Abstract
Purpose of review
Recent efforts to explore the genetic underpinnings of hypertension revealed rare mutations in kidney salt transport genes contribute to blood pressure variation and hypertension susceptibility in the general population. The current review focuses on these latest findings, highlighting a discussion about the rare mutations and how they affect transport function.
Recent findings
Rare mutations that confer a low blood pressure trait and resistance to hypertension have recently been extensively studied. Physiological and biochemical analyses of the effected renal salt transport molecules (NKCC2 (SLC12A1), ROMK (KCNJ1), and NCC (SLC12A3)) revealed that most of the mutations do, in fact, cause a loss of transport function. The mutations disrupt transport by many different mechanisms, including altering biosynthetic processing, trafficking, ion transport, and regulation.
Summary
New insights into the genetic basis of hypertension have recently emerged, supporting a major role of rare, rather than common, gene variants. Many different rare mutations have been found to affect the functions of different salt transporter genes by different mechanisms, yet all confer the same blood pressure phenotype. These studies reinforce the critical roles of the kidney, and renal salt transport in blood pressure regulation and hypertension.
Keywords: Blood pressure, salt, kidney, SLC12A1, NKCC2, ROMK, Kir1.1, KCNJ1, SLC12A3, loop-diuretic, thiazide, hypertension
Introduction
Hypertension is a common disease, affecting over a billion people on the planet. It has complex genetic and environmental underpinnings. Genes play a major role, with heritability of blood pressure (BP) levels estimated to be 30-35%. Environmental triggers, including high dietary sodium and low potassium intake, likely collude with genetic factors to increase disease jeopardy. Recent efforts to explore the genetic underpinnings of the BP trait revealed that rare mutations in kidney salt transport genes contribute to blood pressure variation in the general population, and play an important role in hypertension susceptibility. Here we review these latest advances.
Genetics of the variable BP trait and Hypertension
The “common disease-common variant” hypothesis holds that the genetics of any particular common disease, like hypertension, can be explained by a few common gene variants specific to that disease. According to the theory, common single nucleotide polymorphisms (SNP), the largest source of genetic diversity in the human population, form the basis of most common disease alleles. Approximately 107 different SNP are found at a frequency of 1-5% in the human population, accounting for 90% of human genetic variance. These SNP arose early in human ancestry so that they are found in all modern populations throughout the globe.
In the last several years, numerous large, well-controlled genome wide association studies (GWAS) have been performed (1-5), identifying many different interesting common hypertension susceptibility alleles. For example, a polymorphism in promoter region of the endothelial NO synthase gene was recently indentified (1), reinforcing the contribution of vascular tone regulation in BP. Likewise, the discovery of a functional variant of the SPAK kinase (STK39), which regulates the thiazide-sensitive sodium-chloride co-transporter (6), revealed the potential contribution of a multi-gene kinase network (7) and altered renal salt handling in essential hypertension (6, 8). Nevertheless, GWAS have not supported a major role for common variants in hypertension. Except for a few notable exceptions, like SPAK, it is not clear if and how most variant alleles alter gene function, and their precise link to blood pressure is not usually obvious. More importantly, the common variant alleles have a very small effect size on blood pressure, and their frequency is too low to explain all but a small fraction of the genetic component of the disease.
As a result, the “common disease-rare variant” hypothesis has gained favor (9). This idea holds that common disease has highly heterogeneous underpinnings, attributable to any one of thousands of rare mutations. As the human population began to expand 10,000 years ago with the development of agriculture, the number of rare mutations in the population increased faster (∼100 mutations per generation) than the harmful ones could be eliminated by natural selection. Of course, once a harmful mutation arises, purifying selection keeps it rare in the population. Each individual has approximately half of a million rare variants, ∼13,000 of these are found in gene coding regions, and it is estimated that 1% have functional effects (10). According to the theory, a common disease may have disparate rare genetic origins because a gene or genes in a common pathway may harbor many different severe mutations that produce the same functional effect.
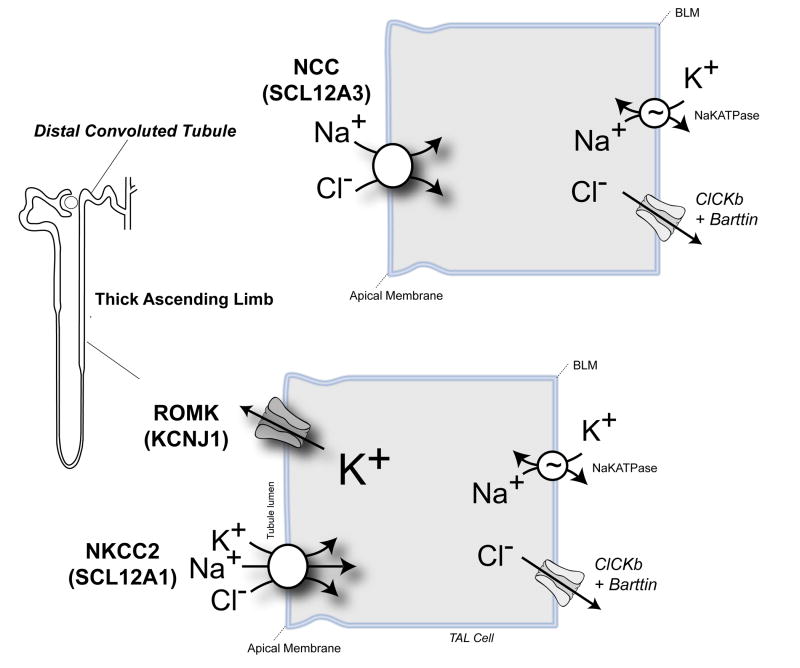
Ji and co-workers explored the rare-mutation/common disease hypothesis for hypertension (11), exploiting advances in high throughput gene sequencing technologies to rapidly and efficiently find rare variants in the general population that might contribute to the blood pressure trait. Their ingenious strategy was guided by insights from Bartter's and Gitelman's syndromes, two rare recessive nephropathies characterized by renal salt-wasting and low blood pressure (12). Bartter's syndrome results from loss-of-function mutations in any one of the key salt transport molecules that are responsible for sodium chloride reabsorption in the thick ascending limb, including the sodium-potassium-two chloride cotransporter (NKCC2, or SLC12A1) and the ROMK channel (KCNJ1 or Kir1.1) (Figure 1). Gitelman's syndrome is caused by mutations in the thiazide-sensitive sodium chloride cotransporter, NCC (SLC12A3) (Figure 1). Because the prevalence of the syndromes predicts ∼1% of the general population should be a carrier, Ji et al reasoned that that the heterozygous disease alleles might commonly impair renal salt reabsorption, and consequently confer a low blood pressure trait.
Figure 1.
Rare Hypertension Resistance Variants were identified in NKCC2 and ROMK, which are essential for salt reabsorption in the thick ascending limb, and NCC, the thiazide sensitive transporter that is responsible for salt reabsorption in the distal convoluted tubule. Homozygous mutations in NKCC2 and ROMK cause Bartter's syndrome. Homozygous mutations in NCC cause Gitelman's syndrome. Carriers of these mutations are protected from hypertension.
They identified rare variants in NKCC2, ROMK and NCC, by exon sequencing in ∼3,000 individuals of the Framingham study cohort. Over a hundred different variants were found in the salt handling genes. The task was to accurately find the few relevant ones. Some of the variants had already been proven to cause inherited salt wasting (i.e. Bartter's or Gitleman's syndromes), making part of the job straightforward. Still, other variants had to be inferred to lead to loss-of-function. Algorithms have been developed to predict the likelihood that a sequence variant might impair function. Most of them, like the one used by Ji, are based on the simple assumption that changes in phylogenetically conserved residues will be damaging. An additional criterion that variants be present at a low frequency was used because harmful alleles are predicted to be under the pressure of purifying selection, and thus rare.
Remarkably, carriers for one of the proven or inferred mutations in NKCC2, NCC, or ROMK had a significantly lower blood pressure compared to the rest of the cohort. The effect size was large, and carriers enjoyed protection against the development of hypertension throughout life. These observations established that rare mutations in renal salt handling genes do, in fact, commonly contribute to blood pressure variation in humans. The study offered a glimpse into the genetic architecture of hypertension, revealing that rare mutations in salt transport genes are likely to play a substantial role in the disease. Just as predicted by the rare mutation-common disease hypothesis, many different rare mutations in a single salt transporting gene or a common pathway of salt transporting genes were found to confer the same blood pressure phenotype.
Of course, interpretation of the Ji et al. study hinged on knowing whether inferred variants actually lead to loss-of-function. The predictive power of the functional variant detection algorithms was, and still is, largely untested. Changes in evolutionarily conserved residues may not always cause loss of function. Conceivably, they might change or, even, increase function. For example, Conn's syndrome (aldosterone-producing adrenal adenomas and hypertension) can be caused by mutations in the highly conserved potassium selectivity filter of the KCNJ5 potassium channel (13), which changes the ion selectivity preference to sodium. This causes membrane depolarization, and stimulates aldosterone production and cell proliferation.
Fortunately, within 18 months of Ji's study, careful analyses of the inferred variants revealed at least 90% have a loss-of-function effect (14-16). Collectively the results of these studies indicate that rare mutations protect against hypertension by reducing the activity of renal salt reabsorptive genes. Genetic discovery followed by careful evaluation of the underlying biochemical and physiological mechanism has driven remarkable progress in nephrology and renal physiology in recent years. Studies on the hypertension resistance (HR) variants in ROMK, NKCC2, and NCC provide new insights into their biology.
HR variants in ROMK
ROMK, the product of the KCNJ1 gene and the founding member of the “inwardly rectifying” (Kir–type) potassium channel family, has two important functions in the thick ascending limb that are essential for salt reabsorption (17). The channel sets a positive transepithelial membrane potential, important for the paracellular reabsorption of cations. It also transports potassium into the TAL lumen to maintain an adequate supply of the ion to preserve the turnover of the potassium-dependent NKCC2 transporter. Without ROMK, the TAL cannot reabsorb salt (18).
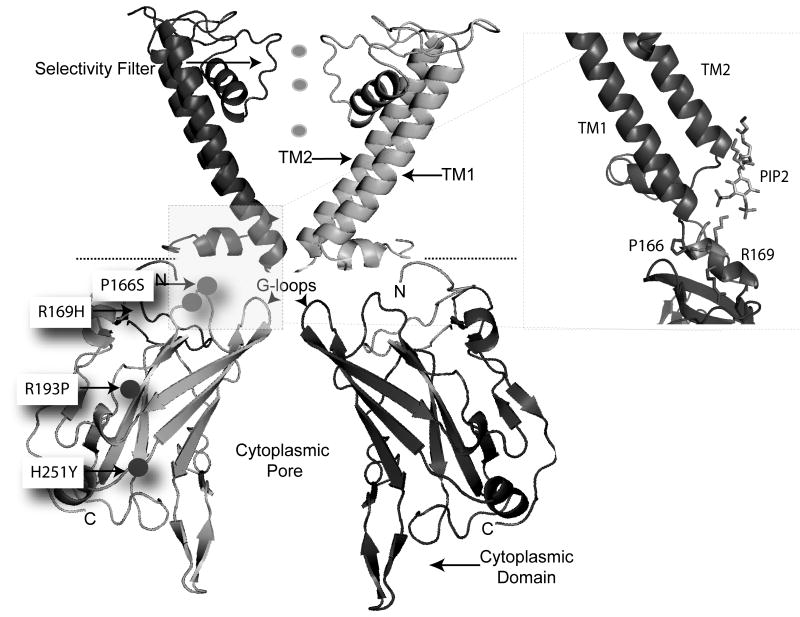
Like other Kir channels, the assembly of four ROMK subunits forms a channel (17). By homology to crystallized family members, each ROMK subunit contains two membrane domains, which flank a potassium selectivity filter. Intracellular N- and C-terminal domains fold to create a large regulatory structure with a long extended pore. The surface of this cytoplasmic domain serves as a site for modulation by kinases, lipids, and trafficking machinery. Interestingly, each of the four inferred functional mutations involve residues within the cytoplasmic domain (Figure 2).
Figure 2.
Atomic Resolution model of ROMK. Two of four subunits are shown for clarity. HR Variants are located in the cytoplasmic domain. Inset on right shows HR variants that affect PIP2-dependent gating reside in the PIP2 binding site.
Fang et al investigated the impact of the four inferred hypertension resistance (HR) variants in ROMK using Xenopus oocytes as an expression system (14). The oocyte system reliably and robustly expresses the wild-type channel, and has been used successfully to evaluate the consequence of many Bartter's disease mutations. Remarkably, all variants affected channel function, but they did so by different mechanisms.
Two of the HR mutations (R193P, H251Y) completely disrupted channel function by inhibiting cell surface membrane expression. Channels bearing these mutations become arrested in their journey through the secretory pathway somewhere between the endoplasmic reticulum (ER) and the Golgi. Such a trafficking defect is commonly observed with misfolded polytopic membrane proteins. This type of mutation, usually referred to as a processing or “trafficking” mutation, is the basis for many genetic diseases including cystic fibrosis (19), Bartter's syndrome, and Gitelman's syndrome (12). Consistent with this idea, R193 and H251 are buried deep within the structure of the cytoplasmic domain (Figure 2), and atomic resolution homology modeling predicts that the mutations should impair proper folding of the cytoplasmic domain. Misfolded proteins are usually retained in the ER or retrieved from the Golgi for anterograde traffic back to the ER, where they are degraded by ER associated degradation (ERAD) machinery (20).
Misfolded proteins are recognized as ERAD substrates in a variety of different ways (20), including by immature glycosylation status, exposure of hydrophobic residues, presence of degradation signals or absence of forward trafficking motifs. Recent studies with a closely related channel, Kir 2.1, suggest a mechanism for coupling protein folding to an early forward trafficking step in the secretory pathway (21). Export of these channels from the Golgi is determined by a conformational signal that is created once the cytoplasmic domains appropriately fold together and adopt a proper tertiary structure. Such a process provides a quality control step in the secretory pathway, ensuring only properly folded channels exit the Golgi for traffic to the cell surface. Misfolded Kir2.1 in the Golgi are targeted for anterograde traffic to the ER, where they are presumably degraded by ERAD. Further studies are required to determine whether a similar process is at play with ROMK and the R193P, H251Y variants.
The other mutations (P166S, R169H) change channel gating, conferring a negative regulatory property. Like the wild-type channel (22), these mutant channels require phosphotidylinositol 4, 5 biphosphate (PIP2) to stay open. But in contrast to the wild-type channel, which binds PIP2 so tightly that it is resistant to regulated changes in PIP2 hydrolysis (23), the P166S and P169H mutant channels bind PIP2 weakly. Remarkably, the crystal structures of related Kir channels have been recently solved with PIP2 bound (24, 25), revealing that the affected residues flank the two critical arginines in the PIP2 binding site (Figure 2). Because the mutations reduce rather than eliminate PIP2 binding, the channels remain open until PIP2 levels are perturbed (14). Indeed, in heterologous expression models, the mutant channels become inhibited upon activation of Gq-protein coupled receptors, which induce PIP2 hydrolysis through activation of Phospholipase C. The observations suggest that the effects of the mutations may be conditionally manifested depending on activation of Gq-protein coupled receptors in the thick ascending limb. For example, activation of the angiotensin II receptor (AT1R), which normally enhances sodium transport in the TAL, may inhibit these mutant ROMK channels and mute the angiotensin II response in the mutant carriers.
HR Variants in NKCC2
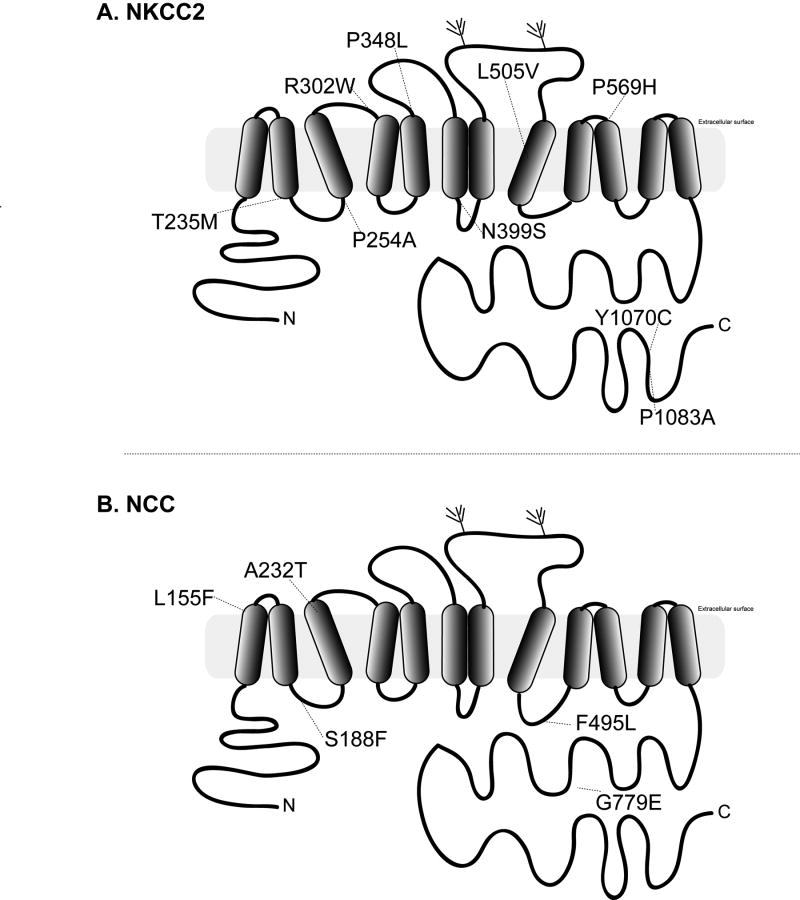
NKCC2, a member of the electroneutral cation-chloride cotransporter family (SLC12A1), is responsible for reabsorbing sodium, chloride and potassium from the tubule lumen of the thick ascending limb, and is the molecular target of loop-diuretic drugs (26). By homology to crystallized APC family members (27), NKCC2 is comprised of 12-transmembrane-helixes, which pack as two homologous domains with inverted symmetry to form the presumed ion translation pathway (Figure 3A). The cytosolic N-terminal domain contains the binding sites for kinases, including OSR-1 and SPAK (28). The large intracellular C terminus folds into an alternating β-strand α-helix bundle (29) that controls dimerization of the protein (30), membrane trafficking (31, 32) and likely serves important roles in regulation of transport activity (33). The nine HR variants affect residues in all of these regions, but most of them occur within the transmembrane domains.
Figure 3.
Location of HR Variants in NKCC2 (A) and NCC (B).
Monette and colleagues (15) and Acunna et al. (16) found that seven of the nine variants reduced transporter function under basal conditions (6 of 9) or activating conditions (1 of 9, P254A). The mutagenic effects were qualitatively similar whether the studies were performed in Xenopus oocytes or HEK cells, providing confidence in the observations. It remains uncertain why two of the mutations (P348L and Y1070C) did not markedly alter function, but results with the gating mutants in the ROMK channel illustrate that deleterious effects of some mutations may be conditionally manifested, depending on cell-specific factors or certain negative regulatory activity.
The most severe loss-of-function mutants (R302W and L505V) exhibit a profound decrease in plasma membrane expression, accumulating within an intracellular compartment, likely the ER (15). These mutants appear to be targeted for degradation by ERAD, similar to the ROMK trafficking mutants. Again, this is a common mechanism for processing misfolded transmembrane proteins, and has been observed with other Bartter's mutations in NKCC2 (12). Interestingly, the two variants exhibit different responses to low temperature-induced adjustments of membrane folding and ER quality control, suggesting that the two mutants are recognized as defective at different steps in the maturation process. Perhaps this shouldn't be so surprising given the different location of the affected residues. R302 is situated at a transmembrane-extracellular interface, while L505V projects into the cytoplasmic domain from the intracellular side of a different transmembrane helix.
The T235M mutation also reduced expression at the cell surface membrane, but effect was much more mild than other trafficking mutants (15). It remains to be determined if T235M is a substrate for ERAD, but the mutant localizes near the cell surface rather than the ER, suggesting other mis-trafficking processes may be at play. A peripheral protein quality-control system for removing unfolded CFTR from the cell surface has recently been reported (34). It will be interesting to learn if the T235M mutant is processed in a similar way.
The other functional mutations affect regulation and ion translocation processes. Two of the mutants, P254A and N399S, exhibited a blunted activation response to low intracellular chloride (15). Normally, low chloride stimulates NKCC2 ion transport by promoting the phosphorylation of three highly conserved threonines in the N- terminus through activation of a WNK-SPAK/OSR-1 kinase network (35). The affected residues, not apart of the kinase docking and phosphorylation sites in NKCC2, are positioned at intracellular ends of two adjacent transmembrane helices. Monette et al speculated that the residues are well positioned to serve as a docking site for the regulatory N-terminus, allowing phosphorylation to be translated to transport-dependent conformational changes in the transmembrane ion translocation pathway (15).
Transport measurements of extracellular ion-dependence revealed that one variant, P569H, exhibited a markedly lower affinity for Na compared to the wild-type NKCC2 (15). Such a change in Na kinetics would cause a strikingly decreased transport flux rate, especially in the cortical thick ascending limb where the tubular fluid becomes dilute. The observation reinforces an important structure function relationship. Homology modeling places the residue in the tenth transmembrane domain, which contributes to the cation translocation pathway in other APC superfamily transporters (36).
HR Variants in NCC
NCC, a member of the electroneutral cation-chloride cotransporter family (gene name SLC12A3), is responsible for reabsorbing sodium and chloride in the distal convoluted tubule, and is the molecular target of thiazide diuretics (26). Comprised of 12-transmembrane-helixes, it shares structural homology with NKCC2 (Figure. The HR variants are scattered throughout the structure. Careful measurements of transport in Xenopus oocytes revealed that all five variants reduce basal activity of NCC to variable degrees (16).
Of the NCC variants, the S186F mutation causes the most severe reduction in activity, and is the only variant to reduce NCC protein abundance (16). Although measurements of cell surface expression have not been performed, this mutant fails to undergo proper N-linked glycosylation, consistent with mistrafficking or misprocessing at an early step in the secretory pathway. Based on the phenotype, it seems likely that the S186F variant is misfolded, and a substrate for ERAD. Recent studies indicate that Gitelman's disease-causing mutations that impair NCC biogenesis also fail to escape ERAD as efficiently as the wild type protein (37, 38). Multichaperone complexes, Hsp70, Hsp40, and CHIP, have recently been implicated in selecting these misfolded NCC for ERAD (38).
The precise mechanisms driving loss-of-function in the four other rare HR variants await further characterization. None of these alter protein abundance. Moreover, each variant is activated by WNK3 or intracellular Cl depletion to the same extent as the wild-type transporter, making it unlikely that faulty phospho-dependent regulation contributes to loss-of-function.
Conclusion
Rare HR mutations in NKCC2, NCC, and ROMK alter transport function by diverse mechanisms, including changing transporter kinetics, or by altering regulation, trafficking, and biogenesis. Nevertheless, they all confer the same low blood pressure phenotype, and resistance to hypertension. The new observations demonstrate that common traits, like blood pressure variation and hypertension susceptibility, can have disparate genetic origins when different rare mutations produce the same functional effect even when vastly different mechanisms are involved.
Key Points.
Carriers for proven or inferred rare mutations in NKCC2, NCC, and ROMK have a significantly lower blood pressure and are resistant to hypertension.
Over 90% of the inferred Hypertension resistance variants were proven to have a functional effect.
HR Mutations alter function by diverse mechanisms, including protein processing, trafficking, transport turnover, regulation.
Blood pressure variation may be explained by disparate genetic origins because different rare mutations produce the same functional effect.
Acknowledgments
This manuscript was funded in part by DK63049 and DK54231 from the National Institutes of Health, National Institute of Diabetes, and Digestive and Kidney Diseases.
Footnotes
Conflicts of Interest: PAW served as a paid consultant for Pfizer and Bristol Myers Squib in the last year.
References
- 1.Salvi E, Kutalik Z, Glorioso N, et al. Genomewide association study using a high-density single nucleotide polymorphism array and case-control design identifies a novel essential hypertension susceptibility locus in the promoter region of endothelial NO synthase. Hypertension. 2012 Feb;59(2):248–55. doi: 10.1161/HYPERTENSIONAHA.111.181990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehret GB, Munroe PB, Rice KM, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011 Oct 6;478(7367):103–9. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wain LV, Verwoert GC, O'Reilly PF, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011 Oct;43(10):1005–11. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newton-Cheh C, Johnson T, Gateva V, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009 Jun;41(6):666–76. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy D, Ehret GB, Rice K, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009 Jun;41(6):677–87. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, O'Connell JR, McArdle PF, et al. From the Cover: Whole-genome association study identifies STK39 as a hypertension susceptibility gene. Proc Natl Acad Sci U S A. 2009 Jan 6;106(1):226–31. doi: 10.1073/pnas.0808358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welling PA, Chang YP, Delpire E, et al. Multigene kinase network, kidney transport, and salt in essential hypertension. Kidney Int. 2010 Jun;77(12):1063–9. doi: 10.1038/ki.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *8.Xi B, Chen M, Chandak GR, et al. STK39 polymorphism is associated with essential hypertension: a systematic review and meta-analysis. PLoS One. 2013;8(3):e59584. doi: 10.1371/journal.pone.0059584. Large metanalysis confirms the significant association of STK39 (SPAK) polymorphism with susceptibility to hypertension, supporting a role of a multi-gene kinase network (7) and altered renal salt handling in essential hypertension. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009 Apr 23;360(17):1696–8. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- **10.Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012 Nov 1;491(7422):56–65. doi: 10.1038/nature11632. This resource enables analysis of common and low-frequency variants in individuals from diverse populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji W, Foo JN, O'Roak BJ, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008 May;40(5):592–9. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seyberth HW, Schlingmann KP. Bartter- and Gitelman-like syndromes: salt-losing tubulopathies with loop or DCT defects. Pediatr Nephrol. 2011 Oct;26(10):1789–802. doi: 10.1007/s00467-011-1871-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi M, Scholl UI, Yue P, et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 2011 Feb 11;331(6018):768–72. doi: 10.1126/science.1198785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang L, Li D, Welling PA. Hypertension resistance polymorphisms in ROMK (Kir1.1) alter channel function by different mechanisms. Am J Physiol Renal Physiol. 2010 Dec;299(6):F1359–64. doi: 10.1152/ajprenal.00257.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monette MY, Rinehart J, Lifton RP, et al. Rare mutations in the human Na-K-Cl cotransporter (NKCC2) associated with lower blood pressure exhibit impaired processing and transport function. Am J Physiol Renal Physiol. 2011 Apr;300(4):F840–7. doi: 10.1152/ajprenal.00552.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Acuna R, Martinez-de-la-Maza L, Ponce-Coria J, et al. Rare mutations in SLC12A1 and SLC12A3 protect against hypertension by reducing the activity of renal salt cotransporters. J Hypertens. 2011 Mar;29(3):475–83. doi: 10.1097/HJH.0b013e328341d0fd. [DOI] [PubMed] [Google Scholar]
- 17.Welling PA, Ho K. A comprehensive guide to the ROMK potassium channel: form and function in health and disease. Am J Physiol Renal Physiol. 2009 Oct;297(4):F849–63. doi: 10.1152/ajprenal.00181.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenz JN, Baird NR, Judd LM, et al. Impaired renal NaCl absorption in mice lacking the ROMK potassium channel, a model for type II Bartter's syndrome. J Biol Chem. 2002;277:37871–80. doi: 10.1074/jbc.M205627200. 2002/// [DOI] [PubMed] [Google Scholar]
- 19.Lukacs GL, Verkman AS. CFTR: folding, misfolding and correcting the DeltaF508 conformational defect. Trends Mol Med. 2012 Feb;18(2):81–91. doi: 10.1016/j.molmed.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012 Dec 7;151(6):1163–7. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma D, Taneja TK, Hagen BM, et al. Golgi Export of the Kir2.1 Channel Is Driven by a Trafficking Signal Located within Its Tertiary Structure. Cell. 2011 Jun 24;145(7):1102–15. doi: 10.1016/j.cell.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang CL, Feng S, Hilgemann DW. Direct activation of inward rectifier potassium channels by PIP2 and its stabilization by Gbetagamma. Nature. 1998;391:803–86. doi: 10.1038/35882. [DOI] [PubMed] [Google Scholar]
- 23.Lopes CM, Zhang H, Rohacs T, et al. Alterations in conserved Kir channel-PIP2 interactions underlie channelopathies. Neuron. 2002 Jun 13;34(6):933–44. doi: 10.1016/s0896-6273(02)00725-0. [DOI] [PubMed] [Google Scholar]
- 24.Whorton MR, MacKinnon R. Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell. 2011 Sep 30;147(1):199–208. doi: 10.1016/j.cell.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen SB, Tao X, MacKinnon R. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature. 2011 Sep 22;477(7365):495–8. doi: 10.1038/nature10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gagnon KB, Delpire E. Physiology of SLC12 transporters: lessons from inherited human genetic mutations and genetically engineered mouse knockouts. Am J Physiol Cell Physiol. 2013 Apr 15;304(8):C693–714. doi: 10.1152/ajpcell.00350.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao X, Lu F, Zhou L, et al. Structure and mechanism of an amino acid antiporter. Science. 2009 Jun 19;324(5934):1565–8. doi: 10.1126/science.1173654. [DOI] [PubMed] [Google Scholar]
- 28.Gagnon KB, England R, Delpire E. A single binding motif is required for SPAK activation of the Na-K-2Cl cotransporter. Cell Physiol Biochem. 2007;20(1-4):131–42. doi: 10.1159/000104161. [DOI] [PubMed] [Google Scholar]
- 29.Warmuth S, Zimmermann I, Dutzler R. X-ray structure of the C-terminal domain of a prokaryotic cation-chloride cotransporter. Structure. 2009 Apr 15;17(4):538–46. doi: 10.1016/j.str.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Parvin MN, Gerelsaikhan T, Turner RJ. Regions in the cytosolic C-terminus of the secretory Na(+)-K(+)-2Cl(-) cotransporter NKCC1 are required for its homodimerization. Biochemistry. 2007 Aug 21;46(33):9630–7. doi: 10.1021/bi700881a. [DOI] [PubMed] [Google Scholar]
- 31.Zaarour N, Demaretz S, Defontaine N, et al. A highly conserved motif at the COOH terminus dictates endoplasmic reticulum exit and cell surface expression of NKCC2. J Biol Chem. 2009 Aug 7;284(32):21752–64. doi: 10.1074/jbc.M109.000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmosino M, Gimenez I, Caplan M, et al. Exon loss accounts for differential sorting of Na-K-Cl cotransporters in polarized epithelial cells. Mol Biol Cell. 2008 Oct;19(10):4341–51. doi: 10.1091/mbc.E08-05-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *33.Monette MY, Forbush B. Regulatory activation is accompanied by movement in the C terminus of the Na-K-Cl cotransporter (NKCC1) J Biol Chem. 2012 Jan 13;287(3):2210–20. doi: 10.1074/jbc.M111.309211. This study provides the first direct evidence that regulation of a SLC12 family member involves large movements of C termini of a dimeric cotransporter, providing a model for understanding the conformational changes required for transport. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okiyoneda T, Barriere H, Bagdany M, et al. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 2010 Aug 13;329(5993):805–10. doi: 10.1126/science.1191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ponce-Coria J, San-Cristobal P, Kahle KT, et al. Regulation of NKCC2 by a chloride-sensing mechanism involving the WNK3 and SPAK kinases. Proc Natl Acad Sci U S A. 2008 Jun 17;105(24):8458–63. doi: 10.1073/pnas.0802966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao X, Zhou L, Jiao X, et al. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature. 2010 Feb 11;463(7282):828–32. doi: 10.1038/nature08741. [DOI] [PubMed] [Google Scholar]
- 37.Needham PG, Mikoluk K, Dhakarwal P, et al. The thiazide-sensitive NaCl cotransporter is targeted for chaperone-dependent endoplasmic reticulum-associated degradation. J Biol Chem. 2011 Dec 23;286(51):43611–21. doi: 10.1074/jbc.M111.288928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *38.Donnelly BF, Needham PG, Snyder AC, et al. Hsp70 and Hsp90 multichaperone complexes sequentially regulate thiazide-sensitive cotransporter endoplasmic reticulum-associated degradation and biogenesis. J Biol Chem. 2013 May 3;288(18):13124–35. doi: 10.1074/jbc.M113.455394. This study provides a new understanding of the mechanisms by which misfolded SLC transport molecules are targeted for degradation by ERAD. [DOI] [PMC free article] [PubMed] [Google Scholar]