Abstract
Recent studies have shown that mesothelial progenitors contribute to mesenchymal lineages of developing organs. To what extent the overlying mesothelium contributes to lung development remains unknown. To rigorously address this question, we employed Wt1CreERT2/+ mice for high-fidelity lineage tracing after confirming that Cre recombinase was mesothelial specific and faithfully recapitulated endogenous Wilms’ tumor 1 (Wt1) gene expression. We visualized WT1+ mesothelial cell entry into the lung by live imaging and identified their progenies in subpopulations of bronchial smooth muscle cells, vascular smooth muscle cells and desmin+ fibroblasts by lineage tagging. Derivation of these lineages was only observed with Cre recombinase activation during early lung development. Using loss-of-function assays in organ cultures, and targeted mesothelial-restricted hedgehog loss-of-function mice, we demonstrated that mesothelial cell movement into the lung requires the direct action of hedgehog signaling. By contrast, hedgehog signaling was not required for fetal mesothelial heart entry. These findings further support a paradigm wherein the mesothelium is a source of progenitors for mesenchymal lineages during organogenesis and indicate that signals controlling mesothelial cell entry are organ specific.
Keywords: Mesothelium, Hedgehog (Hh), Lung mesenchyme, Mouse
INTRODUCTION
The mesothelium is a thin layer of squamous epithelium that surrounds internal organs (visceral mesothelium) and lines body wall cavities (parietal mesothelium) (Mutsaers, 2004). During development, the visceral mesothelium serves as a source of progenitors for differentiated mesenchymal cells within internal organs. Using a mouse that expresses an inducible Cre recombinase from the endogenous Wt1 locus (Zhou et al., 2008), fetal heart mesothelial progenitors were found to undergo an epithelium-to-mesenchyme transition (EMT) before giving rise to cardiomyocytes, vascular smooth muscle (VSM) and endothelial cells. In the gut, the majority of VSM cells arise from the mesothelium (Wilm et al., 2005). In the liver, the mesothelial cells migrate inward and generate hepatic stellate cells and perivascular mesenchyme (Asahina et al., 2011).
Two independent studies using Cre-lox lineage tracing produced conflicting results regarding the contribution of WT1+ fetal mesothelial progenitors to the lung (Que et al., 2008; Greif et al., 2012). One study used a non-inducible Wt1-Cre (WT280Cre YAC) transgenic mouse line and showed that mesothelium gives rise to intrapulmonary artery VSM cells (Que et al., 2008). These results are confounded, however, by uncertainties regarding the strength, timing and specificity of the cellular marking in this transgenic Cre line. The other study, which was focused on lineages in the main pulmonary artery, used an inducible knock-in Wt1CreERT2/+ line and showed that the mesothelium is not a significant source of smooth muscle cells for this structure (Greif et al., 2012). The precise contribution of the early fetal lung mesothelium to lung development thus remains an open question.
Mechanisms underlying mesothelial cell entry into the developing lung are largely unknown. The importance of the hedgehog (Hh) signaling pathway in mesenchymal differentiation and the development of bronchial smooth muscle (BSM), cell migration and EMT suggest a role of Hh signaling in lung mesothelial cell entry (Bellusci et al., 1997; Weaver et al., 2003; Polizio et al., 2011; Yoo et al., 2011). Mammals express three Hh ligands: Indian hedgehog (IHH), desert hedgehog (DHH) and sonic hedgehog (SHH) (Varjosalo and Taipale, 2008). The binding of Hh ligand to the patched family receptor releases the signaling moiety smoothened (SMO) from tonic inhibition, thereby triggering activation of downstream signaling cascades and targets such as Gli1 and patched genes. Cellular sites of active Hh signaling can thus be identified by the expression of these targets.
In this study, we performed a detailed analysis of WT1 expression, and definitively clarified the specific contribution of WT1+ mesothelial lineages to the developing lung parenchyma. Our data show that the mesothelium is a source of distinct subpopulations of BSM, VSM and peri-bronchiolar fibroblasts. We further demonstrated that mesothelial cell entry into the underlying fetal lung requires active Hh signaling whereas this pathway is not operative in the fetal heart. These findings further support a paradigm wherein the mesothelium is a source of mesenchymal progenitors in development and indicate that the signals that control mesothelial cell migration are distinct for each developing internal organ.
MATERIALS AND METHODS
Mice
All original mouse lines were purchased from Jackson Laboratories followed by mating to generate experimental mouse lines used in our study: Wt1CreERT2 (Jackson Laboratories stock 010912), Rosa-CAGfstdTomato (007909), Rosa-lacZ (003474), Ptch1lacZ (003081), Gli1lacZ (008211), Gli1CreERT2 (007913), Smof/f (004526) and ShhCre (005622). For timed pregnancy, identification of the vaginal plug was considered as embryonic day (E) 0.5. To activate CreERT2, 1 mg tamoxifen (TAM) (5 mg/ml, Sigma) was injected intraperitoneally per dose. C-sections were performed on animals that developed dystocia. The pups were fostered with CD1 timed-pregnant mothers. All mouse procedures were performed in accordance with approved protocols by LASC at Boston University School of Medicine.
Immunohistochemistry and detection of β-galactosidase activity
Sections of formalin-fixed rhesus macaque lung tissues (5-6 μm) were kindly provided by Dr Alice Tarantal, National Heart, Lung, and Blood Institute (NHLBI) Center for Fetal Monkey Gene Transfer for Heart, Lung, and Blood Diseases. Dissected mouse lungs were fixed in 4% paraformaldehyde (PFA)/PBS prior to embedding and sectioning. Tissue sections (8 μm) were blocked in 2.5% goat serum or MOM block (Vector Laboratories). High pH antigen retrieval (Vector Laboratories) was used when staining for mesothelin and WT1. Primary antibodies used include: FITC-conjugated anti-α-smooth muscle actin (α-SMA) (1:100, Sigma), anti-WT1 (1:200, Dako), anti-desmin (1:100, Sigma), anti-SNAIL2 (SNAI2 - Mouse Genome Informatics) (1:100, Cell Signaling), anti-KI67 (1:100, BD Biosciences) and anti-mesothelin (1:1000, Abbiotec). Antigen-antibody complex was visualized by fluorescence or DAB.
Staining for β-gal expression (β-gal+) was performed as described (Hogan et al., 1994). After staining, sections were washed in PBS, postfixed with 4% PFA for 4 hours and processed for paraffin embedding. Sections (5 μm) were deparaffinized, dehydrated, and counterstained with Nuclear Fast Red.
In situ hybridization
Wt1 mRNA expression on 8 μm frozen lung sections was assessed by in situ hybridization as described (Ai et al., 2007). The Wt1 antisense probe was generated from a 1.4 kb Wt1 cDNA (Gao et al., 2005).
RT-PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen). cDNA was transcribed using the GoScript reverse transcription system (Promega). All TaqMan probes were obtained from Applied Biosystems and all SYBR primers were from Integrated DNA Technologies. Quantitative real-time PCR (qRT-PCR) was performed using a Step One Plus instrument (Applied Biosystems). Assays were performed in triplicate and normalized to 18S rRNA (Rn18s) or Gapdh. Relative gene expression was calculated by the 2-ΔΔCT method (according to the specifications of the Applied Biosystems protocol). TaqMan primers for 18S rRNA (4319413E), Wt1 (Mm01337048_m1), Gli1 (Mm00494654_m1), Ptch1 (Mm00436026_m1), smoothelin (Mm00449973_m1), Myocd (Mm00455051_m1) and Nkx2.1 (Mm00447558_m1) were purchased from Invitrogen. Primer sequences used for SYBR-based methods were: Acta2, forward GTCCCAGACATCAGGGAGTAA and reverse TCGGATACTT CAGCGTCAGGA; Gapdh, forward AACCAGAAGACTGTGGATGG and reverse CACATTGGGGGTAGGAACAC.
Lung explant cultures and time-lapse live imaging
Wt1CreERT2/+;Rosa(tmRed) pregnant mice were injected with TAM at E10.5 and E11.5 and embryos were harvested at E12.5. Lungs were cultured on Transwell inserts (Corning) in the presence of cyclopamine (0.5 μM, Calbiochem) or DMSO vehicle (Radzikinas et al., 2011). After 24 hours, lungs were processed for analysis. For live imaging, tmRed+ lungs were isolated from mice that had been administered one dose of TAM at E10.5. The lungs were cultured for 24 hours on Transwell inserts and transferred to a glass-bottom culture dish, placed in a 37°C humidified chamber and imaged for 2.5 hours using a Zeiss LSM 710 Live-Duo Scan 2 photon confocal microscope with a 20× objective. For each lung, three different locations were imaged. Transmitted light was used to visualize the edge of the lung. The data were analyzed using Zen 2011 software (Carl Zeiss). Movies were generated for each focal plane and exported to a QuickTime (Apple) format at two frames per second.
Cell quantification
To quantify tmRed+ cells in the parenchyma of lung explants, sections from three vehicle-treated controls and three cyclopamine-treated lungs were analyzed per defined unit area. For in vivo mesothelial loss-of-Hh signaling studies, ten lung sections from each mouse were examined. tmRed+ cells in the lung parenchyma were calculated per defined lung area and quantified using ImageJ [NIH (Schneider et al., 2012)].
Relative quantification of tmRed+ BSM
To measure the relative percentage of the smooth muscle area around bronchi that is tmRed+, lung sections were immunostained for α-smooth muscle actin (α-SMA). After imaging medium-sized airways, ImageJ was used to quantify the total α-SMA immunoreactive area and the area that was concomitantly tmRed+. Ten airways from five mice were examined.
Data analysis
Data are presented as mean ± s.e.m. Statistical analyses were performed using Student’s t-test with P<0.05 considered significant.
RESULTS
Identification of WT1 as a selective fetal lung mesothelial cell marker
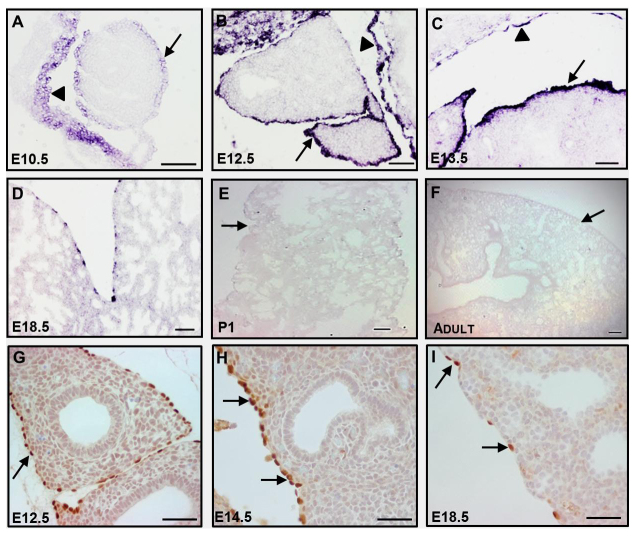
In order to assess whether WT1 can serve as a specific marker of the fetal lung mesothelium, we analyzed the expression of mouse Wt1 mRNA and protein by in situ hybridization and immunohistochemistry. We found that WT1 is localized exclusively and uniformly in visceral mesothelial cells covering the surface of the lung and in parietal mesothelial cells lining the thoracic cavity between E10.5 and E14.5 (Fig. 1A-C,G,H). As development proceeded, expression of WT1 was shut down in an increasing number of visceral mesothelial cells and, ultimately, was not detected in any adult mesothelial cell (Fig. 1D-F,I). The dynamics of Wt1 mRNA expression was corroborated by qRT-PCR of whole mouse lung RNA (including visceral mesothelium) (supplementary material Fig. S1A). At no time point did we find any cells in the lung parenchyma that expressed Wt1 mRNA or protein (Fig. 1A-I). A similar temporal expression pattern for WT1 was found in rhesus macaque lungs (supplementary material Fig. S1B), indicating conserved WT1 expression across mammalian species. By contrast, mesothelin, another putative mesothelial marker, was not expressed in the early lung mesothelium, although it was detected in the heart mesothelium (supplementary material Fig. S1C).
Fig. 1.
Characterization of WT1 expression in lung mesothelial cells. (A-F) Restricted and temporal Wt1 mRNA expression in the mouse lung mesothelium from E10.5 to adult as analyzed by in situ hybridization. Arrows point to the visceral mesothelium and arrowheads point to the parietal mesothelium. (G-I) WT1 immunohistochemistry in the embryonic lung. Arrows point to WT1+ mesothelial cells. Scale bars: 50 μm in A-D,G-I; 100 μm in E,F.
Validation of the Wt1CreERT2/+ mouse for genetic labeling of fetal mesothelial cells and their lineages
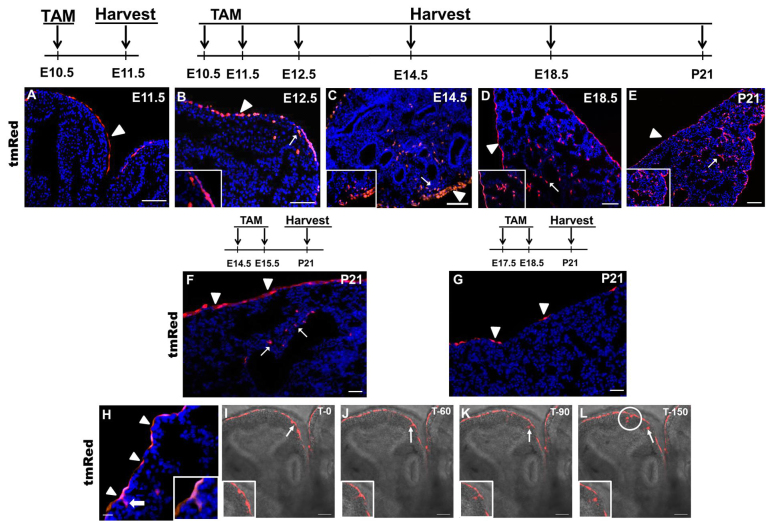
The restricted expression of WT1 to the fetal lung mesothelium indicated that the TAM-inducible knock-in Wt1CreERT2/+ mouse might be an effective tool to study fetal mesothelial cell migration and fate during lung development. To establish this, we crossed the Wt1CreERT2/+ mouse with a Rosa(tmRed) reporter and administered TAM at various time points. Lungs were harvested for evaluation of Cre-dependent expression of the tmRed red fluorescent marker. No tmRed+ cells were detected in Wt1CreERT2/+;Rosa(tmRed) mice that did not receive TAM (data not shown). In addition, tmRed+ cells were not detected in E18.5 lungs from mice that received a single dose of TAM (at E5 or E8) prior to the establishment of the lung primordium. Notably, a single dose of TAM at E10.5 labeled only visceral mesothelial cells in lungs isolated 24 hours later (Fig. 2A), indicating that Cre expression in the Wt1CreERT2/+ fetal mouse lung faithfully recapitulates endogenous Wt1 expression. After two doses of TAM at E10.5 and E11.5, the entire visceral mesothelium was tmRed+ in lungs harvested at E12.5, E14.5, E18.5 and at postnatal day (P) 21 (Fig. 2B,E), showing highly efficient Cre-mediated recombination. In mice that received TAM injections at later gestational time points, however, there was diminished mesothelial labeling (Fig. 2F,G), consistent with the declining expression of WT1 as development advances (Fig. 1). Collectively, these findings demonstrate that WT1 is a marker of early embryonic lung mesothelium and that the Wt1CreERT2/+ mouse is an efficient and faithful tool for characterization of mesothelial cell migration and lineage relationships during lung development.
Fig. 2.
Mesothelial cell entry into the lung parenchyma. (A-E) Examination of tmRed+ cells in the lungs of Wt1CreERT2/+;Rosa(tmRed) mice at various time points after one or two doses of TAM at E10.5 and E11.5. Arrowheads point to tmRed+ mesothelial cells and arrows point to tmRed+ cells in the lung parenchyma, as magnified in insets. (F,G) Analysis of tmRed+ cells in the lungs of Wt1CreERT2/+;Rosa(tmRed) mice at P21 after two doses of TAM at E14.5 and E15.5 (F) or at E17.5 and E18.5 (G). Arrowheads point to scattered tmRed+ cells in the lung mesothelium. Arrows point to tmRed+ cells in the lung parenchyma. (H) tmRed+ mesothelial cells (arrowheads) and tmRed+ cells that straddle the surface and the parenchyma of the lung (arrow) at E14.5 in Wt1CreERT2/+;Rosa(tmRed) mice that received two doses of TAM at E10.5 and E11.5. Inset is an enlargement of the straddling tmRed+ cell. (A-H) Nuclei are counterstained with DAPI (blue). (I-L) Time-lapse series from live confocal imaging of a tmRed+ lung explant. E12.5 lungs were isolated from TAM-treated Wt1CreERT2/+;Rosa(tmRed) mice and cultured on Transwell inserts for 24 hours before imaging. Sequential movie frames from a single z-focal plane show the movement of a tmRed+ cell (arrow and magnified in inset) from the mesothelium into the lung parenchyma over 150 minutes. The appearance of three additional tmRed+ cells in this focal plane over the 150 minutes is indicated by the white circle. The gray contours of the underlying lung are due to transmitted light. Scale bars: 50 μm.
Migration of fetal mesothelial cells into the lung
tmRed+ mesothelial cells appear thin and flat on the surface of the lung in TAM-treated Wt1CreERT2/+;Rosa(tmRed) mice (injection at E10.5 and E11.5) (Fig. 2A,G). However, tmRed+ cells with a rounded morphology were found straddling the surface and parenchyma of the lung at E14.5 (Fig. 2H), suggesting entry into the underlying lung. Consistent with mesothelial cell migration from the surface, tmRed+ cells were also found deeper within the parenchyma at E12.5, E14.5, E18.5 and P21 (Fig. 2B-E). Overall, the tmRed+ mesothelium-derived cells accounted for ∼5% of total lung parenchymal cells at E14.5. With later gestational TAM injections at E14.5 and E15.5, a reduced number of tmRed+ parenchymal cells was observed (compare Fig. 2E with 2F). In addition, no tmRed+ parenchymal cells were found in postnatal lungs after TAM injections at E17.5 and E18.5, although a few scattered visceral mesothelial cells were labeled (Fig. 2G). These observations suggest that WT1+ mesothelial cells enter the fetal lung parenchyma up until ∼E17.
In order to prove that WT1+ mesothelial cells actively migrate into the lung parenchyma, we conducted time-lapse live imaging of lung explants. tmRed+ lungs were harvested from E12.5 Wt1CreERT2/+;Rosa(tmRed) mice after one dose of TAM given at E10.5. The lungs were then cultured for 24 hours in an air-liquid interface prior to live imaging using a two-photon confocal microscope. Images of cells at discrete focal planes were collected every 10 minutes over 2.5 hours. During imaging, we observed thin and flat tmRed+ mesothelial cells moving from the surface into parenchymal lung regions whereupon they attained a rounded morphology (Fig. 2I-L; supplementary material Movie 1). In addition, we also observed tmRed+ cells that were initially out of the focal plane but then appeared during the course of live imaging, consistent with cell movement along a vertical axis.
Lineage analysis of fetal mesothelial cells in the lung
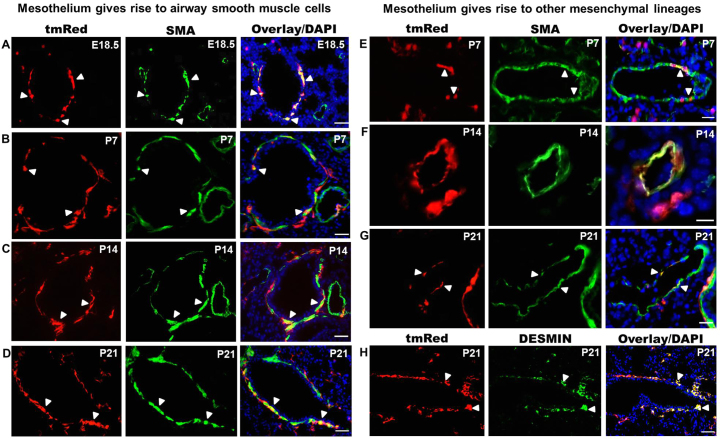
We performed detailed lineage analysis of the early fetal lung mesothelium using Wt1CreERT2/+;Rosa(tmRed) mice. TAM was administered at E10.5 and E11.5 and lungs were harvested at E18.5 and at several postnatal time points (P7, P14 and P21). At all times examined, tmRed+ cells that co-express α-SMA were found circumferentially positioned around airways, indicative of a BSM fate (Fig. 3A-D). These cells comprised a distinct subset of the total BSM compartment. Among five different lungs analyzed, 14% to 60% of airways contained at least one or multiple tmRed+ BSM cells (supplementary material Fig. S3A). The percentage of smooth muscle surface area derived from the mesothelium ranged from 22% to 81% (supplementary material Fig. S3B). Administration of TAM at the time of lung bud formation (E9.5 and E10.5) revealed a similar pattern of tmRed+ cell infiltration and of tmRed+ BSM mesothelial-derived cells at E18.5 (supplementary material Fig. S2). In the postnatal lung only, we observed tmRed+ cells that co-express α-SMA in the walls of pulmonary arteries and veins, indicative of a VSM fate (Fig. 3E-G). We also found tmRed+ peri-bronchiolar cells with a fibroblastic morphology that were α-SMA- and desmin+ (Fig. 3H), indicating a fibroblast cell fate.
Fig. 3.
Lineage tracing of fetal lung mesothelium using Wt1CreERT2/+;Rosa(tmRed) mice. TAM was given at E10.5 and E11.5 and the lungs examined at E18.5, P7, P14 and P21 for tmRed+ cells that co-express α-SMA or desmin. (A-D) Colocalization of tmRed and α-SMA in a subset of bronchial smooth muscle cells (arrowheads) from E18.5 to P21. Vascular smooth muscle cells were not labeled with tmRed at E18.5. (E-G) Colocalization of tmRed and α-SMA in postnatal vascular (E,F) and venous (G) smooth muscle cells (arrowheads). (H) Colocalization of tmRed and desmin in fibroblast cells around the airway at P21 (arrowheads). Nuclei were counterstained with DAPI (blue). Scale bars: 50 μm.
Active Hh signaling in WT1+ fetal mesothelial cells
We next sought to identify signals that might control the entry of fetal visceral mesothelial cells into the lung parenchyma. We focused on the Hh pathway due to its specialized role in cell migration and EMT (Polizio et al., 2011; Yoo et al., 2011) and because of previous work in the fetal kidney in which it was shown that WT1 regulates the expression of Hh pathway constituents (Kreidberg et al., 1993; Hartwig et al., 2010). Using three independent reporter mouse lines, we examined whether the Hh pathway was active in the lung mesothelium between E10.5 and E16.5, a time frame that coincides with Wt1 expression and mesothelial cell entry. The first reporter was a knock-in Ptch1lacZ/+ mouse, in which lacZ expression parallels patched 1 (Ptch1) expression, a direct downstream target of Hh signaling (Bellusci et al., 1997). At E14.5 we found abundant β-gal+ cells in the lung visceral mesothelium, indicative of active Hh signaling (Fig. 4A).
Fig. 4.
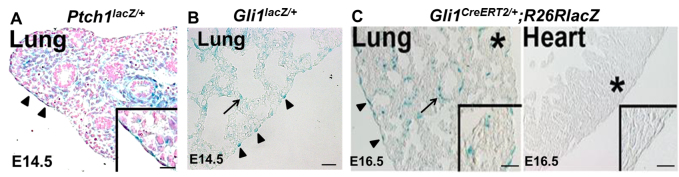
Active Hh signaling in fetal lung mesothelial cells. (A) E14.5 Ptch1lacZ/+ mouse lungs contained β-gal+ visceral mesothelial cells (arrowheads and magnified in insets) and β-gal+ submesothelial and BSM cells. Nuclei were stained with Nuclear Fast Red. (B) E14.5 Gli1lacZ/+ reporter mice contained β-gal+ visceral mesothelial cells (arrowheads) and β-gal+ lung mesenchymal cells (arrow). (C) Analysis of Hh signaling in Gli1CreERT2/+;R26RlacZ reporter mice. TAM was injected at E12.5 and lungs were examined at E16.5. β-gal+ cells were found in the lung mesothelium (arrowheads), BSM (arrow) and lung mesenchyme but not in the heart. Asterisk marks the area enlarged in insets. Scale bars: 50 μm.
To confirm this observation, we sacrificed the Gli1lacZ/+ reporter mouse (Bai et al., 2002) at E14.5 and similarly found β-gal+ visceral mesothelial cells (Fig. 4B). For further validation, we employed the Gli1CreERT2/+;R26RlacZ reporter mouse, which carries a TAM-inducible Cre under the control of the Hh signaling target Gli1 (Ahn and Joyner, 2005). This mouse was given a single dose of TAM between E10.5 and E14.5 and the lungs were analyzed at E16.5. β-gal+ cells were found in the visceral mesothelium (Fig. 4C; supplementary material Fig. S4A) but not in the parietal mesothelium (supplementary material Fig. S4A). Consistent with published findings, active Hh signaling was also observed in other mesenchymal compartments, including the submesothelium and BSM in all three reporter mice. Notably, we did not detect any Hh-responsive cells in the heart mesothelium of TAM-treated Gli1CreERT2/+;R26RlacZ embryos (Fig. 4D). This latter finding is consistent with an earlier role of Hh in heart field specification (Thomas et al., 2008). Collectively, these results demonstrate that the Hh pathway is active in the lung visceral mesothelium during a period that overlaps with WT1 expression and cell entry into the fetal lung parenchyma.
To identify the source and identity of the Hh ligand for mesothelial signaling, we assessed mRNA expression for all three Hh ligands in E14.5 lungs by qRT-PCR. Shh was the most abundantly expressed, whereas Ihh and Dhh were expressed at nearly undetectable levels (supplementary material Fig. S4B). We then performed in situ hybridization and lineage tracing using ShhCre/+;R26RlacZ mice to identify SHH-producing cells. Both assays showed that Shh is only expressed in the lung epithelium (supplementary material Fig. S4C,D), consistent with a paracrine mode of Hh signaling to the fetal visceral mesothelium. Notably, we did not detect Shh expression in the visceral or parietal pleura.
Hh signaling is required for WT1+ fetal mesothelial cell entry into the lung
To further examine the role of Hh signaling in WT1+ mesothelial cell entry, we isolated lungs from E12.5 Wt1CreERT2/+;Rosa(tmRed) embryos after two doses of TAM. These lungs were cultured in an air-liquid interface for 48 hours in the presence or absence of the Hh pathway inhibitor cyclopamine (Radzikinas et al., 2011). Inhibition of Hh signaling was confirmed by qRT-PCR analysis of downstream Hh targets (supplementary material Fig. S5). We assessed the number of tmRed+ cells within the lung parenchyma of three DMSO vehicle-treated control lungs and three cyclopamine-treated lungs; for each lung, tmRed+ parenchymal cells in eight serial sagittal sections were quantified. Cyclopamine-treated lungs had a significant reduction (P<0.05) in the number of tmRed+ cells (2.16±0.71/unit area compared with 7.58±1.18/unit area for controls) within the lung parenchyma (Fig. 5A,B), suggesting that active Hh signaling is required for mesothelial cell entry into the lung parenchyma. Consistent with this, live imaging of cyclopamine-treated lungs revealed no migration of cells into the lung parenchyma (Fig. 5C-F; supplementary material Movie 2).
Fig. 5.
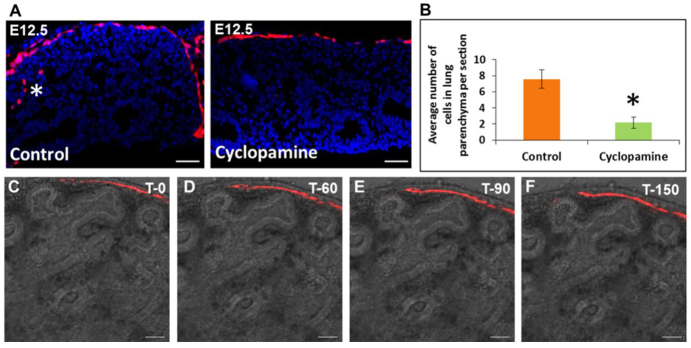
Cyclopamine blocks mesothelial cell entry into the lung parenchyma in lung explants. (A) Effect of Hh pathway inhibition on mesothelial cell entry into cultured fetal lungs. E12.5 lungs were isolated from TAM-treated Wt1CreERT2/+;Rosa(tmRed) mice and cultured for 48 hours with DMSO vehicle (control) or cyclopamine (0.5 μM). Asterisk marks mesothelium-derived tmRed+ cells in the lung parenchyma of a control lung. (B) Quantification of tmRed+ cells in the lung parenchyma of cyclopamine-treated lungs compared with controls. Data are mean ± s.e.m. from three control and three cyclopamine-treated lungs (eight sections each). *P<0.05. (C-F) Time-lapse series from live confocal imaging of a cyclopamine-treated lung explant. Shown are frames of a movie taken from a single z-focal plane over 150 minutes. The gray contours of the underlying lung are due to transmitted light. Scale bars: 50 μm.
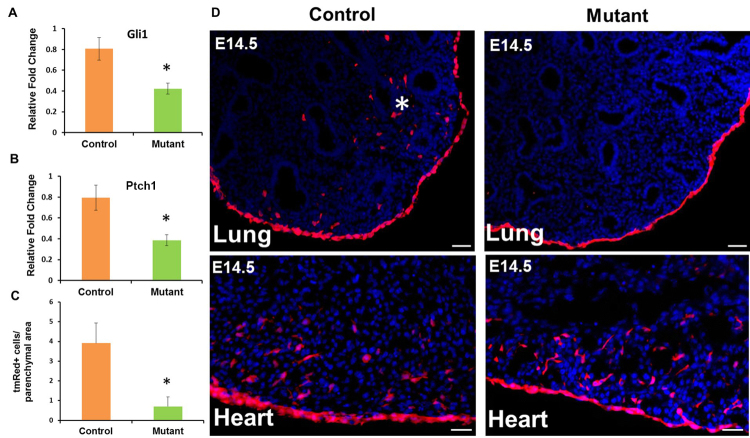
To demonstrate that Shh acts directly on fetal mesothelial cells to induce entry, we generated mesothelial loss-of-Hh signaling embryos by crossing Wt1CreERT2/+ mice with Smof/f mice. To allow simultaneous lineage labeling by tmRed, we took advantage of a rare recombination event that resulted in the localization of the floxed Smo gene and the Rosa locus on the same chromosome 5. Out of 74 embryos from matings between Wt1CreERT2/+;Smof/f males and Smof/+;Rosa(tmRed) females, we identified three Wt1CreERT2/+;Smof/f;Rosa(tmRed) mice. In these mutant mice, TAM administration led to mesothelial-deficient Hh signaling and concurrent activation of the lineage tag tmRed. The Hh target genes Gli1 and Ptch1 were significantly downregulated in loss-of-Hh mutant lungs compared with Wt1CreERT2/+;Smof/+;Rosa(tmRed) littermate controls, confirming disruption of Hh signaling (Fig. 6A,B). Examination of lung sections from mutant mice revealed that the mesothelium was uniformly labeled, similar to controls (Fig. 6D). There was, however, a significant reduction (P<0.05) in the number of tmRed+ cells in the lung parenchyma (0.7±0.4/unit area of mutant lungs compared with 3.91±1.0/unit area for littermate controls) (Fig. 6C,D). This was associated with a significant reduction in the expression of multiple smooth muscle-associated genes in the mutant mouse lung (supplementary material Fig. S6A). By contrast, mRNA expression of Nkx2.1, a key epithelium-specific transcription factor, was not affected in loss-of-Hh mutant lungs (supplementary material Fig. S6A). Grossly, mutant lungs were smaller and had a more rounded morphology at E18.5 (supplementary material Fig. S6B). Importantly, we did not detect any difference in the migration of tmRed+ cells into mutant versus control fetal hearts (Fig. 6D).
Fig. 6.
Mesothelial Hh loss-of-function prevents the appearance of mesothelium-derived cells in the lung parenchyma. Lungs from Wt1CreERT2/+;Smof/f;Rosa(tmRed) mouse embryos (mutant) and Wt1CreERT2/+;Smof/+;Rosa(tmRed) littermates (control) were collected at E14.5 after two doses of TAM at E10.5 and E11.5. (A,B) qRT-PCR analysis for Gli1 and Ptch1 mRNA expression in control and mutant lungs. Results were normalized to 18S rRNA. Data are mean ± s.e.m. from three mice. *P<0.05. (C) Quantification of tmRed+ cells in the lung parenchyma of mesothelial loss-of-Hh signaling mutant lungs compared with controls. The number of tmRed+ cells in the lung parenchyma was normalized by area. Data are the average (± s.e.m.) number of tmRed+ cells per unit area from a total of 30 lung sections, with ten sections per embryo. *P<0.05. (D) Representative images of the lung and heart from mesothelial Hh loss-of-function mutant and control embryos at E14.5. tmRed+ cells were visualized by fluorescent microscopy. Asterisk marks tmRed+ cells in the parenchyma of a control lung. Scale bars: 50 μm.
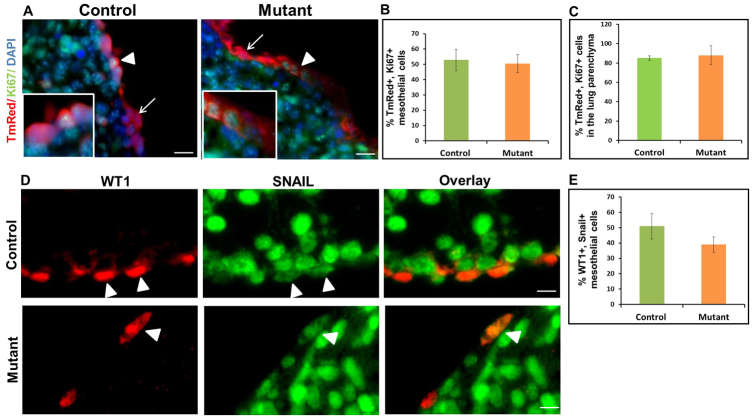
To test whether deficient Hh signaling in mutant lungs alters the proliferation and/or apoptosis of mesothelial lineages leading to a reduced number of tmRed+ lung parenchymal cells, we performed KI67 and activated caspase 3 immunostaining in E14.5 mutant and control lungs. We did not detect any difference in the percentage of surface or parenchymal tmRed+ cells that were KI67+ (Fig. 7A-C). In addition, we did not observe apoptosis in lungs of either genotype at this developmental stage (data not shown: fetal liver served as a positive control). Finally, to determine whether Hh loss-of-function in the visceral mesothelium affected the expression of EMT-related genes, we performed SNAIL2 immunohistochemistry and found no difference between control and mutant lungs (Fig. 7D,E).
Fig. 7.
Mesothelial Hh loss-of-function has no effect on cell proliferation or SNAIL2 expression in mesothelial cells and mesothelium-derived lineages in lung parenchyma. Lungs were dissected from E14.5 Wt1CreERT2/+;Smof/f;Rosa(tmRed) mouse embryos (mutant) and Wt1CreERT2/+;Smof/+;Rosa(tmRed) littermates (control) after two doses of TAM (E10.5 and E11.5). Immunohistochemistry for KI67 and SNAIL2 was performed on lung sections from mutant and control embryos. (A) KI67 immunolabeling of lung sections from mutant and control embryos. Arrowheads mark tmRed+ mesothelial cells that were also KI67+. Arrows point to tmRed+ mesothelial cells that were KI67-. (B,C) The percentage of KI67+ cells among the tmRed+ population in the mesothelium and the lung parenchyma of control and mutant embryos. (D) Double staining for SNAIL2 and WT1 in the mesothelium of mutant and control lungs. Arrowheads point to SNAIL2+ WT1+ cells. (E) The percentage of SNAIL2+ WT1+ cells in the lung mesothelium of control and mutant embryos. Nuclei were counterstained with DAPI. Results are mean ± s.e.m. from three mice. Scale bars: 50 μm.
DISCUSSION
In this study, we employed a rigorous Wt1CreERT2/+ lineage-tracing system whose fidelity was confirmed to identify fetal mesothelium-derived progenies. We showed that fetal WT1+ mesothelial cells give rise to BSM, VSM cells and desmin+ fibroblasts during embryonic and postnatal lung development. Of these cells types, only BSM was found to emerge during the embryonic period, whereas others were identified after birth, suggesting different kinetics for mesothelial progenitor differentiation between lineages. Considering that mesothelial cell entry spans several days of gestation, one possibility is that mesothelial cells that enter the lung early give rise to BSM, whereas cells that migrate at later time points become VSM cells and fibroblast cells. Alternatively, all WT1+ fetal mesothelial cells possess a similar capacity of differentiation, and their fate is determined by the environmental signals that they encounter after entering the lung.
The uniformly high efficiency of Cre recombination in our system and the fact that only subpopulations of differentiated cells were found to be tmRed+ suggest multiple origins for the mesenchymal lineages of the lung. In this context, the primitive fetal lung mesenchyme, including Tbx+ progenitors, has been shown to give rise to smooth muscle (Greif et al., 2012). Whether mesothelial and non-mesothelial progenitors have distinct and specialized roles in the development and maturation of airways, vessels and interstitium is a fundamental issue that will need to be addressed. Another key question relates to whether differentiated mesenchymal progenies with different fetal origins have discrete functional roles in homeostasis and tissue repair in postnatal life.
Using a non-inducible Wt1-Cre transgenic mouse, only VSM cells were found to arise from the surface mesothelium (Que et al., 2008). By contrast, we employed an inducible Cre that was knocked into the Wt1 locus; thereby supporting rigorous lineage tracing. Furthermore, a recent lineage-tracing study using a mesothelin knock-in Cre system found that VSM cells and fibroblasts in many tissues, including lung, arise from the overlying mesothelium (Rinkevich et al., 2012). We, however, did not observe mesothelin expression in the early fetal lung mesothelium, although we did observe expression in the fetal heart mesothelium; this expression pattern is in agreement with GenePaint analysis (Visel et al., 2004). These data argue that mesothelin is not a general marker for the early lung mesothelium. Thus, mesothelin+ mesothelial cells might represent a subset of progenitors whose contribution to lung development follows a timecourse that is different from what we observed with WT1+ progenitors.
Using complementary in vivo and in vitro assays and live imaging, we found that WT1+ mesothelial cell entry into the underlying fetal lung requires the direct action of the Hh signaling pathway. Most notably, Hh signaling was not required for entry of mesothelial cells into the developing fetal heart, which is consistent with previous results showing that the canonical Wnt/β-catenin signaling pathway is a key signal mediating movement of heart mesothelial cells (von Gise et al., 2011). Our data suggest that lung epithelium-derived SHH ligand acts as a paracrine signal to activate Hh signaling in the overlying mesothelium. Interestingly, recent work shows that filopodia can deliver SHH ligand to targets in tissue over a long range (Sanders et al., 2013). In this context, it is important to note that the timing of SHH airway epithelial expression coincides with WT1+ mesothelial cell entry (Bellusci et al., 1997). We confirmed the essential role of this pathway through the targeted disruption of mesothelial SMO function. In these mice, we demonstrated significantly reduced mesothelial cell migration into the fetal lung parenchyma without any accompanying change in the gross morphology of the overlying mesothelium, cell proliferation, expression of EMT-related genes or apoptosis. It is possible that the occurrence of tmRed+ cells in the mutant lung parenchyma is due to incomplete excision of the floxed Smo alleles. Furthermore, we did not observe cells accumulating in the mesothelium, suggesting that Hh loss-of-function mesothelial cells are sloughed from the surface.
Based on our data, we propose two models for how Hh pathway activation controls mesothelial cell migration into the underlying fetal lung parenchyma. In the first model, SHH acts as direct chemoattractant. This model is consistent with several studies showing that SHH is a chemoattractant for multiple cell types, including neural progenitors and macrophages (Charron et al., 2003; Dunaeva et al., 2010; Polizio et al., 2011). In the second model, SHH induces a program that equips mesothelial cells with a facility for movement, which then only occurs following exposure to a second signal. In support of this second model, we did not detect any mesothelial cell movement during time-lapse imaging of cyclopamine-treated lungs. Whatever the mechanism of Hh action, WT1 expression was turned off in all cells after lung entry.
Of interest, Hh pathway activation in fetal visceral mesothelium overlaps with WT1 expression, raising the possibility of a regulatory interaction. Supporting this, WT1 binding sites have been identified within the promoter/enhancer regions of multiple Hh pathway genes, including Smo, patched and Gli genes (Hartwig et al., 2010). In a preliminary ChIP analysis of DNA isolated from early fetal mesothelial cells, we observed binding of WT1 to the promoters of Smo and patched genes. Together, these findings suggest that the distinct embryonic period of mesothelial migration reflects both the timing of WT1-dependent induction of Hh pathway genes and the availability of the SHH ligand.
In conclusion, our study establishes that multiple lung mesenchymal lineages arise from the fetal mesothelium during distinct developmental periods. We were also able to demonstrate that, during this process, active Hh signaling in the mesothelium is required for cells to enter the underlying parenchyma, whereupon they are likely to encounter additional signals that control their fate. The degree to which individual mesothelial cells are multipotent or hard-wired to assume a particular cell fate is an open question that requires further study.
Supplementary Material
Acknowledgments
We gratefully acknowledge the NHLBI Center for Fetal Monkey Gene Transfer for Heart, Lung, and Blood Diseases for providing rhesus macaque lung specimens (A. Tarantal, Principal Investigator; NIH grant #HL08574; and the Primate Center base operating grant #OD011107). We thank Calvin Fong, Kelsi Radzikinas, Melissa Chua, Anneliese Arno and Colleen Keyes for technical assistance, and Dr Jordan Kreidberg for Wt1 in situ probes.
Footnotes
Funding
This work is supported by National Institutes of Health grants [1R21HL112619 and 1R01HL116163]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
R.D. designed and performed experiments, analyzed data and wrote the manuscript. X.A. and A.F. designed the experiments and helped write the manuscript.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.098079/-/DC1
References
- Ahn S., Joyner A. L. (2005). In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature 437, 894–897 [DOI] [PubMed] [Google Scholar]
- Ai X., Do A. T., Kusche-Gullberg M., Lu K., Lindahl U., Emerson C. P., Jr (2006). Conserved domain structures and enzymatic activities of quail heparan sulfate 6-O endosulfatases. J. Biol. Chem. 281, 4969–4976 [DOI] [PubMed] [Google Scholar]
- Asahina K., Zhou B., Pu W. T., Tsukamoto H. (2011). Septum transversum-derived mesothelium gives rise to hepatic stellate cells and perivascular mesenchymal cells in developing mouse liver. Hepatology 53, 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C. B., Auerbach W., Lee J. S., Stephen D., Joyner A. L. (2002). Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129, 4753–4761 [DOI] [PubMed] [Google Scholar]
- Bellusci S., Furuta Y., Rush M. G., Henderson R., Winnier G., Hogan B. L. (1997). Involvement of Sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Development 124, 53–63 [DOI] [PubMed] [Google Scholar]
- Charron F., Stein E., Jeong J., McMahon A. P., Tessier-Lavigne M. (2003). The morphogen sonic hedgehog is an axonal chemoattractant that collaborates with netrin-1 in midline axon guidance. Cell 113, 11–23 [DOI] [PubMed] [Google Scholar]
- Dunaeva M., Voo S., van Oosterhoud C., Waltenberger J. (2010). Sonic hedgehog is a potent chemoattractant for human monocytes: diabetes mellitus inhibits Sonic hedgehog-induced monocyte chemotaxis. Basic Res. Cardiol. 105, 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Chen X., Taglienti M., Rumballe B., Little M. H., Kreidberg J. A. (2005). Angioblast-mesenchyme induction of early kidney development is mediated by Wt1 and Vegfa. Development 132, 5437–5449 [DOI] [PubMed] [Google Scholar]
- Greif D. M., Kumar M., Lighthouse J. K., Hum J., An A., Ding L., Red-Horse K., Espinoza F. H., Olson L., Offermanns S., et al. (2012). Radial construction of an arterial wall. Dev. Cell 23, 482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig S., Ho J., Pandey P., Macisaac K., Taglienti M., Xiang M., Alterovitz G., Ramoni M., Fraenkel E., Kreidberg J. A. (2010). Genomic characterization of Wilms’ tumor suppressor 1 targets in nephron progenitor cells during kidney development. Development 137, 1189–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B., Beddington R., Constantini E., Lacy E. (1994). Manipulating the Mouse Embryo, pp. 344–351 Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Kreidberg J. A., Sariola H., Loring J. M., Maeda M., Pelletier J., Housman D., Jaenisch R. (1993). WT-1 is required for early kidney development. Cell 74, 679–691 [DOI] [PubMed] [Google Scholar]
- Mutsaers S. E. (2004). The mesothelial cell. Int. J. Biochem. Cell Biol. 36, 9–16 [DOI] [PubMed] [Google Scholar]
- Polizio A. H., Chinchilla P., Chen X., Kim S., Manning D. R., Riobo N. A. (2011). Heterotrimeric Gi proteins link Hedgehog signaling to activation of Rho small GTPases to promote fibroblast migration. J. Biol. Chem. 286, 19589–19596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J., Wilm B., Hasegawa H., Wang F., Bader D., Hogan B. L. (2008). Mesothelium contributes to vascular smooth muscle and mesenchyme during lung development. Proc. Natl. Acad. Sci. USA 105, 16626–16630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzikinas K., Aven L., Jiang Z., Tran T., Paez-Cortez J., Boppidi K., Lu J., Fine A., Ai X. (2011). A Shh/miR-206/BDNF cascade coordinates innervation and formation of airway smooth muscle. J. Neurosci. 31, 15407–15415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich Y., Mori T., Sahoo D., Xu P. X., Bermingham J. R., Jr, Weissman I. L. (2012). Identification and prospective isolation of a mesothelial precursor lineage giving rise to smooth muscle cells and fibroblasts for mammalian internal organs, and their vasculature. Nat. Cell Biol. 14, 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders T. A., Llagostera E., Barna M. (2013). Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature 497, 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas N. A., Koudijs M., van Eeden F. J., Joyner A. L., Yelon D. (2008). Hedgehog signaling plays a cell-autonomous role in maximizing cardiac developmental potential. Development 135, 3789–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M., Taipale J. (2008). Hedgehog: functions and mechanisms. Genes Dev. 22, 2454–2472 [DOI] [PubMed] [Google Scholar]
- Visel A., Thaller C., Eichele G. (2004). GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 32, D552–D556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A., Zhou B., Honor L. B., Ma Q., Petryk A., Pu W. T. (2011). WT1 regulates epicardial epithelial to mesenchymal transition through β-catenin and retinoic acid signaling pathways. Dev. Biol. 356, 421–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver M., Batts L., Hogan B. L. (2003). Tissue interactions pattern the mesenchyme of the embryonic mouse lung. Dev. Biol. 258, 169–184 [DOI] [PubMed] [Google Scholar]
- Wilm B., Ipenberg A., Hastie N. D., Burch J. B., Bader D. M. (2005). The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development 132, 5317–5328 [DOI] [PubMed] [Google Scholar]
- Yoo Y. A., Kang M. H., Lee H. J., Kim B. H., Park J. K., Kim H. K., Kim J. S., Oh S. C. (2011). Sonic hedgehog pathway promotes metastasis and lymphangiogenesis via activation of Akt, EMT, and MMP-9 pathway in gastric cancer. Cancer Res. 71, 7061–7070 [DOI] [PubMed] [Google Scholar]
- Zhou B., Ma Q., Rajagopal S., Wu S. M., Domian I., Rivera-Feliciano J., Jiang D., von Gise A., Ikeda S., Chien K. R., et al. (2008). Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454, 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.