Abstract
Juxtacrine signaling interactions between the EphA2 receptor tyrosine kinase and its ephrin-A1 ligand contribute to healthy tissue maintenance and misregulation of this system is observed in at least 40% of human breast cancer. Hybrid live cell – supported membrane experiments, in which membrane-linked ephrin-A1 displayed in supported membranes interacts with EphA2 in living cells, have revealed large scale clustering of EphA2:ephrin-A1 complexes as well as their lateral transport across the cell surface during triggering. Here, we utilize 100nm spaced hexagonally ordered arrays of gold nanodots embedded within supported membranes to present defined obstacles to the movement and assembly of EphA2 clusters. By functionalizing both the supported membrane and the nanodots with ephrin-A1, we perform a type of affinity chromatography on EphA2 signaling clusters in live cell membranes. Analysis of ten different breast cancer cell lines reveals that EphA2 transport is most frustrated by nanodot arrays in the most diseased cell lines. These observations suggest that strong physical association among EphA2 receptors, as well as their assembly into larger clusters, correlates with and may contribute to the pathological misregulation of the EphA2:ephrin-A1 pathway in breast cancer.
Keywords: EphA2, nanoparticles, supported membranes, breast cancer cells, receptor clustering
The Eph receptors are a family of receptor tyrosine kinases (RTK) that bind to ephrin ligands in a juxtacrine configuration.1 They are involved in the positioning, adhesion, repulsion, and migration of cells during development.2 In neural stem cells, Eph receptors have been linked to signature stem cell activities such as self-renewal and differentiation.3 Eph receptors and their ephrin ligands are also points of corruption in cancer cell invasion, metastasis, and angiogenesis.4 EphA2, for example, which binds ephrin-A1 on an apposed cell membrane, is overexpressed in over 40% of all breast cancers and EphA2 overexpression is correlated with tumor metastasis.5-8 As a result, much attention has been directed towards developing therapeutics targeting EphA2.9,10 Despite its role in cancer and its potential as a target, precisely what goes wrong with the EphA2 signaling pathway to contribute to pathological cell behavior remains unclear. In general, the receptor itself is not mutated.5
We have developed a hybrid live cell – supported membrane system in which ephrin-A1, displayed in the supported membrane, interacts with and ultimately triggers EphA2 in the living cell.11, 12 In the absence of physical constraints, triggering of EphA2 by mobile ephrin-A1 in fluid supported membranes results in large-scale clustering of EphA2:ephrin-A1 complexes, followed by their inward radial transport within the plane of the interface, and ultimate endocytosis. Observations across a library of different breast cancer cell lines reveal characteristic differences in EphA2 transport that correlate with cell line tumorigenicity and metastatic potential, thus hinting that the clustering and movement of EphA2 may be misregulated in disease.
Here, we employ ordered arrays of gold nanodots embedded within supported membranes13 to present defined obstacles to the movement and assembly of EphA2 clusters. In these experiments, a supported membrane is formed on a glass substrate with a preformed nanodot array, resulting in a continuous and fluid supported lipid bilayer membrane that surrounds the fixed array of nanodots. Ephrin-A1 ligands can be bound to the mobile membrane (via Ni-polyhistidine interactions),12, 14 to the nanodots (thereby becoming immobile),13 or to both. In the case where both supported membrane and nanodots are functionalized, a type of affinity chromatography is achieved in the membrane of the living cell. The ability of EphA2:ephrin-A1 clusters to percolate through the array is related to the size and internal cohesion of the clusters. The supported membrane embedded gold nanodot arrays enable precision analysis of physical aspects of EphA2 cluster transport. The long range movement of EphA2:ephrin-A1 clusters through the ~100nm spaced arrays of fixed gold nanodots obstacles is readily imaged by fluorescence microscopy. Analysis of ten different breast epithelial cancer cell lines, with similar EphA2 expression levels but spanning a wide range of disease characteristics, reveals that only tumorigenic and metastatic cells exhibit frustrated EphA2 transport through the nanodot arrays. Strong physical association among EphA2 receptors, as well as their assembly into larger clusters, correlates with and may contribute to the pathological misregulation of the EphA2:ephrin-A1 pathway in breast cancer. The nanodot array assay platform introduced here may facilitate the identification of drug compounds that therapeutically modulate this molecular phenotype.
Arrays of gold nanodots are generated on top of a glass coverslip through block copolymer nanolithography.15 The parameters of the nanodot array are adjustable, but generally fixed at an interparticle distance of ~100nm and a particle size of ~7nm for the experiments described here (Fig. 1A). For most experiments, nanodot arrays were formed over half of a glass substrate, leaving the other half free of nanodots as a control surface. A supported lipid bilayer (SLB) is subsequently formed on the entire glass substrate by vesicle fusion.16, 17 Proteins are selectively tethered to either the nanodots or to the phospholipid membrane (or both) via Ni-NTA/His-tag binding (Fig. 1B). The ephrin-A1 fusion protein is composed of the ectodomain of human ephrin-A1, a monomeric fluorescent Eos,18 and a sequence of ten histidines on the C-terminus. This protein, which we hereafter refer to as ephrin-A1, readily binds to supported membranes doped with Ni-NTA lipids, where it diffuses freely on the membrane surface and is monomeric.12 The nanodot arrays present no detectable interference to the mobility of lipids or proteins in the supported membrane,13 as illustrated by fluorescence recovery after photobleaching (FRAP) in Fig. 1C. The membrane-linked ephrin-A1 binds and activates EphA2 in living cells.11, 12
Figure 1.
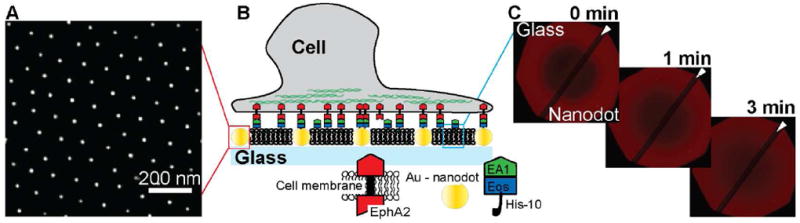
Schematic illustration of the experimental approach. A) SEM of gold nanodots on the surface of a glass coverslip. B) Schematic of the nanodot – SLB platform: cancer cells expressing EphA2 deposited on this surface can interact with molecules of ephrin-A1 tethered to the nanodots or to the lipids within the SLB (or both). C) FRAP images at different time points show the recovery of a Texas Red-DHPE SLB across the entire glass coverslip. This confirms that the membrane fluidity is not affected by the presence of the nanodots as previously reported.13 The dark diagonal stripe (indicated by the white arrow) across the FRAP images is a result of the dipping process for nanodot functionalization and depicts the border between the bare glass side and the nanodot patterned side.
This platform offers the flexibility to generate three different scenarios of ephrin-A1 presentation for live cell studies (Fig. 2): i) mobile ephrin-A1, with ephrin-A1 only anchored to the SLB (nanodots are not functionalized), ii) immobile ephrin-A1, with ephrin-A1 only tethered to the nanodots (SLB is not functionalized), and iii) hybrid ephrin-A1, with mobile and immobile ephrin-A1 tethered to the SLB and nanodots, respectively. Under the conditions of these experiments, mobile ephrin-A1 densities in the supported membrane were several hundreds of molecules/μm2 and nanodot functionalization efficiency was ~24% (~28 molecules/μm2). This functionalization efficiency is a comparison between the number of ephrin-A1 protein molecules, measured with photoactivated localization microscopy and the number of individual nanodots, measured by scanning electron micrograph.13 On the hybrid surfaces, a majority of the total ephrin-A1 is mobile, with a minor fraction of immobile protein anchored to the nanodots.
Figure 2.
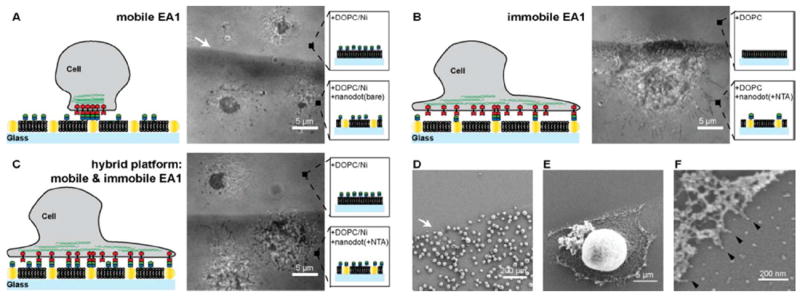
MDA-MB-231 cells are placed on substrates with different ephrin-A1 presentations. A) RICM image of MDA-MB-231 cells placed on a mobile ephrin-A1 surface is shown. Cells form a tight contact area with the SLB displaying mobile ephrin-A1 on both the nanodot patterned side as well as the glass side. B) RICM image of cells placed on immobile ephrin-A1 surface shows a spreading morphology and filopodia protrusions. Essentially no cells attached to a pure 100% DOPC SLB. C) RICM image of cells on hybrid surfaces, consisting of both mobile and immobile ephrin-A1. Cells on the bare glass side that displays only mobile ephrin-A1 form the typical central contact region whereas cells on the hybrid nanodot side spread out, forming protrusions, similar to cells on the immobile ephrin-A1 surface. D-F) SEM images of MDA-MB-231 cells on the immobile ephrin-A1 substrate. Cells are preferentially interacting with the nanodot side since the phospholipids are not functionalized with ephrin-A1. Higher magnification SEM images reveal that individual cell protrusions are interacting with single nanodots (black arrows).
MDA-MB-231 cells, a highly invasive breast epithelial cancer cell line,19 were deposited on surfaces with different presentations of ephrin-A1. After one hour the cells were imaged with reflection interference contrast microscopy (RICM) to observe the regions of tightest adhesion between the cell membrane and the supported bilayer interface, which we have previously confirmed to correspond to regions of EphA2:ephrin-A1 binding and activation.12 In the case of completely mobile ephrin-A1 presentation (no ephrin-A1 bound to nanodots), a tight central contact region between the cell membrane and the SLB was observed by RICM on both the bare glass control surface and on the nanodot array (Fig. 2A). The presence of gold nanodots embedded in the membrane does not lead to any measureable effect on EphA2 transport when all of the ephrin-A1 is linked to the supported membrane. When ephrin-A1 is exclusively linked to the nanodots, cell spreading and the formation of filopodia protrusions were observed on the nanodot array while essentially no nonspecific adhesion to the supported membrane occurs (Fig. 2B). When ephrin-A1 is presented on both the membrane and the nanodots, the minority fraction of immobile ephrin-A1 proved to be dominant. EphA2 transport was stopped completely on the nanodot array (for these MDA-MB-231 cells) while normal junctions were observed on the ephrin-A1-containing supported membrane side (Fig. 2C). Scanning electron micrograph (SEM) images reveal cell protrusions adhering to individual ephrin-A1 coated nanodots (Fig. 2E-F). In a control experiment, cells placed on a surface with Ni-NTA functionalized nanodots but without ephrin-A1 were easily rinsed away, further confirming that ephrin-A1 functionalization of the nanodots is required for adhesion. To verify the functionality of interactions between MDA-MD-231 cells and the ephrin-A1 presentation, we immunostained for the presence of signature downstream signaling molecules (Fig. S1). The results mirror prior published works.11, 12
The most informative aspect of the nanodot array assay is evident in the comparison of behaviors between different cell lines. MCF10A is another breast epithelial cancer cell line that expresses EphA2 but differs from the MDA-MB-231 cell line by being much less invasive and non-tumorigenic. The effects of the nanodot array on EphA2 transport in the MCF10A cells also differ substantially from the effects observed in MDA-MB-231 cells (Fig. 3). On substrates displaying only mobile ephrin-A1, the nanodot array has no effect for either cell type (Fig. 3A). On fully immobile ephrin-A1, MCF10A cells appear to bind only weakly and are easily detached by mild rinsing while MDA-MB-231 cells bind strongly (Fig. 3B). When the two cell lines were individually placed on substrates displaying hybrid ephrin-A1 configurations, consisting of both mobile and immobile ephrin-A1, we observed no change in EphA2 transport by the MCF10A cells in response to ephrin-A1 immobilized nanodot obstacles. This starkly contrasts the response of the MDA-MB-231 cells (Fig. 3C). MCF10A cells can transport ligand-bound EphA2, seemingly unhindered by the presence of immobile ephrin-A1.
Figure 3.
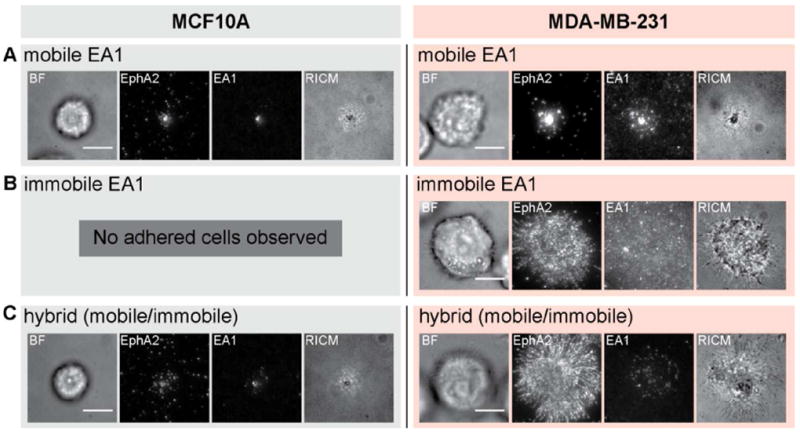
Comparison between MCF10A and MDA-MB-231 cells placed on substrates with three different ephrin-A1 configurations. A) Normal EphA2 transport is observed for both MCF10A and MDA-MB-231 cells on mobile ephrin-A1 surfaces. B) On entirely immobile ephrin-A1 surfaces, and after rinsing steps during fixation and permeabilization processes, only MDA-MB-231 cells adhere. C) MCF10A cells adhere to hybrid surfaces, containing both mobile and immobile ephrin-A1, and EphA2 transport is normal. In contrast, MDA-MB-231 cells exhibit jammed phenotypes on these surfaces. Scale bars are 10 microns.
This dramatic difference in the transport properties of EphA2:ephrin-A1 clusters through the nanodot array between MCF10A and MDA-MB-231 cells provides a readily observable phenotypic parameter, which may correlate with other (pathological) aspects of cellular behavior. To explore this, we extend the above comparative study to a panel of ten cell lines, chosen to have similar EphA2 expression levels but spanning a range of disease characteristics. These experiments were performed using the hybrid (mobile and immobile) ephrin-A1 display configuration. We define a parameter, here referred to as the jamming coefficient, which measures the degree to which the nanodot array frustrates EphA2 transport. This parameter, which is objectively calculated from images of the cells on and off the nanodot arrays (Fig. S2), has a value around 1 for completely jammed states and a value close to 0 for no interference from the nanodot array. The results and corresponding attributes of each cell line are summarized in Fig. 4. There is a visibly obvious correlation between the degree to which EphA2 transport is jammed and the tumorigenicity of the cell lines. EphA2 expression levels themselves are not well correlated with the frustrated transport. This suggests that it is a functional property of EphA2 in the cellular system, rather than simply its expression level, that leads to the jamming phenotype.
Figure 4.
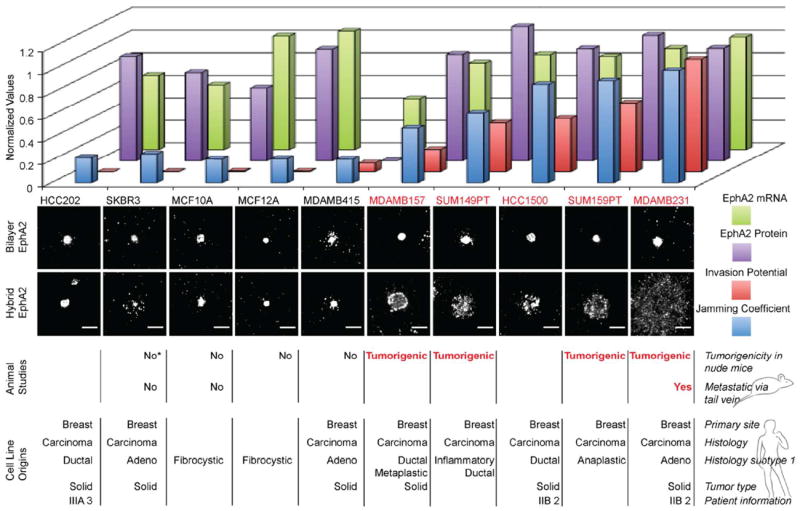
Breast cancer cell line panel study. A panel consisting of 10 human breast cancer cell lines are individually deposited on the hybrid platform. The jamming coefficient of EphA2 is a ratio of the radial profiles measured for cells that were deposited on bilayer only side to the radial profiles of cells on the hybrid side (see supporting information). The cells with the highest normalized jamming coefficient exhibit the most frustrated EphA2 transport. For all cell lines, on the glass side displaying only mobile ephrin-A1, EphA2 is centrally transported. Differential behavior among the cell lines was observed on the nanodot side, on which ephrin-A1 is presented in the hybrid configuration. The animal studies of tumorigenicity and metastasis which correlate with the EphA2 jamming coefficients, the invasion potential values, the EphA2 protein expression levels, and EphA2 mRNA levels were taken from published sources.24-31 The cell line SKBR3 was found to be tumorigenic in one report (*).32 A total of 200 cells were selected and analyzed. Scale bars are 10 microns.
We interpret the results from these experiments to reveal the degree to which EphA2 is clustered in each of the cell lines studied. This hypothesis is summarized schematically in Figure 5. Since the majority of ephrin-A1 is freely mobile on the membrane surface, a corresponding majority of EphA2 receptors are bound to these mobile ephrin-A1 molecules. In order for the transport of these EphA2 receptors to be frustrated, they must be physically coupled, either by direct association or through intracellular structures, to another EphA2 receptor that is bound to one of the immobilized ephrin-A1 ligands. In cells with highly clustered EphA2, one immobilized ephrin-A1 molecule may be sufficient to impede the transport of many EphA2 receptors (Fig. 5A-C). In contrast, for cells with smaller or more dynamic EphA2 clusters, the extent to which a single immobilized ephrin-A1 ligand can impede transport of other EphA2:ephrin-A1 receptor complexes would be reduced (Fig. 5D-F). The observation that the nanodot arrays themselves, without any immobilized ephrin-A1, pose no impedance to EphA2 clustering and transport (Fig. 2A) confirms that this is not a size sieving effect; EphA2:ephrin-A1 clusters easily pass over the bare nanodots. This further confirms that a specific interaction with the ephrin-A1 ligand is required for the impedance of transport caused by the arrays.
Figure 5.
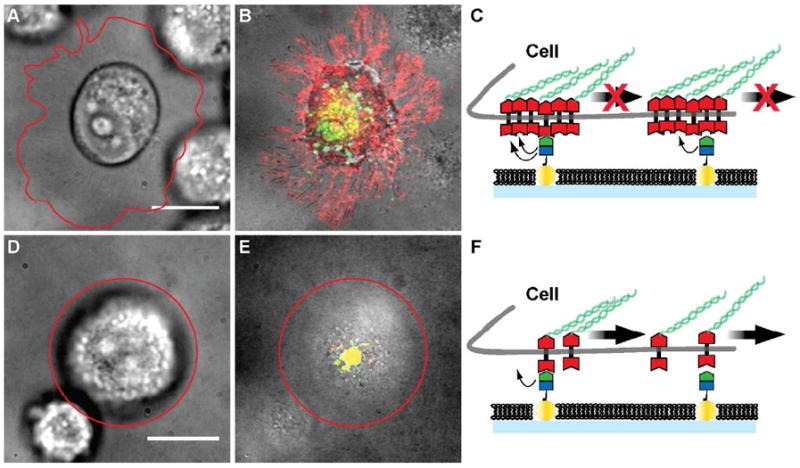
Schematic description of frustrated EphA2 motion on the nanodot array platform. A) – C) Pre-clustered EphA2 on the surface of invasive cell lines are restrained by molecules of immobilized ephrin-A1 linked to the nanodots. If one EphA2 receptor within the cluster unbinds from ephrin-A1, another one in the cluster may re-bind with enhanced probability as a result of proper geometrical juxtaposition. EphA2 clusters (red) are distributed along the cell protrusions imaged in RICM. Only a few ephrin-A1 clusters (green) are transported to the center of the cell – SLB contact area, and are co-localized with EphA2 (yellow). Cell outlines are highlighted by the red line. D) – F) For non-invasive cells, EphA2 may be less tightly clustered and therefore individual EphA2 receptors can bind immobile ephrin-A1 ligands. As the cell applies a pulling force, these receptors can unbind and become centrally transported by re-binding to mobile ephrin-A1. Scale bars are 10 microns.
The differential jamming of EphA2 transport among the various breast cancer cell lines, and its correlation with disease characteristics, suggests that EphA2 clustering itself may contribute to pathological effects. This finding supports previous hypotheses about the roles of RTK cluster formation in invasive cancers.7, 8, 20-23 The nanodot array platform introduced here achieves a type of affinity chromatography in live cell membranes. Molecular-scale physical coupling among EphA2 receptors is transformed into micron-scale spatial patterns, which can be readily quantified by microscopy. This assay could facilitate the identification of molecules that modulate this and other related molecular phenotypes, and potentially offer therapeutic benefit.
Supplementary Material
Acknowledgments
This work was supported by Award U54 CA143836 from the National Cancer Institute (NCI). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NCI or the NIH. Additional support provided by the Director, Office of Science, Office of Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences of the U.S. Department of Energy (DOE) under contract no. DE-AC02-05CH11231. Seed support for biomedical aspects of this work was provided by the U.S. Department of Defense DA Congressionally Directed Medical Research Program Idea Award BC102681 under U.S. Army Medical Research Acquisition Activity no. W81XWH-11-1-0256. T.L. was supported, in part, by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (DFG).
Footnotes
Supporting Information available: Materials and methods and additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pasquale EB. Eph receptor signaling casts a wide net on cell behavior. Nat Rev Mol Cell Biol. 2005;6(6):462–475. doi: 10.1038/nrm1662. [DOI] [PubMed] [Google Scholar]
- 2.Wimmer-Kleikamp SH, Lackmann M. Eph-modulated cell morphology, adhesion and motility in carcinogenesis. IUBMB Life. 2005;57(6):421–31. doi: 10.1080/15216540500138337. [DOI] [PubMed] [Google Scholar]
- 3.Holland SJ, Peles E, Pawson T, Schlessinger J. Cell-contact-dependent signalling in axon growth and guidance: Eph receptor tyrosine kinases and receptor protein tyrosine phosphatase beta. Curr Opin Neurobiol. 1998;8(1):117–27. doi: 10.1016/s0959-4388(98)80015-9. [DOI] [PubMed] [Google Scholar]
- 4.Dodelet VC, Pasquale EB. Eph receptors and ephrin ligands: embryogenesis to tumorigenesis. Oncogene. 2000;19(49):5614–9. doi: 10.1038/sj.onc.1203856. [DOI] [PubMed] [Google Scholar]
- 5.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10(3):165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. EphA2: a determinant of malignant cellular behavior and a potential therapeutic target in pancreatic adenocarcinoma. Oncogene. 2004;23(7):1448–56. doi: 10.1038/sj.onc.1207247. [DOI] [PubMed] [Google Scholar]
- 7.Himanen JP, Yermekbayeva L, Janes PW, Walker JR, Xu K, Atapattu L, Rajashankar KR, Mensinga A, Lackmann M, Nikolov DB, Dhe-Paganon S. Architecture of Eph receptor clusters. Proc Natl Acad Sci U S A. 2010;107(24):10860–5. doi: 10.1073/pnas.1004148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mudali SV, Fu B, Lakkur SS, Luo M, Embuscado EE, Iacobuzio-Donahue CA. Patterns of EphA2 protein expression in primary and metastatic pancreatic carcinoma and correlation with genetic status. Clin Exp Metastasis. 2006;23(7-8):357–65. doi: 10.1007/s10585-006-9045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson D, Gooya J, Mao S, Kinneer K, Xu L, Camara M, Fazenbaker C, Fleming R, Swamynathan S, Meyer D, Senter PD, Gao C, Wu H, Kinch M, Coats S, Kiener PA, Tice DA. A human antibody-drug conjugate targeting EphA2 inhibits tumor growth in vivo. Cancer Res. 2008;68(22):9367–74. doi: 10.1158/0008-5472.CAN-08-1933. [DOI] [PubMed] [Google Scholar]
- 10.Hammond SA, Lutterbuese R, Roff S, Lutterbuese P, Schlereth B, Bruckheimer E, Kinch MS, Coats S, Baeuerle PA, Kufer P, Kiener PA. Selective targeting and potent control of tumor growth using an EphA2/CD3-Bispecific single-chain antibody construct. Cancer Res. 2007;67(8):3927–35. doi: 10.1158/0008-5472.CAN-06-2760. [DOI] [PubMed] [Google Scholar]
- 11.Salaita K, Nair PM, Petit RS, Neve RM, Das D, Gray JW, Groves JT. Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science. 2010;327(5971):1380–1385. doi: 10.1126/science.1181729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Q, Lin W-C, Petit RS, Groves JT. EphA2 Receptor Activation by Monomeric Ephrin-A1 On Supported Membranes. Biophys J. 2011;101(11):2731–9. doi: 10.1016/j.bpj.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohmüller T, Triffo S, O’Donoghue GP, Xu Q, Coyle MP, Groves JT. Supported Membranes Embedded with Fixed Arrays of Gold Nanoparticles. Nano Lett. 2011;11(11):4912–8. doi: 10.1021/nl202847t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nye JA, Groves JT. Kinetic control of histidine-tagged protein surface density on supported lipid bilayers. Langmuir. 2008;24(8):4145–9. doi: 10.1021/la703788h. [DOI] [PubMed] [Google Scholar]
- 15.Lohmüller T, Aydin D, Schweider M, Morhard C, Louban I, Pacholski C, Spatz JP. Nanopatterning by block copolymer micelle nanolithography and bioinspired applications. Biointerphases. 2011;6(1) doi: 10.1116/1.3536839. [DOI] [PubMed] [Google Scholar]
- 16.Groves JT, Ulman N, Boxer SG. Micropatterning fluid lipid bilayers on solid supports. Science. 1997;275(5300):651–3. doi: 10.1126/science.275.5300.651. [DOI] [PubMed] [Google Scholar]
- 17.Lin W-C, Yu C-H, Triffo S, Groves JT. Supported Membrane Formation, Characterization, Functionalization, and Patterning for Application in Biological Science and Technology. Curr Protoc Chem Biol. 2010:235–269. doi: 10.1002/9780470559277.ch100131. [DOI] [PubMed] [Google Scholar]
- 18.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6(2):131–3. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer. 2004;4(5):361–70. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 21.Seiradake E, Harlos K, Sutton G, Aricescu AR, Jones EY. An extracellular steric seeding mechanism for Eph-ephrin signaling platform assembly. Nat Struct Mol Biol. 2010;17(4):398–402. doi: 10.1038/nsmb.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lackmann M, Oates AC, Dottori M, Smith FM, Do C, Power M, Kravets L, Boyd AW. Distinct subdomains of the EphA3 receptor mediate ligand binding and receptor dimerization. J Biol Chem. 1998;273(32):20228–37. doi: 10.1074/jbc.273.32.20228. [DOI] [PubMed] [Google Scholar]
- 23.Casaletto JB, McClatchey AI. Spatial regulation of receptor tyrosine kinases in development and cancer. Nat Rev Cancer. 2012;12(6):387–400. doi: 10.1038/nrc3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moyano JV, Evans JR, Chen F, Lu M, Werner ME, Yehiely F, Diaz LK, Turbin D, Karaca G, Wiley E, Nielsen TO, Perou CM, Cryns VL. AlphaB-crystallin is a novel oncoprotein that predicts poor clinical outcome in breast cancer. J Clin Invest. 2006;116(1):261–70. doi: 10.1172/JCI25888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Pu X, Shi M, Chen L, Qian L, Song Y, Yuan G, Zhang H, Yu M, Hu M, Shen B, Guo N. c-Jun, a crucial molecule in metastasis of breast cancer and potential target for biotherapy. Oncol Rep. 2007;18(5):1207–12. [PubMed] [Google Scholar]
- 26.Mao JH, Kim IJ, Wu D, Climent J, Kang HC, DelRosario R, Balmain A. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321(5895):1499–502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–67. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coletta RD, Christensen KL, Micalizzi DS, Jedlicka P, Varella-Garcia M, Ford HL. Six1 overexpression in mammary cells induces genomic instability and is sufficient for malignant transformation. Cancer Res. 2008;68(7):2204–13. doi: 10.1158/0008-5472.CAN-07-3141. [DOI] [PubMed] [Google Scholar]
- 29.Xie D, Miller CW, O’Kelly J, Nakachi K, Sakashita A, Said JW, Gornbein J, Koeffler HP. Breast cancer. Cyr61 is overexpressed, estrogen-inducible, and associated with more advanced disease. J Biol Chem. 2001;276(17):14187–94. doi: 10.1074/jbc.M009755200. [DOI] [PubMed] [Google Scholar]
- 30.Liang Z, Yoon Y, Votaw J, Goodman MM, Williams L, Shim H. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res. 2005;65(3):967–71. [PMC free article] [PubMed] [Google Scholar]
- 31.Hoenerhoff MJ, Chu I, Barkan D, Liu ZY, Datta S, Dimri GP, Green JE. BMI1 cooperates with H-RAS to induce an aggressive breast cancer phenotype with brain metastases. Oncogene. 2009;28(34):3022–32. doi: 10.1038/onc.2009.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fogh J, Fogh JM, Orfeo T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J Natl Cancer Inst. 1977;59(1):221–6. doi: 10.1093/jnci/59.1.221. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


