Abstract
It has often been speculated that bacterial protein-tyrosine kinases (BY-kinases) evolve rapidly and maintain relaxed substrate specificity to quickly adopt new substrates when evolutionary pressure in that direction arises. Here, we report a phylogenomic and biochemical analysis of BY-kinases, and their relationship to substrates aimed to validate this hypothesis. Our results suggest that BY-kinases are ubiquitously distributed in bacterial phyla and underwent a complex evolutionary history, affected considerably by gene duplications and horizontal gene transfer events. This is consistent with the fact that the BY-kinase sequences represent a high level of substitution saturation and have a higher evolutionary rate compared with other bacterial genes. On the basis of similarity networks, we could classify BY kinases into three main groups with 14 subgroups. Extensive sequence conservation was observed only around the three canonical Walker motifs, whereas unique signatures proposed the functional speciation and diversification within some subgroups. The relationship between BY-kinases and their substrates was analyzed using a ubiquitous substrate (Ugd) and some Firmicute-specific substrates (YvyG and YjoA) from Bacillus subtilis. No evidence of coevolution between kinases and substrates at the sequence level was found. Seven BY-kinases, including well-characterized and previously uncharacterized ones, were used for experimental studies. Most of the tested kinases were able to phosphorylate substrates from B. subtilis (Ugd, YvyG, and YjoA), despite originating from very distant bacteria. Our results are consistent with the hypothesis that BY-kinases have evolved relaxed substrate specificity and are probably maintained as rapidly evolving platforms for adopting new substrates.
Keywords: phylogeny, bacterial protein kinases, kinase evolution, kinase classification, BY-kinases, kinase-substrate coevolution
Introduction
Protein phosphorylation is a widespread posttranslational modification and plays a key role in regulation of cellular functions. Enzymes that perform protein phosphorylation, termed protein kinases, transfers phosphate groups from ATP to reactive side chains of amino acids in proteins. This usually changes the enzyme activity, cellular localization, or interaction with partners of the target protein (Hanks and Hunter 1995). Tyrosine is one of phosphorylatable amino acids. In Eukarya, tyrosine phosphorylation is carried out by Hanks-type kinases (Hanks et al. 1988). In Bacteria, the presence of tyrosine-kinases passed undetected until mid-1990s (Grangeasse et al. 1997). Since then, a number of BY-kinases have been identified, sharing a structure quite distinct from Hanks-type kinases. These tyrosine kinases have been unified in a new, bacteria-specific class of enzymes, named BY-kinases (Grangeasse et al. 1997). A prototype BY-kinase contains an extracellular loop and a cytosolic domain (Grangeasse et al. 2007, 2012). These two domains can appear linked into one large protein encoded by a single gene (e.g., in Escherichia coli) or exist as two proteins: one transmembrane and another cytosolic protein, encoded by two adjacent genes (e.g., in Bacillus subtilis). The cytosolic domain, defined as catalytic domain (CD), contains the catalytic site and performs the phosphorylation on tyrosine. The intracellular juxtamembrane region of the transmembrane domain (following the second transmembrane helix) is essential for the activation of the CD. Thus, this domain was defined as the transmembrane activator domain (TAD) (Jadeau et al. 2008). The CD possesses Walker A, A′ motifs in the N terminus, followed by Walker B motif in the center and a tyrosine-rich cluster in the C terminus. The three Walker motifs constitute an active site required for ATP-Mg binding (the P-loop) (Walker et al. 1982; Doublet et al. 1999; Soulat et al. 2007; Grangeasse et al. 2012), which is significantly different from that used by Hanks-type kinases but usually found in ATP/GTPases (Leipe et al. 2002; Grangeasse et al. 2007, 2012). A nucleotide-binding motif similar to that of BY-kinases has been found in arsenite ATPases (ArsA) and MinD proteins, which lead to the hypothesis that they have all evolved from the same ancestral bacterial ATPase (Grangeasse et al. 2012). The tyrosines in the C-terminal cluster represent the autophosphorylation sites of BY-kinases. The sequence of the C-terminal cluster is not conserved among BY-kinases, it can vary considerably with respect to overall length, number, and position of tyrosines (Grangeasse et al. 2012). It has been shown that no single tyrosine in this region was essential for autophosphorylation (Paiment et al. 2002).
Most of the experimentally validated BY-kinases are encoded by genes located in large operons, which are involved in biosynthesis and export of capsular/extracellular polysaccharides (Whitfield 2006). In E. coli, the first evidenced BY-kinase, Wzc, is encoded by a gene of the cps (or wca) operon that participates in biosynthesis of exopolysaccharides (Vincent et al. 1999). Loss of autophosphorylation of Wzc abolished capsule assembly, inciting the authors to conclude that its autokinase activity is essential for this process (Wugeditsch et al. 2001). BY-kinases were often found to affect virulence or resistance to cationic antimicrobial peptides, which are both associated with capsular polysaccharide synthesis (Stingele et al. 1996; Morona et al. 2004; Morona et al. 2006; Minic et al. 2007). More recently, it was understood that BY-kinases also act via substrate phosphorylation. The first described substrate for BY-kinases was the UDP-glucose dehydrogenase (Ugd, Grangeasse et al. 2003). A recent study indicated that Ugd phosphorylation, mediated by BY-kinases Wzc or Etk, respectively, represents the control element for extracellular polysaccharides production and resistance to polymyxin of E. coli (Lacour et al. 2008). BY-kinases are not only related to polysaccharide biosynthesis, they are also involved in other process such as lysogenization, heat shock response, DNA replication, cell cycle, and others (Klein et al. 2003; Lacour et al. 2006, 2008; Petranovic et al. 2007; Kolot et al. 2008). In E. coli, Etk affects heat shock response through phosphorylation of a heat shock sigma factor RpoH (Klein et al. 2003). In B. subtilis, BY-kinase PtkA phosphorylates the single-stranded DNA-binding protein SsbA and influences DNA replication and the cell cycle (Petranovic et al. 2007). Other substrates of PtkA (such as single-stranded DNA exonuclease YorK, aspartate semialdehyde dehydrogenase Asd, and transcription factor FatR) have been described since defining it clearly as a promiscuous kinase (Jers et al. 2010; Derouiche et al. 2013). When considering the fact that BY-kinases accomplish different tasks by phosphorylating different substrates, one cannot help but wonder how one kinase recognizes different substrates with totally different sequence and structure, and how does the new kinase-substrate couple emerge in terms of evolution.
To better understand the evolution of BY-kinases and the relationship between kinases and their substrates, we performed a phylogenomic analysis of the BY-kinases that led us to conclude that BY-kinases have a complex evolutionary history, mainly driven by horizontal gene transfer (HGT) and duplications, with fast evolution due to higher synonymous substitution rate. Although coevolution was observed in some kinase-substrate pairs (Gildor et al. 2005; Skerker et al. 2008), no evidence of coevolution at the sequence level was detected between BY-kinases and their substrates, in particular the Ugd family proteins. Nevertheless, our results suggest that BY-kinases have the capability to phosphorylate the same set of substrates across very distant bacterial phyla.
Materials and Methods
Identification of BY-Kinases and Ugd Family Proteins
The complete sequences of 1,471 bacterial and 117 archaeal genomes available in March 2012 were downloaded from the National Center for Biotechnology Information FTP website (ftp.ncbi.nih.gov, last accessed April 1, 2014). The use of complete genomes is essential to phylogenomic approaches because it allows determining the exact distribution of homologs. In addition, we compared the taxonomic distribution with those built on sequences from the nr database (data not shown). No important differences were evident between the two analyses, meaning that the use of complete genomes does not bias the interpretation of results. To identify BY-kinase homologs from these genomes, the annotated BY-kinase sequences were first downloaded from the dedicated BY-kinase database (Jadeau et al. 2012), and these are referred to as the BYKdb data set. Because this data set was constructed from UniProt database and was redundant, a nonredundant BY-kinase data set was generated using the CD-hit program (Li and Godzik 2006) with cutoff of 0.7. The resulting sequences were aligned with ClustalW (Larkin et al. 2007), and the HMM profile corresponding to CD region was generated using the HMMER package (Eddy 2011). The HMMER package (Eddy 2011) and self-written scripts were then used to search for BY-kinase homologs in the complete bacterial and archaeal genomes, requiring the presence of the CD. Alignments with E value less than 0.01 were first considered as significant, and the resulting sequences were then filtered with the isBYK algorithm (Jadeau et al. 2012) to identify the BY-kinase homologs. The corresponding sequences were subsequently searched against the Pfam 25.0 database (Finn et al. 2010) to determine the presence of additional known functional domains. For each BY-kinase homolog, the gene context, defined as the 10 neighboring genes (located upstream and downstream the BY-kinase, −5 to +5), was investigated with cluster of orthologous group (COG) (Tatusov et al. 2000) classification using self-written scripts. The identification of the Ugd family protein homologs in complete bacterial and archaeal genomes was performed using the HMMER package (Eddy 2011) requiring the presence of three functional domains in the following order (from N to C-terminal): UDPG_MGDP_dh_N (PF03721), UDPG_MGDP_dh (PF00984), and UDPG_MGDP_dh_C (PF03720). Alignment scores higher than the cut_tc threshold were considered significant.
Phylogenetic Analysis of BY-Kinase and Ugd Family Proteins
The CDs from the retrieved BY-kinase homologous sequences were first aligned using MAFFT v6.860b (Katoh and Toh 2008) with the iterative global alignment option. The resulting alignment was then filtered to remove the positions that contained more than 30% of gaps. A phylogenetic tree of all BY-kinases was constructed using the FastTree version 2.1 (Price et al. 2010) with MinD proteins (AAC74259, CAB14759) as outgroups, and 100 resampled trees were generated to calculate the local bootstrap values. An in-depth phylogenetic analysis using more restricted sequence sampling representative (54 sequences, 187 positions, named BYKsel) of the diversity of BY-kinases was performed using the Bayesian approach implemented in MrBayes 3.2 program (Ronquist and Huelsenbeck 2003). For Bayesian analysis, a mixed substitution model with gamma law (four rate categories) and a proportion of invariant sites were used. The Markov chain Monte Carlo search was run with four chains for 1,000,000 generations. Trees were sampled every 100 generations with the first 1,000 trees were discarded as “burnin.”
For Ugd phylogenetic trees, organisms possessing both Ugd and BY-kinase were selected. Two restricted samples were defined: one (72 sequences, 220 positions) containing Ugd family proteins from organisms in the BYKsel data set and another one (105 sequences, 200 positions) contains Ugd family proteins surrounded by BY-kinase genes (maximum five upstream or five downstream). Each sample was aligned using ClustalW program (Larkin et al. 2007), followed by the selection of unambiguous parts with Gblocks 0.91b program (Castresana 2000). The Bayesian phylogenetic trees were then performed as described above for BY-kinases.
Substitution Saturation and Evolutionary Rate Estimation
The phylogenetic tree using the maximum likelihood (ML) method through the PHYML program (Guindon et al. 2010) was performed with the BYKsel data set. The use of ProtTest program (Abascal et al. 2005) allows to determine the LG model with estimated gamma-distribution parameter (G) and the proportion of invariant sites (I) as the best-fit amino acid substitution model. PHYML with the “LG+I+G” model was then used for ML reconstruction with a 100-replicate bootstrap. The amino acid substitution saturation was evaluated by comparing the number of substitutions inferred by ML method with the number of observed differences. The inferred substitutions were calculated as the sum of all branch lengths between two sequences (Leclere and Rentzsch 2012). The DNA sequences were aligned according to corresponding amino acid alignments. The substitution saturation at the DNA level was assessed by DAMBE according to Xia’s method (Xia and Lemey 2009). PAML’s codeml (Yang 2007) was used to estimate nonsynonymous rate (dN), synonymous rate (dS), and the ratio of these rates (dN/dS). Gapped regions were excluded to avoid spurious rate inference. The phylogenetic trees inferred with FastTree (Price et al. 2010) were used as a constraint tree. For codon sequences from the BYKsel data set and other conserved genes in bacterial lineages (frr, infC, nusA, pyrG, rplB, rplD, rplL, rplM, rplN, rplP, rplS, rplT, rpmA, rpoB, rpsB, rpsE, rpsJ, rpsM, rpsS, smpB, and tsf), only one model of protein evolution was used: model 0 allows a single ω (dN/dS) value throughout the genealogy. To determine whether the BY-kinases were subjected to selection, codon alignment from the BYKsel data set was tested. The presence of positive selection was detected as described by Yang (1998). Likelihood ratio test was used to test the model by comparing model M0 (a single ratio) with M3 (discrete), M1a (nearly neutral) with M2a (positive selection), M7 (β distribution), and M8 (β and ω). P value was calculated using the chi2 program in PAML. Substitution saturation impacts the estimation of dN/dS (Gharib and Robinson-Rechavi 2013). To avoid the influence of substitution saturation, the codon sequences of BY-kinases from three closely related species (Streptococcus pneumoniae TIGR4, Str. suis SC84, and Eubacterium eligens ATCC27750) with low level of substitution saturation were tested for the positive selection using the same method.
Reconstruction of Protein Similarity Network
The protein similarity networks (PSNs) were constructed as described (Zhang et al. 2011). Briefly, pairwise alignments (entire BY-kinase sequences or CDs) were performed with the bl2seq program from the BLAST package (Altschul et al. 1997). Then a series of E-value thresholds were applied for the selection of sequence pairs with significant similarity. Next, the distribution of pairwise alignments E-value was used to define the optimal E-value cutoff. Finally, significant alignments (with E-value less than the optimal E-value) were used to construct the PSN. Each node in the PSN indicates a BY-kinase, and the edge indicates that two nodes share significant similarity with an E-value less than the selected cutoff. The network was visualized using Cytoscape (Shannon et al. 2003) with the yFiles organic layout.
Consensus Sites in BY-Kinases
The consensus sites were determined based on the multiple alignments with “50-10” rule (Carretero-Paulet et al. 2010). A strong conserved site was designed if an amino acid at that site was present in all (100%) sequences. The sites with amino acid present in more than 50% of the sequences were designated as a weakly conserved site. For such a weak conserved site, the amino acid was also added into the 50–10 consensus sequences if it existed in more than 10% of all the sequences. Sequence logos were generated with Weblogo program (Crooks et al. 2004). For each PSN subgroup, a significant conserved motif was defined as an amino acid region with at least two strongly conserved sites spanning a window of 10 amino acids.
Vector Construction and Growth Conditions
Genomic DNA was extracted by DNeasy Blood & Tissue Kit (Qiagen). PCR was performed with specific primers (supplementary table S1, Supplementary Material online) and the corresponding genomic DNA. All PCR products were inserted into pQE-30 Xa (Qiagen) to get the 6xHis-tag fusion proteins. Escherichia coli NM522 was used for cloning, and E. coli M15 (pREP4::GroEL/GroES) strain was used for protein synthesis. Cells were routinely grown in LB medium, with addition of ampicillin (100 µg/ml) and kanamycin (25 µg/ml) when needed; 1 mM IPTG was used to induce expression of cloned genes.
Protein Purification and Phosphorylation Analysis
Protein synthesis and purification were carried out as described previously (Mijakovic et al. 2003). Induction was started at OD600 = 0.6, cells were harvested 3 h later, and sonicated. 6xHis-tagged proteins were purified by Ni-NTA affinity chromatography (Qiagen) from crude extracts and desalted by PD-10 columns (GE Healthcare). For in vitro phosphorylation analysis, reactions were performed in solution containing 50 mM Tris pH 7.5, 100 mM NaCl, 5 mM MgCl2, 5% glycerol, and 50 µM ATP with 20 µCi/mmol [γ-32P]-ATP, incubated at 37 °C for 1 h, then separated by 12% SDS-polyacrylamide gels. Concentration of BY-kinases, substrates, and antimonite in all assays is indicated in figures of this article. The radioactive signals were revealed by autoradiography, using the FUJI phosphoimager as described previously (Mijakovic et al. 2003).
Homology-Based Modeling of 3D-Structures
For six sequences of BY kinases that were used for experimental studies, a template sequence with a known structure was searched using FASTA algorithm against the PDB database on the NPS@ web server (Combet et al. 2000). The molecular models were computed using the Geno3D tools (Combet et al. 2002), following a modeling process under restraints similar to the process used for experimental solving of protein structure by NMR. The Staphylococcus aureus sequence was not modeled as its structure was experimentally solved (PDB code 3BFV [Olivares-Illana et al. 2008]). Molecular models and the experimentally solved structure were analyzed and compared with the DeepView (Guex and Peitsch 1997) and the Matt software (Menke et al. 2008).
Results
Distribution of BY-Kinases in Bacteria
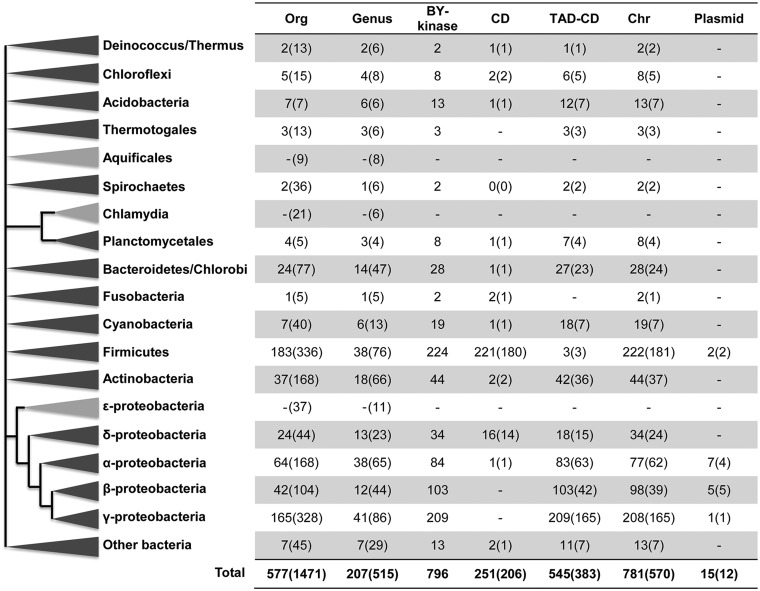
To explore the general occurrence of BY-kinases in bacteria, a large-scale in silico analysis of BY-kinase genes was performed on the available complete bacterial and archaeal genomes. Experimentally characterized BY-kinases contain a CD with or without a TAD (referred as PF02706 in Pfam database). However, the structural core of BY-kinases is defined by three Walker-like motifs (A, A′, and B) and a C-terminal tyrosine cluster (R1), which are all parts of the CD. Furthermore, the TADs do not have particularly conserved distinguishing features besides the transmembrane helices. Thus, in this study, we first defined an HMM profile from the CD. By using the CD profile as a query, we detected 796 BY-kinases homologs (fig. 1 and supplementary table S2, Supplementary Material online) and as their name indicated, all of them were found in bacterial genomes but not in Archaea. These 796 BY-kinas homologs are present in 577 of 1,471 (39.2%) bacterial genomes. The average of BY-kinases per genome is about 1.5 (796/577), and 72.5% of the organisms contain only one copy per genome. However, there can be up to eight copies in Burkholderia strains (supplementary table S2, Supplementary Material online), which indicated the possibility of gene expansion and duplication. BY-kinases are found in members of all bacteria phyla except Chlamydia, Aquificales, and Epsilonproteobacteria (fig. 1). BY-kinase abundance is high in four phyla: Gammaproteobacteria, Firmicutes, Betaproteobacteria, and Alphaproteobacteria, whereas the numbers of organisms harboring BY-kinases are fairly low in some phyla (e.g., 37 out of 168 in Actinobacteria; 2 out of 36 in Spirochaetes). In the four phyla with high BY-kinase abundance, BY-kinases are found in most of the orders (e.g., six out of nine orders in Firmicutes and 10 out of 15 orders in Gammaproteobacteria), whereas in other phyla, the presence of BY-kinases is limited (e.g., three out of seven orders in Cyanobacteria) (supplementary table S2, Supplementary Material online). Extensive variation in distribution of BY-kinases was also observed at the genus level, because less than 50% of genera in each phylum harbor BY-kinases, except in Acidobacteria, Betaproteobacteria, and Planctomycetales (supplementary table S2, Supplementary Material online). The heterogeneous distribution of BY-kinases is consistent with the experimental evidence that their genes are nonessential in bacteria and are often involved in some particular processes in specific organisms or conditions, which include heat-shock response, polymyxin resistance, virulence, and detoxification of polyunsaturated fatty acids (Klein et al. 2003; Lacour et al. 2006; Morona et al. 2006; Derouiche et al. 2013). In addition, the extremely low percentage of BY-kinases in some phyla or genera also indicates the possibility that HGT events occurred during evolution. Interestingly, there are 15 BY-kinase genes (2 in Firmicutes and 13 in Proteobacteria) found in plasmids, whereas the majority (98.1%) of BY-kinase-encoding genes are located within the chromosome (fig. 1), which also supports the notion of HGT acquisition.
Fig. 1.—
The distribution of BY-kinases over the main bacterial phyla. For each of the bacteria main phylum, columns represent: Org, the number of organisms that harbor -kinases (total number of studied organisms); Genus, number of genus with organisms possessing BY-kinases (total number of genus in the phylum); BY-kinase, number of BY-kinases in the phylum; CD and TAD-CD, number of CD and TAD-CD-type BY-kinases, respectively; Chr and plasmid, number of BY-kinases located within the chromosome and plasmid, respectively. For CD, TAD-CD, Chr, and plasmid columns, the numbers in brackets correspond to the number of organisms.
Among the identified putative BY-kinases, 545 (from 383 organisms) belong to TAD-CD type, containing both TAD and CD in single protein, which was encoded by the same gene; and 252 (from 206 organisms) are of the CD type, in which TAD and CD belong to two separate proteins and are encoded by two genes (fig. 1). Previous reports usually described CD type and TAD-CD type as the Firmicutes type and Proteobacteria type, respectively (Jadeau et al. 2008). Our systematic study confirms that 98.7% of BY-kinases from Firmicutes identified in this study are of the CD type and 96% from Proteobacteria are of the TAD-CD type, and yet the distribution of these two types of BY-kinases is not restricted to these two phyla. In most bacterial phyla, a proportion bias is found in favor of one particular BY-kinase type, for example, there is only 1 CD-type versus 83 TAD-CD-type BY-kinases in Alphaproteobacteria (fig. 1). Interestingly, the abundance of CD- and TAD-CD-type BY-kinases is balanced in Deltaproteobacteria, with 16 BY-kinases of the CD type and 18 of TAD-CD type. For the CD type, in most cases (230 over 252), the gene encoding the CD was colocalized with that of encoding the corresponding TAD (supplementary tables S2 and S3, Supplementary Material online), consistent with the notion that the presence of both are necessary to reconstitute a fully functional enzyme. We also found the existence of few orphan CD-type BY-kinases in some organisms (supplementary tables S2 and S3, Supplementary Material online), where there is no corresponding TAD-encoding gene flanking the genes for those orphan CD-type kinases. It has been shown that TAD was essential for CD domain activity (Mijakovic et al. 2003), therefore the mechanism and biological function of those orphan CD-type kinases will be quite interesting to investigate. We speculate that these orphan CD-type kinases may be activated by the kinase-modulator proteins (TAD) from another CD-type kinase in the organisms harboring both, for example, Clostridium acetobutylicum ATCC 824.
Evolutionary History of BY-Kinases
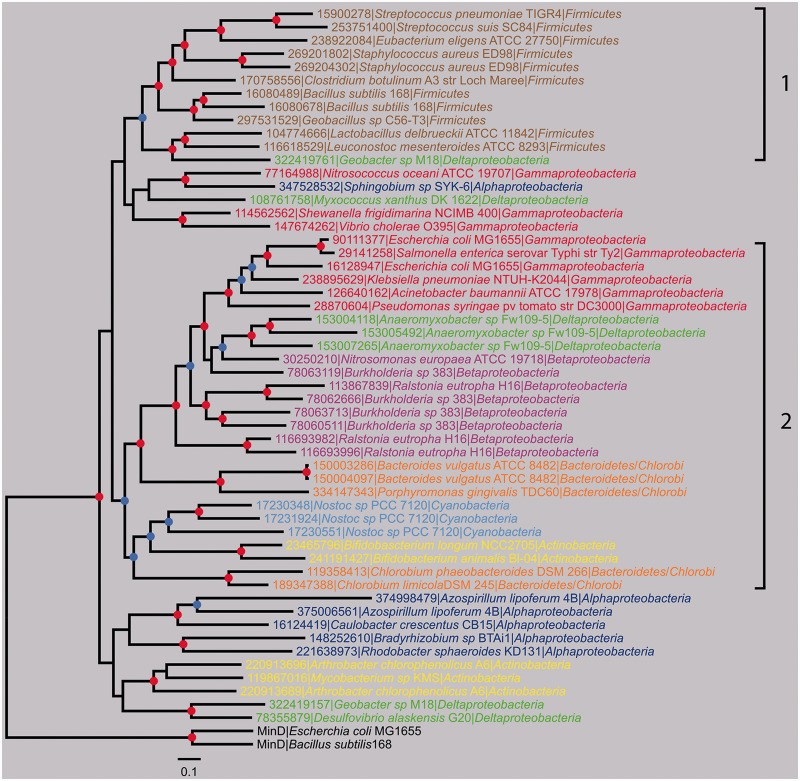
For the same reason mentioned previously, we performed the phylogenetic analysis of BY-kinases based on the CD. According to the large number of BY-kinase sequences and the limited number of unambiguously aligned positions (178 amino acids) available for phylogenetic analysis, the rooted tree of 796 BY-kinases is only partially resolved, especially at the most basal nodes (supplementary fig. S1, Supplementary Material online). The exhaustive phylogenetic tree showed that all the BY-kinases formed a monophyletic clade with MinD from E. coli and B. subtilis as outgroups, which suggested that all these BY-kinases have evolved from a common origin. Although the order of emergence of the main bacterial phyla is not fully resolved, the overall BY-kinase tree does not fit the reference bacterial tree, because several phyla were split into different clusters. The resulting tree is composed of four major clades, as well as several minor ones. Interestingly, several phyla were split into different clusters (e.g., two clusters for Firmicutes, three clusters for Gammaproteobacteria, three clusters for Alphaproteobacteria, and three clusters of Betaproteobacteria). To gain more insight in the evolutionary history of BY-kinases, a subset of 54 sequences representative of their phylogenetic diversity was used to perform an in-depth phylogenetic analysis. The overall topology of the resulting Bayesian tree was in agreement with the general BY-kinas tree (fig. 2), with equally limited resolution in some basal nodes. Two main well-supported clades (labeled 1 and 2) and several smaller groups were identified (with posterior probability >0.5), corresponding to major bacterial phyla. Clade 1 is mainly composed of BY-kinases from Firmicutes, for which all of the members are exclusively of the CD type. Clade 2 mainly contains BY-kinases of the TAD-CD type, from Gammaproteobacteria, Betaproteobacteria, Cyanobacteria, Bacteroidetes, Actinobacteria, and Deltaproteobacteria. Moreover, BY-kinases from Cyanobacteria formed a well-supported small clade, BY-kinases from Deltaproteobacteria were split into clade 2 and other unclassified groups, and Actinobacteria BY-kinases fell into two-separated clusters, which was in agreement with the previous global analysis (supplementary fig. S1, Supplementary Material online).
Fig. 2.—
Bayesian phylogenetic tree of 54 BY-kinase sequences representative of the BY-kinase diversity. MinD proteins (AAC74259, CAB14759) were used as outgroups. Leafs are colored according to bacterial main phyla. The colored dots at the nodes indicate the posterior probabilities. Nodes supported with a posterior probability ≥0.9 are indicated by a red dot, whereas nodes supported with a posterior probability ≥0.5 are indicated by a blue dot. The scale bar represents the average number of substitutions per site. The well-supported major clades are marked with 1 and 2.
As mentioned before, gene expansion and duplication of BY-kinases has been observed, and the perfect show case for these events is the genus Burkoholderia (from Betaproteobacteria). Twenty-five sequenced Burkholderia strains harbor 78 BY-kinases of the TAD-CD type. BY-kinases are particularly abundant in Burkholderia phytofirmans PsJN (6 BY-kinases), Burkholderia sp. CCGE1002 (6 BY-kinases), and Bu. xenovorans LB400 (8 BY-kinases) (supplementary tables S2 and S4, Supplementary Material online). To determine the origin of multiple BY-kinases in the Burkoholderia common ancestor, additional phylogenetic analysis based on CD were performed and resulted in six distinct well-supported subclades (Burk 1–6) (supplementary fig. S2 and table S4, Supplementary Material online). Some organisms, such as Bu. xenovorans LB400, contain two or more copies of BY-kinases within the same subclade (supplementary table S4, Supplementary Material online), but most of the organisms (19 out of 25) have multiple copies of BY-kinases from different subclades. It suggests that the Burkoholderia common ancestor probably already contained an expansion of BY-kinases, which was followed by recent duplications in some strains. Moreover, the presence of only one copy of BY-kinase in most organisms from the Burk6 subclade (23 out of 25) suggests that one of the BY-kinases ancestor genes is likely to be from this subclade. It is worth mentioning that Burkholderia usually have multiple chromosomes, and the extra copies of BY-kinase genes are usually located in different chromosomes and even in plasmids. Considering the genome size variation and high genomic plasticity in the genus Burkholderia (Chain et al. 2006), the accumulation of BY-kinases may be the result of genome scale expansions.
To check if the CD region of two types of BY-kinases, CD and TAD-CD, have similar evolutionary histories, we performed an in-depth phylogenetic analysis of the BY-kinases from Deltaproteobacteria, which possess a balanced set of BY-kinases of both types. The phylogenetic analysis of Deltaproteobacteria BY-kinases based on CD domain and the full sequences clearly shows that all BY-kinases from this phylum fall into two well-supported clades (posterior probabilities of 1), one corresponding to the CD type and the other to the TAD-CD type (supplementary fig. S3, Supplementary Material online). Moreover, the speciation between CD-type and TAD-CD-type kinases from the same organism (e.g., Geobacter sp. M18 and Desulfobacca acetoxidans DSM) suggests that CDs of each BY-kinase type have different evolutionary histories, even in the same organism. In addition, because most Geobacter species harbor only one CD-type BY-kinase gene (supplementary fig. S3, Supplementary Material online), the occurrence of both BY-kinase types in Geobacter sp. M18 and Geobacter uraniireducens Rf4 suggests possible HGT acquisition of the TAD-CD-type protein in those strains during evolution.
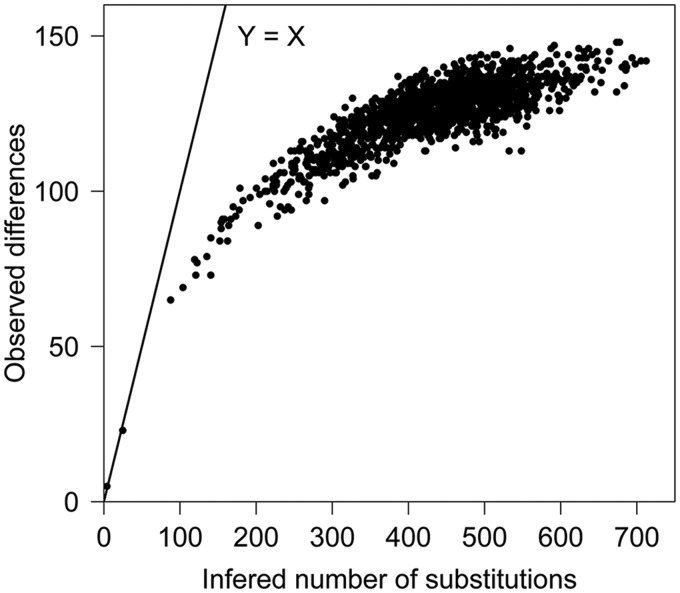
As shown earlier, the less-well supported deeper nodes of our phylogenetic trees restricted the tracking of the BY-kinase evolutionary history to full extent. Previous analyses have suggested that for the proteins with a high degree of mutational saturation, such as AAA+ superfamily of P-loop NTPase, which contains Walker A and Walker B motifs, accumulation of mutations at the same positions throughout time generate more noise than signal for molecular phylogeny (Gribaldo and Philippe 2002; Leclere and Rentzsch 2012). Because BY-kinases belong to this superfamily, we examined the mutational saturation of these enzymes by amino acid substitution analysis. Amino acid substitution is considered as saturated if the number of observed differences keeps constant while increasing the inferred number of substitutions. As shown in figure 3, most of the sequence pairs are located within the plateau, therefore inferring that the substitution has already reached saturation in a number of BY-kinases. Moreover, the nucleotide substitution saturation was also detected among these species (data not shown). Although we cannot exclude alternative explanations at present, this finding suggests that BY-kinases may evolve fast, and the substitution level in BY-kinase genes has reached saturation even between some closely related species. Faster evolution may explain the weak confidence levels in the deeper branches of our phylogenetic trees and supports the idea suggested previously that BY-kinases are open to fast evolution to adopt new substrates (Mijakovic and Macek 2012). To further test the hypothesis of fast evolution of BY-kinases, the synonymous and nonsynonymous substitution rates were estimated with PAML. As shown in supplementary table S5, Supplementary Material online, BY-kinases have a significantly higher nonsynonymous substitution rate (dN = 0.1607) compared with 21 conserved genes in bacterial lineages (from 0.0358 to 0.1034) chosen as reference. Most of proteins are dominated by purifying selection (i.e., the removal of functionally deleterious mutations), and their nonsynonymous rate will be less than the synonymous rate (dN/dS < 1), independently of the evolution model. To investigate the evolutionary forces that shaped BY-kinases, the dN/dS ratio was estimated. As shown in supplementary table S6A, Supplementary Material online, strong purifying selection pressure leads to the low dN/dS value. Because the substitution saturation could bias the estimation of the dN/dS, we restricted our analysis to three BY-kinases (Str. pneumoniae TIGR4, Str. suis SC84, and Eu. eligens ATCC27750, three closely related strains with low level of substitution saturation). Low dN/dS value (ω = 0.00283 for model M0) were observed, and no positively selected sites were identified in those three kinases (supplementary table S6B, Supplementary Material online). All these evidences indicated that the purifying selection, and not the positive selection, was the major driving force in the evolution of the BY-kinase family.
Fig. 3.—
Amino acid substitution analysis of BY-kinases. The level of substitution saturation is evaluated by the ratio between inferred number of substitutions (X axis) and the observed difference (Y axis). Dots in the straight line Y = X correspond to the completely unsaturated sequence pairs.
Classification of BY-Kinases
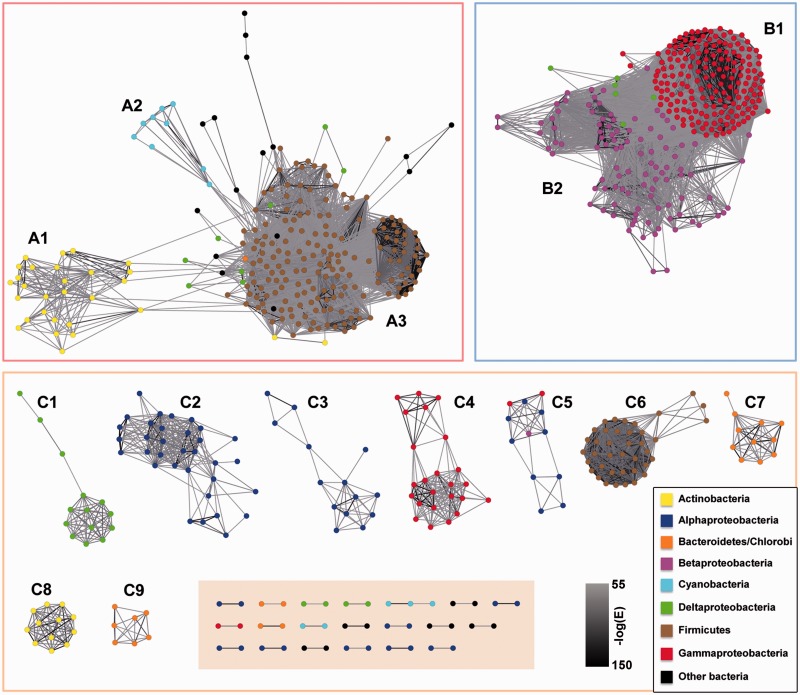
Because the phylogenetic approach could not provide an exhaustive classification of all BY-kinase sequences, as an alternative strategy, the PSN, which was shown recapitulate much of the information present in phylogenetic trees (Atkinson et al. 2009), was used. The emergence of connections between putative clusters was examined with different alignment E-value cutoffs from 10−20 to 10−100. Permissive cutoffs (e.g., 10−20) collapsed all sequences into one single cluster without any outliers (supplementary fig. S4A, Supplementary Material online), whereas more stringent cutoffs (e.g., 10−100) broke the data set into small-disconnected groups (supplementary fig. S4B, Supplementary Material online). Through the distribution of pairwise alignment numbers with decreasing E-value cutoffs (from 10−20 to 10−100) (supplementary fig. S4C, Supplementary Material online), an optimal E-value cutoff of 10−55 was determined, and the resulting pairwise alignments kept the majority (92%) of BY-kinases. As shown in figure 4, the analysis produced two major groups and also a number of peripheral clusters. Most of the sequences grouped together according to taxonomic relationship, which was consistent with previous phylogenetic results. We proposed to define them as groups A, B, and C. In Group A, three subgroups were identified: subgroups A1, A2, and A3 mainly include BY-kinases from Actinobacteria, Cyanobacteria, and Firmicutes, respectively. The Group B mainly includes proteins from Gammaproteobacteria (subgroup B1) and Betaproteobacteria (subgroup B2). The Group C contains nine subgroups (C1–C9) representing seven bacterial phyla and a miscellaneous ungroup cluster (shaded box in fig. 4). In summary, numerous phyla including Alphaproteobacteria (C2, C3, and C5), Gammaproteobacteria (B1 and C4), Actinobacteria (A1 and C8), Firmicutes (A3 and C6), and Bacteroidetes (C7 and C9) are mainly represented by at least two subgroups. Furthermore, through the overall taxonomic information of each subgroup (supplementary table S7, Supplementary Material online), some of these subgroups are mainly related to a particular genus (e.g., C6 from Streptococcus genus; C8, from Bifidobacterium genus), whereas in some of the subgroups, there is a small number of BY-kinases belonging to other phyla.
Fig. 4.—
The PSN reconstructed from the CD of the BY-kinases. Sequences are represented by nodes, and the nodes from the same taxonomic groups are in the same color. The edges are colored with a gray scale, and the darker the color is, the more significant similarity is. A1 to C9 represent the BY-kinases subgroups determined by the PSN reconstruction. Miscellaneous BY-kinases are shown in the shaded area.
Consensus Sites in BY-Kinase Sequences
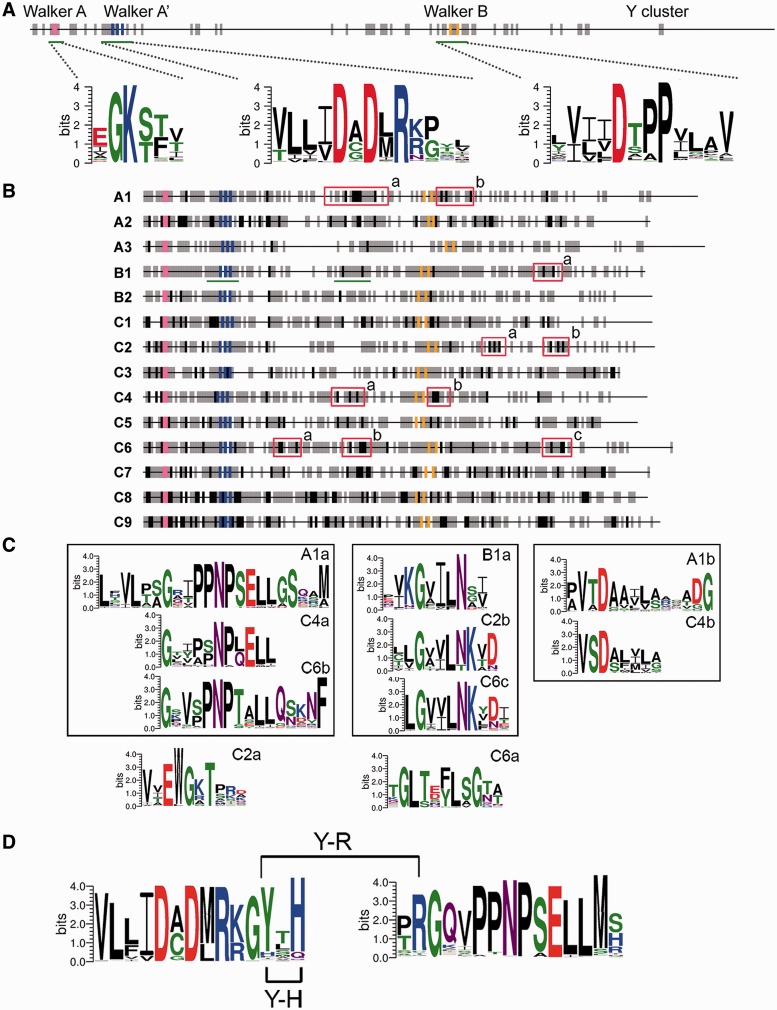
To examine the conservation of residues in BY-kinase sequences, the consensus sites based on the multiple alignments with “50-10” rule (Carretero-Paulet et al. 2010) were determined. As shown in figure 5A, only seven strong conserved sites (100% identical at the position) are found in the entire data set of CD domains from 796 BY-kinases, and they are all situated in the three Walker motifs. These sites include “GK” in Walker A motif, “DXDXR” in Walker A′ motif, and “DXXPX” in Walker B motif, which was not surprising because those three motifs are included in the isBYK algorithm and also have been experimentally shown to participate directly in ATP hydrolysis and ATP-Mg interaction (Soulat et al. 2007; Lee et al. 2008; Olivares-Illana et al. 2008). This result is consistent with amino acid substitution analysis of BY-kinases and supports the notion that these enzymes evolve fast and only conserve at essential sites to maintain core enzymatic activity. In addition, we found that several regions (marked with gray bars in fig. 5A) with a relatively high degree of conservation. These include regions between the Walker A and Walker A′, regions around the Walker B motif, and the Y cluster.
Fig. 5.—
Motif and amino acid consensus sites in BY-kinases and related subgroups. (A) Schematic view of amino acid conserved sites in all 796 BY-kinases. Black and gray ticks represent the strong and relatively strong conserved sites in BY-kinases, respectively (see Materials and Methods). “GK” in Walker A motif, “DXDXR” in Walker A′ motif, and “DXXPX” in Walker B motif are indicated by pink, orange, and blue ticks, respectively. The three motifs Walker A, A′, and B are marked with green underlines, and the corresponding sequence logos are shown. (B) Schematic maps of the conserved sites of BY-kinase subgroups (A1 to C9). Strong and relatively strong conserved sites over the protein subgroups are indicated by black and gray ticks. Except for Walker A, A′, and B motifs, the signature motifs in each subgroup are indicated within a red box. The region marked with green underlines contained the conserved Y-R and Y-H pairs, and their weblogs are indicated in (D). (C) Sequence logos of signature motifs (defined in B) are shown. Motifs located in the same region of the BY-kinase are displayed in the same box. (D) Sequence logos of motifs are related to the Y-R and Y-H interactions in subgroup B1 (also underlined in green lines in B).
For each BY-kinase subgroup, we performed the same analysis as described earlier. As shown in figure 5B, more strong conserved sites can be found within each subgroup, compared with only seven such sites in the total of 796 BY-kinases. For example, subgroup B1 contains 13 strictly conserved sites and subgroup A3 has 10. From these strong conserved sites, signature motifs were defined (supplementary fig. S5, Supplementary Material online), and the 10 most significant of them are illustrated (red boxes in fig. 5B) with the corresponding sequence logos in figure 5C. Besides strong conserved sites, more sites with a relatively high degree of conservation (marked with gray bars in fig. 5B) were also identified within each subgroup. All these sites are seem to be of functional importance. The functional importance of these sites is highlighted by the example of Y574 of Etk and Y569 of Wzc from E. coli, which have been suggested to mediate the intraphosphorylation in a two-step activation process (Grangeasse et al. 2002; Lu et al. 2009). It has been reported that H576 (located 5 amino acids downstream the DXDXR motif) and R614 (located between the DXDXR and DXXP motifs) of Etk can interact with phosphorylated Y574 (located three amino acids downstream the DXDXR motif) (Lee et al. 2008), and R614 was critical for Etk kinase activity. On the basis of this, we looked for correlation in presence of these amino acids in each subgroup (supplementary table S8, Supplementary Material online) and found that Y574-R614 and Y574-H576 pairs of amino acids were only conserved in subgroup B1 (fig. 5D). Within this subgroup, 87% of the sequences harbor the conserved Y, 89% contained the conserved R, 89% contained the conserved H, 80% and 78% possessed Y-R and Y-H pairs, respectively, and 72% harbor both Y-R and Y-H. The high conservation of both Y-R and Y-H pairs suggests that it may play an important role in BY-kinases intraphosphorylation within the subgroup B1, which mainly include organisms from Gammaproteobacteria.
Conserved Functional Genomic Context of BY-Kinases
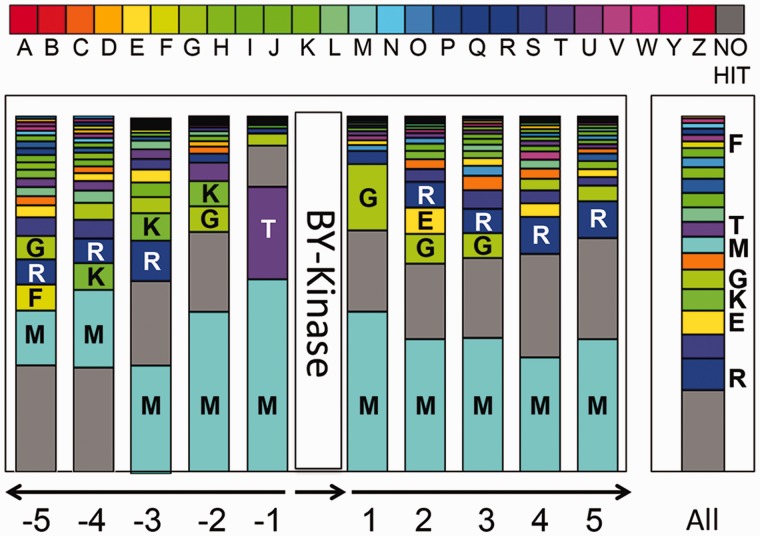
Experimentally characterized BY-kinases are usually encoded by genes located in operons involved in synthesis and export of capsular or extracellular polysaccharides and have been described as polysaccharide copolymerases in previous studies (Cuthbertson et al. 2009; Grangeasse et al. 2012). We sought to shed more light on the function of BY-kinases by screening the genomic neighborhoods of their genes. As shown in figure 6, the genomic context of BY-kinase genes was not conserved and varied considerably in different genomes. However, by comparing the COG classification between the immediate neighbor genes of BY-kinase genes and the entire genome, we found the location of BY-kinase genes was not entirely random. For the proteins encoded by the three immediate neighbor genes, 45.4% in average were involved in cell wall/membrane/envelop biogenesis (COG category M), 9.4% in average were involved in carbohydrate transport and metabolism (COG category G), and about 20% in average were belong to proteins without hits in COG, independently of the neighbor position from −3 to +3 except −1. The high frequency of neighbor genes encode proteins for cell wall/membrane/envelop biogenesis (COG category M) is not related to their proportion within the genomes, because the overall distribution of COG category M genes from all genomes is lower than 5%. This finding suggests a functional enrichment of BY-kinase regions in capsular biosynthesis proteins, which is consistent with the initial definition of BY-kinases as a component of the polysaccharide biosynthesis pathway. The same conclusion can be achieved by the analysis of genomic-context network associated to BY-kinase genes (supplementary fig. S6, Supplementary Material online), for example, BY-kinase (COG0489) was strongly linked to COG category M (COG3944), which corresponds to capsular biosynthesis proteins. Moreover, at position −1, proteins related to signal transduction (COG category T) appear to be among the most frequent neighbors of BY-kinase genes (fig. 6), and protein tyrosine phosphatases (COG0394) were linked with high frequency to BY-kinases (COG3206, another COG which BY-kinases were defined to) (supplementary fig. S6, Supplementary Material online). This result was consistent with previous report that BY-kinase genes are often colocalized with phosphatase-encoding genes (Mijakovic et al. 2003) and indicates that dephosphorylation performed by these phosphatases also plays a major role in the regulatory mechanisms mediated by BY-kinases. However, the genes flanking BY-kinases genes are also involved in many other biological processes (supplementary fig. S6, Supplementary Material online), which was not surprising because it was known that BY-kinases Wzc from E. coli and PtkA from B. subtilis participate in many processes besides capsular biosynthesis (Klein et al. 2003; Lacour et al. 2006, 2008). The variety of the immediate genomic environment supports the notion that BY-kinases may play a complex role in bacterial physiology and may contribute to several distinct signaling pathways in the same bacterium.
Fig. 6.—
Distribution and functional classification of genes surrounding the BY-kinases. For each of the five neighboring genes (located upstream and downstream of the BY-kinase), the functional COG category was determined. For each surrounded position, the bar indicates the frequency of each functional gene type (represented here by COG category) among the overall categories of the same position. Bar in the right part represents the COG distribution in all genomes harboring BY-kinases. COG categories are shown in different colors (see the COG color legend) and are associated with the corresponding capital letters: A, RNA processing and modification; B, chromatin structure and dynamics; C, energy production and conversion; D, cell cycle control, cell division, and chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; J, translation, ribosomal structure, and biogenesis; K, transcription; L, replication, recombination, and repair; M, cell wall/membrane/envelope biogenesis; N, cell motility; O, posttranslational modification; protein turnover, chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism; R, general function prediction only; S, function unknown; T, signal transduction mechanisms; U, intracellular trafficking, secretion, and vesicular transport; V, defense mechanisms; W, extracellular structures; Y, nuclear structure; Z, cytoskeleton.
Evolutionary Relationship between BY-Kinases and Their Substrates
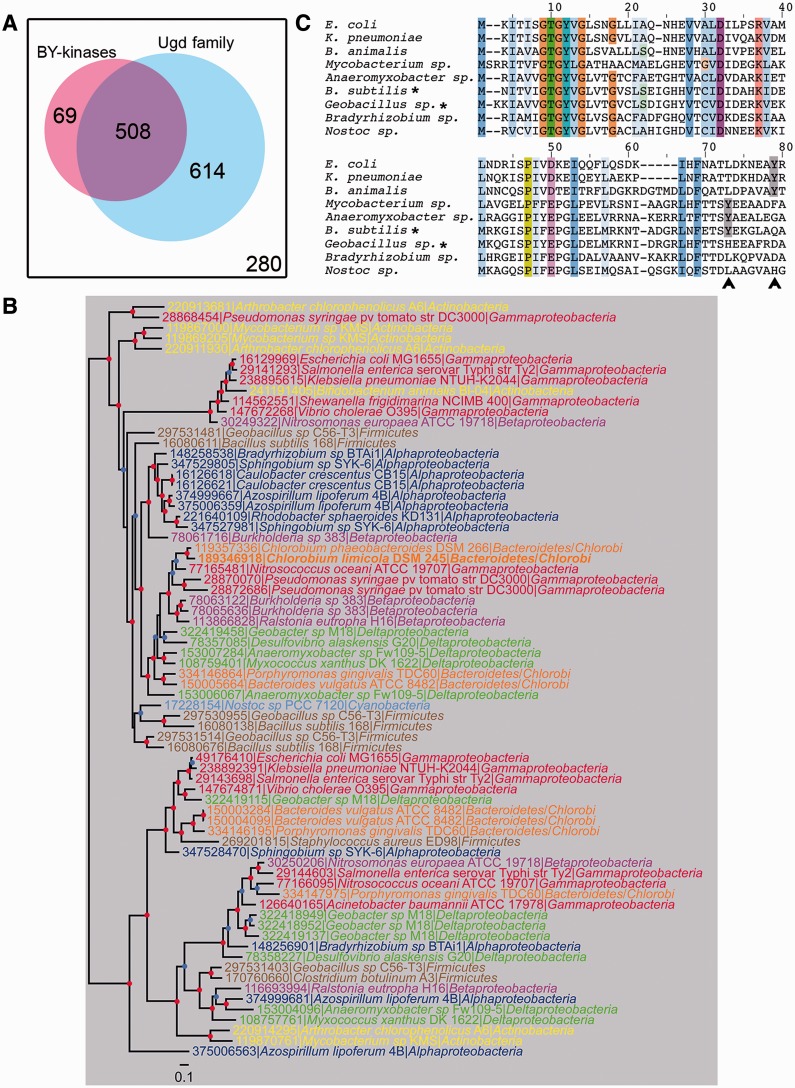
Ugds are the first discovered and the best characterized substrates of BY-kinases (Mijakovic et al. 2003; Lacour et al. 2008; Egger et al. 2010). It has been shown that cognate Ugds are phosphorylated in vitro by BY-kinases PtkA from B. subtilis and Wzc from E. coli, which significantly increased the Ugd dehydrogenase activity (Grangeasse et al. 2003; Mijakovic et al. 2003). The activation of Ugd depends on phosphorylation of a specific tyrosine residue which in its nonphosphorylated form hinders the binding of substrates in the active site (Petranovic et al. 2009). We sought to elucidate the evolutionary past of the relationship between BY-kinases and their substrates Ugds. As shown in supplementary table S9, Supplementary Material online, 2,346 homologs of Ugd family proteins are present in 1,122 bacterial genomes and 93 archaeal genomes, which means that they are more widespread than BY-kinases. Furthermore, only 508 organisms possess both BY-kinases and Ugd family proteins, whereas 614 organisms, distributed in almost all bacterial phyla, only contain Ugd family proteins without BY-kinases (fig. 7A). The different phylogenetic profiles suggest that Ugd proteins are not universally regulated by BY-kinases. Organisms with both Ugds and BY-kinases are mainly members of Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Firmicutes (supplementary tables S2 and S9, Supplementary Material online), the four phyla with high BY-kinase abundance. There are also 69 organisms without an Ugd, but with BY-kinases, they are mainly members of Firmicutes (e.g., organisms from Lactobacillus and Streptococcus genera) and are found in both A3 and C6 subgroups (fig. 7A and supplementary table S2, Supplementary Material online). Because previous studies have shown that some kinases coevolved with their substrates, that is, eukaryal cyclin Pcl5 and its substrate Gcn4 (Gildor et al. 2005), or the bacterial histidine kinases and response regulator pairs (Skerker et al. 2008), we performed phylogenetic analysis of Ugds from organisms harboring both Ugds and BY-kinases and observed that the topology of the Ugd phylogenetic tree is very different from the tree of BY-kinases (fig. 7B). The incongruence between those trees suggested a complex evolutionary relationship between BY-kinases and Ugds. Because BY-kinase and Ugd encoding genes are located in the same gene cluster in some organisms, we narrowed down our focus on 105 of such BY-kinase and Ugd co-occurrence pairs. Phylogenetic analysis of these pairs resulted once again in very different trees for BY-kinases and Ugd proteins (supplementary fig. S7, Supplementary Material online). Thus, we could detect no evidence of coevolution between BY-kinases and Ugds at the sequence level, despite the fact that their genes are located in the same gene clusters (operons) and probably highly correlated at the expression level.
Fig. 7.—
Relationship of BY-kinases and their substrates Ugds. (A) Distribution of BY-kinases and Ugd family proteins in bacteria. The white square with the black frame indicates all analyzed genomes; organisms containing BY-kinases are in pink; and organisms harboring proteins from Ugd family are in blue. (B) Bayesian tree of a sample of 72 Ugd sequences representative of the Ugd family diversity. Leafs are colored according to bacterial phyla. The colored dots at the nodes indicate the posterior probabilities. Nodes supported with a posterior probability ≥0.9 are indicated by a red dot, whereas nodes supported with a posterior probability ≥0.8 are indicated by a blue dot. The scale bar represents the number of substitutions per site. (C) Multiple sequence alignment of Ugd family proteins. Proteins from Escherichia coli, Klebsiella pneumoniae, Bifidobacterium animalis, Mycobacterium sp. Anaeromyxobacter sp, Bacillus subtilis, Geobacillus sp., Bradyrhizobium sp., and Nostoc sp. were aligned. Only the N-terminal extremity of helix α4 including the possible phosphorylation site (Egger et al. 2010) were shown. Conserved amino acids are highlighted. The CD-type BY-kinases were indicated by asterisk (*). The positions of phosphorylation site (Y) are marked with gray and indicated with an arrow.
The tyrosine 70 of Ugd is phosphorylated in the B. subtilis, and homology-based structure modeling reveals that this phosphorylated tyrosine residue locate in the positioned at the N-terminal extremity of helix α4 and is in close proximity to the N-terminal NAD-binding site (Petranovic et al. 2009; Egger et al. 2010). To examine whether the Ugd tyrosine phosphorylation site was conserved, the Ugd proteins were aligned. As shown in figure 7C, the tyrosine phosphorylation site of Ugd, which had been characterized in E. coli and B. subtilis, is present in Klebsiella pneumoniae, Bifidobacterium animalis, Anaeromyxobacter sp., and Mycobacterium sp.; however, the position of the phosphorylated tyrosine is not conserved and varies in helix α4 region. One possible explanation is that the tyrosine phosphorylation may occur within a particular region but not at a particular position in a protein and therefore may shift position during evolution (Moses et al. 2007; Nguyen Ba and Moses 2010). Another explanation is that proteins with multiple sites may lose or gain some sites without affecting the regulation of the protein (Moses et al. 2007; Nguyen Ba and Moses 2010). Moreover, the Ugd phosphorylation site seems to be lost in some organisms (e.g., Geobacillus sp. and Nostoc sp. 7120 in fig. 7C), despite the presence of a BY-kinase in the genome. In such cases, it is probable that the Ugd activity is no longer under control of BY-kinase.
The BY-Kinase Substrate Specificity across Species Boundaries
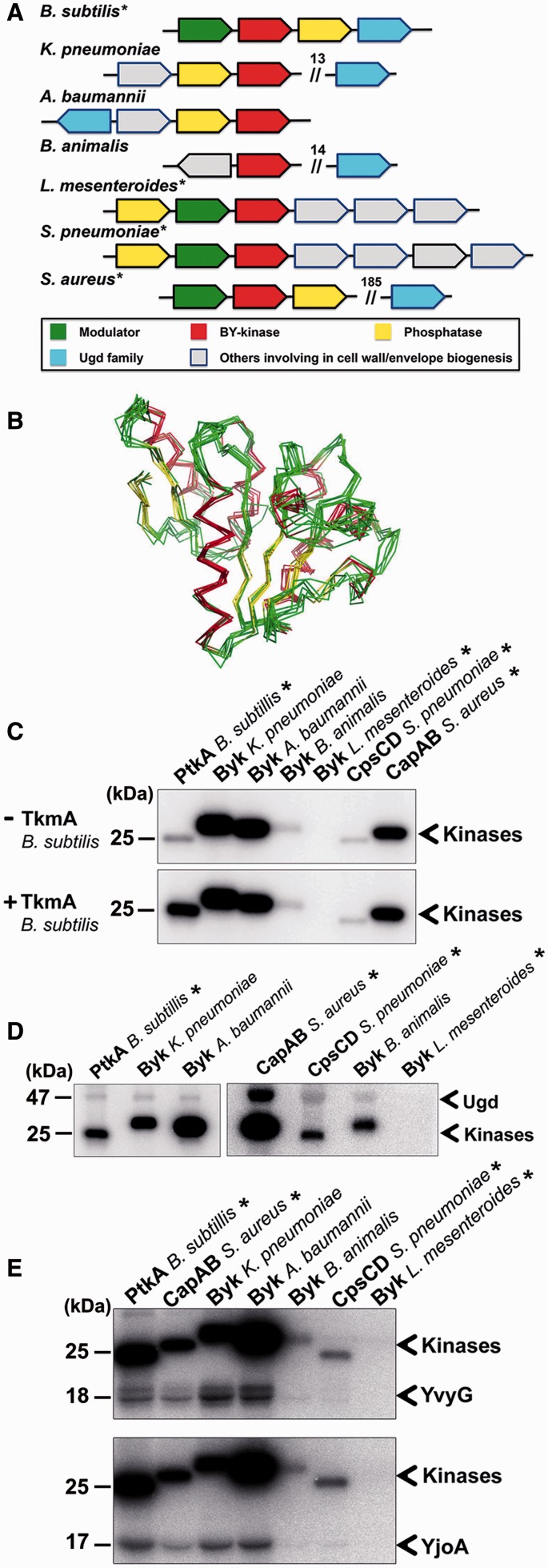
To investigate the relationship between BY-kinases and substrates, we decided to probe experimentally the ability of a number of evolutionary distant BY-kinases to phosphorylate some substrates (including Ugd) of the canonical BY-kinase PtkA from B. subtilis. In the set of kinases, we included three well characterized ones: PtkA from B. subtilis (Mijakovic et al. 2003), CapB from Sta. aureus (Olivares-Illana et al. 2008), CpsD from S. pneumoniae (Morona et al. 2004), and four putative BY-kinases from K. pneumoniae, Acinetobacter baumannii, Bi. animalis, and Leuconostoc mesenteroides identified by the BY-kinase database. Among these kinases, PtkA, CapB, and Byk from L. mesenteroides belong to group A (subgroup A3); Byk from K. pneumoniae and Byk from A. baumannii belong to group B (subgroup B1); and CpsD and Byk from Bi. animalis belong to group C (subgroups C6 and C8, respectively). Additionally, PtkA, CapB, CpsD, and Byk from L. mesenteroides are CD type, whereas the other three are TAD-CD type (fig. 8A). For the CD-type kinases, PtkA was carried out with experiments in the presence of its modulator TkmA (TAD) (Mijakovic et al. 2003), Byk from L. mesenteroides was also assayed in the presence of B. subtilis TkmA with the attempt to supplement its function, whereas CapB and CpsD were expressed as translational fusions with the activator region of their corresponding modulators CapA and CpsC (TAD) and purified as CapAB and CpsCD (Olivares-Illana et al. 2008). For the other three TAD-CD-type kinases, we cloned the intacellular CDs from the end of the second transmembrane helix to the stop codon to maintain the activator region of TAD. First, to get a global structural overview of the putative kinases, homology-based modeling was used to generate their structural models. All models were superimposed with the resolved structure of CapAB (Olivares-Illana et al. 2008) (fig. 8B). The extensive overlap between models (Core RMSD : 1.371 A) suggests that these uncharacterized proteins also exhibit the conserved BY-kinase fold, in spite of the high sequence divergence. Autophosphorylation activity of all proteins was examined by autoradiography. The experiment was carried out first without TkmA, and we found autophosphorylation activity for three TAD-CD-type putative BY-kinases (fig. 8C), as well as for three previously characterized BY-kinases CpsCD, CapAB, and PtkA. However, there was no detectable autophosphorylation activity for the CD-type putative Byk from L. mesenteroides. PtkA exhibited very weak autophosphorylation due to the absence of its modulator TkmA (Mijakovic et al. 2003). Thus, the same experiment was repeated in the presence of B. subtilis TkmA. Although TkmA increased autophosphorylation of PtkA, Byk from L. mesenteroides still did not show any autophosphorylation. This finding suggested that Byk from L. mesenteroides probably requires its cognate modulator (encoded by the same operon as the kinase) (fig. 8A) or may not be a functional kinase at all. We also found that TkmA did not change the autophosphorylation of any of the other kinases (fig. 8C). Our data set is too small for drawing definite conclusions, but these findings do suggest that the kinase-modulator relationship is probably species specific.
Fig. 8.—
In vitro phosphorylation of the BY-kinases. Asterisk (*) indicates BY-kinase of CD type. (A) Schematic diagram of gene organization of BY-kinases and Ugd in Staphylococcus aureus, Streptococcus pneumoniae, Klebsiella pneumoniae, Acinetobacter baumannii, Bifidobacterium animalis, Leuconostoc mesenteroides, and Bacillus subtilis. The number above the double slash indicates the number of interval genes. (B) The superposition of all BY-kinase structural models (S. pneumoniae, K. pneumoniae, A. baumannii, B. animalis, L. mesenteroides, and B. subtilis) with the structure from S. aureus, colored by secondary structure (truncated from N-ter and C-ter, Core Residues: 164, Core RMSD: 1.371 Å). (C) In vitro autophosphorylation of BY-kinases; 1-µM BY-kinases were incubated without or with 1 µM TkmA. Bands corresponding to autophosphorylated BY-kinases are indicated. Molecular weight of PtkA is shown on the left. (D) Phosphorylation of Ugd by seven BY-kinases; 10-µM Ugd was incubated individually with following kinases: 2 µM CapAB from S. aureus, 2 µM CpsCD from S. pneumoniae, 2 µM Byk from B. animalis, 2 µM Byk from L. mesenteroides, 1 µM of Byk from K. pneumoniae, 1.5 µM of Byk from A. baumannii, or 0.8 µM of PtkA and TkmA from B. subtilis. Bands corresponding to autophosphorylated BY-kinases and phosphorylated Ugd are indicated. Molecular weights of Ugd and PtkA are shown on the left. (E) Phosphorylation of YvyG and YjoA by the seven BY-kinases; 10 µM of YvyG or YjoA were incubated with the following kinases: 2 µM Byk from A. baumannii, 2 µM Byk from L. mesenteroides, 1 µM of CapAB from S. aureus, 1 µM CpsCD from S. pneumoniae, 1 µM Byk from K. pneumoniae, 1 µM Byk from B. animalis, 1 µM PtkA, and TkmA from B. subtilis. Bands corresponding to autophosphorylated BY-kinases and phosphorylated YvyG and YjoA are indicated. Molecular weights of YvyG, YjoA, and PtkA are shown on the left.
Next, we examined whether all these kinases can phosphorylate Ugd from B. subtilis. Previous work showed that Ugd from B. subtilis can be phosphorylated by BY-kinases from E. coli (Mijakovic et al. 2003). Our results here suggest that this was not an isolated phenomenon. Six out of seven kinases examined here phosphorylated B. subtilis Ugd (fig. 8D). Byk from L. mesenteroides was the only one that has shown no activity (fig. 8D), which was not surprising because the kinase was already inactive for autophosphorylation (fig. 8C). This may explain why BY-kinase and Ugd do not need coevolution—they retain the capacity to recognize each other even across very distant evolutionary boundaries. To turn to some more idiosyncratic and far less widely distributed substrates, we also performed the same analysis with two newly identified substrates of B. subtilis PtkA: YvyG and YjoA (Jers et al. 2010), which are only present in Firmicutes. The result was identical to the one obtained for Ugd, six out of seven kinases (all except the Byk from L. mesenteroides) phosphorylated YvyG and YjoA (fig. 8E). In conclusion, we propose that BY-kinases maintain the capability to phosphorylate the same set of substrates across very distant phyla, probably by keeping the ability to recognize global structural motifs and not specific sequences around the target sites.
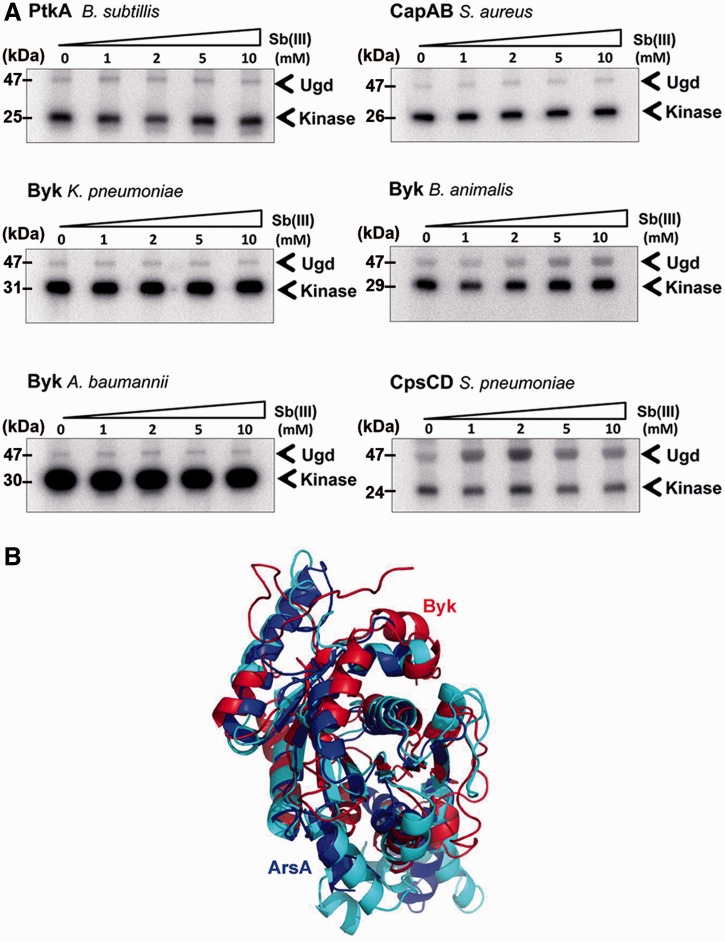
Regulation by Antimonite as a Shared Mechanism between ArsA Proteins and Some BY-Kinases
Previous studies have revealed that BY-kinases exhibit significant sequence similarity with ArsA proteins and suggested that they may have evolved from the same ancestor (Grangeasse et al. 2012). ArsA ATPase is the cytosolic subunit of the Ars pump protein in E. coli, which provides resistance to arsenic or antimony. ArsA contains two nucleotide-binding sites and an allosteric-binding site for antimonite Sb(III). Binding of Sb(III) increases the ArsA ATPase activity (Li et al. 1996; Zhou et al. 2000; Walmsley et al. 2001). Presumably, the ATP-binding domain in ArsA and the CD of BY-kinases come from the same origin, so we decided to examine whether BY-kinase active site could be affected by antimonite. Interestingly, the activity of Byk from Bi. animalis and CpsCD from S. pneumoniae increased slightly with the addition of Sb(III) to the reaction (fig. 9A). The phosphorylation of Ugd by Byk from Bi. animalis reached the maximum at 5 mM Sb(III), whereas for CpsCD, the optimum concentration of Sb(III) was 2 mM, and further increase in antimonite reduced the kinase activity. Other BY-kinases were not affected by Sb(III). It is noteworthy that the structures of ArsA and BY-kinase ATP-binding domains overlap extensively (fig. 9B). However, beyond that domain, there is a low sequence identity (about 20%) between the two enzymes. In particular, the Sb(III)-binding site of ArsA is outside this region, and no homolog can be find in BY-kinases. Thus, the molecular mechanism of activation of BY-kinases by antimonite remains puzzling, even though it supports the notion of an evolutionary link between BY-kinases and ArsA ATPases.
Fig. 9.—
Phosphorylation of Ugd by BY-kinases in presence of Sb(III). (A) Phosphorylation of Ugd by the seven BY-kinases in presence of different concentrations of Sb(III); 10 µM Ugd was incubated individually with the following kinases: 2 µM of CapAB from Staphylococcus aureus, 2 µM CpsCD from Streptococcus pneumoniae, 2 µM Byk from Bifidobacterium animalis, 2 µM Byk from Leuconostoc mesenteroides; 1 µM of Byk from Klebsiella pneumoniae; 1.5 µM of Byk from Acinetobacter baumannii; and 0.8 µM of PtkA and TkmA from Bacillus subtilis. Concentration of Sb(III) is indicated above each lane. Bands corresponding to autophosphorylated BY-kinases and phosphorylated Ugd are indicated by arrows. Molecular weights of Ugd and BY-kinase are shown on the left. (B) Comparison of monomeric ArsA (domains 1 and 2) and monomeric Wzc. Domains 1 and 2 are colored with blue and cyan. Wzc is colored with red. Core residues: 139; Core RMSD: 3.311.
Discussion
In this study, we performed a large scale in silico analysis of BY-kinases and its well-known substrates, the Ugd family proteins. Phylogenomic analysis of these proteins reveals that BY-kinases ubiquitously distributed in most bacterial phyla, except Aquificales, Chlamydia, and Epsilonproteobacteria. In spite of the partially resolved nodes at the basal position of the phylogenetic trees, it still can be asserted that the evolutionary history of BY-kinases has been affected considerably by HGT and duplications. With an alternative clustering method, the BY-kinases were classified into three main groups and 14 subgroups, and we provide in silico evidence that each subgroup contains specific signatures. Our results indicated that BY-kinases underwent fast evolution and maintain a high conservation only for the residues directly involved in structure formation and catalytic activity. Analysis of the relationship between BY-kinases and their substrates showed that there is no coevolution between the two, and BY-kinases retain the ability to phosphorylate substrates from very distant bacterial relatives. In Eukarya, prereplicative complex (RC) is regulated by the cyclin-dependent kinase (CDK) through phosphorylation on CDK consensus sites. Interestingly, those CDK consensus sites are not conserved in position or number. By consequence, the regulation does not require the precise phosphorylation sites (Moses et al. 2007). We observed the similar “turnover” event with respect to phosphorylation of Ugd (Chen et al. 2011), suggesting that this phenomenon is not restricted to Eukarya.
Bacteria are widely distributed in most habitats on earth, even in some extreme conditions. They can adapt to frequent and rapid changed environment through adjusting their physiology and behavior, chemotaxis, phototaxis, dormancy, biofilm formation, etc. Signal sensing and transduction are the first steps of adaptative processes, and it has been shown that protein phosphorylation play a major role in signaling pathways (Cohen 2000). Previous studies suggested that BY-kinases were expressed as stress responds and modulate cellular processes to survive (Vincent et al. 2000). Moreover, BY-kinases were involved in different processes via phosphorylation different substrates (Klein et al. 2003; Mijakovic et al. 2003). It therefore leads to a question how those substrates emerged and keep to be modulated during evolution. Our study suggests that BY-kinases evolve very rapidly and may develop a new substrate by recognizing its overall structure and not a particular short sequence or phosphorylation site. Phosphorylation would then occur within a particular region of the substrate and be selected for if it conveys any regulatory potential.
Supplementary Material
Supplementary figures S1–S7 and tables S1–S9 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to the following colleagues: Sylvain Brisse, Gérman Bou, Philippe Horvath, and Agusting Lopez-Munguia for providing genomic DNA of the following bacteria, respectively: Klebsiella pneumoniae, Acinetobacter baumanii, Bifidobacterium animalis, and Leuconostoc mesenteroides. This work was supported by grants from the “Agence Nationale de la Recherche” (2010-BLAN-1303-01 to I.M. and C.G.; the “Fondation pour la Recherche Médicale” (SPF20111223359) to L.S.; and the “Chinese Scholarship Council Fellowship” to B.J.). The BYKdb database used in this study was supported by the Agence Nationale de la Recherche (ANR-07-JCJC0125-01 BACTYRKIN) and the Pôle Rhône-Alpes de BioInformatique (PRABI) platform by the Groupement d’Interêt Scientifique Infrastructures en Biologie, Santé et Agronomie (GIS IBiSA).
Literature Cited
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L, et al. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153:1398–1412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chain PSG, et al. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci U S A. 2006;103:15280–15287. doi: 10.1073/pnas.0606924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YY, Ko TP, Lin CH, Chen WH, Wang AH. Conformational change upon product binding to Klebsiella pneumoniae UDP-glucose dehydrogenase: a possible inhibition mechanism for the key enzyme in polymyxin resistance. J Struct Biol. 2011;175:300–310. doi: 10.1016/j.jsb.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Cohen P. The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- Combet C, Jambon M, Deleage G, Geourjon C. Geno3D: automatic comparative molecular modelling of protein. Bioinformatics. 2002;18:213–214. doi: 10.1093/bioinformatics/18.1.213. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson L, Mainprize IL, Naismith JH, Whitfield C. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol Mol Biol Rev. 2009;73:155–177. doi: 10.1128/MMBR.00024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derouiche A, et al. Interaction of bacterial fatty-acid-displaced regulators with DNA is interrupted by tyrosine phosphorylation in the helix-turn-helix domain. Nucleic Acids Res. 2013;41:9371–9381. doi: 10.1093/nar/gkt709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublet P, Vincent C, Grangeasse C, Cozzone AJ, Duclos B. On the binding of ATP to the autophosphorylating protein, Ptk, of the bacterium Acinetobacter johnsonii. FEBS Lett. 1999;445:137–143. doi: 10.1016/s0014-5793(99)00111-8. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger S, Chaikuad A, Kavanagh KL, Oppermann U, Nidetzky B. UDP-glucose dehydrogenase: structure and function of a potential drug target. Biochem Soc Trans. 2010;38:1378–1385. doi: 10.1042/BST0381378. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharib WH, Robinson-Rechavi M. The branch-site test of positive selection is surprisingly robust but lacks power under synonymous substitution saturation and variation in GC. Mol Biol Evol. 2013;30:1675–1686. doi: 10.1093/molbev/mst062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildor T, Shemer R, Atir-Lande A, Kornitzer D. Coevolution of cyclin Pcl5 and its substrate Gcn4. Eukaryot Cell. 2005;4:310–318. doi: 10.1128/EC.4.2.310-318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeasse C, Cozzone AJ, Deutscher J, Mijakovic I. Tyrosine phosphorylation: an emerging regulatory device of bacterial physiology. Trends Biochem Sci. 2007;32:86–94. doi: 10.1016/j.tibs.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Grangeasse C, Doublet P, Cozzone AJ. Tyrosine phosphorylation of protein kinase Wzc from Escherichia coli K12 occurs through a two-step process. J Biol Chem. 2002;277:7127–7135. doi: 10.1074/jbc.M110880200. [DOI] [PubMed] [Google Scholar]
- Grangeasse C, Nessler S, Mijakovic I. Bacterial tyrosine kinases: evolution, biological function and structural insights. Philos Trans R Soc Lond B Biol Sci. 2012;367:2640–2655. doi: 10.1098/rstb.2011.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeasse C, et al. Characterization of a bacterial gene encoding an autophosphorylating protein tyrosine kinase. Gene. 1997;204:259–265. doi: 10.1016/s0378-1119(97)00554-4. [DOI] [PubMed] [Google Scholar]
- Grangeasse C, et al. Autophosphorylation of the Escherichia coli protein kinase Wzc regulates tyrosine phosphorylation of Ugd, a UDP-glucose dehydrogenase. J Biol Chem. 2003;278:39323–39329. doi: 10.1074/jbc.M305134200. [DOI] [PubMed] [Google Scholar]
- Gribaldo S, Philippe H. Ancient phylogenetic relationships. Theor Popul Biol. 2002;61:391–408. doi: 10.1006/tpbi.2002.1593. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Jadeau F, et al. Identification of the idiosyncratic bacterial protein tyrosine kinase (BY-kinase) family signature. Bioinformatics. 2008;24:2427–2430. doi: 10.1093/bioinformatics/btn462. [DOI] [PubMed] [Google Scholar]
- Jadeau F, et al. BYKdb: the Bacterial protein tYrosine Kinase database. Nucleic Acids Res. 2012;40:D321–D324. doi: 10.1093/nar/gkr915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jers C, et al. Bacillus subtilis BY-kinase PtkA controls enzyme activity and localization of its protein substrates. Mol Microbiol. 2010;77:287–299. doi: 10.1111/j.1365-2958.2010.07227.x. [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Klein G, Dartigalongue C, Raina S. Phosphorylation-mediated regulation of heat shock response in Escherichia coli. Mol Microbiol. 2003;48:269–285. doi: 10.1046/j.1365-2958.2003.03449.x. [DOI] [PubMed] [Google Scholar]
- Kolot M, Gorovits R, Silberstein N, Fichtman B, Yagil E. Phosphorylation of the integrase protein of coliphage HK022. Virology. 2008;375:383–390. doi: 10.1016/j.virol.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Lacour S, Bechet E, Cozzone AJ, Mijakovic I, Grangeasse C. Tyrosine phosphorylation of the UDP-glucose dehydrogenase of Escherichia coli is at the crossroads of colanic acid synthesis and polymyxin resistance. PLoS One. 2008;3:e3053. doi: 10.1371/journal.pone.0003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacour S, Doublet P, Obadia B, Cozzone AJ, Grangeasse C. A novel role for protein-tyrosine kinase Etk from Escherichia coli K-12 related to polymyxin resistance. Res Microbiol. 2006;157:637–641. doi: 10.1016/j.resmic.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leclere L, Rentzsch F. Repeated evolution of identical domain architecture in metazoan netrin domain-containing proteins. Genome Biol Evol. 2012;4:883–899. doi: 10.1093/gbe/evs061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DC, Zheng J, She YM, Jia Z. Structure of Escherichia coli tyrosine kinase Etk reveals a novel activation mechanism. EMBO J. 2008;27:1758–1766. doi: 10.1038/emboj.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- Li J, Liu S, Rosen BP. Interaction of ATP binding sites in the ArsA ATPase, the catalytic subunit of the Ars pump. J Biol Chem. 1996;271:25247–25252. doi: 10.1074/jbc.271.41.25247. [DOI] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lu T, Tan H, Lee D, Chen G, Jia Z. New insights into the activation of Escherichia coli tyrosine kinase revealed by molecular dynamics simulation and biochemical analysis. Biochemistry. 2009;48:7986–7995. doi: 10.1021/bi900811p. [DOI] [PubMed] [Google Scholar]
- Menke M, Berger B, Cowen L. Matt: local flexibility aids protein multiple structure alignment. PLoS Comput Biol. 2008;4:e10. doi: 10.1371/journal.pcbi.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijakovic I, Macek B. Impact of phosphoproteomics on studies of bacterial physiology. FEMS Microbiol Rev. 2012;36:877–892. doi: 10.1111/j.1574-6976.2011.00314.x. [DOI] [PubMed] [Google Scholar]
- Mijakovic I, et al. Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J. 2003;22:4709–4718. doi: 10.1093/emboj/cdg458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minic Z, et al. Control of EpsE, the phosphoglycosyltransferase initiating exopolysaccharide synthesis in Streptococcus thermophilus, by EpsD tyrosine kinase. J Bacteriol. 2007;189:1351–1357. doi: 10.1128/JB.01122-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona JK, Miller DC, Morona R, Paton JC. The effect that mutations in the conserved capsular polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J Infect Dis. 2004;189:1905–1913. doi: 10.1086/383352. [DOI] [PubMed] [Google Scholar]
- Morona JK, Morona R, Paton JC. Attachment of capsular polysaccharide to the cell wall of Streptococcus pneumoniae type 2 is required for invasive disease. Proc Natl Acad Sci U S A. 2006;103:8505–8510. doi: 10.1073/pnas.0602148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses AM, Liku ME, Li JJ, Durbin R. Regulatory evolution in proteins by turnover and lineage-specific changes of cyclin-dependent kinase consensus sites. Proc Natl Acad Sci U S A. 2007;104:17713–17718. doi: 10.1073/pnas.0700997104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba AN, Moses AM. Evolution of characterized phosphorylation sites in budding yeast. Mol Biol Evol. 2010;27:2027–2037. doi: 10.1093/molbev/msq090. [DOI] [PubMed] [Google Scholar]
- Olivares-Illana V, et al. Structural basis for the regulation mechanism of the tyrosine kinase CapB from Staphylococcus aureus. PLoS Biol. 2008;6:e143. doi: 10.1371/journal.pbio.0060143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiment A, Hocking J, Whitfield C. Impact of phosphorylation of specific residues in the tyrosine autokinase, Wzc, on its activity in assembly of group 1 capsules in Escherichia coli. J Bacteriol. 2002;184:6437–6447. doi: 10.1128/JB.184.23.6437-6447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petranovic D, et al. Bacillus subtilis strain deficient for the protein-tyrosine kinase PtkA exhibits impaired DNA replication. Mol Microbiol. 2007;63:1797–1805. doi: 10.1111/j.1365-2958.2007.05625.x. [DOI] [PubMed] [Google Scholar]
- Petranovic D, et al. Activation of Bacillus subtilis Ugd by the BY-kinase PtkA proceeds via phosphorylation of its residue tyrosine 70. J Mol Microbiol Biotechnol. 2009;17:83–89. doi: 10.1159/000206635. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerker JM, et al. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulat D, et al. Tyrosine-kinase Wzc from Escherichia coli possesses an ATPase activity regulated by autophosphorylation. FEMS Microbiol Lett. 2007;274:252–259. doi: 10.1111/j.1574-6968.2007.00841.x. [DOI] [PubMed] [Google Scholar]
- Stingele F, Neeser JR, Mollet B. Identification and characterization of the eps (Exopolysaccharide) gene cluster from Streptococcus thermophilus Sfi6. J Bacteriol. 1996;178:1680–1690. doi: 10.1128/jb.178.6.1680-1690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent C, et al. Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J Bacteriol. 1999;181:3472–3477. doi: 10.1128/jb.181.11.3472-3477.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent C, et al. Relationship between exopolysaccharide production and protein-tyrosine phosphorylation in gram-negative bacteria. J Mol Biol. 2000;304:311–321. doi: 10.1006/jmbi.2000.4217. [DOI] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley AR, Zhou T, Borges-Walmsley MI, Rosen BP. Antimonite regulation of the ATPase activity of ArsA, the catalytic subunit of the arsenical pump. Biochem J. 2001;360:589–597. doi: 10.1042/0264-6021:3600589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- Wugeditsch T, et al. Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J Biol Chem. 2001;276:2361–2371. doi: 10.1074/jbc.M009092200. [DOI] [PubMed] [Google Scholar]
- Xia X, Lemey P. Assessing substitution saturation with DAMBE. In: Lemey P, Salemi P, Vandamme AM, editors. The phylogenetic handbook: a practical approach to DNA and protein phylogeny. Cambridge University Press. 2009:615–630. [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zagnitko O, Rodionova I, Osterman A, Godzik A. The FGGY carbohydrate kinase family: insights into the evolution of functional specificities. PLoS Comput Biol. 2011;7:e1002318. doi: 10.1371/journal.pcbi.1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Radaev S, Rosen BP, Gatti DL. Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. EMBO J. 2000;19:4838–4845. doi: 10.1093/emboj/19.17.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.