Abstract
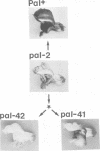
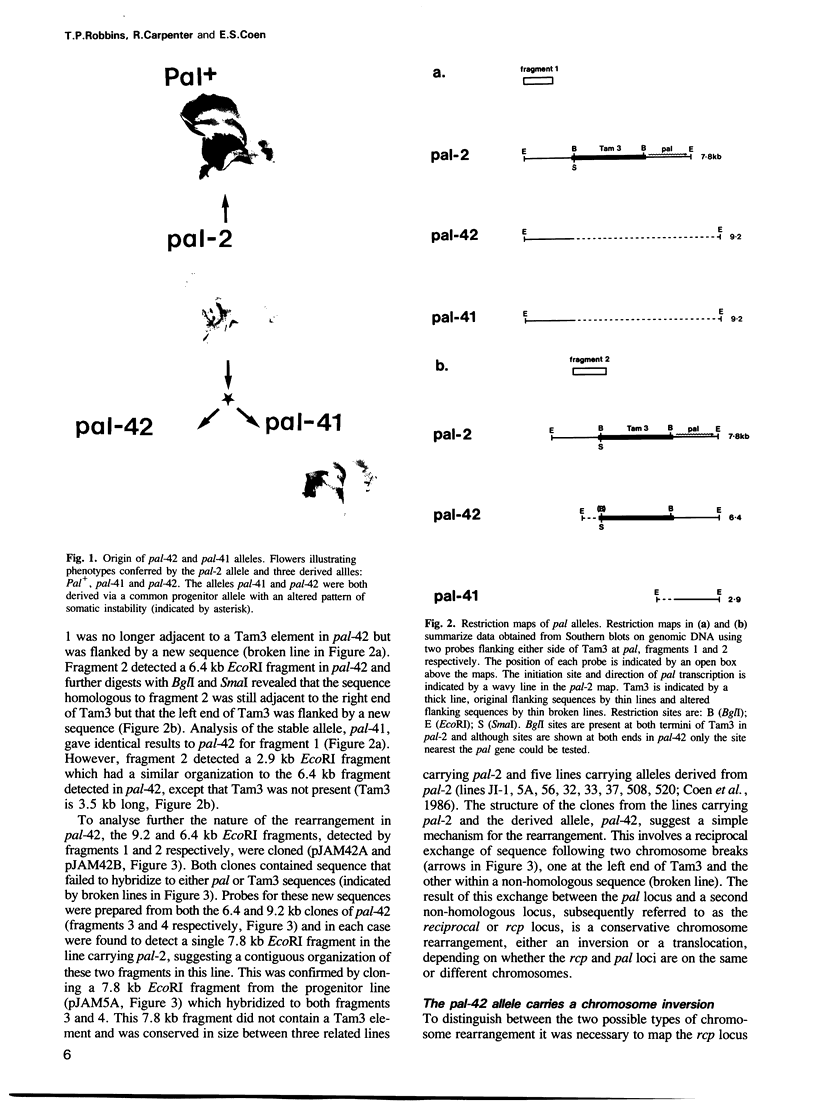
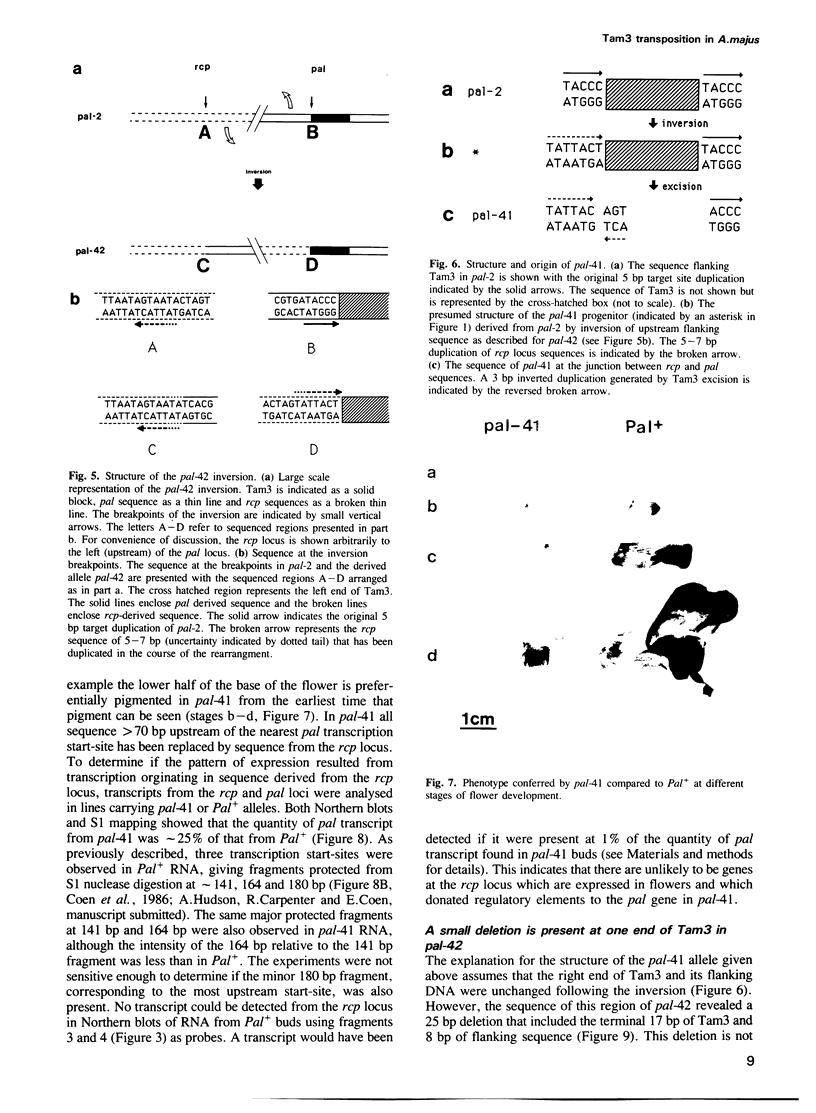
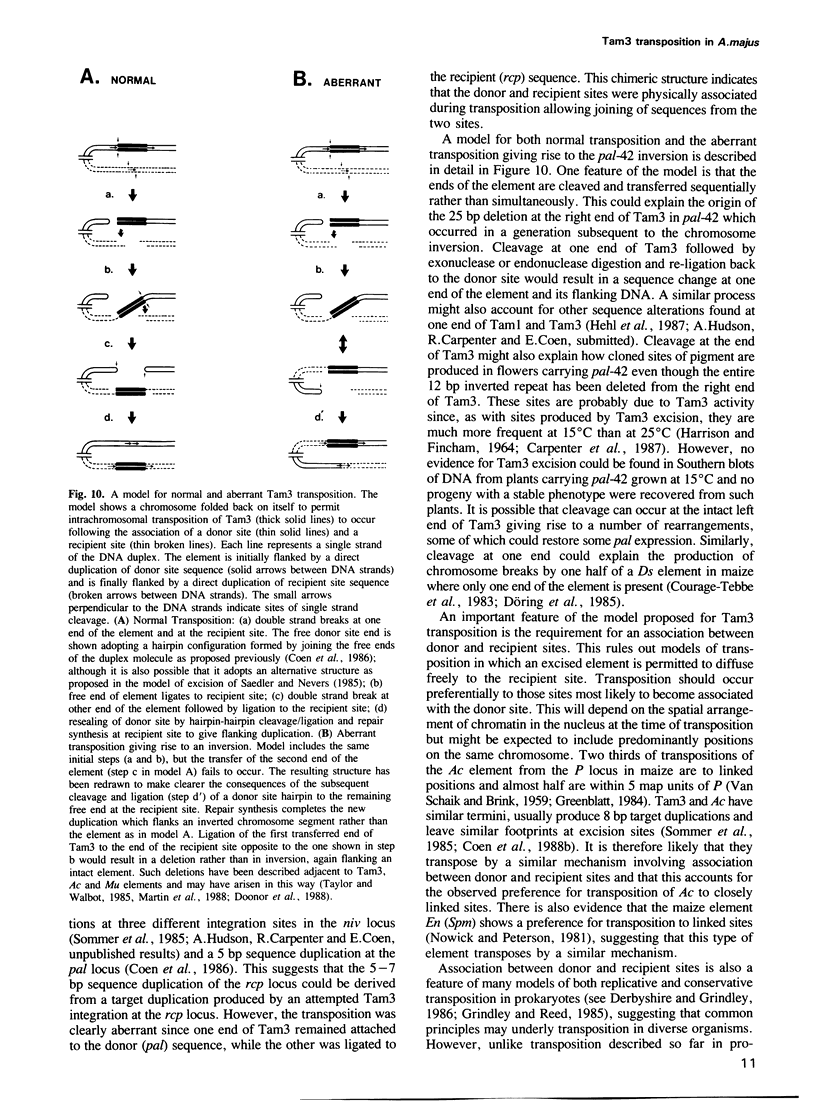
We describe the structure of a chromosome rearrangement which changes the spatial pattern of expression of the pallida gene of Antirrhinum majus. The rearrangement involves a chromosome inversion of ~6 map units with one breakpoint at the end of a copy of the transposable element Tam3 located in the promoter region of the pallida locus. The sequence at the breakpoints shows that 5-7 bp, present once in the progenitor, has been duplicated and flanks both ends of the inversion. We propose that this structure arose from an aberrant Tam3 transposition, suggesting a model for normal transposition which involves physical association between donor and recipient sites. This may explain why transposition of plant transposable elements such as Ac in maize occurs preferentially to recipient sites closely linked to the donor site. Excision of the Tam3 copy located at the end of the chromosome inversion, results in a unique spatial pattern of pallida gene expression as a consequence of replacing all sequence 70 bp upstream of transcription by a new sequence. This pattern may be the result of deleting specific upstream components which regulate pallida expression and/or of changing the relative chromosome position of the pallida gene.
Keywords: Tam3, Antirrhinum majus, transposable elements
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Coen E. S., Carpenter R. A semi-dominant allele, niv-525, acts in trans to inhibit expression of its wild-type homologue in Antirrhinum majus. EMBO J. 1988 Apr;7(4):877–883. doi: 10.1002/j.1460-2075.1988.tb02891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E. S., Carpenter R., Martin C. Transposable elements generate novel spatial patterns of gene expression in Antirrhinum majus. Cell. 1986 Oct 24;47(2):285–296. doi: 10.1016/0092-8674(86)90451-4. [DOI] [PubMed] [Google Scholar]
- Courage-Tebbe U., Döring H. P., Fedoroff N., Starlinger P. The controlling element Ds at the Shrunken locus in Zea mays: structure of the unstable sh-m5933 allele and several revertants. Cell. 1983 Sep;34(2):383–393. doi: 10.1016/0092-8674(83)90372-0. [DOI] [PubMed] [Google Scholar]
- Davis P. S., Shen M. W., Judd B. H. Asymmetrical pairings of transposons in and proximal to the white locus of Drosophila account for four classes of regularly occurring exchange products. Proc Natl Acad Sci U S A. 1987 Jan;84(1):174–178. doi: 10.1073/pnas.84.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire K. M., Grindley N. D. Replicative and conservative transposition in bacteria. Cell. 1986 Nov 7;47(3):325–327. doi: 10.1016/0092-8674(86)90586-6. [DOI] [PubMed] [Google Scholar]
- Dooner H. K., English J., Ralston E. J. The frequency of transposition of the maize element Activator is not affected by an adjacent deletion. Mol Gen Genet. 1988 Mar;211(3):485–491. doi: 10.1007/BF00425705. [DOI] [PubMed] [Google Scholar]
- Engels W. R., Preston C. R. Formation of chromosome rearrangements by P factors in Drosophila. Genetics. 1984 Aug;107(4):657–678. doi: 10.1093/genetics/107.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels W. R., Preston C. R. Identifying P factors in Drosophila by means of chromosome breakage hotspots. Cell. 1981 Nov;26(3 Pt 1):421–428. doi: 10.1016/0092-8674(81)90211-7. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Ish-Horowicz D. Extrachromosomal circular copies of the eukaryotic transposable element copia in cultured Drosophila cells. Nature. 1981 Aug 13;292(5824):591–595. doi: 10.1038/292591a0. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Ish-Horowicz D. The origin of extrachromosomal circular copia elements. Cell. 1983 Sep;34(2):415–419. doi: 10.1016/0092-8674(83)90375-6. [DOI] [PubMed] [Google Scholar]
- Frischer L. E., Hagen F. S., Garber R. L. An inversion that disrupts the Antennapedia gene causes abnormal structure and localization of RNAs. Cell. 1986 Dec 26;47(6):1017–1023. doi: 10.1016/0092-8674(86)90816-0. [DOI] [PubMed] [Google Scholar]
- Greenblatt I M, Brink R A. Twin Mutations in Medium Variegated Pericarp Maize. Genetics. 1962 Apr;47(4):489–501. doi: 10.1093/genetics/47.4.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt I. M. A chromosome replication pattern deduced from pericarp phenotypes resulting from movements of the transposable element, modulator, in maize. Genetics. 1984 Oct;108(2):471–485. doi: 10.1093/genetics/108.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Reed R. R. Transpositional recombination in prokaryotes. Annu Rev Biochem. 1985;54:863–896. doi: 10.1146/annurev.bi.54.070185.004243. [DOI] [PubMed] [Google Scholar]
- Levis R., Hazelrigg T., Rubin G. M. Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science. 1985 Aug 9;229(4713):558–561. doi: 10.1126/science.2992080. [DOI] [PubMed] [Google Scholar]
- Martin C., Carpenter R., Sommer H., Saedler H., Coen E. S. Molecular analysis of instability in flower pigmentation of Antirrhinum majus, following isolation of the pallida locus by transposon tagging. EMBO J. 1985 Jul;4(7):1625–1630. doi: 10.1002/j.1460-2075.1985.tb03829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C., Mackay S., Carpenter R. Large-scale chromosomal restructuring is induced by the transposable element tam3 at the nivea locus of antirrhinum majus. Genetics. 1988 May;119(1):171–184. doi: 10.1093/genetics/119.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock W. J., Dennis E. S., Gerlach W. L., Sachs M. M., Schwartz D. Insertion and excision of Ds controlling elements in maize. Cold Spring Harb Symp Quant Biol. 1984;49:347–354. doi: 10.1101/sqb.1984.049.01.041. [DOI] [PubMed] [Google Scholar]
- Roiha H., Rubin G. M., O'Hare K. P element insertions and rearrangements at the singed locus of Drosophila melanogaster. Genetics. 1988 May;119(1):75–83. doi: 10.1093/genetics/119.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A. M., Snutch T. P. Isolation of the closed circular form of the transposable element Tc1 in Caenorhabditis elegans. Nature. 1984 Oct 4;311(5985):485–486. doi: 10.1038/311485a0. [DOI] [PubMed] [Google Scholar]
- Ruan K., Emmons S. W. Extrachromosomal copies of transposon Tc1 in the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4018–4022. doi: 10.1073/pnas.81.13.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedler H., Nevers P. Transposition in plants: a molecular model. EMBO J. 1985 Mar;4(3):585–590. doi: 10.1002/j.1460-2075.1985.tb03670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneuwly S., Kuroiwa A., Gehring W. J. Molecular analysis of the dominant homeotic Antennapedia phenotype. EMBO J. 1987 Jan;6(1):201–206. doi: 10.1002/j.1460-2075.1987.tb04739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Gierl A., Cuypers H., Peterson P. A., Saedler H. Plant transposable elements generate the DNA sequence diversity needed in evolution. EMBO J. 1985 Mar;4(3):591–597. doi: 10.1002/j.1460-2075.1985.tb03671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V., Freeling M. An extrachromosomal form of the Mu transposons of maize. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4924–4928. doi: 10.1073/pnas.84.14.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor L. P., Walbot V. A deletion adjacent to the maize transposable element Mu-1 accompanies loss of Adh1 expression. EMBO J. 1985 Apr;4(4):869–876. doi: 10.1002/j.1460-2075.1985.tb03712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schaik N W, Brink R A. Transpositions of Modulator, a Component of the Variegated Pericarp Allele in Maize. Genetics. 1959 Jul;44(4):725–738. doi: 10.1093/genetics/44.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]