Abstract
Forward genetic approaches in zebrafish have provided invaluable information about developmental processes. However, the relative difficulty of mapping and isolating mutations has limited the number of new genetic screens. Recent improvements in the annotation of the zebrafish genome coupled to a reduction in sequencing costs prompted the development of whole genome and RNA sequencing approaches for gene discovery. Here we describe a whole exome sequencing (WES) approach that allows rapid and cost-effective identification of mutations. We used our WES methodology to isolate four mutations that cause kidney cysts; we identified novel alleles in two ciliary genes as well as two novel mutants. The WES approach described here does not require specialized infrastructure or training and is therefore widely accessible. This methodology should thus help facilitate genetic screens and expedite the identification of mutants that can inform basic biological processes and the causality of genetic disorders in humans.
Keywords: Kidney cyst, Exome sequencing, Cilia
INTRODUCTION
Forward genetics is a major strength of the zebrafish system. The relative ease of inducing mutations affecting a variety of organ systems has been exploited in numerous genetic screens, leading to new insights for a range of biological processes and assisting the modeling of human genetic disorders (Patton and Zon, 2001; Bakkers, 2011; Lawson and Wolfe, 2011). However, a limitation to the continuous use of this approach has been the relative difficulty in isolating mutated loci by traditional genetic approaches, such as bulk segregant analysis and chromosome walking (Zhou and Zon, 2011). This limitation has been partially overcome by the implementation of whole genome sequencing (WGS) (Bowen et al., 2012; Leshchiner et al., 2012; Obholzer et al., 2012; Voz et al., 2012) and RNA-seq-based approaches (Hill et al., 2013; Miller et al., 2013). Although both mapping strategies expedite linkage analysis, the direct identification of candidate pathogenic mutations remains a challenge. In WGS-based gene discovery approaches, the size of the zebrafish genome forces relatively low coverage due to cost considerations (typically 4× or lower). RNA-seq, although not burdened by target size, is also limited with regard to: (1) distinguishing nonsense mutations that cause mRNA decay from gene expression changes irrelevant to the phenotype; (2) the relative difficulty in uncovering genomic lesions that affect splice sites; and (3) substantial differences in the relative abundance of transcripts and thus the possibility of missing the causal gene by leaving it with low or no coverage.
Whole exome sequencing (WES) is now used routinely to isolate mutations in humans (Bamshad et al., 2011), and this technology has already been adopted by other model systems, such as the mouse (Fairfield et al., 2011). Recently, the newly released version of the zebrafish genome [Zv9 (Howe et al., 2013)] enabled the design of an exon-capture library that allowed large-scale genotyping of ENU-induced mutations in zebrafish (Kettleborough et al., 2013). Here we describe a WES-based methodology for the mapping and rapid isolation of mutations using commercially available exon-capture reagents and the freely available software SNPtrack (Leshchiner et al., 2012). We have validated our WES-based approach on an existing mutant and have used it subsequently to rapidly isolate four ENU-induced mutations that cause kidney cyst phenotypes. These include missense, nonsense and splice site mutations, demonstrating the robustness of the method. We report new alleles for ift172 and lrrc6, as well as novel mutants for the ciliary components kif3a and dync2h1.
Our WES-based method allows rapid and direct identification of both coding and splice site mutations. Importantly, the use of free and intuitive software and a robust, commercially available library preparation technology make this approach accessible to the average laboratory and considerably more efficient than the current alternatives.
MATERIALS AND METHODS
Fish stocks
Zebrafish were maintained at 28°C and raised as described (Westerfield, 2000). The zebrafish lines used for this work were: AB/TL, hdac1s436 (Noël et al., 2008), ift172hi2211Tg (Sun et al., 2004), dx36.18pd1084, aa65.6pd1086, ccn1.9pd1085 and dx33.9pd1087.
Mutagenesis and genetic screen
We used the AB/TL background and followed a classic three generation scheme using N-ethyl-N-nitrosourea (ENU) as a mutagen. Adult AB/TL males were exposed to 3 mM ENU in fish water for 1 hour at 22°C and then bathed in fresh fish water three times. The ENU treatment was repeated six times over 12 weeks. Mutagenized males were mated to AB/TL females to generate 17 F1 families with ∼500 fish in total. The F1 fish were either in-crossed or out-crossed (∼50% each) to generate 450 F2 families with at least ten pairs each. F3 embryos from crosses of F2 siblings were screened at 2 days post-fertilization (dpf) and 5 dpf.
DNA extraction, exome capture and sequencing
Genomic DNA was isolated from pools of 5-dpf larvae using the blood and tissue DNA isolation kit from Qiagen according to the manufacturer’s instructions. For library preparation, 1-3 μg genomic DNA was sheared to 150-250 bp fragments using a Covaris sonicator. These products were then run on an Agilent chip to confirm the size range.
Next, sequencing platform-specific adaptors (Illumina HiSeq) were ligated using the Kapa Biosystems library preparation kit following the manufacturer’s instructions and purified with Agencourt AMPure XP beads (Beckman-Coulter). Two different bar-coded adaptors were used for wild-type (WT) and mutant DNA to allow for pooled exon enrichment and subsequent sequencing in a single lane. Up to three separate WT/mutant pairs were sequenced in a single lane.
Exon capture was performed using the Agilent SureSelect solution-based zebrafish exon-capture kit, following the manufacturer’s protocol with the following modifications: (1) blocking oligonucleotides (5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTC CGATCT-3′; 5′-GATCGGAAGAGCACACGTCTGAACTCCAGTCACI*I*I*I*I*I*ATCTCGTATGCCGTCTTCTGCTTG-3′) were added during the hybridization step to a final concentration of 4 μM (I* indicates deoxyinosine); (2) post-capture amplification was performed using the Kapa Biosystems library amplification kit. The resulting library was then analyzed using an Agilent BioAnalyzer chip. Sequencing (100 bp paired-end run) was performed on an Illumina HiSeq 2000 instrument.
Linkage analysis
Linkage analysis was performed with SNPtrack software (http://genetics.bwh.harvard.edu/snptrack/) (Leshchiner et al., 2012).
Genetic mapping
Rough mapping of the dx36.18pd1084 and aa65.6pd1086 loci was achieved using 80 mutant embryos using HpyCH4IV, EarI and HincII for dx36.18pd1084; BsrGI and HinfI (lesion site) for aa65.6pd1086.
Chemicals and antibodies
Anti-acetylated tubulin antibody and Alcian Blue were obtained from Sigma. Phalloidin conjugated to Alexa 568 and secondary antibodies were obtained from Invitrogen.
Histology and microscopy
Tissue sections were generated from 4% paraformaldehyde-fixed specimens that were embedded in 4% low-melt agarose using a vibratome (Leica) and mounted in Vectashield (Vector Labs) (Bagnat et al., 2010). Whole-mount embryo staining for acetylated tubulin was performed as described (Navis et al., 2013). Tissue sections and whole-mount embryos were imaged with a Leica SP5 laser-scanning confocal microscope. Live embryos were imaged with a Zeiss Discovery stereo microscope. Alcian Blue staining was performed as described (Ellis et al., 2013).
RNA injection
Human KIF3A and zebrafish lrrc6 open reading frames were cloned into the pCS2 vector. RNA was injected into the yolk of one-cell stage embryos.
Morpholino knockdown
The antisense splice morpholino for dync2h1 (5′-AAACTGTGAGTTTGGCCTCAGTGTC-3′) was designed and synthesized by Gene Tools. The efficacy of the morpholino was evaluated by RT-PCR using the following primers: dync2h1-ex1, 5′-CAAAATCTCCCGCTCGTACT-3′; DYNC2H1-ex2,3-R, 5′-CCTGGAACTTCACCTGAGGA-3′ (supplementary material Fig. S1A).
RESULTS AND DISCUSSION
Development of a WES and mapping approach
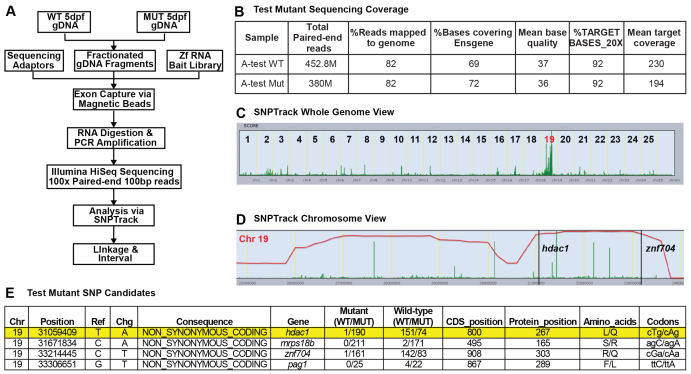
To facilitate the isolation of mutated loci in zebrafish, we sought to assemble a simple WES-based paradigm to map and identify candidate pathogenic genetic lesions (Fig. 1A). To test the exon-capture technology and mapping approach, we first used a previously isolated mutation, hdac1s436 (Noël et al., 2008). We selected this allele to develop our protocols because it is a point mutation that causes a relatively mild missense change (L to Q), thus making the affected locus inconspicuous in a blind experiment.
Fig. 1.
Development of a pipeline for the isolation of zebrafish mutations via whole exome sequencing (WES). (A) Flow diagram of our WES approach. (B) Sequencing statistics. (C) Genome view of linkage analysis using SNPtrack. (D) Homozygosity interval showing two candidate mutations. (E) The four candidate mutations for the test mutant. The T-to-A change encoding an L-to-Q missense mutation in hdac1 is highlighted in yellow.
DNA from pools of ten homozygous mutant (Mut) and ten wild-type (WT) siblings was sequenced using an Illumina HiSeq 2000 100 bp paired-end (PE) run. The WT pool was sequenced in one lane, yielding 226 million (M) PE reads with 92% of targeted bases covered ≥20×. The Mut pool was sequenced in two-thirds of a lane yielding 190M PE reads, also with 92% of targeted bases covered ≥20× (Fig. 1B). Linkage analysis was performed using the mapping tool SNPtrack (Leshchiner et al., 2012) (http://genetics.bwh.harvard.edu/snptrack/), which allows the identification of single nucleotide polymorphisms (SNPs) and regions of homozygosity. Mapping analysis yielded a single peak on chromosome 19 that was then interrogated manually by focusing on the homozygosity interval (Fig. 1C,D). Using a cut-off of >5× coverage to minimize the analysis of sequencing artifacts, we generated a list of candidate mutations. Using the ‘Find SNPs’ function of the SNPtrack software we then excluded variants found previously in the SNPtrack database. Filtering was then set to homozygous mutant/heterozygous WT only, allowing for up to one reference read in the mutant pool. Prioritized lists were generated for each of the following: stop changes, non-synonymous, and splice-site variants, resulting in 0, 4, and 0 variants, respectively. Two of these four variants had a disproportionate number of non-reference reads in the WT pool, making them unlikely candidates (Fig. 1E). The two remaining variants, znf704 R303Q and hdac1 L267Q, had an appropriate ratio of non-reference/reference reads in the WT pool, and were homozygous in the mutant pool (Fig. 1E). Although each of these variants had one reference read in the mutant pool, this was an acceptable margin of error, given the high number of total reads. Thus, WES combined with analysis using the user-friendly SNPtrack software allowed us to quickly narrow down to two candidate mutations, including the previously known causal mutation in hdac1. Importantly, this type of analysis would not have been possible with a low coverage WGS approach.
Implementation of the WES method for the identification of mutations causing renal cysts
Encouraged by the efficiency of the method, we turned our attention to its implementation in novel mutants. We recently completed a forward genetic screen to identify mutations affecting fluid regulation in zebrafish (S.R., L.M., J.B., J.M. and M.B.). Among the 67 mutants identified, we found four recessive mutant lines with large kidney cysts. Defects in primary cilia have been causally linked to cyst formation in humans (Badano et al., 2006; Hildebrandt et al., 2011) and zebrafish (Sun et al., 2004). However, the in vivo function of many key cilia components has not yet been elucidated. Further, cilia are proposed to contain over 1000 proteins (Gherman et al., 2006) and the majority of the genetic burden of ciliopathies in humans is not yet known.
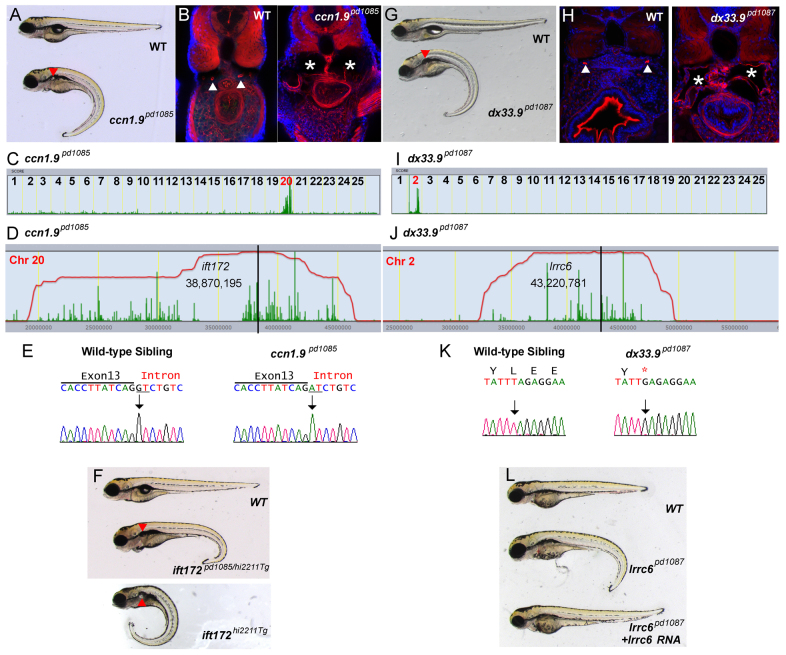
We first analyzed two mutants (ccn1.9pd1085 and dx33.9pd1087) presenting large, mostly bilateral kidney cysts and a downward body curvature (Fig. 2). Subsequent to sequencing Mut/WT, we mapped ccn1.9pd1085 and dx33.9pd1087 to chromosomes 20 and 2, respectively (Fig. 2C,I). Examination of the homozygosity interval for ccn1.9pd1085 revealed the presence of a G-to-A mutation in an essential splice donor site on exon 13 of ift172 (Fig. 2D,E), which encodes a known intraflagellar transport particle component (Ocbina et al., 2011). To validate this mutation, we first sequenced cDNA from WT and Mut and confirmed the absence of exon 13 in the mutant pool (data not shown). Next, we obtained a previously described insertional mutant, ift172hi2211tg (Sun et al., 2004), and performed a complementation test; we confirmed that ccn1.9pd1085 is a mutated allele of ift172. Similarly, dx33.9pd1087 mapped to an interval containing lrrc6, which encodes a dynein arm ciliary component (Horani et al., 2013), that bears a T-to-G nonsense mutation (Fig. 2J,K). Validation of the causality of this allele was performed by rescuing the phenotype by injecting WT lrrc6 RNA (Fig. 2L). Injection of 50 pg WT lrrc6 cRNA into one-cell stage embryos generated from dx33.9pd1087+/- parents led to ∼70% rescue (n=464).
Fig. 2.
Isolation of new mutant alleles for ift172 and lrrc6 that cause kidney cyst formation. (A) Brightfield image of 5-dpf WT sibling and ccn1.9pd1085 homozygous mutant. Red arrowhead indicates a kidney cyst. (B) Confocal images of transverse sections of 5-dpf WT sibling and ccn1.9pd1085 mutant stained with phalloidin (red) and DAPI (blue). Arrowheads point to the pronephric ducts and asterisks mark kidney cysts. (C) Linkage analysis for ccn1.9pd1085 using SNPtrack. (D) The homozygosity interval for ccn1.9pd1085. (E) Sequencing of genomic DNA of WT and ccn1.9pd1085 mutant larvae. Mutants bear a G-to-A mutation at an essential splice site in exon 13 of ift172 (underlined). (F) Brightfield image of 5-dpf WT sibling, ift172hi2211Tg/pd1085 and ift172hi2211Tg larvae. (G) Brightfield image of 5-dpf WT sibling and dx33.9pd1087 mutant larvae. (H) Confocal images of transverse sections of 5-dpf WT sibling and dx33.9pd1087 mutant larvae stained with phalloidin and DAPI as in B. (I) Linkage analysis for dx33.9pd1087 using SNPtrack. (J) The homozygosity interval for dx33.9pd1087. (K) Sequencing of genomic DNA of WT and dx33.9pd1087 mutant larvae. Mutants are homozygous for a T-to-G nonsense mutation. (L) Rescue of the dx33.9pd1087 phenotype via injection of WT lrrc6 cRNA.
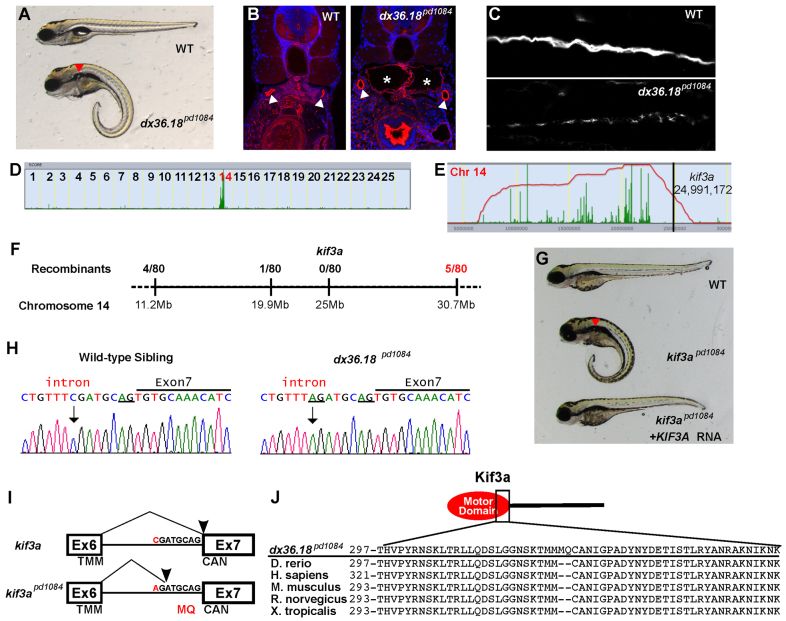
We next analyzed dx36.18pd1084, a mutant with bilateral kidney cysts and short cilia (Fig. 3A-C). This mutation mapped to a 21 Mb interval on chromosome 14 (Fig. 3D). Within this interval we found six genes with non-synonymous changes or splice site mutations. To reduce the list of candidates, we genotyped a group of 80 homozygous mutants; we defined a 10 Mb critical genomic interval that excluded four of the candidate genes. The experimentally defined critical region contained kif3a and rnf20 (supplementary material Table S3C). Given the phenotype, the known ciliary component kif3a was the most likely candidate (Kodani et al., 2013). We therefore injected cRNA for human KIF3A to try to rescue the phenotype. Injection of 100 pg human cRNA almost completely (98%, n>500 embryos) rescued the mutant phenotype. Examination of the genomic sequence revealed a C-to-A change six bases upstream of the splice acceptor site of exon 7 (Fig. 3H). Upon sequencing cDNA, we found a six nucleotide insertion between exons 6 and 7 causing an MQ insertion within the motor domain of Kif3a (Fig. 3I). Sequence alignment revealed that this region is 100% conserved in vertebrates (Fig. 3J). Taken together, these data demonstrate that dx36.18pd1084 is a mutated allele of kif3a.
Fig. 3.
kif3apd1084 mutants present defects in cilia formation and kidney cysts. (A) Brightfield image of 5-dpf WT sibling and dx36.181084 mutant larvae. Red arrowhead indicates a kidney cyst. (B) Confocal images of transverse sections of 5-dpf WT sibling and dx36.181084 mutant larvae stained with phalloidin (red) and DAPI (blue). The arrowheads point to the pronephric ducts and the asterisks mark kidney cysts. (C) Confocal image of 3-dpf WT and dx36.181084 mutant in whole-mount, stained for acetylated tubulin to mark cilia. (D) Linkage analysis for dx36.181084 using SNPtrack. (E) The homozygosity interval for dx36.181084. (F) Linkage analysis of the dx36.181084 locus. (G) Rescue of the dx36.18pd1084 phenotype with WT human KIF3A cRNA. Rescue was confirmed by genotyping (not shown). (H) Sequencing of genomic DNA of WT and kif3apd1084 mutant larvae showing a six nucleotide insertion in kif3a that adds two extra codons between exons 6 and 7 (underlined). (I) Sequencing of cDNA confirms the insertion of codons coding for MQ in kif3apd1084 mutants. TMM, threonine, methionine, methionine; CAN, cysteine, alanine, asparagine. (J) Alignment of Kif3a protein sequences from zebrafish, human, mouse, rat and frog.
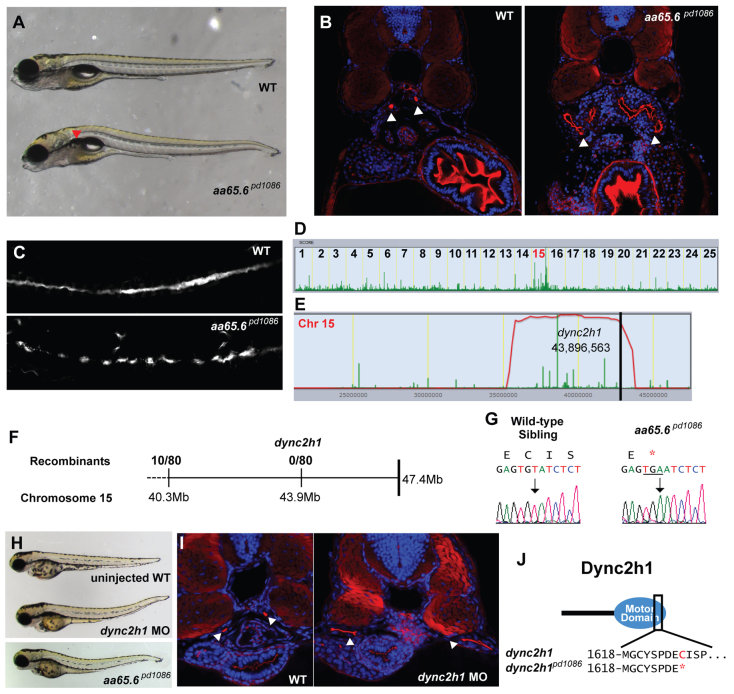
The final mutation that we studied, aa65.6pd1086, shows a relatively mild bilateral kidney cyst phenotype with no other obvious anatomical defects (Fig. 4A). Confocal imaging of transverse sections revealed expanded and collapsed pronephric ducts (Fig. 4B). We also observed short cilia along the length of the pronephros (Fig. 4C). SNPtrack mapping linked the mutation to chromosome 15 (Fig. 4D), with a homozygosity interval near the telomere (Fig. 4E). This interval contained five candidate mutations in three genes: cd28, numa1 and dync2h1. We confirmed this interval by genotyping 80 homozygous mutants (Fig. 4F) and were also able to exclude cd28 (supplementary material Table S3D). Because mutations in DYNC2H1 cause thoracic dysplasia and ciliary defects in humans (Dagoneau et al., 2009; Merrill et al., 2009; El Hokayem et al., 2012; Schmidts et al., 2013) we first confirmed the nonsense mutation by sequencing single WT and homozygous aa65.6pd1086 mutant larvae (Fig. 4G). The aa65.6pd1086 mutation produces a truncation of the motor domain of Dync2h1 (Fig. 4J). In this case, RNA rescue was not possible due to the large size of the transcript. Therefore, we pursued an alternative strategy in which we injected antisense morpholinos against a splice donor site within the same domain as the mutation; we were able to phenocopy the mutation (Fig. 4H,I), suggesting that aa65.6pd1086 is an allele of dync2h1. Of note, while analyzing the genomic locus we noticed that in Ensembl dync2h1 is currently annotated as four separate transcripts (CABZ01072601.1, CABZ01072338.1, CABZ010728921.1 and CABZ010728919.1) that correspond to the different domains of the full-length transcript (supplementary material Fig. S1C). In humans, mutations in DYNC2H1A are associated with skeletal defects (Dagoneau et al., 2009); however, we did not detect signs of skeletal abnormalities (supplementary material Fig. S1B). A previous study of cytoplasmic dynein function in zebrafish using morpholinos targeted against the region encoding the cargo-binding domain reported the formation of kidney cysts, a pronounced body curvature and cardiac edemas (Krock et al., 2009). Although it is formally possible that some aspects of that phenotype might be due to non-specific toxicity of the morpholino, it is also likely that differences in the genetic background may lead to significant phenotypic variation (Davis and Katsanis, 2012).
Fig. 4.
A truncation at the end of the motor domain of Dync2h1 leads to a cystic kidney phenotype. (A) Brightfield image of 5-dpf WT sibling and aa65.6pd1086 mutant larvae. Red arrowhead indicates a kidney cyst. (B) Confocal images of transverse sections of 5-dpf WT sibling and aa65.6pd1086 mutant larvae stained with phalloidin (red) and DAPI (blue). The arrowheads point to pronephric ducts and asterisks mark kidney cysts. (C) Confocal image of 3-dpf WT and aa65.6pd1086 mutant embryos in whole-mount, stained for acetylated tubulin to mark cilia. (D) Linkage analysis for aa65.6pd1086 using SNPtrack. (E) The homozygosity interval for aa65.6pd1086. (F) Linkage analysis using single embryos. (G) Sequencing of genomic DNA of WT and aa65.6pd1086 mutant larvae. (H) Injection of a morpholino directed against a splice site within the motor domain of dync2h1 phenocopies the aa65.6pd1086 mutation. (I) Confocal images of transverse sections 3-dpf WT and dync2h1 morphants stained with phalloidin and DAPI, showing distended pronephric ducts (arrowheads). (J) Schematic of the Dync2h1 protein illustrating that the mutation removes a portion of the motor domain.
Technical considerations
In this study, we used two different exome-capture bait libraries: v1, with a 96.7 Mb target region that was designed to contain 3′ and 5′ untranslated regions; and v2, with a 70.6 Mb target region. The number of reads mapping to exons was increased by 12-16% using v2 when compared with the v1 version, reducing cost and computational time. Another factor affecting exon coverage was prelibrary shearing size. Our data indicate that DNA sheared larger than 150-200 bp results in more off-target reads. Sequencing quality scores and the number of reads mapping to the genome remained stable within a range of 32M to 111M total PE reads. Thus, the smaller target region of v2 and a number of PE reads between 60M and 100M is optimal for achieving sufficient coverage (80% at ∼20×) for candidate mutation discovery (supplementary material Table S1).
Increasing the number of individual fish per Mut/WT pool has been shown to decrease interval size (Leshchiner et al., 2012). Nonetheless, our results suggest that when using WES even a small pool can be sufficient, since smaller sample pools with DNA from 10-40 individuals showed comparable mapping interval size (supplementary material Table S2). Outcrossing is another factor that affects interval determination; panels that have been outcrossed multiple times consistently show less noise throughout the genome and tighter linkage intervals. However, we found that panels that had been outcrossed only once were still viable for determining a linkage interval, yielding one predominate peak in the genome that in each case harbored the driver mutation.
Concluding remarks
In summary, using a WES-based approach we were able to map and efficiently identify candidate mutations in a single step. The high number of quality on-target reads obtained also provided a reliable set of SNPs that allowed us to determine critical genomic intervals accurately and reduce the number of candidate mutations to be evaluated. The approach described here represents a straightforward and versatile method for identifying and isolating all types of mutations in zebrafish. Because all reagents are commercially available and no particular expertise in bioinformatics is required to use the SNPtrack software, this method is readily accessible to the community. Finally, the scalability of the method suggests that it is now possible, within a reasonable time frame and cost, to interrogate systematically all mutants that emerge from screens.
Supplementary Material
Acknowledgments
We thank Agilent and Derek Stemple for providing early access to the exome-capture arrays; the zebrafish facility at Duke University; Voot Yin for help with mutagenesis; Zhaoxia Sun and Elke Ober for providing fish lines; and Ken Poss for critical reading of the manuscript.
Footnotes
Funding
This work was funded by the National Institutes of Health (NIH) [innovator grant DP2OD006486 to M.B., R01 DK095721 to W.G., and a pilot project of P50-DK096415 to M.B.]. Support for L.M. was provided by an NIH training grant [5T32HL098099-02]. N.K. is a Distinguished Brumley Professor. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
M.B. and N.K. concieved the study. S.R., L.M., J.B., J.M. and M.B. performed the forward genetic screen designed by M.B. and characterized mutant zebrafish. S.R. isolated genomic DNA from mutant embryos and J.W. performed exome capture and compiled data from Illumina sequencing from this genomic DNA. S.R. and J.W. performed linkage analysis with help from I.L. and W.G. using SNPtrack. S.R. performed the gene mapping for mutants with L.M. and J.W. providing additional help. M.B. and N.K. wrote the manuscript with input from the other authors.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.101170/-/DC1
References
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N. (2006). The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7, 125–148 [DOI] [PubMed] [Google Scholar]
- Bagnat M., Navis A., Herbstreith S., Brand-Arzamendi K., Curado S., Gabriel S., Mostov K., Huisken J., Stainier D. Y. (2010). Cse1l is a negative regulator of CFTR-dependent fluid secretion. Curr. Biol. 20, 1840–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkers J. (2011). Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc. Res. 91, 279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M. J., Ng S. B., Bigham A. W., Tabor H. K., Emond M. J., Nickerson D. A., Shendure J. (2011). Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 12, 745–755 [DOI] [PubMed] [Google Scholar]
- Bowen M. E., Henke K., Siegfried K. R., Warman M. L., Harris M. P. (2012). Efficient mapping and cloning of mutations in zebrafish by low-coverage whole-genome sequencing. Genetics 190, 1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagoneau N., Goulet M., Geneviè ve D., Sznajer Y., Martinovic J., Smithson S., Huber C., Baujat G., Flori E., Tecco L., et al. (2009). DYNC2H1 mutations cause asphyxiating thoracic dystrophy and short rib-polydactyly syndrome, type III. Am. J. Hum. Genet. 84, 706–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis E. E., Katsanis N. (2012). The ciliopathies: a transitional model into systems biology of human genetic disease. Curr. Opin. Genet. Dev. 22, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hokayem J., Huber C., Couvé A., Aziza J., Baujat G., Bouvier R., Cavalcanti D. P., Collins F. A., Cordier M. P., Delezoide A. L., et al. (2012). NEK1 and DYNC2H1 are both involved in short rib polydactyly Majewski type but not in Beemer Langer cases. J. Med. Genet. 49, 227–233 [DOI] [PubMed] [Google Scholar]
- Ellis K., Bagwell J., Bagnat M. (2013). Notochord vacuoles are lysosome-related organelles that function in axis and spine morphogenesis. J. Cell Biol. 200, 667–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairfield H., Gilbert G. J., Barter M., Corrigan R. R., Curtain M., Ding Y., D’Ascenzo M., Gerhardt D. J., He C., Huang W., et al. (2011). Mutation discovery in mice by whole exome sequencing. Genome Biol. 12, R86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherman A., Davis E. E., Katsanis N. (2006). The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat. Genet. 38, 961–962 [DOI] [PubMed] [Google Scholar]
- Hildebrandt F., Benzing T., Katsanis N. (2011). Ciliopathies. N. Engl. J. Med. 364, 1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. T., Demarest B. L., Bisgrove B. W., Gorsi B., Su Y. C., Yost H. J. (2013). MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq. Genome Res. 23, 687–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horani A., Ferkol T. W., Shoseyov D., Wasserman M. G., Oren Y. S., Kerem B., Amirav I., Cohen-Cymberknoh M., Dutcher S. K., Brody S. L., et al. (2013). LRRC6 mutation causes primary ciliary dyskinesia with dynein arm defects. PLoS ONE 8, e59436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K., Clark M. D., Torroja C. F., Torrance J., Berthelot C., Muffato M., Collins J. E., Humphray S., McLaren K., Matthews L., et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettleborough R. N., Busch-Nentwich E. M., Harvey S. A., Dooley C. M., de Bruijn E., van Eeden F., Sealy I., White R. J., Herd C., Nijman I. J., et al. (2013). A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 496, 494–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A., Salomé Sirerol-Piquer M., Seol A., Garcia-Verdugo J. M., Reiter J. F. (2013). Kif3a interacts with Dynactin subunit p150 Glued to organize centriole subdistal appendages. EMBO J. 32, 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krock B. L., Mills-Henry I., Perkins B. D. (2009). Retrograde intraflagellar transport by cytoplasmic dynein-2 is required for outer segment extension in vertebrate photoreceptors but not arrestin translocation. Invest. Ophthalmol. Vis. Sci. 50, 5463–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson N. D., Wolfe S. A. (2011). Forward and reverse genetic approaches for the analysis of vertebrate development in the zebrafish. Dev. Cell 21, 48–64 [DOI] [PubMed] [Google Scholar]
- Leshchiner I., Alexa K., Kelsey P., Adzhubei I., Austin-Tse C. A., Cooney J. D., Anderson H., King M. J., Stottmann R. W., Garnaas M. K., et al. (2012). Mutation mapping and identification by whole-genome sequencing. Genome Res. 22, 1541–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill A. E., Merriman B., Farrington-Rock C., Camacho N., Sebald E. T., Funari V. A., Schibler M. J., Firestein M. H., Cohn Z. A., Priore M. A., et al. (2009). Ciliary abnormalities due to defects in the retrograde transport protein DYNC2H1 in short-rib polydactyly syndrome. Am. J. Hum. Genet. 84, 542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. C., Obholzer N. D., Shah A. N., Megason S. G., Moens C. B. (2013). RNA-seq-based mapping and candidate identification of mutations from forward genetic screens. Genome Res. 23, 679–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navis A., Marjoram L., Bagnat M. (2013). Cftr controls lumen expansion and function of Kupffer’s vesicle in zebrafish. Development 140, 1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël E. S., Casal-Sueiro A., Busch-Nentwich E., Verkade H., Dong P. D., Stemple D. L., Ober E. A. (2008). Organ-specific requirements for Hdac1 in liver and pancreas formation. Dev. Biol. 322, 237–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obholzer N., Swinburne I. A., Schwab E., Nechiporuk A. V., Nicolson T., Megason S. G. (2012). Rapid positional cloning of zebrafish mutations by linkage and homozygosity mapping using whole-genome sequencing. Development 139, 4280–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocbina P. J., Eggenschwiler J. T., Moskowitz I., Anderson K. V. (2011). Complex interactions between genes controlling trafficking in primary cilia. Nat. Genet. 43, 547–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton E. E., Zon L. I. (2001). The art and design of genetic screens: zebrafish. Nat. Rev. Genet. 2, 956–966 [DOI] [PubMed] [Google Scholar]
- Schmidts M., Arts H. H., Bongers E. M., Yap Z., Oud M. M., Antony D., Duijkers L., Emes R. D., Stalker J., Yntema J. B., et al. ; UK10K (2013). Exome sequencing identifies DYNC2H1 mutations as a common cause of asphyxiating thoracic dystrophy (Jeune syndrome) without major polydactyly, renal or retinal involvement. J. Med. Genet. 50, 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Amsterdam A., Pazour G. J., Cole D. G., Miller M. S., Hopkins N. (2004). A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 131, 4085–4093 [DOI] [PubMed] [Google Scholar]
- Voz M. L., Coppieters W., Manfroid I., Baudhuin A., Von Berg V., Charlier C., Meyer D., Driever W., Martial J. A., Peers B. (2012). Fast homozygosity mapping and identification of a zebrafish ENU-induced mutation by whole-genome sequencing. PLoS ONE 7, e34671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. (2000) The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). Eugene, OR: University of Oregon Press; [Google Scholar]
- Zhou Y., Zon L. I. (2011). The Zon laboratory guide to positional cloning in zebrafish. Methods Cell Biol. 104, 287–309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.