Abstract
Background
Japanese encephalitis virus (JEV) is classified into the genus Flavivirus in the family Flaviviridae. JEV can cause febrile illness and encephalitis mainly in humans and horses, and occasionally in cattle.
Case presentation
In late September 2010, a 114-month-old cow showed neurological symptoms similar to the symptoms observed in previous bovine cases of Japanese encephalitis (JE); therefore, we conducted virological and pathological tests on the cow. As a result, JEV was isolated from the cerebrum of the affected cow. We determined the complete genome sequence of the JEV isolate, which we named JEV/Bo/Aichi/1/2010, including the envelope (E) gene region and 3’ untranslated region (3’UTR). Our phylogenetic analyses of the E region and complete genome showed that the isolate belongs to JEV genotype 1 (G1). The isolate, JEV/Bo/Aichi/1/2010, was most closely related to several JEV G1 isolates in Toyama Prefecture, Japan in 2007–2009 by the phylogenetic analysis of the E region. In addition, the nucleotide alignment revealed that the deletion in the 3’UTR was the same between JEV/Bo/Aichi/1/2010 and several other JEV G1 isolates identified in Toyama Prefecture in 2008–2009. A hemagglutination inhibition (HI) test was conducted for the detection of anti-JEV antibodies in the affected cow, and the test detected 2-mercaptoethanol (2-ME)-sensitive HI antibodies against JEV in the serum of the affected cow. The histopathological investigation revealed nonsuppurative encephalomyelitis in the affected cow, and the immunohistochemical assay detected JEV antigen in the cerebrum.
Conclusion
We diagnosed the case as JE of a cow based on the findings of nonsuppurative encephalomyelitis observed in the central nervous system, JEV antigen detected in the cerebrum, JEV isolated from the cerebrum, and 2-ME-sensitive HI antibodies against JEV detected in the serum. This is the first reported case of JE in a cow over 24 months old.
Keywords: Japanese encephalitis, Cow, Arthropod-borne virus, Viral encephalomyelitis
Background
Japanese encephalitis virus (JEV) is a member of the genus Flavivirus in the family Flaviviridae, and it is distributed throughout eastern, southern and southeastern Asia, Papua New Guinea, and the Torres Strait of northern Australia [1]. JEV is transmitted by mosquitoes and amplified in pigs and wading birds in nature. It can also infect humans, horses, cattle and many other vertebrates [2,3]. Although the titers of viremia in JEV-infected humans, horses and cattle seem not to be high enough for infecting mosquitoes [2,4,5], JEV can cause febrile illness and encephalitis, mainly in humans and horses [1-3,6], and occasionally in cattle [7-9]. A case of nonsuppurative encephalomyelitis occurred recently in a 141-day-old calf in Miyazaki Prefecture, Japan, and JEV genotype 1 (G1) was isolated from the cerebrum of the affected calf, suggesting that JEV G1 can cause neurological disorders in cattle [10]. In late summer of the following year, a 114-month-old cow in Aichi Prefecture, Japan showed clinical symptoms similar to those of the calf; we therefore conducted virological and pathological tests on the cow.
Case presentation
Sample collection
In late September 2010, a 114-month-old Japanese Black cow on a beef farm in Aichi Prefecture, Japan showed decreased appetite and fever. Four days after the onset of these symptoms, the cow showed astasia. Despite palliative treatments, no improvement was seen, and the cow then became recumbent and was euthanized. A serum sample was collected after the onset of the initial symptoms, and organs were collected at necropsy for virological and pathological investigations including the brain, spinal cord, heart, lung, liver, kidney, spleen, rumen, reticulum, omasum, abomasum, intestines, right axillary lymph node, and right popliteal lymph node. Serum samples were collected from the other 14 cows on the farm for anti-JEV hemagglutination inhibition (HI) antibody.
Virological, serological and pathological investigations
Homogenate was prepared from the cerebrum of the affected cow, and RNA was extracted from the homogenate and the cow’s serum sample with a QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA, USA). The RNA was tested for the presence of JEV by reverse transcription-polymerase chain reaction (RT-PCR) [11]. For virus isolation, the homogenate was inoculated onto Vero cells [6], and the isolated virus was subjected to RT-PCR and direct sequencing using both primer sets targeting the envelope (E) gene region and the 3’ untranslated region (3’UTR), respectively [12]. The complete genome sequence was determined by a direct sequencing using the products of RT-PCR, 5’RACE, and 3’RACE as sequencing templates [10]. All of the primers used in the present study are shown in Table 1.
Table 1.
Oligonucleotide primers used for the cDNA amplification and sequencing of JEV/Bo/Aichi/1/2010
| Primer | Sequence (5'-3') | Position | Purpose | References |
|---|---|---|---|---|
| JF1 |
TTACTCAGCGCAAGTAGGAGCGTCTCAAG |
1440-1468 |
RT-PCR (E region) |
Yeh et al. (2010) |
| JF2 |
TTACTCAGCGCAAGTTGGGGCGTC |
1440-1463 |
|
|
| JR1 |
ATGCCGTGCTTGAGGGGGACG |
1675-1655 |
|
|
| JR2 |
CAYGCTGTGCTCGAAGGGGACG |
1675-1654 |
|
|
| JE955f |
TGYTGGTCGCTCCGGCTTA |
956-974 |
RT-PCR (E region) |
Nerome et al. (2007) |
| JE2536r |
AAGATGCCACTTCCACAYCTC |
2537-2517 |
|
|
| JE10141f |
TGGATTGAAGAAAATGAATGGATG |
10141-10164 |
RT-PCR (3'UTR) |
Nerome et al. (2007) |
| JE10965r |
AGATCCTGTGTTCTTCCTCTC |
10965-10945 |
|
|
| JEV9-2193F |
ATCTGTGTGAACTTCTTGGC |
9-28 |
RT-PCR and sequencing |
Katayama et al. (2013) |
| JEV9-2193R |
TTTACCCAGCGTGCTTCCAGC |
2193-2173 |
|
|
| JEV1850-3845F |
TGGACAAACTGGCTCTGAAGGG |
1850-1871 |
|
|
| JEV1850-3845R |
TTCTCTTGGTTCGTCCATCTCG |
3845-3824 |
|
|
| JEV3655-5606F |
CTACTTGTGCTGATGCTTGG |
3655-3674 |
|
|
| JEV3655-5606R |
ATTGGGGCATTTGAGTC |
5606-5590 |
|
|
| JEV5409-7958F |
CCATAGACTAATGTCACCAAAC |
5409-5430 |
|
|
| JEV5409-7958R |
AGAGTTGCTGCGTAGTAG |
7958-7941 |
|
|
| JEV7543-9421F |
GACAATGGAGCCAGTGC |
7543-7559 |
|
|
| JEV7543-9421R |
CCTTGACCACTTTGTGCCTG |
9421-9402 |
|
|
| JEV9234-10965F |
CATTCTCCGTGACATAGCAGG |
9234-9254 |
|
|
| JEV9234-10965R |
AGATCCTGTGTTCTTCCTCACC |
10965-10944 |
|
|
| JEV1108R |
GGACATCTAGTGTTGGTTTG |
1108-1089 |
5'RACE and sequencing |
Katayama et al. (2013) |
| JEV1030R |
CTCCACTGGCTCCTTCTATG |
1030-1011 |
|
|
| JEV157R |
ATCCGCGTTTCAGCATATTGATGG |
157-134 |
|
|
| JEV9568F |
GTCATCGGACCACAACACTTG |
9568-9588 |
3'RACE and sequencing |
Katayama et al. (2013) |
| JEV10757F |
CCGTGGAAACAACATTATGC |
10757-10776 |
|
|
| JEV3305R |
TCAAGGACAATGCCGTTCTC |
3305-3286 |
Sequencing |
Katayama et al. (2013) |
| JEV5124R |
CTCTTGACGGTCGCCTTGC |
5124-5106 |
|
|
| JEV7367R |
TCCACGACGGCATTCTTCATTAT |
7367-7345 |
|
|
| JEV7819F |
AACATAGTGGGAGGACATC |
7819-7837 |
|
|
| JEV8993R |
GCATTGACCATCTCCCAGAAC |
8993-8973 |
|
|
| JEV10306R |
CCCTCACTTGGTTTATTGCCG |
10306-10286 |
|
|
| JEV970R |
CTGGAGCGACCAATAGCAAG |
970-951 |
Sequencing |
This study |
| JEV2240F |
GTGACACAGCCTGGGACTTC |
2240-2259 |
|
|
| JEV3906F |
AGTCCACGGAATCCTGAATG |
3906-3925 |
|
|
| JEV6041F |
CCACCAGTGAAGATGACAGCAAC |
6041-6063 |
|
|
| JEV7539R |
CAAGGTGAGTGTGGCTGCCGTC |
7539-7518 |
|
|
| JEV8090F |
AGCCTAGTGACACCCTGTTC |
8090-8109 |
|
|
| JEV9249R | TATGTCACGGAGGATGTATC | 9249-9230 |
To analyze the phylogenetic relationships among virus isolates, we aligned sequences using the Clustal W program [13], and we constructed phylogenetic trees with MEGA5 using the neighbor-joining method. The reliability of the branching orders was evaluated by the bootstrap test (n = 1,000) [14]. The nucleotide sequence data reported in this study were deposited in the DNA Data Bank of Japan (DDBJ) with the accession numbers AB797319 (E region), AB797320 (3’UTR) and AB853904 (complete genome). We also tested the affected cow for bovine spongiform encephalopathy (BSE) by enzyme-linked immunosorbent assay (ELISA) (NippiBL, Nippi, Tokyo).
A hemagglutination inhibition (HI) test was conducted to determine the anti-JEV HI antibody titers of the affected cow and all other cows at the farm [6]. Prior to the test, all of the serum samples were treated with acetone and then mixed with goose red blood cells. The HI antibody titers were determined with the use of JEV antigen prepared from the Nakayama strain (Kyoto Biken, Kyoto, Japan). JEV-positive swine serum was used as a positive control. We also treated the serum sample of the affected cow with 2-mercaptoethanol (2-ME) and then subjected it to the HI test, to test for anti-JEV immunoglobulin M antibodies.
We examined the affected cow for gross and histological lesions. The collected organs were fixed in 10% neutral buffered formalin. Fixed organs were then embedded in paraffin wax, sectioned at 3 μm, and stained with hematoxylin and eosin. An immunohistochemical (IHC) assay was performed using a Histofine Simple Stain MAX-PO (MULTI) Kit (Nichirei, Tokyo). Anti-JEV (AS-6 strain [15]) polyclonal rabbit serum was used as the primary antibody for the detection of JEV antigen.
Isolation and genetic characterization of JEV
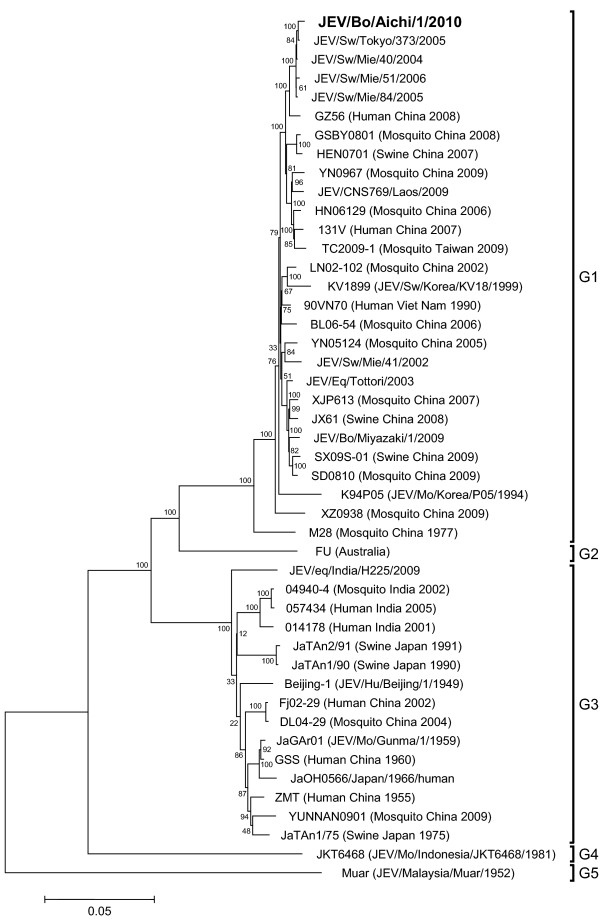
The cerebrum of the affected cow tested positive for JEV, but the serum tested negative. We then subjected the cerebrum to virus isolation, and JEV was isolated after the second passage. From the JEV isolate, which we named JEV/Bo/Aichi/1/2010, cDNAs containing the E region and the 3’UTR were successfully amplified and sequenced. Our phylogenetic analysis of the E region showed that the isolate clustered with other isolates belonging to JEV G1 (Figure 1), and it was most closely related to several isolates from Toyama Prefecture, Japan in 2007–2009 [16], including JEV/Sw/Toyama/08253c/2008, JEV/Sw/Toyama/08253c/2008, JEV/Sw/Toyama/07296c/2007 and JEV/Mo/Toyama/3140c/2009.
Figure 1.
Phylogenetic profile showing the relationships among JEV isolates based on a comparison of the E gene. The percentage bootstrap values calculated from 1,000 replications are indicated around the internal nodes. The scale represents 0.05% sequence divergence. G1–G5: genotypes 1–5.
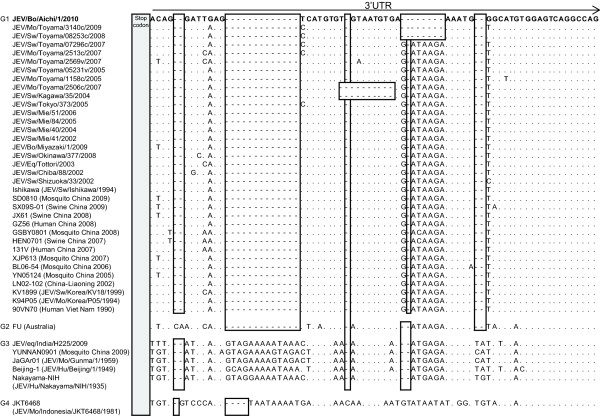
The newly identified JEV isolate, JEV/Bo/Aichi/1/2010, was found to contain 10,958 nucleotides, and our phylogenetic analysis of the complete genome also showed that the isolate was closely related with other isolates of JEV G1 (Figure 2). The nucleotide sequence of the 3’UTR was then compared between JEV/Bo/Aichi/1/2010 and other JEV isolates belonging to genotypes 1 - 5 available in GenBank. The nucleotide alignment revealed that JEV/Bo/Aichi/1/2010 has the same deletion in the 3’UTR as several other isolates of JEV G1 in Toyama Prefecture, such as JEV/Mo/Toyama/3140c/2009 and JEV/Mo/Toyama/08253c/2008, although their sequences were not completely identical (Figure 3). The affected cow was negative for BSE by the ELISA.
Figure 2.
Phylogenetic profile showing the relationships among JEV isolates based on a comparison of the complete genome. The percentage bootstrap values calculated from 1,000 replications are indicated around the internal nodes. The scale represents 0.05% sequence divergence. G1–G5: genotypes 1–5.
Figure 3.
Nucleotide sequence alignment of the 5’ distal region of the 3′ UTRs. The nucleotide sequences of the 3′ UTR were compared between the JEV isolate characterized in this study (JEV/Bo/Aichi/1/2010) and other isolates of genotypes 1–5 collected elsewhere, available from GenBank. The sequence of JEV/Bo/Aichi/1/2010 is shown in bold, and only differences from the sequence are indicated for other JEV isolates. Sequence gaps were introduced to optimize the alignment, and deletions are indicated by hyphens and open boxes.
JEV seroprevalence
We conducted HI tests for the affected cow and the other healthy cows on the farm to determine the serological prevalence of JEV infection. All 15 of the cows tested positive for anti-JEV HI antibodies, and the titers were 1:160 in the affected cow and ranged from 1:160 to 1:2560 in the other cows (Table 2). In addition, the affected cow had 2-ME sensitive HI antibody, and the titer was 1:20.
Table 2.
Anti-JEV HI titers of serum samples collected from all the cows in the farm, located in Aichi Prefecture, Japan
| Cow no. | Age (month) | HI titer |
|---|---|---|
| 1 |
29 |
1:160 |
| 2 |
33 |
1:160 |
| 3 |
35 |
1:640 |
| 4 |
58 |
1:320 |
| 5 |
68 |
1:640 |
| 6 |
80 |
1:640 |
| 7 |
81 |
1:640 |
| 8 |
90 |
1:640 |
| 9 |
100 |
1:1280 |
| 10* |
114 |
1:160 |
| 11 |
115 |
1:640 |
| 12 |
143 |
1:640 |
| 13 |
147 |
1:640 |
| 14 |
148 |
1:320 |
| 15 | 188 | 1:2560 |
*The affected cow.
Pathological findings
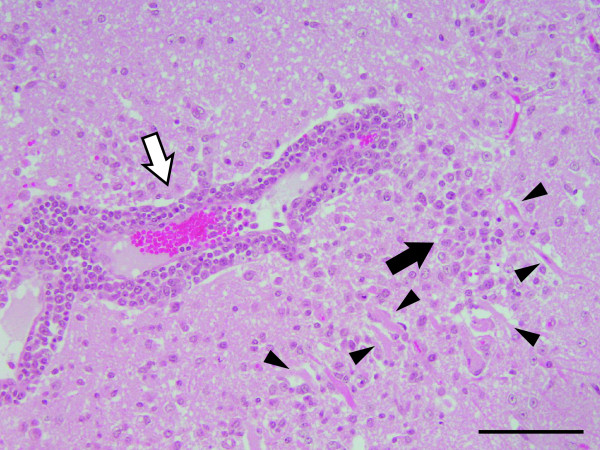
The gross examination of the affected cow revealed hemorrhage and congestion of the brain, pulmonary emphysema, rough surface of the kidney, swelling of the right axillary and right popliteal lymph nodes, and a wound and muscle necrosis of the right upper limb. The histological examination revealed typical nonsuppurative encephalomyelitis. Perivascular infiltration of lymphocytes was widely distributed throughout the cerebrum (Figure 4), and neuronal necrosis, neuronophagia and glial nodules were observed in the cerebral cortex (Figures 4 and 5).
Figure 4.
Neuronal necrosis, a glial nodule and perivascular infiltration of lymphocytes in the cerebral cortex of the affected cow. Arrowheads point out the neuronal necrosis. Black arrows: the glial nodules; white arrows: the perivascular infiltration of lymphocytes. Hematoxylin and eosin (H&E) stain. Bar = 100 μm.
Figure 5.
Neuronophagia in the cerebral cortex of the affected cow. The arrow indicates the neuronophagia. H&E stain. Bar = 100 μm.
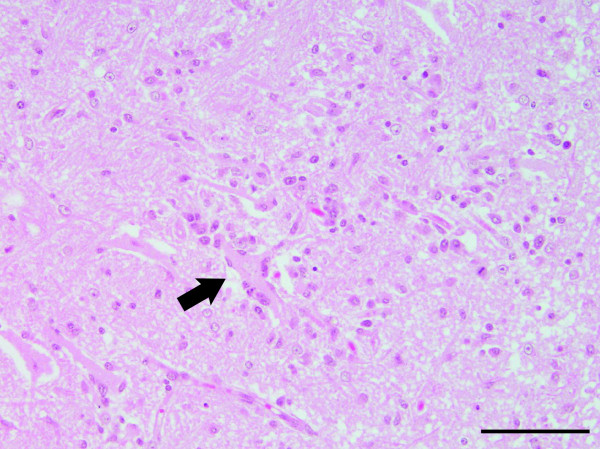
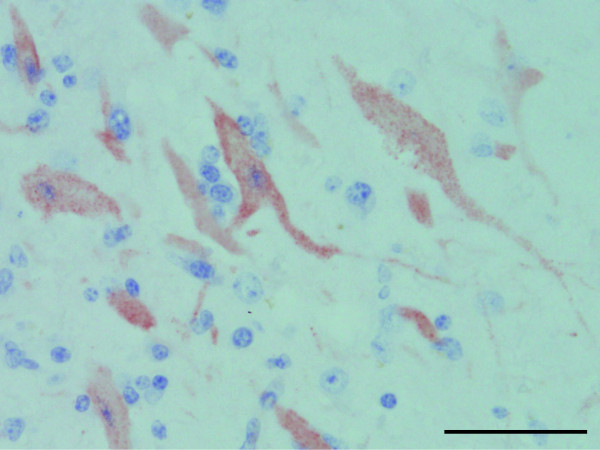
These histological lesions were also observed in the medulla oblongata and spinal cord, and neuronal necrosis was observed in the diencephalon. The lesions were most severe in the hippocampus and were also severe in the frontal lobe among the affected sites in the central nervous system (CNS). Histological findings in other organs included Sarcocystis spp. infection in the myocardium, mild abomasal ulceration, mild nonsuppurative interstitial nephritis, severe diffuse hemosiderosis in the spleen, and infiltration of neutrophils in the right axillary and right popliteal lymph nodes. The IHC assay detected JEV antigens in the cerebrum. JEV-positive granules were observed in the cytoplasm of neurons and in the nerve fibers (Figure 6).
Figure 6.
The IHC assay results. Detection of JEV antigen in the cerebrum of the affected cow. JEV-positive granules (brown color) were observed in the cytoplasm of neurons and in the nerve fibers. Bar = 50 μm.
Discussion
We diagnosed the case as JE of a cow based on the following findings: nonsuppurative encephalomyelitis observed in the CNS, JEV antigen detected in the cerebrum, JEV isolated from the cerebrum and 2-ME sensitive HI antibodies against JEV in the serum. The histopathological findings observed in the CNS of the affected cow were similar to those observed in the 141-day-old calf in the previous JE case [10], including perivascular infiltration of lymphocytes, neuronal necrosis, neuronophagia and glial nodules.
However, two other lesions observed in the present case were not observed in the 141-day-old calf but were seen in other bovine JE cases: pulmonary emphysema was observed in a 24-month-old milking cow in 1950, and congestion of the brain was seen in an 18-month-old Japanese Black cow in 1996 [7,9]. It is possible that the pulmonary emphysema—as well as the mild abomasal ulceration and mild nonsuppurative interstitial nephritis—observed in the present study influenced the immune status of the affected cow, and contributed to the invasion of JEV into the CNS. The congestion of the brain may have been caused by the onset of encephalomyelitis, since similar gross lesions were observed in some human JE cases [17,18].
The affected cow’s wound and muscle necrosis of the right upper limb seemed to have resulted from the onset of astasia and recumbency, and the infiltration of neutrophils in the right axillary and right popliteal lymph nodes may have been immune responses to wound infections. However, it is unclear whether the other pathological findings in the affected cow had relevance to the onset of JE, such as the severe diffuse hemosiderosis in the spleen and the infection of Sarcocystis spp. in the myocardium, although some species of Sarcocystis protozoan parasites are highly pathogenic to ruminants [19].
Our genetic analyses revealed that JEV/Bo/Aichi/1/2010 was most closely related to several other JEV G1 isolates in Toyama Prefecture, which is located approx. 100 miles away from Aichi Prefecture. The JEV isolate JEV/Bo/Aichi/1/2010 was most closely related to several JEV G1 isolates in Toyama Prefecture identified in 2007–2009 by phylogenetic analysis of the E region [16]. In addition, the deletion patterns in the 3’UTR were the same between JEV/Bo/Aichi/1/2010 and some of the JEV G1 isolates found in Toyama Prefecture in 2008–2009.
There were some differences in the phylogenetic characteristics between JEV/Bo/Aichi/1/2010 and another JEV isolate from an affected calf in Miyazaki Prefecture [10], which is located approx. 400 miles away from Aichi Prefecture. Our present findings suggest that JEV/Bo/Aichi/1/2010 may be a part of the JEVs overwintering in the Chubu region of Japan [16], a region that consists of Aichi, Toyama and several other prefectures.
In the bovine JE cases reported to date, the ages of onset were 141 days, 6 months, 18 months and 24 months [7-10]; the 114-month-old affected cow in the present study was the oldest cow affected by JEV infection. The advanced age may have influenced the immune status of the affected cow. In addition, heat stress may have triggered the onset of JE in the extraordinarily hot summer of 2011. However, none of the other cows (including a 188-month-old cow on the same farm) showed any neurological signs, even though the HI test provided evidence of JEV infection in the herd. Therefore, advanced age seems not to be an important factor in the onset after JEV infection in cattle.
Conclusions
A 114-month-old cow showed several clinical signs including decreased appetite, fever, astasia and recumbency in the summer of 2010 in Aichi Prefecture, Japan. The histopathological investigation revealed nonsuppurative encephalomyelitis in the affected cow, and JEV antigen was observed in the lesions. We isolated JEV from the cerebrum, and we thus diagnosed the case as JE of a cow. Our genetic analyses revealed that the isolate was closely related to other JEV G1 isolates from pigs and mosquitoes in Toyama Prefecture, which is approx. 100 miles away from Aichi Prefecture. Although our data clearly indicated that the causative agent was JEV in the cow’s case, the causes of the onset are still unclear, since JEV infection usually results in an asymptomatic infection in cattle. Further study is needed to clarify the actual causes of the onset after JEV infection in cattle.
Additional information
Alignments and phylogenetic trees created in the present study are available from the Dryad Digital Repository (https://datadryad.org/resource/doi:10.5061/dryad.s0c34).
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
NK and SS performed the pathological and virological investigations, respectively. NK, SS and NS made the diagnosis of the cow’s case. TK, TY, MY and HS performed the genetic and phylogenetic analyses of the JEV isolate. NK and HS wrote the manuscript, and all authors read and approved the final manuscript.
Contributor Information
Naomi Kako, Email: naomi_kako@pref.aichi.lg.jp.
Seiji Suzuki, Email: seiji_4_suzuki@pref.aichi.lg.jp.
Norie Sugie, Email: norie_sugie@pref.aichi.lg.jp.
Tomoko Kato, Email: katoto@affrc.go.jp.
Tohru Yanase, Email: tyanase@affrc.go.jp.
Makoto Yamakawa, Email: yamato@affrc.go.jp.
Hiroaki Shirafuji, Email: shirah@affrc.go.jp.
Acknowledgements
We thank Dr. Masanori Kubo (National Institute of Animal Health, National Agriculture and Food Research Organization) for providing technical advice and for conducting the IHC assay.
References
- Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10(12 Suppl):S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- Gubler DJ, Kuno G, Markoff L. In: Fields VIROLOGY. Kinipe DM, Howley PM, editor. Philadelphia: Lippin Cott Williams & Wilkins, a Wolters Kluwer Business; 2007. Flaviviruses; pp. 1153–1252. [Google Scholar]
- Endy TP, Nisalak A. Japanese encephalitis virus: ecology and epidemiology. Curr Top Microbiol Immunol. 2002;267:11–48. doi: 10.1007/978-3-642-59403-8_2. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Mochizuki H, Sugawa Y, Okazaki K, Matumoto M. Studies on Japanese encephalitis of cattle - 2. Experimental infection of calves with virus of Japanese encephalitis. Nat Inst Anim Hlth Quart. 1951;23:119–128. [Google Scholar]
- Ilkal MA, Dhanda V, Rao BU, George S, Mishra AC, Prasanna Y, Gopalkrishna S, Pavri KM. Absence of viraemia in cattle after experimental infection with Japanese encephalitis virus. Trans R Soc Trop Med Hyg. 1988;82:628–631. doi: 10.1016/0035-9203(88)90536-6. [DOI] [PubMed] [Google Scholar]
- OIE. OIE Terrestrial Manual 2010. Paris: Office International des Epizooties; 2010. Japanese encephaliis. [Google Scholar]
- Katayama K, Ikeda A, Saika K, Ishizaka M, Onodera M, Ito T. A case of Japanese encephalitis in a black Japanese Heifer. J Jpn Vet Med Assoc. 2000;53:293–296. [Google Scholar]
- Yamamoto S, Tsubahara H, Yoshida T, Harada K. First observation on case of Japanese B encephalitis of cattle. Bull Nat Inst Anim Hlth. 1949;22:197–203. [Google Scholar]
- Shimizu T, Mochizuki H, Sugawa Y, Okazaki K, Matumoto M. Studies on Japanese encephalitis of cattle - 1. Bovine encephalitis caused by natural infection with Japanese encephalitis virus. Nat Inst Anim Hlth Quart. 1951;23:111–118. [Google Scholar]
- Katayama T, Saito S, Horiuchi S, Maruta T, Kato T, Yanase T, Yamakawa M, Shirafuji H. Nonsuppurative encephalomyelitis in a calf in Japan and isolation of Japanese encephalitis virus genotype 1 from the affected calf. J Clin Microbiol. 2013;51:3448–3453. doi: 10.1128/JCM.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh JY, Lee JH, Seo HJ, Park JY, Moon JS, Cho IS, Lee JB, Park SY, Song CS, Choi IS. Fast duplex one-step reverse transcriptase PCR for rapid differential detection of West Nile and Japanese encephalitis viruses. J Clin Microbiol. 2010;48:4010–4014. doi: 10.1128/JCM.00582-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerome R, Tajima S, Takasaki T, Yoshida T, Kotaki A, Lim CK, Ito M, Sugiyama A, Yamauchi A, Yano T, Kameyama T, Morishita I, Kuwayama M, Ogawa T, Sahara K, Ikegaya A, Kanda M, Hosoya Y, Itokazu K, Onishi H, Chiya S, Yoshida Y, Tabei Y, Katsuki K, Tabata K, Harada S, Kurane I. Molecular epidemiological analyses of Japanese encephalitis virus isolates from swine in Japan from 2002 to 2004. J Gen Virol. 2007;88:2762–2768. doi: 10.1099/vir.0.82941-0. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisaki Y, Miura Y, Sugimori T, Murakami Y, Miura K. Experimental studies on vertical infection of mice with Japanese encephalitis virus. IV. Effect of virus strain on placental and fetal infection. Nat Inst Anim Hlth Quart. 1983;23:21–26. [PubMed] [Google Scholar]
- Obara M, Yamauchi T, Watanabe M, Hasegawa S, Ueda Y, Matsuno K, Iwai M, Horimoto E, Kurata T, Takizawa T, Kariwa H, Takashima I. Continuity and change of Japanese encephalitis virus in Toyama Prefecture, Japan. Am J Trop Med Hyg. 2011;84:695–708. doi: 10.4269/ajtmh.2011.10-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unni SK, Ruzek D, Chhatbar C, Mishra R, Johri MK, Singh SK. Japanese encephalitis virus: from genome to infectome. Microbes Infect. 2011;13:312–321. doi: 10.1016/j.micinf.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Totsuka S, Matsumoto Y, Kato A. On the Subacute and Chronic Japanese B Encephalitis: Neuropathological Studies on the Nervous and Mental Diseases (1) Chiba Med J. 1968;44:329–343. [Google Scholar]
- Tenter AM. Current research on Sarcocystis species of domestic animals. Int J Parasitol. 1995;25:1311–1330. doi: 10.1016/0020-7519(95)00068-D. [DOI] [PubMed] [Google Scholar]