Fig. 2.
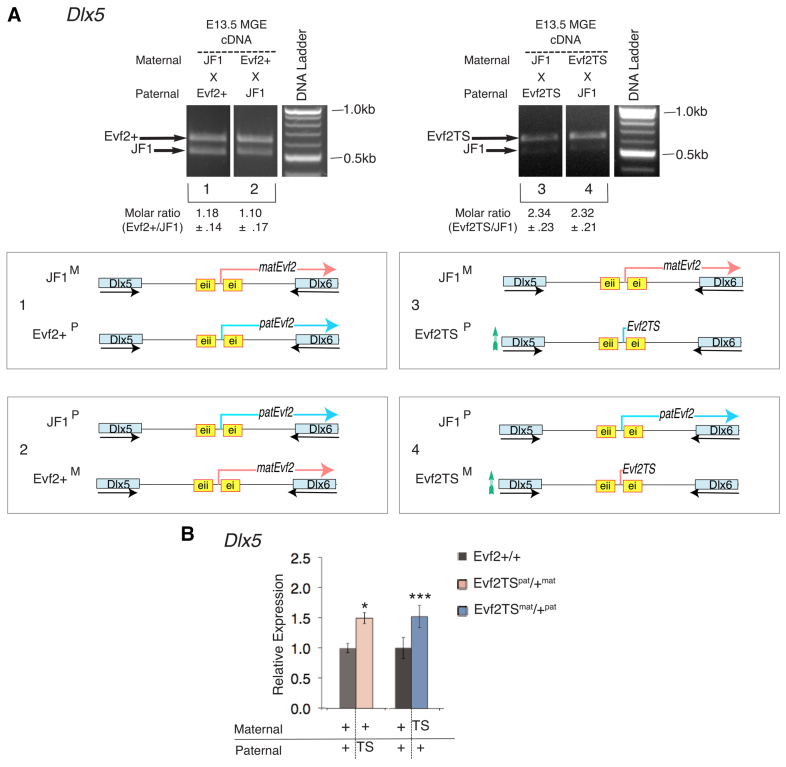
Evf2 represses Dlx5 equally on maternal and paternal alleles. (A) Imprinting analysis of Dlx5 RNA in E13.5 MGE. A SNP within Dlx5 generates a HindIII site in JF1 (Horike et al., 2005), distinguishing parental origin of Dlx5 transcripts in crosses of JF1 and Evf2+ (mixed 129/Bl6) mice. Evf2TS/+ mice are on a mixed 129/Bl6 background. Wild types are referred to as Evf2+ or Evf2+/+ to indicate littermate controls. Crosses of (1) JF1mat × Evf2+pat or (2) JF1pat × Evf2+mat indicate equal expression of Dlx5 from maternal and paternal alleles, showing that Dlx5 is not imprinted [ratios (1) Evf2+/JF1=1.18±0.14, (2) Evf2+/JF1=1.10±0.17; n=4, P>0.05]. Analysis of Dlx5 expression in (3) JF1mat × Evf2TSpat and (4) JF1pat × Evf2TSmat shows that Dlx5 expressed adjacent to transcription stop site insertion is increased (∼2.3-fold) for both maternal and paternal alleles; n=3 for each genotype, P>0.05. Schematics of the genotypes of crosses (1-4) corresponding to gel lanes are shown. M, maternal; P, paternal; pink, maternal Evf2 transcript; blue, paternal Evf2 transcript; TS, transcription stop; Evf2TS, truncated transcript from TS insertion; green arrow, increased Dlx5 expression adjacent to TS insertion. (B) Dlx5 expression increases to the same level upon maternal Evf2 or paternal Evf2 loss. Crosses of Evf2TS/+ with Evf2+/+ generate Evf2TSpat/+mat (pink) and Evf2TSpat/+mat (blue), depending on Evf2TS parental origin, as well as Evf2+/+ littermates (black). n=5 for Evf2TSpat/+mat, and n=5 Evf2+/+ littermates (*P=0.03), n=6 Evf2TSmat/+pat, n=6 Evf2+/+ littermates (***P=3.4×10-4). P values are generated by Student’s two-tailed t-test.