Abstract
The ortholog conjecture (OC), which is central to functional annotation of genomes, posits that orthologous genes are functionally more similar than paralogous genes at the same level of sequence divergence. However, a recent study challenged the OC by reporting a greater functional similarity, in terms of Gene Ontology (GO) annotations and expression profiles, among within-species paralogs compared with orthologs. These findings were taken to indicate that functional similarity of homologous genes is primarily determined by the cellular context of the genes, rather than evolutionary history. However, several subsequent studies suggest that GO annotations and microarray data could artificially inflate functional similarity between paralogs from the same organism. We sought to test the OC using approaches distinct from those used in previous studies. Analysis of a large RNAseq data set from multiple human and mouse tissues shows that expression similarity (correlations coefficients, rank‘s, or Z-scores) between orthologs is substantially greater than that for between-species paralogs with the same sequence divergence, in agreement with the OC and the results of recent detailed analyses. These findings are further corroborated by a fine-grain analysis in which expression profiles of orthologs and paralogs were compared separately for individual gene families. Expression profiles of within-species paralogs are more strongly correlated than profiles of orthologs but it is shown that this is caused by high background noise, that is, correlation between profiles of unrelated genes in the same organism. Z-scores and rank scores show a nonmonotonic dependence of expression profile similarity on sequence divergence. This complexity of gene expression evolution after duplication might be at least partially caused by selection for protein dosage rebalancing following gene duplication.
Keywords: duplicated genes, selection, neutral evolution, rebalancing dosage effect model, duplication–degeneration–complementation model, neofunctionalization model, subfunctionalization model
Introduction
The evolutionary history of genomes combines vertical descent with numerous gene duplications and lineage-specific losses. The genes that are related via vertical descent (speciation) are known as orthologs whereas genes that evolved via duplication in a certain lineage are called paralogs (Fitch 1970, 2000). These definitions are straightforward in principle but the actual evolutionary relationships between genes are often complex and involve not only one-to-one but also one-to-many and many-to-many correspondence due to the complicated combinations of lineage-specific duplication and gene loss (Koonin 2005; Kristensen et al. 2011; Gabaldon and Koonin 2013).
Beyond the reconstruction of evolutionary scenarios, robust identification of orthologs is of central importance for comparative and functional genomics due to a rarely stated but almost universally implied concept that recently has been denoted ortholog conjecture (OC) (Nehrt et al. 2011). The OC holds that orthologous genes perform equivalent functions in the respective organisms and accordingly, experimentally determined functions of a gene can be transferred to its experimentally uncharacterized orthologs in other species (certainly, taking into account the biological differences between the organisms involved) (Koonin 2005; Gabaldon and Koonin 2013). Thus, the OC effectively forms the foundation of all functional annotation of sequenced genomes given that experimental characterization of the functions of any sizable fraction of the genes in the numerous sequenced genomes remains unrealistic for the foreseeable future.
A key corollary of the OC is that orthologous genes are more functionally similar to each other that any of them is to its paralogs at the same level of sequence divergence, in the same or other species. The functional diversification of paralogs is a subject with a long, rather circuitous history of theoretical and experimental study (Lynch 2002, 2007; Lynch and Katju 2004; Conant and Wolfe 2008; Zhang 2013). With the increasing availability of genomic data, it became clear that numerous gene families have diverged in function through series of duplications, including many lineage-specific expansions identified in each of the sequenced genomes (Hughes 1994; Force et al. 1999; Kondrashov et al. 2002; Lespinet et al. 2002; Koonin 2005; Conant and Wolfe 2008; Innan and Kondrashov 2010; Zhang 2013). Gene duplications are considered to be a major evolutionary source of new protein functions (Ohno 1970; Hughes 1994; Force et al. 1999; Conant and Wolfe 2008; Innan and Kondrashov 2010; Zhang 2013).
The importance of appropriately designed tests to disambiguate models of gene evolution between orthologs and paralogs was emphasized by Studer and Robinson-Rechavi (2009) who suggested that functional changes between orthologs might be as common as between paralogs (the uniform model), and that more studies should be designed to test the impact of different models. Recently, an attempt has been undertaken to directly and systematically test the OC by quantifying the functional similarity between orthologs and paralogs (Nehrt et al. 2011). Two independent metrics, namely experiment-based annotations in the Gene Ontology (GO) database (Ashburner et al. 2000) and microarray gene expression data (Su et al. 2004), were used as proxies for gene function, that is, to compare the functional and expression similarities of orthologs and paralogs in human and mouse (Nehrt et al. 2011). Unexpectedly, this comparison has shown that at the same level of protein sequence divergence, orthologs and between-species paralogs are much less functionally similar than within-species paralogs (Nehrt et al. 2011). Furthermore, it has been shown that functional and expression similarity between orthologs is (virtually) independent of the protein sequence identity. These results appeared inconsistent with the OC which prompted Nehrt et al. (2011) to conclude that the primary determinant of the evolutionary rate of gene function and expression is the cellular and organismal context in which the genes act. This cellular context hypothesis could explain why within-species paralogs were found to be more similar in function and expression than between-species paralogs and orthologs (Nehrt et al. 2011).
The purported refutation of the OC by Nehrt et al. (2011) stimulated further reassessment of the functional relationships among orthologs and paralogs. Several studies have suggested that GO annotations should be used to test the OC with extreme caution (Altenhoff et al. 2012; Thomas et al. 2012) or even are unsuitable for this purpose (Chen and Zhang 2012). However, all the crudity and likely biases of the GO annotations notwithstanding, the emerging consensus appears to be that these annotations are generally compatible with the OC (Chen and Zhang 2012; Thomas et al. 2012) or with the uniform model (Altenhoff et al. 2012). In addition, Chen and Zhang (2012) analyzed a large RNAseq data set from multiple tissues and found that the similarity of expression profiles between orthologs is significantly higher than that between within-species paralogs, supporting the OC and conversely refuting the cellular context hypothesis (Chen and Zhang 2012). These results are consistent with an earlier analysis of the differences in the tissue-specific expression among between-species paralogs in human and mouse (Huerta-Cepas et al. 2011). The results of this work indicate that expression divergence is most pronounced among relatively young paralogs, that is, those originating from duplications that postdate the primary divergence of mammals but antedate the divergence of primates and rodents, consistent with the notion that tissue specificity evolves shortly after duplication (Huerta-Cepas et al. 2011).
We decided to reanalyze these controversial results and hypotheses using approaches as different as possible from those used before and further to attempt to reconcile them with the existing models of paralogous gene evolution. Our analysis of a large RNAseq data set for multiple tissues from human and mouse shows that rank and Z-score measures of the expression similarity indicate significantly greater similarity between orthologs than that for either between-species or within-species paralogs, in agreement with the OC and the results of Chen and Zhang (2012). In contrast, correlation coefficients between expression profiles of orthologs were found to be greater than those for between-species paralogs but lower than those for within-species paralogs. We conclude that the human/mouse gene expression data generally support the OC but deeper understanding of the evolution of expression also should invoke explicit models of gene evolution after duplication.
Materials and Methods
Human and Mouse Protein-Coding Genes
The protein sets for human and mouse were from the genome division of the National Center for Biotechnology Information (NCBI) /am/ftp-refseq/H_sapiens/mRNA_Prot/human.protein.faa/am/ftp-refseq/M_musculus/mRNA_Prot/mouse.protein.faa (last accessed March 28, 2014), gene names /am/ftp-gene/DATA/gene2refseq.gz (last accessed March 28, 2014). The ortholog–paralog cluster construction protocol included: first, all-against-all comparison of protein sequences from the analyzed human and genomes using the BlastP program, with masking of low sequence complexity regions using the SEG program. At the second step, we identified orthologs using symmetrical best hits (fig. 1). Paralogs were delineated using within-species and between-species BlastP hits (e value < 10–20) using the single linkage clustering procedure (the 50% identity score was used as a thresholds, supplementary table S1, Supplementary Material online). We chose this straightforward, simple approach for orthology identification over more sophisticated, phylogenetically based procedures (Altenhoff et al. 2012) given the results of benchmarking indicating high efficiency of similarity based methods, at least in pairwise comparisons of closely related organisms (Altenhoff and Dessimoz 2009; Kristensen et al. 2011). In addition, we deliberately sought to use a methodology distinct from that used in the previous analysis of the OC (Nehrt et al. 2011). Kimura distance for protein alignments as implemented in the EMBOSS package was used to estimate sequence divergence. Clusters of orthologs and paralog are available at ftp://ftp.ncbi.nlm.nih.gov/pub/managdav/paper_suppl/ortholog_conjecture/ (last accessed March 28, 2014).
Fig. 1.—

Construction of clusters of orthologous and paralogous genes from human and mouse. Solid lines show symmetrical best BlastP hits between human and mouse proteins (predicted orthologs). Dotted lines illustrate the identification of paralogs using single linkage clustering of within-species and between-species BlastP hits.
Expression of Protein-Coding Genes
Expression of the human and mouse genes was assessed by analysis of the available RNAseq data given the indications that RNAseq is superior to microarrays for comparing the expression levels of different genes or in different species (e.g., Chen and Zhang 2012). For human, the run files of the Illumina Human Body Map 2.0 project for adipose, adrenal, brain, breast, colon, heart, kidney, liver, lung, lymph node, ovary, prostate, skeletal muscle, testis, thyroid, and white blood cells, were downloaded from The NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra, last accessed March 28, 2014; Study ERP000546; runs ERR030888 to ERR030903). For mouse, RNAseq data of the ENCODE project for tissues: bone marrow, cerebellum, cortex, ES-Bruce4, heart, kidney, liver, lung, mouse embryonic fibroblast cells, and spleen, were downloaded from the UCSC Table Browser FTP site (ftp://hgdownload.cse.ucsc.edu/goldenPath/mm9/encodeDCC/wgEncodeLicrRNAseq/, last accessed March 28, 2014).
Prebuilt Bowtie indices of human and mouse, based on UCSC hg19 and mm9, were downloaded from Bowtie FTP site (ftp://ftp.cbcb.umd.edu/pub/data/bowtie_indexes/hg19.ebwt.zip and ftp://ftp.cbcb.umd.edu/pub/data/bowtie_indexes/mm9.ebwt.zip, respectively [last accessed March 28, 2014]). The reads were aligned with the cognate genomic sequences using TopHat (Trapnell et al. 2012).
The TopHat-generated alignments were analyzed using an ad hoc Python script that accepts alignments and genomic coordinates in SAM and BED formats, respectively, and uses the HTSeq Python package (http://www-huber.embl.de/users/anders/HTSeq, last accessed March 28, 2014) to calculate the number of aligned reads (counts). The RPKM values, that is, reads per kilobase of exon model per million mapped reads (Mortazavi et al. 2008), were calculated from the counts values for each of four tissues shared by human and mouse (heart, kidney, liver, and lung). These expression data are available at ftp://ftp.ncbi.nlm.nih.gov/pub/managdav/paper_suppl/ortholog_conjecture/ (last accessed March 28, 2014).
Comparison of Expression Profiles
Four measures of expression similarity were used for pairwise comparison of gene expression profiles: Pearson linear correlation coefficient, Kendall τ rank correlation coefficient, Z-score, and rank score. The linear correlation coefficient was used by Nehrt et al. (2011). We also used the Kendall τ correlation coefficient because this measure of expression similarity is expected to be more robust with respect to the small sample size (four tissues) compared with the linear correlation coefficient (Newson 2002). The Z-score and rank score measures were used by Chen and Zhang (2012). Briefly, log 2(RPKM) values were transformed into Z-scores within each tissue of each species forcing expression values within a tissue to have a mean of 0 and a standard deviation of 1 (Nehrt et al. 2011; Chen and Zhang 2012). The expression similarity between genes i and j was calculated as ES(i,j) = 1 − |Zi − Zj| which has the maximum value of 1 (Chen and Zhang 2012). To calculate the rank scores, the genes in each tissue of each species were ranked according to their expression level and the ranks were converted to percentile ranks. The expression similarity was estimated as for the Z-scores (Chen and Zhang 2012). All genes without RNAseq hits were excluded from the analysis.
Results and Discussion
Clusters of Orthologs and Paralogs
An all-against-all comparison of the human and mouse protein sequences using Blast, followed by single-linkage clustering, was used to construct clusters of orthologs and paralogs (fig. 1; see Materials and Methods for details). The number of orthologous gene pairs was similar to the numbers used in previous studies (e.g., 14,815 vs. 15,588 pairs of orthologs in the study of Chen and Zhang [2012]) (supplementary table S1, Supplementary Material online). To streamline the analysis and following the approach of Nehrt et al. (2011), we merged in-paralogs (i.e., paralogs produced by recent duplications represented only in human or only in mouse) and out-paralogs (i.e., paralogs resulting from older duplications and represented in both species) (Koonin 2005) in a single data set of paralogs; where appropriate, however, within-species and between-species comparisons were performed separately. We tried to implement approaches as different as possible from those used in previous studies, that is, an independent RNAseq data set, the simple, BlastP-based approach for the delineation of orthologs and paralog, and pooling all paralogs in a single data set for some of the statistical tests.
Expression Profiles of Orthologs and Paralogs Compared Using Different Measures of Expression Similarity and the Background Noise
Gene expression certainly does not equal gene function but similarity of the expression levels and the profiles of expression across different tissues is expected to reflect functional similarity between the respective gene products and so can be used to test the OC (Nehrt et al. 2011; Chen and Zhang 2012). Nehrt et al. (2011) used microarray data to analyze the correlations between the profiles of normalized gene expression across 25 tissues and found a generally greater similarity between within-species paralogs compared with human–mouse orthologs with the same amount of sequence divergence. Chen and Zhang (2012) analyzed RNAseq data instead of microarrays under the premise that RNAseq is the preferred method of expression quantification that is not prone to probe biases that are inherent in expression array data and has a wider dynamic range of detection of low-expressed genes (Wang et al. 2009; Xiong et al. 2010; Brawand et al. 2011). Chen and Zhang also argued against the use of the correlation between across-tissue expression profiles as a measure of expression similarity because it substantially underestimates the similarity between genes with uniform patterns of expression (house-keeping genes) (Pereira et al. 2009; Chen and Zhang 2012; Piasecka et al. 2012). Instead, Z-scores and ranking measures for each individual tissue were compared separately (Chen and Zhang 2012).
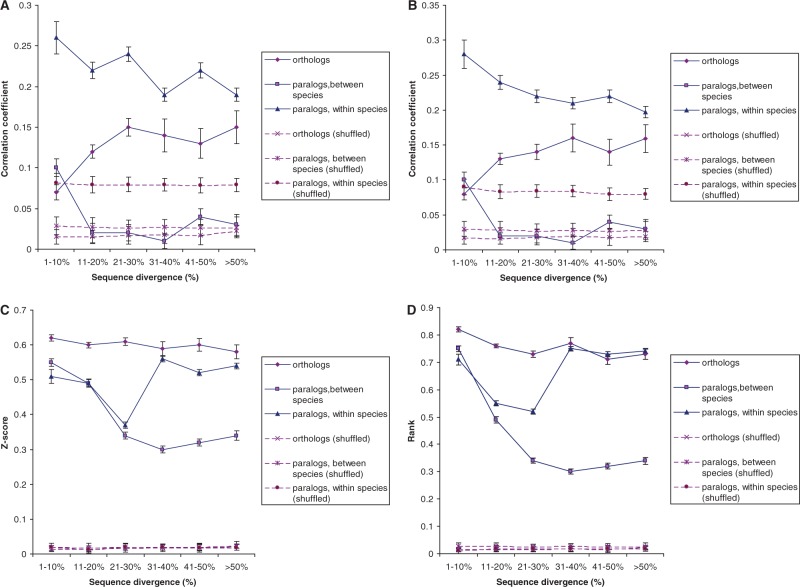
We first examined the potential biases introduced by the correlation measures when applied to the RNAseq data. Random samples of orthologs and paralogs (one randomly chosen orthologous gene pair and one pair of paralogs from each cluster of orthologs and paralogs) were generated, the expression profiles were randomly shuffled as proposed by Piasecka et al. (2012), and the procedure was repeated 1,000 times. Low but significant correlations (Kendall τ rank correlation coefficient or Pearson linear correlation coefficient of approximately 0.08) were detected among randomly shuffled within-species paralogs (fig. 2A and B). A weaker positive correlation (correlation coefficients of ∼0.04) was found among shuffled between-species paralogs and no significant correlation after shuffling the orthologs was observed (fig. 2A and B). The shuffled genes showed no appreciable dependency of the expression profile correlation on the sequence divergence (fig. 2A and B). By default, the significant positive correlation among the shuffled within-species paralogs should be taken as resulting from conditions of sample preparation and other experimental conditions that are identical (or closely similar) for all genes from the same species but differ between the species. However, it cannot be ruled out that the identical cellular context, as per the hypothesis of Nehrt et al. (2011), also results in some similarity between the expression profiles of within-species paralogs. In contrast to the correlation coefficients, no substantial background was observed for Z-scores and ranking scores (fig. 2C and D).
Fig. 2.—
Expression similarity of orthologous and paralogous genes (solid lines) and background noise for randomly shuffled profiles of orthologs and paralogs (dashed lines). (A) Kendall τ rank correlation coefficient, (B) Pearson linear correlation coefficient, (C) Z-score similarity averaged across four tissues, and (D) rank-based similarity averaged across four tissues.
Thus, the analysis of the randomized expression profiles indicates that Z-scores and ranking scores are more robust measures of the similarity between genes in terms of expression than correlation coefficients because the latter produce nonnegligible noise that appears to be biased toward certain categories of genes. This conclusion is consistent with recent studies showing that the comparison of across-tissue expression-profile similarities of different gene pairs using correlation can be problematic because it substantially underestimates the expression similarity between orthologous (or paralogous) genes with a conserved, uniform pattern of expression (Pereira et al. 2009; Chen and Zhang 2012; Piasecka et al. 2012). Nevertheless, we reasoned that different measures of expression similarity could reflect different salient features of gene expression, namely tissue-specificity in the case of the correlation coefficients, and relative abundance of individual mRNAs in the case of Z-scores and ranking scores. Therefore, here we employed each of these measures of expression similarity, with all the caveats that are associated with the use of correlation coefficients.
The plots of expression similarity measured using linear or rank correlation coefficients were qualitatively similar to the analogous plots reported by Nehrt et al. (2011) (fig. 2A and B) in that the strongest correlation was observed among within-species paralogs, followed by orthologs and then by between-species paralogs. This result suggested that the details of orthology and paralogy identification had no major effect on the results. For the between-species paralogs, significant expression similarity was observed only at low sequence divergence whereas at higher divergence, the correlation coefficient values were indistinguishable from the noise (fig. 2A and B). Although the correlation among within-species paralogs was well above the noise level for all values of sequence divergence, it also dropped with increasing divergence. In a sharp contrast, highly similar orthologs tend to produce low values of correlation coefficients (fig. 2A and B). This artifact has been noticed previously, the likely explanation being that many of the highly conserved orthologs are housekeeping genes that are broadly and (nearly) uniformly expressed across tissues which results in a severe underestimation of the expression similarity (Pereira et al. 2009; Chen and Zhang 2012; Piasecka et al. 2012). A dramatic difference was observed between the within-species and between-species measurements: the correlation among between-species paralogs was much lower than that between orthologs but within-species paralogs showed a greater correlation than orthologs, along with the greater background noise (fig. 2A and B). Thus, the OC appears to hold when the experimental conditions are comparable (between-species comparisons for orthologs or paralogs). However, the high positive correlation between paralogs for within-species comparisons cannot be explained by the background noise alone (discussed earlier) (fig. 2A and B) and is likely to reflect some features of evolution of duplicated genes.
For the Z-scores and ranking scores, our results (fig. 2C and D) agreed with those of Chen and Zhang (2012), with the greatest similarity consistently observed between orthologs, followed by within-species paralogs and then by between-species paralogs. However, the results obtained with Z-scores and ranking scores also showed an anomaly, namely the drop in the Z-scores and ranking scores for within-species paralog comparisons in the intermediate sequence divergence range and the seemingly paradoxical high scores in the high divergence range (fig. 2C and D). Collectively, these observations reveal the complexity of the dependencies between sequence and expression divergence, and further imply that different expression similarity measures capture distinct aspects of the functions of orthologous and paralogous genes. Later, we attempt to reconcile all these observations in the light of different models of the evolution of gene duplications.
Comparison of Expression Profiles of Orthologs and Paralogs within the Same Gene Family
The functional characteristics of orthologous genes and duplicated genes are not necessarily expected to be the same because paralogs are not a random subset of genes that have readily identifiable human–mouse orthologs (Kondrashov et al. 2002; Pal et al. 2003; Yang et al. 2003; Marland et al. 2004; He and Zhang, 2005, 2006; Prachumwat and Li, 2006; Amoutzias et al. 2010). The set of recently duplicated paralogs appears to be enriched in genes coding for proteins involved in different aspects of the organism’s interaction with the environment. In particular, a substantial fraction of these paralogs encode (predicted) membrane or secreted proteins (Kondrashov et al. 2002; Yang et al. 2003; Pal et al. 2003; Marland et al. 2004; He and Zhang 2005, 2006; Prachumwat and Li 2006; Amoutzias et al. 2010). To avoid this and other potential functional biases (He and Zhang 2006; Prachumwat and Li 2006; Vinogradov 2013; Amoutzias et al. 2010), we compared expression of paralogs and orthologs from the same gene clusters (gene families). For the purposes of this analysis, we merged all paralogous gene pairs into one set, to increase the power of statistical analysis, and considering that the OC does not explicitly differentiate between within-species paralogs and other paralogs. The distances between orthologs and between paralogs were required to be similar (no statistically significant difference at the 0.95 level according to the χ2 test). Altogether 14,242 pairs of orthologs and paralogs (N = 14,242) were analyzed; the numbers of within-species and between-species comparisons were approximately the same (7,020 and 7,222, respectively). For all measures of expression similarity, a greater similarity between orthologs than between paralogs was observed (fig. 3). Although the standard error of these analyses was much higher compared with the initial data sets (fig. 2) due to the smaller sample sizes, the overall expression similarity between orthologs was significantly greater (P < 0.0001 for all expression similarity measures according to the Student’s t-test). These findings indicate that the OC holds when comparisons between orthologs and paralogs are done with the family specificity of orthologs and paralogs taken into account.
Fig. 3.—
Expression similarity of orthologous and paralogous genes from the same clusters. The distance between each pair of orthologs and each pair of paralogs from the same cluster was chosen to be the same or similar (according to the χ2 test, the 0.95 level of significance). (A) Kendall τ rank correlation coefficient, (B) Pearson linear correlation coefficient, (C) Z-score similarity averaged across four tissues, and (D) rank-based similarity averaged across four tissues.
This test does not address another important potential bias in the data set, namely the substantial background noise observed for both correlation coefficients (fig. 2A and B). A similar level of the background noise was detected in the original microarray data of Nehrt et al. (2011) (supplementary fig. S1, Supplementary Material online). Thus, this background noise is a general property of expression data when a correlation coefficient (linear or τ) is used as a measure of the expression similarity. We examined this issue in greater detail by incorporating the background noise directly into the model. To this end, the following sampling procedure was used.
A subset of size L is randomly sampled from the set of pairs of orthologs and paralogs described earlier (the size of the set is N = 14,242; L << N). Various L values (L = 100, 300, and 500) were tested, and given that the results were similar (not shown), L = 300 was used for further analysis.
For the selected subset of size L, the expression profiles of orthologous gene pairs and paralogous gene pairs were separately, randomly shuffled 1,000 times.
Background correlation coefficients between expression profiles were calculated for each randomly shuffled version of the selected subset, and the mean value and variance of these coefficients were calculated for the 1,000 shuffled subsets. Mean and variance values for orthologs were compared with the respective values for paralogs using Student’s t-test (0.95 significance level). If no significant difference was found, that is, the background noise was approximately the same for orthologs and paralogs, the given subset was accepted for further analysis (balanced subsets).
Steps 1–3 were repeated 1,000 times, and the resulting balanced subsets (72 for the Pearson linear correlation coefficient and 139 for the Kendall τ correlation coefficient) were analyzed (table 1). Two approaches were used to compare the lists of balanced subsets. First, the number of cases when the mean expression similarity was greater for orthologs than for paralogs was compared with the number of reverse cases (table 1). Second, we tallied the cases when the mean expression similarity for orthologs was significantly greater (Student’s t-test, 0.95 significance level) than that for paralogs and vice versa (table 1). This analysis showed that the number of cases when the mean expression similarity was greater for orthologs than that for paralogs for both correlation coefficients was substantially and significantly greater than the number of reverse cases (table 1). Thus, the results of balanced subset analysis are fully compatible with the OC.
Table 1.
Analysis of Balanced Subsets of Orthologs and Paralogs (the Background Noise Is Approximately the Same for Orthologs and Paralogs According to the Student’s t-Test, the 0.95 Significance Level)
| Correlation Coefficient | Balanced Subsets | Expression Similarity |
P (Sign Test) | |
|---|---|---|---|---|
| Ortholog > Paralog | Ortholog < Paralog | |||
| Pearson linear correlation coefficient | All (72) | 55 | 17 | <0.001 |
| Significant only (16) | 13 | 3 | 0.011 | |
| Kendall τ rank correlation coefficient | All (139) | 137 | 2 | <0.001 |
| Significant only (38) | 38 | 0 | <0.001 | |
Note.—Expression similarity Ortholog > Paralog is the number of cases when the mean expression similarity was greater (All) or significantly greater (Significant only, Student’s t-test, 0.95 significance level) for orthologs than for paralogs. Expression similarity Ortholog < Paralog is the number of cases when the mean expression similarity was greater (All) or significantly greater (Significant only) for paralogs than for orthologs. The significance of the difference for each pair of these numbers was estimated using the sign test.
Discussion
In general, the results of gene expression analysis presented here as well as the results of Chen and Zhang (2012) are compatible with the OC. Even apart from additional tests that take into account gene families, all used measures of expression indicate that human–mouse orthologs are more similar to each other than between-species paralogs at the same level of sequence divergence (fig. 2). This is the most relevant comparison for testing the OC because two between-species measurements are compared. In addition, when expression similarity is measured using Z-scores or ranking scores, orthologs appear more similar than within-species paralogs as well (fig. 2C and D). Arguably, the comparison of expression profiles of orthologs and paralogs within gene families provide the strongest available argument in support of the OC because in this case only comparisons of functionally similar genes are taken into account.
In contrast, within-species paralogs show a significantly stronger correlation between expression profiles than orthologs at the same sequence distance, in agreement with the observations of Nehrt et al. (2011) that have been interpreted as a challenge to the OC (fig. 2A and B). The strong correlation between the expression profiles of within-species paralogs is largely caused by the high background noise that becomes apparent when correlations are measured among randomly shuffled expression profiles (fig. 2A and B; supplementary fig. S1, Supplementary Material online). When this noise is taken into account, a greater similarity between the expression profiles of orthologs compared with those of paralogs is observed (table 1), in accord with the OC.
The comparisons between expression profiles of orthologs and paralogs described here take account of multiple sources of bias and show that, once these biases are dealt with, profiles of orthologs are significantly more similar than profiles of paralogs, whether within or between species, in agreement with the OC. As shown here, the high similarity among within species paralogs reported by Nehrt et al. (2011) stems from the correlation between unrelated genes (background noise). Such correlation could be caused by the same experimental conditions that are used for sample preparation from the same organism, in contrast to unescapable differences between experimental procedures when different organisms are involved, by the same cellular context as suggested by Nehrt et al. (2011) or by a combination of both factors. In the absence of direct evidence, the appropriate null hypothesis is that experimental conditions are the main factor.
The dependency of the expression similarity on sequence divergence, regardless of the measure used, appears nonmonotonic for the within-species paralogs such that, somewhat paradoxically, the difference between orthologs and within-species paralogs nearly disappears at high divergence, in particular when Z-scores or ranking scores are used as measures of expression similarity (fig. 2C and D). This nonmonotonic dependency potentially can be explained by selection for balanced expression of recently duplicated genes in different tissues and environmental conditions. This scenario is consistent with several previous studies suggesting that rebalancing of expression after duplication, at least for some genes, could be subject to selection (Makova and Li 2003; Qian et al. 2010; Colbourne et al. 2011; Huerta-Cepas et al. 2011; Liu et al. 2011; Pegueroles et al. 2013). For example, Qian et al. (2010) have shown that yeast and mammalian genes often experienced a significant drop of the level of expression after duplication. Although the majority of the expression reduction is likely to be neutral, for some of the duplicated genes, it could be beneficial through the rebalanced gene dosage.
Taken together, the results of this work are fully compatible with the OC. However, the OC, all its importance notwithstanding, reflects only one aspect of gene evolution. The complete picture must integrate vertical descent encapsulated in the OC with the lineage-specific aspects of the evolution of paralogs.
Supplementary Material
Supplementary figure S1 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Koonin group members for useful discussions and Jean and Danielle Thierry-Mieg for helpful advice on RNAseq data analysis. This work was supported by the Intramural Research Program of the National Library of Medicine at National Institutes of Health (US Department Health and Human Services).
Literature Cited
- Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput Biol. 2009;5:e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenhoff AM, Studer RA, Robinson-Rechavi M, Dessimoz C. Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs. PLoS Comput Biol. 2012;8:e1002514. doi: 10.1371/journal.pcbi.1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias GD, et al. Posttranslational regulation impacts the fate of duplicated genes. Proc Natl Acad Sci U S A. 2010;107:2967–2971. doi: 10.1073/pnas.0911603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang J. The ortholog conjecture is untestable by the current gene ontology but is supported by RNA sequencing data. PLoS Comput Biol. 2012;8:e1002784. doi: 10.1371/journal.pcbi.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe HK. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–106. [PubMed] [Google Scholar]
- Fitch WM. Homology a personal view on some of the problems. Trends Genet. 2000;16:227–231. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T, Koonin VE. Functional and evolutionary implications of gene orthology. Nat Rev Genet. 2013;14:360–366. doi: 10.1038/nrg3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J. Gene complexity and gene duplicability. Curr Biol. 2005;15:1016–1021. doi: 10.1016/j.cub.2005.04.035. [DOI] [PubMed] [Google Scholar]
- He X, Zhang J. Higher duplicability of less important genes in yeast genomes. Mol Biol Evol. 2006;23:144–151. doi: 10.1093/molbev/msj015. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Dopazo J, Huynen AM, Gabaldon T. Evidence for short-time divergence and long-time conservation of tissue-specific expression after gene duplication. Brief Bioinform. 2011;12:442–448. doi: 10.1093/bib/bbr022. [DOI] [PubMed] [Google Scholar]
- Hughes AL. The evolution of functionally novel proteins after gene duplication. Proc Biol Sci. 1994;256:119–124. doi: 10.1098/rspb.1994.0058. [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Rogozin IB, Wolf IY, Koonin VE. Selection in the evolution of gene duplications. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-research0008. RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- Kristensen DM, Wolf YI, Mushegian RA, Koonin VE. Computational methods for Gene Orthology inference. Brief Bioinform. 2011;12:379–391. doi: 10.1093/bib/bbr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lespinet O, Wolf YI, Koonin VE, Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–1059. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Baute JG, Adams KL. Organ and cell type-specific complementary expression patterns and regulatory neofunctionalization between duplicated genes in Arabidopsis thaliana. Genome Biol Evol. 2011;3:1419–1436. doi: 10.1093/gbe/evr114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Genomics. Gene duplication and evolution. Science. 2002;297:945–947. doi: 10.1126/science.1075472. [DOI] [PubMed] [Google Scholar]
- Lynch M. The evolution of genetic networks by non-adaptive processes. Nat Rev Genet. 2007;8:803–813. doi: 10.1038/nrg2192. [DOI] [PubMed] [Google Scholar]
- Lynch M, Katju V. The altered evolutionary trajectories of gene duplicates. Trends Genet. 2004;20:544–549. doi: 10.1016/j.tig.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Makova KD, Li HW. Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res. 2003;13:1638–1645. doi: 10.1101/gr.1133803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marland E, Prachumwat A, Maltsev N, Gu Z, Li HW. Higher gene duplicabilities for metabolic proteins than for nonmetabolic proteins in yeast and E. coli. J Mol Evol. 2004;59:806–814. doi: 10.1007/s00239-004-0068-x. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nehrt NL, Clark WT, Radivojac P, Hahn WM. Testing the ortholog conjecture with comparative functional genomic data from mammals. PLoS Comput Biol. 2011;7:e1002073. doi: 10.1371/journal.pcbi.1002073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newson R. Parameters behind “nonparametric” statistics: Kendall’s tau, Somers’ D and median differences. Stata J. 2002;2:45–63. [Google Scholar]
- Ohno S. Evolution by gene duplication. Berlin-Heidelberg-New York: Springer-Verlag; 1970. [Google Scholar]
- Pal C, Papp B, Hurst DL. Genomic function: rate of evolution and gene dispensability. Nature. 2003;421:496–497. doi: 10.1038/421496b. discussion 497–498. [DOI] [PubMed] [Google Scholar]
- Pegueroles C, Laurie S, Alba MM. Accelerated evolution after gene duplication: a time-dependent process affecting just one copy. Mol Biol Evol. 2013;30:1830–1842. doi: 10.1093/molbev/mst083. [DOI] [PubMed] [Google Scholar]
- Pereira V, Waxman D, Eyre-Walker A. A problem with the correlation coefficient as a measure of gene expression divergence. Genetics. 2009;183:1597–1600. doi: 10.1534/genetics.109.110247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecka B, Robinson-Rechavi M, Bergmann S. Correcting for the bias due to expression specificity improves the estimation of constrained evolution of expression between mouse and human. Bioinformatics. 2012;28:1865–1872. doi: 10.1093/bioinformatics/bts266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prachumwat A, Li HW. Protein function, connectivity, and duplicability in yeast. Mol Biol Evol. 2006;23:30–39. doi: 10.1093/molbev/msi249. [DOI] [PubMed] [Google Scholar]
- Qian W, Liao BY, Chang YA, Zhang J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010;26:425–430. doi: 10.1016/j.tig.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer RA, Robinson-Rechavi M. How confident can we be that orthologs are similar, but paralogs differ? Trends Genet. 2009;25:210–216. doi: 10.1016/j.tig.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Wood V, Mungall CJ, Lewis ES, Blake AJ. On the use of gene ontology annotations to assess functional similarity among orthologs and paralogs: a short report. PLoS Comput Biol. 2012;8:e1002386. doi: 10.1371/journal.pcbi.1002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinogradov AE. Density peaks of paralog pairs in human and mouse genomes. Gene. 2013;527:55–61. doi: 10.1016/j.gene.2013.05.039. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, et al. RNA sequencing shows no dosage compensation of the active X-chromosome. Nat Genet. 2010;42:1043–1047. doi: 10.1038/ng.711. [DOI] [PubMed] [Google Scholar]
- Yang J, Lusk R, Li HW. Organismal complexity, protein complexity, and gene duplicability. Proc Natl Acad Sci U S A. 2003;100:15661–15665. doi: 10.1073/pnas.2536672100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Gene duplication. In: Losos J, editor. 2013. Princeton guide to evolution. Princeton (NJ): Princeton University Press. p. 397–405. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.