posttranslational modification, protein turnover, chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport and catabolism; R, general function prediction only; S, function unknown; T, signal transduction mechanisms; U, intracellular trafficking, secretion, and vesicular transport; V, defense mechanisms; W, extracellular structures.
Reannotation of Start Codons
The TSSs indicated start codon prediction errors for more than 250 annotated genes, which is important for further genetic or biochemical studies of the genes/proteins as well as for analysis of the upstream sequences. For example, various TSSs were found downstream of the annotated start, often at an internal ATG or GTG codon. Guided by these TSSs, start codon positions of 152 genes were reannotated, resulting in proteins that are 1–59 amino acid residues (aa) shorter (average 11 aa). TSSs were also found upstream of annotated genes at ATG or GTG codons in frame with the gene. This allowed 105 start codons reannotations that result in longer proteins (ranging from 1 to 231 aa longer; average 27 aa). The modifications include several important DNA repair proteins: RecF (Deide_14250) (fig. 5), RecN (Deide_12310), RarA (Deide_04980), RuvA (Deide_09360), RuvC (Deide_20630), and UvrA2 (Deide_2p02060) (supplementary fig. S4, Supplementary Material online). Sequence comparisons with homologous proteins from other species supported the start codon reannotations in D. deserti (see supplementary fig. S5, Supplementary Material online, for examples). Moreover, these analyses showed that start codon reannotation, including that of several DNA repair proteins, may also be required for various homologs in other bacteria (fig. 5C; supplementary fig. S5, Supplementary Material online).
Additional Leaderless mRNAs for Novel Small Peptides and Proteins in D. deserti
Various studies on translation initiation of leaderless mRNA strongly suggest that an AUG triplet at the 5′-end of an mRNA is a distinct signal required and sufficient for ribosome binding and expression (Brock et al. 2008; Benelli and Londei 2009; Hering et al. 2009; Malys and McCarthy 2011). In D. deserti, we have detected protein expression from leaderless genes possessing an ATG or GTG start codon (table 2; supplementary table S3, Supplementary Material online). Therefore, besides the gTSSs of the leaderless annotated genes described in the previous subsections, we inspected all other identified TSSs in D. deserti for the presence of an ATG or GTG (AUG and GUG in RNA) at the 5′-end of the transcripts. Interestingly, a 5′-ATG (65 cases) or 5′-GTG (25 cases) triplet was found for 90 orphan transcripts, suggesting that they could be new translatable leaderless mRNAs for peptides and proteins ranging from 4 to 219 amino acid residues (average 46 aa) (supplementary table S4, Supplementary Material online). The number of cDNA reads that were mapped to the TSSs of these orphan transcripts varies between dozens (indicating low expression) to thousands (indicating high expression), as found for annotated genes (supplementary tables S4 and S8, Supplementary Material online). An ATG (60 cases) or GTG (23 cases) was also found at the 5′-end of leadered mRNAs of annotated genes. The 5′-ATG or 5′-GTG of these 83 leader sequences, which were thought to be 5′-UTRs, could direct synthesis of peptides ranging from 4 to 86 residues (average 22 aa) (supplementary table S8, Supplementary Material online). The stop codons for the latter peptides are present upstream of, or overlap with, the start codon of the annotated “leadered” gene. Together, the data indicate the presence of many additional leaderless mRNAs that do not correspond to known genes, predicting that the genome of D. deserti codes for much more peptides and small proteins than previously thought. These potential novel peptides and proteins are further analyzed in the next two subsections.
Conserved New Proteins and Identification of Putative Leader Peptides Involved in Transcription Attenuation
For 17 novel small proteins deduced from the predicted additional leaderless transcripts, one or more homologous proteins were found, mostly only from Deinococcus species and including several nonannotated homologs from D. radiodurans, D geothermalis, and Deinococcus gobiensis (supplementary fig. S6, Supplementary Material online). The conservation of these proteins strongly suggests that the corresponding transcripts are indeed translatable leaderless mRNAs. Labels (Deide) were attributed to these 17 new genes, which all code for peptides and proteins of unknown function (25 to 92 aa). We noticed a remarkable amino acid composition for five of these proteins, revealing low complexity regions that cover almost the entire proteins (supplementary fig. S6, Supplementary Material online). Deide_15148 (91 aa) has a predicted cytoplasmic region rich in Gly (28%), Ser (19%), Arg (15%), and His (10%) followed by a transmembrane helix at the C-terminus. Deide_00694 (63 aa) is rich in Glu, Gly, and Thr (16% each). Deide_04426 (58 aa) and Deide_12656 (70 aa) possess a signal peptide for, respectively, type II and type I signal peptidase, and the mature proteins are particularly rich in Thr (45% and 19%). Deide_2p00483 (49 aa), rich in Lys (25%) and Ala (20%), is coded by a gene located adjacent to a partial integrase gene. Deide_2p00483 homologs of several other species are also located next to phage-associated genes, suggesting that Deide_2p00483 might be of phage origin.
We observed that several leadered mRNAs for tRNA synthetase and amino acid biosynthesis genes contain an AUG at the 5′-end that would direct synthesis of small peptides rich in specific residues. For example, the ten amino acid-long peptide encoded by the 5′-end of the mRNA leader of the cysteinyl-tRNA synthetase gene cysS contains two Cys residues (fig. 6). These peptides could play a role in transcription attenuation control of the downstream gene (Naville and Gautheret 2010). Transcription attenuation involving leader peptides was first observed for the tryptophan biosynthesis operon in E. coli (Yanofsky 1981). Briefly, the 5′-leader of the trp operon mRNA codes for a leader peptide of 14 residues, two of which are Trp. Efficient translation of this peptide results in formation of a transcription terminator structure in the mRNA between this leader peptide-coding region and the first trp gene. When Trp-tRNA levels are low in the cell, the ribosome will stall at the Trp codons for the leader peptide and not the terminator but an alternative secondary RNA structure (antiterminator) is formed, resulting in transcription elongation into the trp operon.
In E. coli and others, the start codon of leader peptides is generally preceded by an SD sequence present in the mRNA leader. Therefore, other leadered mRNAs of D. deserti were inspected for leader peptides that do not have their start codon at the extreme 5′-end. One additional putative leader peptide was found, encoded by trpEGD mRNA. Also here translation occurs from a leaderless transcript, because the TSS is at only one nucleotide upstream of the leader peptide’s start codon. In total, 14 putative leader peptides predicted to be involved in transcription attenuation were found in D. deserti (supplementary table S9, Supplementary Material online).
New Gene Predictions and Proteome Data Correlate with Novel Leaderless mRNAs
For many of the deduced translation products of the additional leaderless transcripts, no protein homologs were identified using sequence- or structure-based homology searches. These putative peptides and proteins may be specific for D. deserti, or failure to detect homologs may be related to the small peptide sizes. To see whether the additional leaderless transcripts correspond to CDSs that were missed previously, new gene predictions were obtained and analyzed. In addition, a new proteome analysis after enrichment of small proteins was performed.
For the initial D. deserti genome annotation, FrameD and MED were used (de Groot et al. 2009). Here, we applied AMIGene for identification of potential new CDSs (Bocs et al. 2003). After analysis of these new predictions, 142 CDSs were added and annotated (including 49 partial genes) (supplementary table S10, Supplementary Material online). Identification of these new genes and reannotation of several start codons also allowed removal of 49 erroneously annotated hypothetical genes (supplementary table S10, Supplementary Material online). It is worth noting that the software did not predict the 14 leader peptide genes proposed to be involved in transcription attenuation and 3 of the 17 conserved genes described in the previous subsection, that is, Deide_15148 (encoding Gly-, Ser-, Arg-, and His-rich protein), Deide_2p00483 (Lys- and Ala-rich), and Deide_11672 (gene fragment). Homology with one or more proteins in databases was detected for 43 of the new CDSs, but a putative function could be assigned to only two (Deide_11194, putative excisionase; Deide_2p02235, putative transposase). Twenty-seven of the new additional leaderless transcripts appeared to correlate with new predicted genes (supplementary table S10, Supplementary Material online).
For the new proteome analysis, fractions were proteolyzed with trypsin on one hand, and chymotrypsin on the other, with the intention to increase the global polypeptide sequence coverage. The peptides were analyzed by orbitrap-based tandem mass spectrometry resulting in a data set of 556,375 MS/MS spectra. A total of 233,301 spectra could be assigned to peptide sequences after querying a six-frame translation database, revealing the presence of 21,232 peptide sequences (supplementary table S11, Supplementary Material online) and pointing at a total of 1,481 proteins detected with two or more tryptic or chymotryptic peptides (supplementary table S12, Supplementary Material online). Of these, the products of 160 previously annotated genes were detected here for the first time, including 14 proteins smaller than 100 residues (supplementary table S3, Supplementary Material online). Eight of these small proteins appeared to be translated from a leaderless mRNA. For the 160 newly detected proteins, a TSS for leaderless and leadered mRNA was observed for 74 and 26 cases, respectively.
With the detection of two or more tryptic or chymotryptic peptides, the new proteome analysis also validated the expression of seven previously nonannotated proteins of 69–158 residues: Deide_05864, Deide_13059, Deide_15253, Deide_1p00482, Deide_2p01542, and Deide_3p02615/Deide_23165 (the latter two are indistinguishable by tandem mass spectrometry) (supplementary fig. S7 and table S12, Supplementary Material online). Two additional new proteins, namely Deide_12656 (70 aa) and Deide_11207 (70 aa), were detected with only one peptide but with a high confidence score. The nine corresponding genes were also found among the new CDSs predicted by AMIGene. Five of these newly detected proteins correspond to additional leaderless mRNAs described above, and their mass spectrometry detection thus validates the translation of these new mRNAs. A single peptide was also found for the products of two other predicted new genes (Deide_14224 and Deide_3p02814) that retained our attention. Remarkably, three peptides were detected for an ORF located at the reverse strand of Deide_04940, suggesting that RNA antisense to Deide_04940 could be translated (supplementary fig. S7, Supplementary Material online). The protein coded by Deide_04940, glycine dehydrogenase, was also found expressed (supplementary table S12, Supplementary Material online).
For five of the newly detected proteins, homologs were found in other bacteria, mainly from the genus Deinococcus (supplementary fig. S7, Supplementary Material online). In addition, Deide_05864 (76 aa) showed 70% identity with the product of another predicted new gene, Deide_05654 (74 aa), and with that of a new gene that was not predicted but whose expression correlated with RNA-seq data (Deide_11206, 76 aa). Moreover, a peptide was detected for Deide_11206. Deide_23165 (78 aa) and Deide_3p02615 (78 aa) are almost identical (76 identical residues). Both genes are downstream of putative phytanoyl-CoA dioxygenase genes (Deide_23170 and Deide_3p02610; 99% identity), suggesting duplication of this gene pair. RNA-seq data indicated better expression of Deide_23165 (and Deide_23170) than Deide_3p02615 (and Deide_3p02610). Deide_15253 (158 aa) and Deide_12656 (70 aa) both contain a predicted signal peptide followed by a threonine-rich part and a remarkably conserved glycine and leucine-rich region of about 18 residues (supplementary fig. S7, Supplementary Material online).
Taken together, the data from the analyses (gene prediction, proteomics, and protein/peptide conservation) in this and the previous subsection indicate the existence of at least 44 novel polypeptides that support the identification of new leaderless mRNAs. In total, 160 new CDSs have been added and annotated (table 3; supplementary table S10, Supplementary Material online).
Table 3.
New Annotated CDSs and Their Prediction and/or Detected Expression
| New Annotated CDSs | 160 |
| Predicted by AMIGene | 142 |
| Transcripts with 5′-AUG or 5′-GUG | 43a |
| Detected by mass spectrometryb | 9 |
aOne additional putative leader peptide with TSS at −1.
bWith >1 (chymo)tryptic peptide or with 1 peptide with high confidence score.
Noncoding RNA
In addition to new leaderless transcripts, many other orphan TSSs and transcripts were found, which may correspond to noncoding RNAs (supplementary table S4, Supplementary Material online). It is also possible that one or more are bifunctional, that is, functioning as a regulatory RNA and also coding for a peptide (Dinger et al. 2008). As suggested by the read numbers, many potential noncoding RNAs are well expressed in D. deserti (supplementary table S4, Supplementary Material online). For three of the transcripts, homology was found with noncoding RNAs belonging to conserved RNA families present in the Rfam database (Burge et al. 2013), that is, signal recognition particle RNA, transfer-messenger RNA and bacterial RNase P class A (supplementary table S13, Supplementary Material online). Besides these noncoding RNAs, the long 5′-UTR of 13 genes appeared to correspond to cis-regulatory elements (supplementary table S13, Supplementary Material online). These include TPP, FMN, SAM, cyclic di-GMP-I, cyclic di-GMP-II, and cobalamin riboswitches for genes involved in processes such as thiamine biosynthesis and transport, riboflavin biosynthesis, and methionine and vitamin B12 transport. A T-box leader in the 5′-UTR of the valine- and glycine tRNA ligase genes was also found. Therefore, although many genes entirely lack a 5′-UTR in D. deserti, expression of various others is likely regulated by structured elements formed by 5′-UTRs.
Radiation-Induced Genes
After addition of new genes and correction of start codons, gene expression levels in RD19 NI and RD19 IR were compared. Table 4 lists the genes for which an induction of at least 10-fold was found after irradiation. Fold changes for all genes are present in supplementary table S14, Supplementary Material online. Several expected and novel genes were found among the highly upregulated genes after exposure to radiation (table 4). In previous microarray experiments with D. radiodurans, the five most highly radio-induced genes were the Deinococcus-specific genes ddrA, ddrB, ddrC, ddrD, and pprA (Tanaka et al. 2004). Their homologs in D. deserti were also among the most highly induced, showing that not only their presence but also their strong upregulation in response to radiation damage is conserved. Deinococcus deserti possesses two homologs of ddrO, namely Deide_20570 (ddrOC) and Deide_3p02170 (ddrOP3), encoding XRE family transcriptional regulators sharing 84% identity. Both genes were found induced. Deide_20580 is another highly induced gene, specifying a small protein of unknown function (supplementary fig. S8, Supplementary Material online). It is located adjacent and divergent to Deide_20570 (ddrOC).
Table 4.
Most Highly Induced Genes in D. deserti After Irradiation
| Gene | Product | Fold Changea | TSSb |
|---|---|---|---|
| Deide_09150 | DNA damage response protein DdrA | +++ | 1 |
| Deide_01090 | DinB family protein | +++ | −25 |
| Deide_09148 | Putative protein of unknown function (30 aa) | +++ | |
| Deide_23280 | DNA damage response protein DdrC | +++ | 1 |
| Deide_04721 | Conserved hypothetical protein (74 aa) | +++ | 1 |
| Deide_01160 | DNA damage response protein DdrD | +++ | 1 |
| Deide_02990 | DNA damage response protein DdrB | ++ | −17 |
| Deide_18350 | Holliday junction DNA helicase RuvB | ++ | 1 |
| Deide_3p02170 | XRE family transcriptional regulator DdrOP3 | ++ | −2 |
| Deide_19490 | Hypothetical protein (53 aa) | ++ | 1 |
| Deide_11446 | Putative protein of unknown function (57 aa) | ++ | −26 |
| Deide_2p00980 | Conserved hypothetical protein (64 aa) | + | 1 |
| Deide_05260 | Conserved hypothetical protein (62 aa) | + | −12 |
| Deide_20580 | Conserved hypothetical protein (83 aa) | + | −78 |
| Deide_15340 | Bug family protein, precursor | + | −59 |
| Deide_18730 | Putative SWIM zinc finger domain protein | + | −30 |
| Deide_20570 | XRE family transcriptional regulator DdrOC | + | −131 |
| Deide_01100 | DinB family protein | + | |
| Deide_2p01380 | DNA repair protein PprA | + | −97 |
| Deide_03320 | Conserved hypothetical protein, precursor | + | −44 |
| Deide_08010 | ABC transporter permease | + | −34 |
| Deide_1p00730 | Putative peptidase S8, precursor | + | −49 |
| Deide_1p01880 | Y-family DNA polymerase ImuY | + | |
| Deide_3p00210 | Recombinase A | + | |
| Deide_00100 | 50S ribosomal protein L9 | + | |
| Deide_00110 | 30S ribosomal protein S18 | + | |
| Deide_14940 | Putative N-acetyltransferase | + | |
| Deide_19440 | 2′–5′ RNA ligase | + | |
| Deide_19965 | Conserved hypothetical protein (63 aa) | + | −85 |
| Deide_20140 | Putative N-acetyltransferase | + | 1 |
| Deide_21600 | RtcB family protein | + | −73 |
| Deide_15490 | DNA gyrase, subunit B (GyrB) | + | −88 |
| Deide_1p01870 | Repressor LexA | + | −42 |
| Deide_21420 | Conserved hypothetical protein (96 aa) | + | −12 |
| Deide_19450 | Recombinase A | + | |
| Deide_1p01260 | Recombinase A | + | −57 |
| Deide_1p01890 | Hypothetical protein | + | |
| Deide_07900 | Conserved hypothetical protein (63 aa) | + | −22 |
| Deide_13590 | Conserved hypothetical protein (77 aa) | + | −37 |
a+++, >50-fold; ++, >20-fold; +, >10-fold.
bTSS relative to the translation initiation codon.
Table 4 contains several DNA repair genes that were previously found radiation-induced in D. deserti and/or D. radiodurans (Liu et al. 2003; Tanaka et al. 2004; Dulermo et al. 2009). These include three recA and genes from the operon encoding LexA and mutagenic translesion DNA polymerases (Deide_1p01870, Deide_1p01880, and Deide_1p01890 in table 4). Deide_19440 (ligT) is located upstream and in operon with recAC. Deide_00100 and Deide_00110 encoding ribosomal proteins are located downstream and in operon with the 9-fold-induced ssb gene (Deide_00120) (fig. 1).
Deide_01100 is located downstream of and in the same orientation as Deide_01090. Both encode a protein of unknown function belonging to the DinB family, which includes DNA damage-inducible DinB from Bacillus subtilis. Two genes encoding putative N-acetyltransferases were found among the highly induced genes. One of these, Deide_20140, was also found induced at the protein level using a 2DE approach, which led us to speculate that induced N-acetyltransferase might be responsible for the N-terminal acetylation observed on upregulated DNA gyrase GyrA (Dedieu et al. 2013). Deide_18730 is the first gene of an operon encoding five proteins, including a MoxR-like AAA+ ATPase (Deide_18710), which could function as a chaperon system for the folding/activation of specific substrate proteins (Snider and Houry 2006). Deide_21600 is an RtcB family protein. Recent work on the RNA ligase RtcB from E. coli led the authors to speculate that RtcB might afford bacteria a means to recover from stress-induced RNA damage (Tanaka and Shuman 2011). Interestingly, Deide_3p01893 codes for a second RtcB homolog (62% identity with Deide_21600).
Several of the highly induced genes code for (mostly small) proteins of unknown function (supplementary fig. S8, Supplementary Material online). Deinococcus-specific Deide_04721 has two pairs of conserved CXXC residues. Deide_05260 contains a conserved Domain of Unknown Function (DUF1540), which also has four conserved cysteine residues, suggestive of a metal-binding function. Homologs of Deide_19965, with one conserved cysteine, were only found in several Deinococcus species and in Meiothermus ruber. TBLASTN analysis indicated nonannotated homologs of Deide_05260 and Deide_19965 in D. geothermalis (supplementary fig. S8, Supplementary Material online). Deinococcus radiodurans does not possess homologs of Deide_04721, Deide_05260, and Deide_19965. Deide_2p00980 and its D. radiodurans homolog DR_A0234 share limited similarity with stress-induced proteins YciG from E. coli and GsiB from B. subtilis. Deide_09148 is a putative gene for a peptide of only 30 residues. It is located directly downstream and in the same orientation as highly induced ddrA (Deide_09150). TBLASTN analysis revealed that potential homologs could be present downstream of ddrA in D. radiodurans and D. geothermalis. Deide_11446 is located downstream of and in the opposite orientation of uvrC (Deide_11450) and may code for a protein of 57 residues that has a low level of homology only with DGo_CA1576 (55 aa) from D. gobiensis (DGo_CA1576 is also located downstream of uvrC). Alternatively, Deide_11446 may correspond to a novel noncoding RNA (no homology was detected using BLASTN and the Rfam database).
A 17-base pair palindromic motif called RDRM (radiation and desiccation response motif) has been found upstream of about 20 radiation-induced genes in D. radiodurans and their homologs in D. geothermalis (Makarova et al. 2007), and found to be conserved in D. deserti (de Groot et al. 2009). Here, we analyzed the location of the RDRM with respect to the TSS of the radiation-induced genes (supplementary fig. S9, Supplementary Material online). There was clearly no conserved distance between the TSS and the RDRM. TSSs were found up to 20 bp upstream, within, or up to 50 bp downstream of the RDRM. These data would be compatible with a potential repressor protein binding to the RDRM and blocking initiation of transcription in standard growth conditions.
Discussion
In this work, we showed that RNA-seq, complemented with proteomics, was a powerful method that strongly improved our knowledge of radiation-tolerant bacterium D. deserti. The RNA-seq data and identified TSSs revealed several new highly radiation-induced genes, many novel genes, and allowed numerous start codon reannotations. Importantly, hundreds of efficiently translated leaderless mRNAs with either an AUG or GUG start codon were identified. Analysis of the new genes and start codon reannotations indicated that nonannotated genes and start reannotations could also be proposed for other bacteria. Similarly, it is plausible that translation from leaderless mRNAs is also a major translation initiation mechanism in other Deinococcus species. Bioinformatic analysis predicted more than 40% leaderless mRNAs in D. radiodurans (Zheng et al. 2011), but these results were not confirmed by experimental evidence and are dependent on the quality of start codon predictions.
Among the highly radiation-induced genes in D. deserti are several unknown small proteins of less than 100 aa. Three of these contain conserved cysteine residues, which are probably structurally and/or functionally important, for example, for metal binding or antioxidant defence (Netto et al. 2007; Requejo et al. 2010). Another small protein, Deide_20580, is encoded by a gene located adjacent and divergent to Deide_20570. The latter codes for DdrO, a radiation-induced transcriptional regulator protein highly conserved in Deinococcus and therefore proposed to be implicated in the radiation response (Makarova et al. 2007). An identical genetic organization is present in other sequenced Deinococcus genomes, with D. radiodurans, which does not have a Deide_20580 homolog, as a remarkable exception. A homologous gene pair is also present in four other sequenced members of the Deinococcus/Thermus phylum: M. ruber, M. silvanus, Marinithermus hydrothermalis, and Oceanithermus profundus. The coinduction of Deide_20570 and Deide_20580 and the conservation of this gene pair in other species indicate that their gene products might function together.
Leaderless mRNAs are rare in most organisms characterized so far, but in the last years their number is steadily increasing. In recent studies, 4–505 leaderless genes (up to 27%) have been identified in several bacterial species (Qiu et al. 2010; Sharma et al. 2010; Mitschke et al. 2011; Vockenhuber et al. 2011; Dötsch et al. 2012; Kröger et al. 2012; Schmidtke et al. 2012; Seo et al. 2012; Cortes et al. 2013; Schlüter et al. 2013). Thus, the very high number and proportion of leaderless mRNAs in D. deserti (1,174 cases, 60%) is unprecedented for a bacterial species. A high abundance of leaderless mRNAs has also been found in some archaea, for example, 69% in Sulfolobus solfataricus (Wurtzel et al. 2010), whereas only few leaderless transcripts were found in other archaeal species (Jäger et al. 2009; Toffano-Nioche et al. 2013).
Translation initiation on canonical leadered mRNAs has specific features in Archaea, Bacteria, and Eukarya (Benelli and Londei 2009; Malys and McCarthy 2011). Leaderless mRNAs, however, can be universally translated by archaeal, bacterial, and eukaryotic ribosomes (Grill et al. 2000). It has been suggested that leaderless mRNAs may represent the ancestral form of messenger for a less complex and less regulated translation initiation mechanism (Benelli and Londei 2009; Malys and McCarthy 2011). This is supported by data obtained with E. coli, where the antibiotic kasugamycin induced the formation of 61S ribosomes lacking several functionally important proteins. These 61S particles, which might reflect ancient bacterial protoribosomes, were proficient in selectively translating leaderless mRNA (Kaberdina et al. 2009). Leaderless initiation is different from the mechanisms on leadered mRNAs. Unlike canonical bacterial mRNAs, which first bind to the small 30S ribosomal subunit prior to joining of the 50S subunit, leaderless mRNAs can be efficiently bound and read by nondissociated 70S ribosomes (O'Donnell and Janssen 2002; Moll et al. 2004; Udagawa et al. 2004). Obviously, as leaderless mRNAs have no or very short 5′-UTR, they lack an SD sequence or other signals present in the 5′-UTR for ribosome recruitment. The only identified translation signal in a leaderless mRNA is the 5′-terminal start codon. The AUG start codon, and not codon-anticodon complementarity, is required for translation of leaderless mRNA (Van Etten and Janssen 1998). Although GUG and UUG can be efficiently used as start codons on leadered mRNAs, the presence of a 5′-AUG on leaderless mRNAs is much more important for efficient translation in two studied model bacteria. Using a natural leaderless reporter gene in the haloarchaeon Haloferax volcanii, changing the native AUG start to GUG or UUG totally inhibited translation (Hering et al. 2009). Similar studies in E. coli showed that UUG and CUG start codons did not support expression of leaderless mRNAs, but low levels of expression and binding to 70S ribosomes were observed when a 5′-terminal GUG was present (Van Etten and Janssen 1998; O'Donnell and Janssen 2001, 2002). Importantly, addition of a 5′-terminal AUG to random RNA fragments made these competent for ribosome binding and translation in E. coli and H. volcanii (Brock et al. 2008; Hering et al. 2009). Taken together, the data strongly suggest that an AUG (or GUG) triplet at the 5′-end of an mRNA is a distinct signal required and sufficient for ribosome binding and expression, which prompted us to inspect all TSSs for the presence of a 5′-AUG or 5′-GUG.
Besides the leaderless annotated genes, an AUG or GUG triplet was found at the 5′-end of more than 170 additional transcripts, which suggested that these are translatable leaderless mRNAs for novel peptides and low molecular weight proteins. After having used a combination of homology search, sequence analysis, new gene predictions, and new proteome analysis, we annotated new CDSs that correspond to 44 of these new leaderless transcripts. Fourteen of these code for putative leader peptides predicted to be involved in transcription attenuation control of tRNA synthetase or amino acid biosynthesis genes. For 17 other products, one or more homologs (annotated or nonannotated) were identified, mainly in Deinococcus genomes. One of these conserved proteins, Deide_15148, is predicted to contain a hydrophilic cytoplasmic domain of low complexity followed by a C-terminal transmembrane helix. Unstructured hydrophilic low complexity proteins have a role in desiccation tolerance by stabilizing membranes and by limiting aggregation of cellular proteins (Chakrabortee et al. 2007, 2012; Krisko et al. 2010). Deide_15148 may thus contribute to membrane protection and prevention of protein aggregation during desiccation of D. deserti. Three other new small proteins, Deide_04426, Deide_12656, and Deide_15253, might have a similar function in the periplasm. They have a signal peptide followed by the mature part of the protein consisting almost entirely of a hydrophilic region of low complexity. Lipoprotein Deide_04426 would be anchored to the periplasmic side of the cytoplasmic membrane. Noteworthy, Deide_12656 and Deide_15253 were detected by tandem mass spectrometry.
No homology was found for the deduced peptides from many other new leaderless transcripts in D. deserti. As previous data have established that an AUG or GUG at the 5′-end of RNA is generally sufficient for ribosome binding and translation initiation (Brock et al. 2008), many of these transcripts could also be translated. Interestingly, recent studies have revealed that peptides are abundant components of protein-free cell extracts of D. radiodurans and the radiation-tolerant archaeon Halobacterium salinarum, and important for protection of proteins against radiation-induced oxidation (Daly et al. 2010; Robinson et al. 2011). Antioxidant properties of peptides of various length and amino acid composition, which have substantially higher antioxidant activity than intact proteins, have also been reported in protein hydrolysates (Elias et al. 2008). It has been suggested that the accumulated cellular peptides in D. radiodurans may be derived from proteolysis and/or peptide import (Daly et al. 2010; Krisko and Radman 2013). As D. deserti efficiently translates many leaderless genes (table 2), we propose that translation of many newly identified leaderless transcripts, and of 5′-leaders of leadered mRNAs that were thought to be 5′-UTR but which possess an AUG or GUG at the 5′-end, provides an alternative explanation for the enrichment in the cell of small peptides important for protection of proteins against oxidation and thus for radiation- and desiccation tolerance. Moreover, in addition to the transcripts with a 5′-AUG or 5′-GUG, it is likely that even more peptides could be translated from transcripts that contain only one or few nucleotides upstream of an AUG or GUG triplet. Such leaderless mRNAs with a very short 5′-UTR were also found to be efficiently translated (table 2) (Krishnan et al. 2010). As a high number of leaderless mRNAs has also been reported for Halobacterium salinarum (Brenneis et al. 2007), a correlation between radiation tolerance and leaderless initiation may also exist in this archaeon and possibly in other radiation tolerant species.
Only a single-nucleotide change can result in generation of a -10 consensus motif TAnnnT and associated new TSS, as observed in Mycobacterium tuberculosis (Rose et al. 2013). And if appropriate sequences are present in such a novel transcript, like a start codon at the 5′-end, it may direct translation initiation. Similarly, mutations in already existing (and possibly highly expressed) leadered mRNAs could result in a start codon at or very near the 5′-end of a transcript. For example, Deide_02390 has a TSS at -59 of the start codon. A single C to G mutation at the 5′-AUC of this transcript would result in a leaderless mRNA with a 5′-AUG for a peptide of 20 residues. We therefore suggest that such mutations resulting in synthesis of small peptides has contributed to adaptation to extreme environmental conditions such as present in dry deserts, and thus to radiation tolerance.
Supplementary Material
Supplementary figures S1–S9 and tables S1–S14 are available at Genome Biology and Evolution online.
Acknowledgments
This work was supported by the Commissariat à l’énergie atomique et aux energies alternatives (CEA) and the Agence Nationale de la Recherche (grant number ANR-07-BLAN-0106-02). The authors also thank J. Vogel and F. Thümmler for discussion, and J. Vicente for access to gamma irradiation facilities.
Literature Cited
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Battista JR. Against all odds: the survival strategies of Deinococcus radiodurans. Annu Rev Microbiol. 1997;51:203–224. doi: 10.1146/annurev.micro.51.1.203. [DOI] [PubMed] [Google Scholar]
- Baudet M, et al. Proteomics-based refinement of Deinococcus deserti genome annotation reveals an unwonted use of non-canonical translation initiation codons. Mol Cell Proteomics. 2010;9:415–426. doi: 10.1074/mcp.M900359-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benelli D, Londei P. Begin at the beginning: evolution of translational initiation. Res Microbiol. 2009;160:493–501. doi: 10.1016/j.resmic.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Blasius M, Hubscher U, Sommer S. Deinococcus radiodurans: what belongs to the survival kit? Crit Rev Biochem Mol Biol. 2008;43:221–238. doi: 10.1080/10409230802122274. [DOI] [PubMed] [Google Scholar]
- Bocs S, Cruveiller S, Vallenet D, Nuel G, Medigue C. AMIGene: annotation of microbial genes. Nucleic Acids Res. 2003;31:3723–3726. doi: 10.1093/nar/gkg590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouthier de la Tour C, et al. Comparative proteomics reveals key proteins recruited at the nucleoid of Deinococcus after irradiation-induced DNA damage. Proteomics. 2013;13:3457–3469. doi: 10.1002/pmic.201300249. [DOI] [PubMed] [Google Scholar]
- Brenneis M, Hering O, Lange C, Soppa J. Experimental characterization of Cis-acting elements important for translation and transcription in halophilic archaea. PLoS Genet. 2007;3:e229. doi: 10.1371/journal.pgen.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock JE, Pourshahian S, Giliberti J, Limbach PA, Janssen GR. Ribosomes bind leaderless mRNA in Escherichia coli through recognition of their 5'-terminal AUG. RNA. 2008;14:2159–2169. doi: 10.1261/rna.1089208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge SW, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41:D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, et al. GenomicFeatures: tools for making and manipulating transcript centric annotations. 2011. R package version 1.4.3. [Google Scholar]
- Chakrabortee S, et al. Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proc Natl Acad Sci U S A. 2007;104: 18073–18078. doi: 10.1073/pnas.0706964104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortee S, et al. Intrinsically disordered proteins as molecular shields. Mol Biosyst. 2012;8:210–219. doi: 10.1039/c1mb05263b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie-Oleza JA, Miotello G, Armengaud J. Proteogenomic definition of biomarkers for the large roseobacter clade and application for a quick screening of new environmental isolates. J Proteome Res. 2013;12:5331–5339. doi: 10.1021/pr400554e. [DOI] [PubMed] [Google Scholar]
- Cortes T, et al. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 2013;5:1121–1131. doi: 10.1016/j.celrep.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM, Battista JR. Deinococcus radiodurans—the consummate survivor. Nat Rev Microbiol. 2005;3:882–892. doi: 10.1038/nrmicro1264. [DOI] [PubMed] [Google Scholar]
- Croucher NJ, Thomson NR. Studying bacterial transcriptomes using RNA-seq. Curr Opin Microbiol. 2010;13:619–624. doi: 10.1016/j.mib.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly MJ. Death by protein damage in irradiated cells. DNA Repair (Amst). 2012;11:12–21. doi: 10.1016/j.dnarep.2011.10.024. [DOI] [PubMed] [Google Scholar]
- Daly MJ, et al. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science. 2004;306:1025–1028. doi: 10.1126/science.1103185. [DOI] [PubMed] [Google Scholar]
- Daly MJ, et al. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 2007;5:e92. doi: 10.1371/journal.pbio.0050092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly MJ, et al. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS One. 2010;5:e12570. doi: 10.1371/journal.pone.0012570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot A, et al. Deinococcus deserti sp. nov., a gamma-radiation-tolerant bacterium isolated from the Sahara Desert. Int J Syst Evol Microbiol. 2005;55:2441–2446. doi: 10.1099/ijs.0.63717-0. [DOI] [PubMed] [Google Scholar]
- de Groot A, et al. Alliance of proteomics and genomics to unravel the specificities of Sahara bacterium Deinococcus deserti. PLoS Genet. 2009;5:e1000434. doi: 10.1371/journal.pgen.1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedieu A, et al. Major soluble proteome changes in Deinococcus deserti over the earliest stages following gamma-ray irradiation. Proteome Sci. 2013;11:3. doi: 10.1186/1477-5956-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol. 2008;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dötsch A, et al. The Pseudomonas aeruginosa transcriptome in planktonic cultures and static biofilms using RNA sequencing. PLoS One. 2012;7:e31092. doi: 10.1371/journal.pone.0031092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulermo R, Fochesato S, Blanchard L, de Groot A. Mutagenic lesion bypass and two functionally different RecA proteins in Deinococcus deserti. Mol Microbiol. 2009;74:194–208. doi: 10.1111/j.1365-2958.2009.06861.x. [DOI] [PubMed] [Google Scholar]
- Dupierris V, Masselon C, Court M, Kieffer-Jaquinod S, Bruley C. A toolbox for validation of mass spectrometry peptides identification and generation of database: IRMa. Bioinformatics. 2009;25:1980–1981. doi: 10.1093/bioinformatics/btp301. [DOI] [PubMed] [Google Scholar]
- Earl AM, Mohundro MM, Mian IS, Battista JR. The IrrE protein of Deinococcus radiodurans R1 is a novel regulator of recA expression. J Bacteriol. 2002;184:6216–6224. doi: 10.1128/JB.184.22.6216-6224.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias RJ, Kellerby SS, Decker EA. Antioxidant activity of proteins and peptides. Crit Rev Food Sci Nutr. 2008;48:430–441. doi: 10.1080/10408390701425615. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Fredrickson JK, et al. Protein oxidation: key to bacterial desiccation resistance? ISME J. 2008;2:393–403. doi: 10.1038/ismej.2007.116. [DOI] [PubMed] [Google Scholar]
- Georg J, Hess WR. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev. 2011;75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill S, Gualerzi CO, Londei P, Blasi U. Selective stimulation of translation of leaderless mRNA by initiation factor 2: evolutionary implications for translation. EMBO J. 2000;19:4101–4110. doi: 10.1093/emboj/19.15.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering O, Brenneis M, Beer J, Suess B, Soppa J. A novel mechanism for translation initiation operates in haloarchaea. Mol Microbiol. 2009;71:1451–1463. doi: 10.1111/j.1365-2958.2009.06615.x. [DOI] [PubMed] [Google Scholar]
- Hua Y, et al. PprI: a general switch responsible for extreme radioresistance of Deinococcus radiodurans. Biochem Biophys Res Commun. 2003;306:354–360. doi: 10.1016/s0006-291x(03)00965-3. [DOI] [PubMed] [Google Scholar]
- Jäger D, et al. Deep sequencing analysis of the Methanosarcina mazei Go1 transcriptome in response to nitrogen availability. Proc Natl Acad Sci U S A. 2009;106:21878–21882. doi: 10.1073/pnas.0909051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaberdina AC, Szaflarski W, Nierhaus KH, Moll I. An unexpected type of ribosomes induced by kasugamycin: a look into ancestral times of protein synthesis? Mol Cell. 2009;33:227–236. doi: 10.1016/j.molcel.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Krishnan KM, Van Etten WJ, 3rd, Janssen GR. Proximity of the start codon to a leaderless mRNA's 5' terminus is a strong positive determinant of ribosome binding and expression in Escherichia coli. J Bacteriol. 2010;192:6482–6485. doi: 10.1128/JB.00756-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisko A, Leroy M, Radman M, Meselson M. Extreme anti-oxidant protection against ionizing radiation in bdelloid rotifers. Proc Natl Acad Sci U S A. 2012;109:2354–2357. doi: 10.1073/pnas.1119762109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisko A, Radman M. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc Natl Acad Sci U S A. 2010;107:14373–14377. doi: 10.1073/pnas.1009312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisko A, Radman M. Biology of extreme radiation resistance: the way of Deinococcus radiodurans. Cold Spring Harb Perspect Biol. 2013;5:a012765. doi: 10.1101/cshperspect.a012765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisko A, Smole Z, Debret G, Nikolic N, Radman M. Unstructured hydrophilic sequences in prokaryotic proteomes correlate with dehydration tolerance and host association. J Mol Biol. 2010;402:775–782. doi: 10.1016/j.jmb.2010.08.012. [DOI] [PubMed] [Google Scholar]
- Kröger C, et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A. 2012;109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin-Zaidman S, et al. Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science. 2003;299:254–256. doi: 10.1126/science.1077865. [DOI] [PubMed] [Google Scholar]
- Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc Natl Acad Sci U S A. 2003;100:4191–4196. doi: 10.1073/pnas.0630387100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Chen H, Xu G, Shah AM, Hua Y. DNA binding is essential for PprI function in response to radiation damage in Deinococcus radiodurans. DNA Repair (Amst). 2012;11:139–145. doi: 10.1016/j.dnarep.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Lu H, et al. Deinococcus radiodurans PprI switches on DNA damage response and cellular survival networks after radiation damage. Mol Cell Proteomics. 2009;8:481–494. doi: 10.1074/mcp.M800123-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, et al. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, et al. Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks. PLoS One. 2007;2:e955. doi: 10.1371/journal.pone.0000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malys N, McCarthy JE. Translation initiation: variations in the mechanism can be anticipated. Cell Mol Life Sci. 2011;68:991–1003. doi: 10.1007/s00018-010-0588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattimore V, Battista JR. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J Bacteriol. 1996;178:633–637. doi: 10.1128/jb.178.3.633-637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitschke J, et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci U S A. 2011;108:2124–2129. doi: 10.1073/pnas.1015154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll I, Hirokawa G, Kiel MC, Kaji A, Blasi U. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acids Res. 2004;32:3354–3363. doi: 10.1093/nar/gkh663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naville M, Gautheret D. Transcription attenuation in bacteria: theme and variations. Brief Funct Genomics. 2010;9:178–189. doi: 10.1093/bfgp/elq008. [DOI] [PubMed] [Google Scholar]
- Netto LE, et al. Reactive cysteine in proteins: protein folding, antioxidant defense, redox signaling and more. Comp Biochem Physiol C Toxicol Pharmacol. 2007;146:180–193. doi: 10.1016/j.cbpc.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norais CA, Chitteni-Pattu S, Wood EA, Inman RB, Cox MM. DdrB protein, an alternative Deinococcus radiodurans SSB induced by ionizing radiation. J Biol Chem. 2009;284:21402–21411. doi: 10.1074/jbc.M109.010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell SM, Janssen GR. The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5' untranslated leader. J Bacteriol. 2001;183:1277–1283. doi: 10.1128/JB.183.4.1277-1283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell SM, Janssen GR. Leaderless mRNAs bind 70S ribosomes more strongly than 30S ribosomal subunits in Escherichia coli. J Bacteriol. 2002;184:6730–6733. doi: 10.1128/JB.184.23.6730-6733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, et al. Structural and operational complexity of the Geobacter sulfurreducens genome. Genome Res. 2010;20:1304–1311. doi: 10.1101/gr.107540.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requejo R, Hurd TR, Costa NJ, Murphy MP. Cysteine residues exposed on protein surfaces are the dominant intramitochondrial thiol and may protect against oxidative damage. FEBS J. 2010;277:1465–1480. doi: 10.1111/j.1742-4658.2010.07576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CK, et al. A major role for nonenzymatic antioxidant processes in the radioresistance of Halobacterium salinarum. J Bacteriol. 2011;193:1653–1662. doi: 10.1128/JB.01310-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G, et al. Mapping of genotype-phenotype diversity among clinical isolates of Mycobacterium tuberculosis by sequence-based transcriptional profiling. Genome Biol Evol. 2013;5:1849–1862. doi: 10.1093/gbe/evt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubiano-Labrador C, et al. Proteogenomic insights into salt tolerance by a halotolerant alpha-proteobacterium isolated from an Andean saline spring. J Proteomics. 2014;97:36–47. doi: 10.1016/j.jprot.2013.05.020. [DOI] [PubMed] [Google Scholar]
- Schlüter JP, et al. Global mapping of transcription start sites and promoter motifs in the symbiotic alpha-proteobacterium Sinorhizobium meliloti 1021. BMC Genomics. 2013;14:156. doi: 10.1186/1471-2164-14-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtke C, et al. Genome-wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Res. 2012;40:2020–2031. doi: 10.1093/nar/gkr904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JH, et al. Multiple-omic data analysis of Klebsiella pneumoniae MGH 78578 reveals its transcriptional architecture and regulatory features. BMC Genomics. 2012;13:679. doi: 10.1186/1471-2164-13-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- Shine J, Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade D, Radman M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev. 2011;75:133–191. doi: 10.1128/MMBR.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Snider J, Houry WA. MoxR AAA+ ATPases: a novel family of molecular chaperones? J Struct Biol. 2006;156:200–209. doi: 10.1016/j.jsb.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Sorek R, Cossart P. Prokaryotic transcriptomics: a new view onregulation, physiology and pathogenicity. Nat Rev Genet. 2010;11:9–16. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Shuman S. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J Biol Chem. 2011;286:7727–7731. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, et al. Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics. 2004;168:21–33. doi: 10.1534/genetics.104.029249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillett D, Burns BP, Neilan BA. Optimized rapid amplification of cDNA ends (RACE) for mapping bacterial mRNA transcripts. Biotechniques. 2000;28:448. doi: 10.2144/00283st01. 450, 452–453, 456. [DOI] [PubMed] [Google Scholar]
- Toffano-Nioche C, et al. RNA at 92 degrees C: the non-coding transcriptome of the hyperthermophilic archaeon Pyrococcus abyssi. RNA Biol. 2013;10:1211–1220. doi: 10.4161/rna.25567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toueille M, et al. A comparative proteomic approach to better define Deinococcus nucleoid specificities. J Proteomics. 2012;75:2588–2600. doi: 10.1016/j.jprot.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Udagawa T, Shimizu Y, Ueda T. Evidence for the translation initiation of leaderless mRNAs by the intact 70 S ribosome without its dissociation into subunits in eubacteria. J Biol Chem. 2004;279:8539–8546. doi: 10.1074/jbc.M308784200. [DOI] [PubMed] [Google Scholar]
- Vallenet D, et al. MicroScope—an integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 2013;41:D636–D647. doi: 10.1093/nar/gks1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten WJ, Janssen GR. An AUG initiation codon, not codon-anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli. Mol Microbiol. 1998;27:987–1001. doi: 10.1046/j.1365-2958.1998.00744.x. [DOI] [PubMed] [Google Scholar]
- van Vliet AH. Next generation sequencing of microbial transcriptomes: challenges and opportunities. FEMS Microbiol Lett. 2010;302:1–7. doi: 10.1111/j.1574-6968.2009.01767.x. [DOI] [PubMed] [Google Scholar]
- Vockenhuber MP, et al. Deep sequencing-based identification of small non-coding RNAs in Streptomyces coelicolor. RNA Biol. 2011;8:468–477. doi: 10.4161/rna.8.3.14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujicic-Zagar A, et al. Crystal structure of the IrrE protein, a central regulator of DNA damage repair in Deinococcaceae. J Mol Biol. 2009;386:704–716. doi: 10.1016/j.jmb.2008.12.062. [DOI] [PubMed] [Google Scholar]
- Wurtzel O, et al. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–141. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981;289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- Zheng X, Hu GQ, She ZS, Zhu H. Leaderless genes in bacteria: clue to the evolution of translation initiation mechanisms in prokaryotes. BMC Genomics. 2011;12:361. doi: 10.1186/1471-2164-12-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JM, Battista JR. A ring-like nucleoid is not necessary for radioresistance in the Deinococcaceae. BMC Microbiol. 2005;5:17. doi: 10.1186/1471-2180-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.