Abstract
Chlorarachniophyte and cryptophyte algae have complex plastids that were acquired by the uptake of a green or red algal endosymbiont via secondary endosymbiosis. The plastid is surrounded by four membranes, and a relict nucleus, called the nucleomorph, remains in the periplastidal compartment that is the remnant cytoplasm of the endosymbiont. Thus, these two algae possess four different genomes in a cell: Nuclear, nucleomorph, plastid, and mitochondrial. Recently, sequencing of the nuclear genomes of the chlorarachniophyte Bigelowiella natans and the cryptophyte Guillardia theta has been completed, and all four genomes have been made available. However, the copy number of each genome has never been investigated. It is important to know the actual DNA content of each genome, especially the highly reduced nucleomorph genome, for studies on genome evolution. In this study, we calculated genomic copy numbers in B. natans and G. theta using a real-time quantitative polymerase chain reaction approach. The nuclear genomes were haploid in both species, whereas the nucleomorph genomes were estimated to be diploid and tetraploid, respectively. Mitochondria and plastids contained a large copy number of genomic DNA in each cell. In the secondary endosymbioses of chlorarachniophytes and cryptophytes, the endosymbiont nuclear genomes were highly reduced in size and in the number of coding genes, whereas the chromosomal copy number was increased, as in bacterial endosymbiont genomes. This suggests that polyploidization is a general characteristic of highly reduced genomes in broad prokaryotic and eukaryotic endosymbionts.
Keywords: chlorarachniophyte, cryptophyte, endosymbiosis, nucleomorph, plastid
Introduction
Plastids (chloroplasts) have been acquired through multiple endosymbiotic events. Land plants and three algal groups (green, red, and glaucophyte algae) acquired plastids via a primary endosymbiosis of a photosynthetic cyanobacterium (Rodríguez-Ezpeleta et al. 2005; Price et al. 2012). In contrast, other algae have more complex plastids, called secondary plastids, which originated from the ingestion of primary plastid-bearing algae such as green and red algae (Archibald 2009; Keeling 2010). Secondary plastids are thus bound by one or two additional membranes in comparison with primary plastids which are bound by two membranes. Chlorarachniophytes and cryptophytes have secondary plastids derived from a green and red algal endosymbiont, respectively (Curtis et al. 2012). These two algal groups are of special interest, because the relict nucleus, called the nucleomorph, of engulfed algae still exists in the periplastidal compartment (PPC), which is the space between the inner and outer pair of plastid membranes, whereas many other algae have lost this organelle (Archibald 2007; Moore and Archibald 2009).
Nucleomorphs contain a greatly reduced genome that is the smallest known eukaryotic nuclear genome. Complete nucleomorph genomic sequences have been reported in a chlorarachniophyte (Gilson et al. 2006) and four cryptophyte species (Douglas et al. 2001; Lane et al. 2007; Tanifuji et al. 2011; Moore et al. 2012). The nucleomorph genomes sequenced to date consist of three linear chromosomes with ribosomal DNA operons on all six chromosome ends. The genomes are extremely small and compact ranging in size from 373 to 703 kb with only several hundred genes. The structural similarities of nucleomorph genomes between chlorarachniophytes and cryptophytes are striking when one considers that they are of independent origin. All nucleomorph genomes encode a small number of plastid proteins and hundreds of housekeeping proteins, suggesting that nucleomorphs are essential to maintain plastids. However, numerous fundamental proteins for nucleomorph biogenesis are absent from nucleomorph genomes (e.g., DNA polymerase genes). Recently, complete nuclear genomes have been sequenced in the chlorarachniophyte Bigelowiella natans and the cryptophyte Guillardia theta (Curtis et al. 2012). These revealed that thousand(s) of nucleus-encoded proteins are imported into the PPC to compensate for the lacking proteins. In secondary endosymbioses, numerous genes were lost and transferred from the endosymbiont nuclei to the host nuclei, and consequently the highly reduced nucleomorph genomes were generated.
Two major organelles, chloroplasts and mitochondria, were derived from bacterial endosymbionts, and they generally contain a large copy number of genomic DNA (plastid DNA [ptDNA] and mitochondrial DNA [mtDNA]). For instance, the copy numbers of ptDNA and mtDNA in the unicellular red alga Cyanidioschyzon merolae have been estimated to be 24 and 9 per organelle, respectively (Moriyama et al. 2010), and approximately 80 ptDNA and several hundred mtDNA copies have been predicted to exist in a single cell of the green alga Chlamydomonas reinhardtii (Nishimura et al. 1998). In contrast, many bacterial species generally possess a single copy or a small number of chromosome(s); the model proteobacterium, Escherichia coli, contains a single copy of the chromosome (Lau et al. 2003), and the cyanobacterium, Synechococcus elongatus, is known to possess one to eight copies of the circular chromosome (Chen et al. 2012). These imply that bacterial endosymbiont genomes tend to increase their copy numbers to be polyploid organelle genomes. This feature has also been seen in the bacterial endosymbiont of insects, Buchnera, which has more than 100 copies of a highly reduced genome (Komaki and Ishikawa 1999). However, reports of polyploidization in endosymbiotically derived genomes have been limited to prokaryotic genomes. To investigate the generality of polyploidization in broad endosymbiotic genomes, we analyzed the ploidy of endosymbiotically derived nuclear genomes in two algal groups, chlorarachniophytes and cryptophytes.
In this study, we calculated copy numbers of the four genomes in the chlorarachniophyte, B. natans, and the cryptophytes, G. theta, using a real-time quantitative polymerase chain reaction (qPCR) approach. These species have a single nucleus, nucleomorph, plastid, and mitochondrion in a cell, which is suitable for estimating genomic copy numbers (Hill and Wetherbee 1990; Moestrup and Sengco 2001). Our data indicate that the nuclear genomes are haploid, whereas the nucleomorphs are diploid or tetraploid. The mitochondrion and plastid contained a large copy number (20 to over 200 copies) of genomic DNA. We also analyzed relative transcript levels of homologous ribosomal protein genes in the four genomes of B. natans and the three genomes of G. theta by reverse transcription qPCR. The mRNA levels were increased in the nuclear, nucleomorph, mitochondrial, and plastid genes in the order of representation, which correlated with genomic copy numbers. Together, our data suggest that endosymbiotically derived genomes tend to be polyploid even in the case of eukaryotic nuclear genomes and that gene transcription appears to be increased according to the degree of polyploidy.
Materials and Methods
DNA and RNA Extraction from Cell Culture
Bigelowiella natans (CCMP621) and G. theta (CCMP2712) cells were grown at 20 °C under white illumination on a 12 h/12 h light/dark cycle in ESM medium (Kasai et al. 2009). Fresh cultures in midlight phase (predicted G1-phase) were used for DNA and RNA extractions, as most nuclear DNA replication and cell division of B. natans are performed in the dark phase (Hirakawa et al. 2011). DNA was purified using a DNeasy Plant Mini Kit (Qiagen) and a Plant DNA Preparation Kit (Jena Bioscience) for real-time qPCR. To quantify the amount of DNA per cell, total DNA was extracted from cells by a high-yield purification method using the Plant DNA Preparation Kit (without phenol/chloroform and spin columns). Cultured cell density (cells/ml) was calculated using a hemocytometer. DNA amount was measured using a microvolume spectrophotometer (NanoDrop 1000, Thermo Scientific). Total RNA was extracted using Trizol Reagent (Invitrogen) according to the manufacturer’s protocol. cDNA was synthesized from 3 µg total RNA using a ReverTra Ace qPCR RT Kit (Toyobo) with random 9-mer primers in a total reaction volume of 40 µl.
Real-Time Quantitative PCR
PCR primers for single-copy genes from each genome were designed using Primer3Plus software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/primer3plus.cgi, last accessed April 17, 2014), based on the sequences of nuclear genomes (Curtis et al. 2012), nucleomorph genomes (Douglas et al. 2001; Gilson et al. 2006), plastid genomes (Douglas and Penny 1999; Rogers et al. 2007), and mitochondrial genomes (GenBank: HQ840955; GQ896379) for B. natans and G. theta; target genes and primer sequences are shown in table 1 and supplementary table S1, Supplementary Material online. Each fragment was amplified and cloned into a pGEM-T easy vector (Promega), and serial dilutions of the plasmids were used to create standard curves. Real-time qPCR was carried out using a Thermal Cycler Dice Real Time System II (Takara) under the following conditions: 10 ng of DNA/0.5 µl of cDNA, 0.4 µM of each primer, 12.5 µl of SYBR Premix Ex Taq II (Takara), and DNase/RNase-free water up to 25 µl. The cycling conditions were as follows: 3 min of denaturation at 95 °C followed by 40 cycles of 10 s at 95 °C and 30 s at 60 °C, and a melting curve program for detection of nonspecific products. The absolute copy number of target DNA/cDNA were calculated by the Ct values (2nd derivative maximum) and the standard curves of serial dilutions. The qPCR analysis was repeated at least three times for estimating standard deviations.
Table 1.
Primer Sequences for Quantitative PCR
| Target Genome | Coding Gene | Forward Primer | Reverse Primer |
|---|---|---|---|
| BnN #1 | rbcS2 | TCGCTAACCTGCTGACTCTTTG | TTGACATTGCTGCTCTGGTTG |
| BnN #2 | atpD | ACTTCAAACTTCTTGAACCTTCTCC | CATCTTCATCACTTCTTCCTCAAAC |
| BnNm #1 | rpL8 (Ch 1) | TACTTACCGCCACCTGCTACAAC | GCCTTTGTCGGTGTTCTTTCTC |
| BnNm #2 | cdc48 (Ch 2) | TTCTCCTACGCCCACATACTTACTC | TGGCATTTTACTTTACGGACCAC |
| BnNm #3 | eef2 (Ch 3) | GGAGGGGAAAAGTGTATGAGGAG | AATGCCTGCCCTGATGTAGAAG |
| BnM #1 | nad9 | TGGGATTTGCTTGGTGTGTTC | GTCTTTTCGTAGTGGGTGTCCTTC |
| BnM #2 | nad3 | AGAAAAGCTATCCGCCTATGAGTG | AAACGGCTCAAGGAAATAGAAGG |
| BnP #1 | psbD | CAAGATCAACCGCACGAGAAG | GAGCATTTCCAGACCACCAAG |
| BnP #2 | petA | TCAAGAAGGGCAACAGGTAAAAG | CAGTTTCAGATTGACCAAATCCAC |
| GtN #1 | rpL8 | GTCGTGGTGCTGGTTCTGTC | CCATTCCTCTCAGCAAAGTCC |
| GtN #2 | rpL15 | CAACGATCCTCGCCTCAAC | ACCTGTGATTGCCGTGTCC |
| GtNm #1 | rpb2 (Ch 1) | TTGCCTTTAGCGGATGGTG | CGGAAACTGGGGAACAGATAAG |
| GtNm #2 | gidA (Ch 2) | AGCCATTTCTCCTGACCTTCC | GGATTTTACCGATGCCGTTC |
| GtNm #3 | kin (Ch 3) | GAAGCGGATCATTTGGAGTTG | CTCTGTTTTCATACCTGTCGTCTTG |
| GtM #1 | cox1 | GTCAACCCTTGGGCATTTTC | AACGTGTGAAATGATGGTGGAG |
| GtP #1 | atpH | TCTTCCTTCTGCTTCTGGTTGG | CTGCTTCTGTTGTTGCTTCTGG |
| GtP #2 | rpoB | TTAATGCGTGCGTGTGTTG | CGTGATAGAGCAGGTTTTGGTG |
Note.—Bn, Bigelowiella natans; Gt, Guillardia theta; N, nucleus; Nm, nucleomorph; M, mitochondrion; P, plastid; Ch, chromosome.
Results
Copy Numbers of the Four Genomes in a Chlorarachniophyte and a Cryptophyte
To estimate copy numbers of the four genomes (nuclear, nucleomorph, mitochondrial, and plastid) in the chlorarachniophyte B. natans and the cryptophyte G. theta, we used an absolute quantification method by real-time qPCR with genome-specific primers. The primers were designed on single copy genes of each genome; two primer sets were designed specific to the nuclear/plastid/mitochondrial genomes except for a single set in the G. theta mitochondrial genome, and three primer sets were designed for the three nucleomorph chromosomes (table 1). All target fragments were amplified by PCR, cloned into plasmid vectors, and serial dilutions of the plasmids were used to create standard curves for calculating exact DNA copy numbers. Total DNA was extracted from different batches of cell cultures in three independent experiments (samples A–C) and used for qPCR analyses.
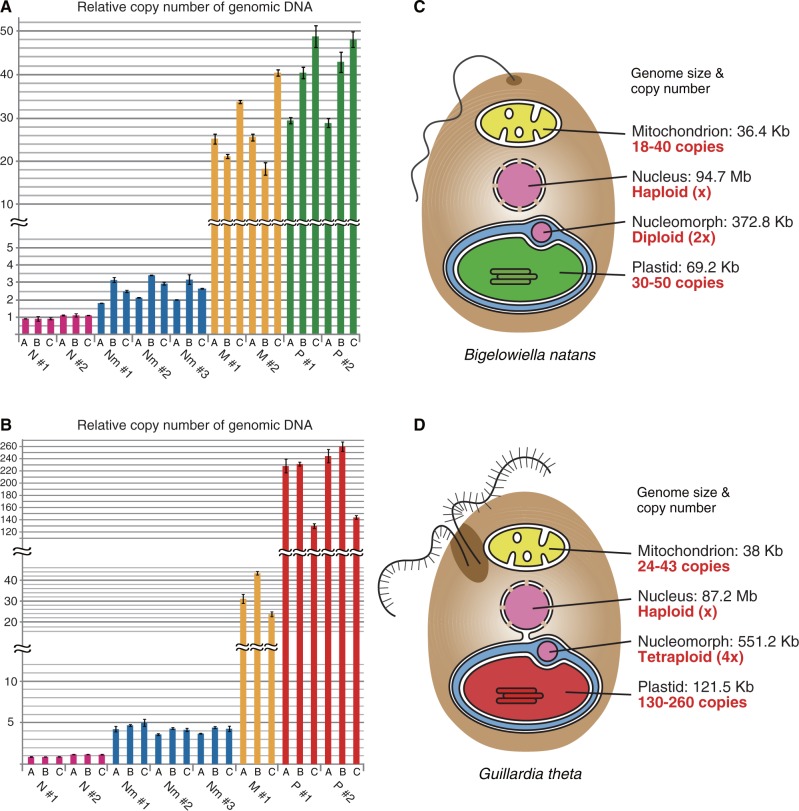
The copy numbers of nucleomorph chromosomes in B. natans and G. theta were predicted to be double to triple and quadruple that of nuclear chromosomes, respectively (fig. 1A and B). A stable copy number of nucleomorph chromosomes in G. theta was seen in three different DNA samples (fig. 1B), whereas the nucleomorph chromosome copy number of B. natans varied (fig. 1A). This variability was due to contamination by cells with divided plastids and nucleomorphs in the cultures; double plastids were detected by confocal laser microscopy in approximately 50% of cells (39 out of 75 cells) in sample C that showed a triple copy number of nucleomorph chromosomes (fig. 1A, supplementary fig. S1, Supplementary Material online). This suggests that the chromosome copy number of a single nucleomorph in B. natans is double that of nucleus. Three nucleomorph chromosomes indicated the same copy number in both species (fig. 1A and B), as previously seen by Southern blot analyses of pulsed-field gel separation that showed identical signal intensities of the three nucleomorph chromosomes (McFadden et al. 1994; Lane and Archibald 2006). Copy numbers of the B. natans mitochondrial and plastid genomes were approximately 18–40 and 30–50 times greater than that of the nuclear genome, respectively (fig. 1A). For G. theta, the mitochondrion and plastid contained approximately 24- to 43-fold and 130- to 260-fold more genomic copies than the nucleus, respectively (fig. 1B).
Fig. 1.—
Copy numbers of the four genomes in the chlorarachniophyte, Bigelowiella natans, and the cryptophyte, Guillardia theta. (A, B) Each bar shows the relative copy numbers of genomes (N, nuclear; Nm, nucleomorph; M, mitochondrial; P, plastid) calculated by real-time quantitative PCR in three independent experiments (samples A–C). The average value of the nuclear genome was set to 1.0, and other values were normalized. Error bars represent the standard deviation (SD) (n = 3–4). (C, D) Size and copy number of each genome are summarized in the cellular schemes.
Ploidy of the nuclear genomes of B. natans and G. theta was calculated from the amount of DNA per cell. Total DNA was extracted from cell cultures by a high-yield solution-based purification method without using phenol/chloroform and spin columns, and its weight was quantified by the absorbance at 280 nm (NanoDrop 1000). It was estimated that a single cell contained from 8.6 × 10−2 to 9.1 × 10−2 pg DNA in B. natans, and from 10.0 × 10−2 to 11.4 × 10−2 pg DNA in G. theta, based on experiments performed in triplicate. Assuming that the nuclear genomes are haploid, the total DNA content was calculated to be 10.4 × 10−2 and 11.8 × 10−2 pg/cell for B. natans and G. theta, respectively, based on the estimated total nucleotides of four genomes (genome size multiplied by the copy number) and a factor used to convert DNA weight to the number of base pairs (0.978 × 10−9) (Dolezel et al. 2003), using the following formulas: (94.7 × 106 × 1 + 372.8 × 103 × 2 + 69.2 × 103 × 40 + 36.4 × 103 × 27)/0.978 × 10−9 = 10.4 × 10−2 (pg/cell) for B. natans, and (87.2 × 106 × 1 + 551.2 × 103 × 4 + 121.5 × 103 × 206 + 38 × 103 × 33)/0.978 × 10−9 = 11.8 × 10−2 (pg/cell) for G. theta. The above data indicate that the nuclear genomes consist of single copy chromosomes in both species. Altogether, these results suggest that the nucleomorph genomes are diploid (2 n = 6) in B. natans and tetraploid (4 n = 12) in G. theta and that the plastids and mitochondria contain a large copy number of genomic DNA.
Transcript Levels of Homologous Genes in the Four Genomes
Both B. natans and G. theta possess multiple copies of organelle genomes (nucleomorph, mitochondrial, and plastid), and these genomes show different copy numbers. To determine the relationship between genomic copy number and mRNA expression in each genome, we examined transcription of homologous genes in each genome of B. natans and G. theta. Several ribosomal protein genes, rpL4 and rpL8 of B. natans and rpL8 and rpL15 of G. theta, were used to calculate the relative transcript levels of nuclear and nucleomorph genes, and rpL5 and rpL16 were used for plastid and/or mitochondrial genes of both species. cDNA was synthesized from total RNA using random primers (9 mers), and the relative transcription of each gene was quantified by real-time qPCR with gene-specific primers (supplementary table S1, Supplementary Material online). Transcript levels of the nucleomorph rpL genes were 3–4-fold higher than those of their nuclear counterparts with the exception of G. theta rpL8. (fig. 2A and B). Although we cannot compare the transcription of exact homologous genes between eukaryotic and prokaryotic type genomes, transcript levels of the plastid and/or mitochondrial rpL genes were generally greater than those of the nuclear and nucleomorph rpL genes in both B. natans and G. theta (fig. 2A and B). Transcript levels of rpL genes largely increased from the nuclear, nucleomorph, mitochondrial, to plastid genomes in that order, which seems to be positively correlated with the copy number of each genome.
Fig. 2.—
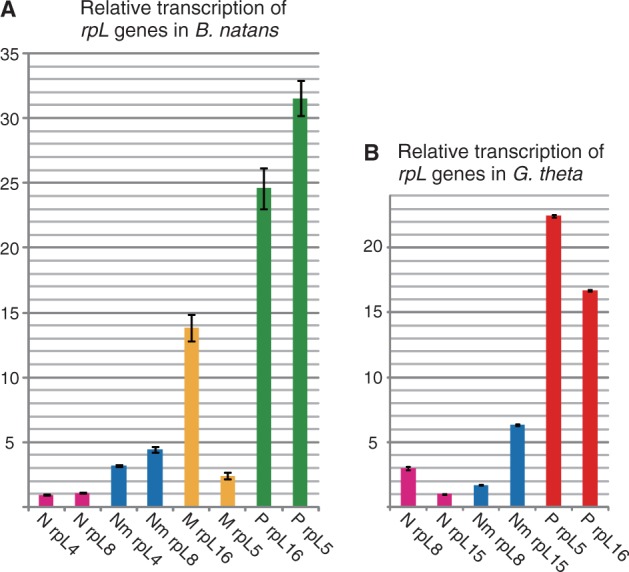
Relative transcription levels of homologous genes residing in the four genomes of Bigelowiella natans and the three genomes of Guillardia theta. (A, B) Each bar shows the relative transcript levels of ribosomal protein genes calculated by real-time quantitative PCR (N, nuclear; Nm, nucleomorph; M, mitochondrial; P, plastid). The value of nuclear rpL8 genes was set to 1.0, and other values were normalized. Error bars represent the standard deviation (SD) (n = 3).
Discussion
In this study, we show that the nucleomorphs of the chlorarachniophyte B. natans and the cryptophyte G. theta contained a diploid and tetraploid genome, respectively, whereas their nuclear genomes were haploid. The nucleomorphs are a relict nucleus of a green or red algal endosymbiont (Douglas et al. 2001; Gilson et al. 2006). Although in algae the chromosome copy numbers can vary at different stages of the life cycles, the genomes of Chlorophyta and Rhodophyta gametophytes have been typically reported as haploid (Kapraun 2007; Kapraun and Freshwater 2012). Thus, copy numbers of chromosomes in nucleomorph genomes seem to have multiplied after the endosymbiotic events. Interestingly, the nucleomorph genomes of B. natans and G. theta are highly reduced in size (373 and 551 kb) and coding genes (284 and 487 genes), whereas increased in their chromosomal copy numbers. This feature has been observed in other endosymbiotically derived genomes of plastids and mitochondria. Both organelles contain greatly reduced multiple copy genomes, though the relatives of their origins, proteobacteria and cyanobacteria, have a single or a few genomic copies. Furthermore, the bacterial symbiont of aphids, Buchnera, has more than a hundred copies of a highly reduced genome, despite a single genome copy in the closely related bacterium E. coli (Komaki and Ishikawa 1999). Therefore, we hypothesize that genomic polyploidization is a general characteristic in the highly reduced genomes of both prokaryotic and eukaryotic endosymbionts.
The evolutionary advantages of genomic polyploidy in flowering plants and animals have been discussed in several reviews (Comai 2005; Sémon and Wolfe 2007; Arrigo and Barker 2012); for example, gene expression changes, buffering of deleterious mutations, and sub- and neofunctionalization of duplicated genes. Although the functional significance of polyploidy for nucleomorph genomes is unclear, the increased gene dosage would allow for higher productivity in the PPC. In fact, transcript levels of homologous genes were higher in the diploid/tetraploid nucleomorph genomes than in the haploid nuclear genomes of B. natans and G. theta (fig. 2). Very recently, a high level of mRNA transcription for B. natans and G. theta nucleomorph genes was reported based on RNA-seq transcriptome data (Tanifuji et al. 2014). The authors hypothesized that the high gene expression compensates for an inefficiency of protein complexes functioning in the PPC, because part of the fundamental proteins were absent from the PPC (e.g., 17 of 79 ribosomal protein subunits are not present). Although there is no proof for this hypothesis, polyploidy of the nucleomorph genome appears to be related to the high expression of nucleomorph genes.
Nucleomorph genomes of both chlorarachniophytes and cryptophytes have a gene-dense structure with shrinking intergenic regions (Douglas et al. 2001; Gilson et al. 2006). Many syntenic regions with the same gene order among different species were found in cryptophyte nucleomorph genomes (Lane et al. 2007; Tanifuji et al. 2011; Moore et al. 2012), as well as draft genomes of chlorarachniophyte nucleomorphs. Overlapping gene transcription has also been reported in nucleomorph genomes (Williams et al. 2005). It is known that gene-dense genomes generally show a low rate of viable genetic rearrangements, because nonhomologous recombination is likely to disrupt coding sequences. Indeed, nucleomorph genomes are composed of many syntenic regions; however, many intra- and interchromosomal recombination of syntenic blocks have also been reported in nucleomorph genomes (Lane et al. 2007). This implies that the multiple copies of nucleomorph genomes would lead to an increase in the recombination frequency.
The replication and segregation systems of nucleomorph DNA are at present unknown, and chromosome structure has never been observed by electron microscopy. We previously reported that expression of nucleomorph histone genes was controlled during the cell cycle of the chlorarachniophyte B. natans; transcript levels of the histones increased in S-phase, which would correlate to nucleomorph chromosome replication (Hirakawa et al. 2011). This study indicates that nucleomorphs contain stable copy numbers of three chromosomes. Though there is no direct evidence, replication of nucleomorph chromosomes is likely tightly controlled by the cell cycle, and duplicated chromosomes are segregated to daughter plastids nonrandomly.
In this study, we investigated copy numbers of the four genomes in a chlorarachniophyte and cryptophyte and found that the nucleomorph genomes were diploid and tetraploid, respectively. It is interesting that two distantly related algal groups have individually evolved multiple copied genomes in endosymbiotically derived organelles. We hypothesize that genomic polyploidization may be a general characteristic of highly reduced genomes in endosymbionts. To test this hypothesis, genomic copy numbers should be investigated in other endosymbiotically derived genomes from a broad variety of organisms.
Supplementary Material
Supplementary figure S1 and table S1 are available at Genome Biology and Evolution online.
Acknowledgments
This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Number 23117004.
Literature Cited
- Archibald JM. Nucleomorph genomes: structure, function, origin and evolution. BioEssays. 2007;29:392–402. doi: 10.1002/bies.20551. [DOI] [PubMed] [Google Scholar]
- Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19:R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Arrigo N, Barker MS. Rarely successful polyploids and their legacy in plant genomes. Curr Opin Plant Biol. 2012;15:140–146. doi: 10.1016/j.pbi.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Chen AH, Afonso B, Silver PA, Savage DF. Spatial and temporal organization of chromosome duplication and segregation in the cyanobacterium Synechococcus elongatus PCC 7942. PloS One. 2012;7:e47837. doi: 10.1371/journal.pone.0047837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Curtis BA, et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature. 2012;492:59–65. doi: 10.1038/nature11681. [DOI] [PubMed] [Google Scholar]
- Dolezel J, Bartos J, Voglmayr H, Greilhuber J. Nuclear DNA content and genome size of trout and human. Cytometry A. 2003;51:127–128. doi: 10.1002/cyto.a.10013. [DOI] [PubMed] [Google Scholar]
- Douglas SE, Penny SL. The plastid genome of the cryptophyte alga, Guillardia theta: complete sequence and conserved synteny groups confirm its common ancestry with red algae. J Mol Evol. 1999;48:236–244. doi: 10.1007/pl00006462. [DOI] [PubMed] [Google Scholar]
- Douglas SE, et al. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410:1091–1096. doi: 10.1038/35074092. [DOI] [PubMed] [Google Scholar]
- Gilson PR, et al. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature’s smallest nucleus. Proc Natl Acad Sci U S A. 2006;103:9566–9571. doi: 10.1073/pnas.0600707103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D, Wetherbee R. Guillardia theta gen. et sp. nov. (Cryptophyceae) Can J Bot. 1990;68:1873–1876. [Google Scholar]
- Hirakawa Y, Burki F, Keeling PJ. Nucleus- and nucleomorph-targeted histone proteins in a chlorarachniophyte alga. Mol Microbiol. 2011;80:1439–1449. doi: 10.1111/j.1365-2958.2011.07643.x. [DOI] [PubMed] [Google Scholar]
- Kapraun DF. Nuclear DNA content estimates in green algal lineages: chlorophyta and streptophyta. Ann Bot. 2007;99:677–701. doi: 10.1093/aob/mcl294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapraun DF, Freshwater DW. Estimates of nuclear DNA content in red algal lineages. AoB Plants. 2012;2012:pls005. doi: 10.1093/aobpla/pls005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai F, Kawachi M, Erata M, Yumoto K, Sato M. NIES-collection list of strains, 8th edition. Jpn J Phycol (Sosui). 2009;57:220. [Google Scholar]
- Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 2010;365:729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki K, Ishikawa H. Intracellular bacterial symbionts of aphids possess many genomic copies per bacterium. J Mol Evol. 1999;48:717–722. doi: 10.1007/pl00006516. [DOI] [PubMed] [Google Scholar]
- Lane CE, Archibald JM. Novel nucleomorph genome architecture in the cryptomonad genus Hemiselmis. J Eukaryot Microbiol. 2006;53:515–521. doi: 10.1111/j.1550-7408.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- Lane CE, et al. Nucleomorph genome of Hemiselmis andersenii reveals complete intron loss and compaction as a driver of protein structure and function. Proc Natl Acad Sci U S A. 2007;104:19908–19913. doi: 10.1073/pnas.0707419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau IF, et al. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- McFadden GI, Gilson PR, Hofmann CJ, Adcock GJ, Maier UG. Evidence that an amoeba acquired a chloroplast by retaining part of an engulfed eukaryotic alga. Proc Natl Acad Sci U S A. 1994;91:3690–3694. doi: 10.1073/pnas.91.9.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moestrup Ø, Sengco M. Ultrastructural studies on Bigelowiella natans, gen. et sp. nov., a chlorarachniophyte flagellate. J Phycol. 2001;37:624–646. [Google Scholar]
- Moore CE, Archibald JM. Nucleomorph genomes. Ann Rev Genet. 2009;43:251–264. doi: 10.1146/annurev-genet-102108-134809. [DOI] [PubMed] [Google Scholar]
- Moore CE, Curtis B, Mills T, Tanifuji G, Archibald JM. Nucleomorph genome sequence of the cryptophyte alga Chroomonas mesostigmatica CCMP1168 reveals lineage-specific gene loss and genome complexity. Genome Biol Evol. 2012;4:1162–1175. doi: 10.1093/gbe/evs090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama T, et al. Characterization of cell-cycle-driven and light-driven gene expression in a synchronous culture system in the unicellular rhodophyte Cyanidioschyzon merolae. Microbiology. 2010;156:1730–1737. doi: 10.1099/mic.0.037754-0. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Higashiyama T, Suzuki L, Misumi O, Kuroiwa T. The biparental transmission of the mitochondrial genome in Chlamydomonas reinhardtii visualized in living cells. Eur J Cell Biol. 1998;77:124–133. doi: 10.1016/S0171-9335(98)80080-0. [DOI] [PubMed] [Google Scholar]
- Price DC, et al. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science. 2012;335:843–847. doi: 10.1126/science.1213561. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Ezpeleta N, et al. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol Biol Evol. 2007;24:54–62. doi: 10.1093/molbev/msl129. [DOI] [PubMed] [Google Scholar]
- Rogers MB, et al. Plastid-targeting peptides from the chlorarachniophyte Bigelowiella natans. J Eukaryot Microbiol. 2004;51:529–535. doi: 10.1111/j.1550-7408.2004.tb00288.x. [DOI] [PubMed] [Google Scholar]
- Sémon M, Wolfe KH. Consequences of genome duplication. Curr Opin Genet Dev. 2007;17:505–512. doi: 10.1016/j.gde.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Tanifuji G, Onodera NT, Moore CE, Archibald JM. Reduced nuclear genomes maintain high gene transcription levels. Mol Biol Evol. 2014;31:625–635. doi: 10.1093/molbev/mst254. [DOI] [PubMed] [Google Scholar]
- Tanifuji G, et al. Complete nucleomorph genome sequence of the nonphotosynthetic alga Cryptomonas paramecium reveals a core nucleomorph gene set. Genome Biol Evol. 2011;3:44–54. doi: 10.1093/gbe/evq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BA, Slamovits CH, Patron NJ, Fast NM, Keeling PJ. A high frequency of overlapping gene expression in compacted eukaryotic genomes. Proc Natl Acad Sci U S A. 2005;102:10936–10941. doi: 10.1073/pnas.0501321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.