Abstract
Background
Tumor endothelial transdifferentiation and VEGFR1/2 expression by cancer cells have been reported in glioblastoma but remain poorly documented for many other cancer types.
Methods
To characterize vasculature of patient-derived tumor xenografts (PDXs), largely used in preclinical anti-angiogenic assays, we designed here species-specific real-time quantitative RT-PCR assays. Human and mouse PECAM1/CD31, ENG/CD105, FLT1/VEGFR1, KDR/VEGFR2 and VEGFA transcripts were analyzed in a large series of 150 PDXs established from 8 different tumor types (53 colorectal, 14 ovarian, 39 breast and 15 renal cell cancers, 6 small cell and 5 non small cell lung carcinomas, 13 cutaneous melanomas and 5 glioblastomas) and in two bevacizumab-treated non small cell lung carcinomas xenografts.
Results
As expected, mouse cell proportion in PDXs -evaluated by quantifying expression of the housekeeping gene TBP- correlated with all mouse endothelial markers and human VEGFA RNA levels. More interestingly, we observed human PECAM1/CD31 and ENG/CD105 expression in all tumor types, with higher rate in glioblastoma and renal cancer xenografts. Human VEGFR expression profile varied widely depending on tumor types with particularly high levels of human FLT1/VEGFR1 transcripts in colon cancers and non small cell lung carcinomas, and upper levels of human KDR/VEGFR2 transcripts in non small cell lung carcinomas. Bevacizumab treatment induced significant low expression of mouse Pecam1/Cd31, Eng/Cd105, Flt1/Vegfr1 and Kdr/Vefr2 while the human PECAM1/CD31 and VEGFA were upregulated.
Conclusions
Taken together, our results strongly suggest existence of human tumor endothelial cells in all tumor types tested and of both stromal and tumoral autocrine VEGFA-VEGFR1/2 signalings. These findings should be considered when evaluating molecular mechanisms of preclinical response and resistance to tumor anti-angiogenic strategies.
Keywords: Tumor vasculature, Patient-derived xenografts, Species-specific PCR assays, Endothelial markers, VEGFA-VEGFR1/2 signalings
Background
Tumor vasculature, a crucial feature in cancer development and progression, is based on angiogenesis and vasculogenesis driven by VEGF signalings [1-3] but also on tumor endothelial transdifferentiation and vascular mimicry [4]. The VEGFR1 and VEGFR2 tyrosine kinase receptors are primarily expressed by endothelial cells. Recent studies, however, suggest that tumor-derived VEGF provides not only paracrine survival cues for endothelial cells, but may also autocrine processes in tumor cells expressing VEGFRs and play a role in tumor resistance to existing anti-angiogenic therapies [5-7].
Growth of patient tumor fragments into immunodeficient mice allows an accurate depiction of human tumor biological characteristics and are considered to represent the heterogeneity of human cancers (for review [8]). These patient-derived tumor xenografts (PDX) are greatly helpful to evaluate fundamental issues in cancer and chemosensitivity response, including characteristics of angiogenesis, tumor-stroma interactions and response to antiangiogenic therapies. As real-time quantitative RT-PCR is highly specific, species-specific primer sets can allow to discriminating between mouse/stromal and human/cancer gene expression in PDX models.
To obtain further insight into tumor vascularization and VEGFR expression by cancer and non-tumor cells, we used real-time qRT-PCR to quantify species-specific mRNAs of PECAM1/CD31, ENG/CD105, FLT1/VEGFR1, KDR/VEGFR2 and VEGFA genes in a large series of 150 xenografts from different tumor types. We also validated clinical relevance of species-specific PCR assays for in vivo evaluation of anti-angiogenesis therapy in two non small cell lung carcinoma models. We showed human PECAM1/CD31 and ENG/CD105 expression in all tumor types, supporting existence of human tumor endothelial cells in all tumor types. In addition, the VEGFR expression profiles led to involvement of both stromal and tumoral autocrine VEGFA-VEGFR1/2 signalings in tumors.
Results and discussion
First, the proportion of mouse cells was estimated in a panel of 8 different PDX types, using a real-time qRT-PCR assay combining primers specific for mouse Tbp RNA and primers able to amplify a common sequence on both human and mouse TBP transcripts. (Additional file 1: Table S1). As this gene encoding the TATA box-binding protein is a robust house-keeping gene [9] with similar amplification efficiency for the 2 primer sets, the ratio reflects the percentage of mouse cells within xenograft as validated in a standard curve of mouse and human cDNA mixtures (data not shown).
In an initial series of 157 human xenografts, the proportion of mouse cells was 100% in 7 tumors. These 7 tumor samples probably originated from spontaneous mouse lymphoma, frequently observed in immunodeficient mice [10].
In the 150 other xenografts, mouse host cells were found in all specimens with a median proportion of mouse cells of 9%, ranged between 3.3% in SCLC and 20% in NSCLC (p < 0.05, Table 1). To note, all the xenografts used here, have been passaged at least 5 times in mice, leading to a replacement of human stroma by mouse components [8].
Table 1.
Normalized gene expression for each of the 150 PDX samples, classified by tumor type (noted in bold)
|
Sample nature |
Derived from primary tumor or metastatis |
% of mouse cells |
PECAM1 |
ENG |
VEGFR1 |
VEGFR2 |
VEGFA |
% of
mVegfa
vs human + mouse VEGFA transcripts |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hs | Mm | Hs | Mm | Hs | Mm | Hs | Mm | Hs | Mm | ||||
| Pure human control |
|
0% |
1265 |
0 |
796 |
0 |
2610 |
0 |
157 |
0 |
287 |
0 |
|
| Pure mouse control |
|
100% |
0 |
1176 |
0 |
736 |
0 |
303 |
0 |
879 |
0 |
790 |
|
|
Colorectal carcinoma PDX |
|
|
|
|
|
|
|
|
|
|
|
|
|
| CRC#1 |
Primary |
11% |
0 |
894 |
2 |
492 |
23 |
453 |
0 |
405 |
4010 |
212 |
5% |
| CRC#2 |
Primary |
5% |
0 |
917 |
3 |
398 |
9 |
383 |
0 |
309 |
4912 |
51 |
1% |
| CRC#3 |
Metastasis |
21% |
1 |
2380 |
34 |
893 |
14 |
843 |
0 |
803 |
4642 |
628 |
12% |
| CRC#4 |
Primary |
17% |
0 |
836 |
<1 |
285 |
0 |
368 |
0 |
299 |
2876 |
302 |
10% |
| CRC#5 |
Metastasis |
8% |
0 |
813 |
0 |
492 |
3 |
337 |
0 |
374 |
3552 |
109 |
3% |
| CRC#6 |
Primary |
9% |
46 |
458 |
217 |
326 |
77 |
196 |
<1 |
176 |
1866 |
84 |
4% |
| CRC#7 |
Metastasis |
8% |
17 |
553 |
27 |
272 |
65 |
292 |
0 |
210 |
5230 |
251 |
5% |
| CRC#8 |
Primary |
14% |
0 |
1193 |
469 |
614 |
3 |
349 |
0 |
689 |
2999 |
92 |
3% |
| CRC#9 |
Primary |
8% |
0 |
967 |
8 |
550 |
3 |
475 |
0 |
379 |
7973 |
204 |
2% |
| CRC#10 |
Primary |
10% |
0 |
733 |
<1 |
409 |
176 |
246 |
0 |
284 |
3463 |
124 |
3% |
| CRC#11 |
Metastasis |
9% |
1 |
1083 |
<1 |
481 |
300 |
567 |
0 |
410 |
5461 |
135 |
2% |
| CRC#12 |
Metastasis |
4% |
48 |
479 |
0 |
182 |
26 |
274 |
0 |
230 |
4937 |
106 |
2% |
| CRC#13 |
Metastasis |
4% |
3 |
356 |
5 |
135 |
289 |
163 |
0 |
168 |
3606 |
145 |
4% |
| CRC#14 |
Primary |
2% |
<1 |
260 |
7 |
139 |
305 |
119 |
0 |
143 |
5085 |
76 |
1% |
| CRC#15 |
Primary |
17% |
<1 |
1287 |
<1 |
715 |
51 |
530 |
0 |
419 |
6541 |
311 |
5% |
| CRC#16 |
Metastasis |
5% |
<1 |
477 |
44 |
237 |
89 |
197 |
0 |
219 |
3406 |
196 |
5% |
| CRC#17 |
Primary |
17% |
21 |
1067 |
49 |
539 |
42 |
382 |
0 |
323 |
3674 |
555 |
13% |
| CRC#18 |
Primary |
14% |
4 |
1078 |
81 |
550 |
33 |
370 |
<1 |
356 |
2016 |
262 |
12% |
| CRC#19 |
Primary |
4% |
3 |
288 |
<1 |
162 |
<1 |
120 |
0 |
135 |
4258 |
111 |
3% |
| CRC#20 |
Metastasis |
22% |
4 |
1580 |
19 |
754 |
10 |
584 |
<1 |
684 |
5604 |
391 |
7% |
| CRC#21 |
Metastasis |
17% |
10 |
1336 |
373 |
749 |
10 |
656 |
0 |
639 |
4894 |
432 |
8% |
| CRC#22 |
Primary |
18% |
0 |
2315 |
322 |
1081 |
32 |
908 |
0 |
1262 |
4671 |
1244 |
21% |
| CRC#23 |
Metastasis |
8% |
0 |
446 |
407 |
406 |
42 |
202 |
0 |
173 |
2360 |
155 |
6% |
| CRC#24 |
Primary |
12% |
0 |
981 |
5 |
581 |
13 |
508 |
0 |
331 |
4773 |
233 |
5% |
| CRC#25 |
Primary |
5% |
0 |
622 |
36 |
329 |
0 |
246 |
0 |
285 |
2643 |
68 |
3% |
| CRC#26 |
Primary |
11% |
0 |
1245 |
569 |
480 |
112 |
375 |
0 |
296 |
3607 |
237 |
6% |
| CRC#27 |
Primary |
14% |
4 |
1789 |
3 |
895 |
83 |
682 |
0 |
581 |
3101 |
891 |
22% |
| CRC#28 |
Carcinosis |
5% |
3 |
526 |
1 |
326 |
1 |
215 |
0 |
268 |
2545 |
29 |
1% |
| CRC#29 |
Primary |
11% |
5 |
1000 |
2 |
541 |
0 |
364 |
0 |
344 |
3172 |
391 |
11% |
| CRC#30 |
Primary |
7% |
0 |
753 |
11 |
332 |
22 |
282 |
0 |
258 |
2247 |
231 |
9% |
| CRC#31 |
Metastasis |
10% |
<1 |
629 |
1 |
294 |
29 |
241 |
0 |
216 |
2896 |
210 |
7% |
| CRC#32 |
Primary |
16% |
0 |
1073 |
304 |
556 |
28 |
357 |
0 |
469 |
1731 |
166 |
9% |
| CRC#33 |
Primary |
7% |
4 |
563 |
<1 |
277 |
7 |
202 |
0 |
218 |
1253 |
129 |
9% |
| CRC#34 |
Primary |
13% |
2 |
749 |
379 |
530 |
15 |
306 |
0 |
390 |
4293 |
157 |
4% |
| CRC#35 |
Primary |
9% |
0 |
958 |
3 |
484 |
9 |
329 |
0 |
318 |
2206 |
212 |
9% |
| CRC#36 |
Primary |
21% |
1 |
991 |
0 |
504 |
32 |
388 |
<1 |
436 |
3296 |
140 |
4% |
| CRC#37 |
Primary |
19% |
6 |
1978 |
16 |
840 |
10 |
391 |
0 |
668 |
2692 |
182 |
6% |
| CRC#38 |
Primary |
8% |
2 |
1114 |
8 |
446 |
2 |
320 |
0 |
367 |
1889 |
218 |
10% |
| CRC#39 |
Metastasis |
12% |
0 |
1156 |
478 |
523 |
40 |
366 |
0 |
418 |
4034 |
214 |
5% |
| CRC#40 |
Primary |
10% |
<1 |
547 |
94 |
356 |
49 |
199 |
0 |
242 |
1848 |
142 |
7% |
| CRC#41 |
Carcinosis |
16% |
0 |
1552 |
3 |
762 |
7 |
325 |
0 |
457 |
918 |
228 |
20% |
| CRC#42 |
Primary |
31% |
0 |
1786 |
<1 |
922 |
94 |
447 |
0 |
599 |
2710 |
493 |
15% |
| CRC#43 |
Primary |
10% |
0 |
1024 |
75 |
459 |
249 |
358 |
2 |
431 |
4126 |
272 |
6% |
| CRC#44 |
Carcinosis |
15% |
<1 |
938 |
159 |
565 |
1 |
285 |
0 |
364 |
2523 |
269 |
10% |
| CRC#45 |
Primary |
12% |
1654 |
807 |
512 |
388 |
9 |
215 |
1 |
332 |
969 |
124 |
11% |
| CRC#46 |
Primary |
3% |
<1 |
412 |
3 |
158 |
2 |
139 |
0 |
168 |
1865 |
61 |
3% |
| CRC#47 |
Metastasis |
6% |
0 |
521 |
2 |
252 |
<1 |
173 |
0 |
195 |
1662 |
68 |
4% |
| CRC#48 |
Carcinosis |
10% |
0 |
843 |
<1 |
417 |
0 |
252 |
0 |
252 |
1705 |
227 |
12% |
| CRC#49 |
Metastasis |
6% |
1 |
379 |
426 |
274 |
11 |
248 |
0 |
267 |
4587 |
149 |
3% |
| CRC#50 |
Metastasis |
18% |
31 |
1697 |
0 |
690 |
23 |
485 |
0 |
421 |
5271 |
299 |
5% |
| CRC#51 |
Primary |
23% |
0 |
1294 |
2 |
662 |
67 |
476 |
0 |
375 |
6660 |
583 |
8% |
| CRC#52 |
Primary |
38% |
14 |
3265 |
398 |
1126 |
640 |
736 |
0 |
836 |
7517 |
953 |
11% |
| CRC#53 |
Metastasis |
19% |
0 |
1657 |
0 |
566 |
15 |
430 |
0 |
430 |
4014 |
209 |
5% |
|
Median
|
|
10.6%
|
0.7
|
958
|
7
|
484
|
22
|
349
|
0
|
344
|
3463
|
210
|
6%
|
|
Ovarian carcinoma PDX |
|
|
|
|
|
|
|
|
|
|
|
|
|
| OVC#1 |
Metastasis |
28% |
42 |
2575 |
0 |
1498 |
89 |
1191 |
159 |
867 |
10390 |
459 |
4% |
| OVC#2 |
Metastasis |
5% |
4 |
565 |
99 |
350 |
2 |
439 |
34 |
259 |
6391 |
88 |
1% |
| OVC#3 |
Metastasis |
21% |
1 |
1427 |
304 |
809 |
69 |
406 |
<1 |
583 |
3133 |
710 |
18% |
| OVC#4 |
Primary |
6% |
26 |
709 |
9 |
474 |
0 |
259 |
4 |
272 |
1528 |
144 |
9% |
| OVC#5 |
Primary |
7% |
16 |
974 |
81 |
807 |
0 |
802 |
45 |
525 |
14226 |
95 |
1% |
| OVC#6 |
Primary |
12% |
3 |
2052 |
97 |
593 |
19 |
734 |
101 |
528 |
2628 |
427 |
14% |
| OVC#7 |
Primary |
8% |
0 |
762 |
4 |
470 |
0 |
270 |
32 |
278 |
6156 |
266 |
4% |
| OVC#8 |
Primary |
3% |
2 |
219 |
30 |
119 |
6 |
88.8 |
3 |
59.2 |
652 |
37 |
5% |
| OVC#9 |
Primary |
8% |
5 |
1795 |
2 |
674 |
3 |
518 |
5 |
372 |
2981 |
184 |
6% |
| OVC#10 |
Primary |
4% |
1 |
444 |
16 |
288 |
0 |
204 |
22 |
141 |
2812 |
52 |
2% |
| OVC#11 |
Primary |
20% |
24 |
1586 |
54 |
1036 |
0 |
482 |
2 |
648 |
2781 |
493 |
15% |
| OVC#12 |
Primary |
13% |
3 |
877 |
177 |
487 |
0 |
259 |
12 |
285 |
1720 |
127 |
7% |
| OVC#13 |
Primary |
3% |
17 |
550 |
207 |
263 |
2 |
196 |
<1 |
224 |
1134 |
16 |
1% |
| OVC#14 |
Primary |
5% |
0 |
332 |
<1 |
255 |
21 |
238 |
<1 |
164 |
19239 |
62 |
0% |
|
Median
|
|
7%
|
3.7
|
819
|
42
|
480
|
2
|
338
|
9
|
281
|
2896
|
136
|
5%
|
|
Glioblastoma PDX |
|
|
|
|
|
|
|
|
|
|
|
|
|
| GBM#1 |
Primary |
8% |
22 |
712 |
2051 |
457 |
378 |
559 |
8 |
186 |
18822 |
241 |
1% |
| GBM#2 |
Primary |
13% |
1 |
1351 |
1143 |
819 |
0 |
799 |
378 |
328 |
17084 |
296 |
2% |
| GBM#3 |
Primary |
13% |
1 |
2372 |
422 |
1184 |
0 |
1325 |
0 |
1237 |
8452 |
131 |
2% |
| GBM#4 |
Primary |
5% |
55 |
870 |
321 |
328 |
0 |
503 |
0 |
372 |
5923 |
78 |
1% |
| GBM#5 |
Primary |
15% |
0 |
2600 |
268 |
1389 |
28 |
1361 |
294 |
1413 |
15443 |
100 |
1% |
|
Median
|
|
13%
|
1.4
|
1351
|
422
|
819
|
0
|
799
|
8
|
372
|
15443
|
131
|
1%
|
|
Breast cancer carcinoma PDX |
|
|
|
|
|
|
|
|
|
|
|
|
|
| BC#1 |
Primary |
2% |
<1 |
222 |
204 |
113 |
3 |
89.4 |
2 |
80.7 |
637 |
114 |
15% |
| BC#2 |
Primary |
8% |
0 |
666 |
177 |
335 |
19 |
289 |
0 |
162 |
2997 |
310 |
9% |
| BC#3 |
Metastasis |
10% |
0 |
679 |
286 |
447 |
5 |
539 |
36 |
259 |
4961 |
334 |
6% |
| BC#4 |
Primary |
15% |
46 |
803 |
91 |
498 |
5 |
366 |
0 |
222 |
3547 |
447 |
11% |
| BC#5 |
Primary |
1% |
<1 |
116 |
0 |
61.9 |
33 |
89.1 |
4 |
31.9 |
15066 |
77 |
1% |
| BC#6 |
Metastasis |
15% |
2 |
1351 |
289 |
634 |
29 |
719 |
150 |
439 |
17360 |
357 |
2% |
| BC#7 |
Primary |
22% |
8 |
1908 |
442 |
887 |
0 |
1370 |
0 |
739 |
27659 |
365 |
1% |
| BC#8 |
Primary |
6% |
1 |
810 |
149 |
412 |
18 |
327 |
9 |
280 |
8360 |
160 |
2% |
| BC#9 |
Metastasis |
10% |
0 |
713 |
6 |
322 |
13 |
420 |
0 |
279 |
1020 |
294 |
22% |
| BC#10 |
Primary |
6% |
3 |
370 |
460 |
233 |
0 |
325 |
17 |
134 |
7447 |
154 |
2% |
| BC#11 |
Primary |
6% |
6 |
993 |
347 |
403 |
29 |
461 |
68 |
325 |
14282 |
256 |
2% |
| BC#12 |
Primary |
8% |
6 |
1005 |
466 |
543 |
28 |
664 |
3 |
391 |
25794 |
363 |
1% |
| BC#13 |
Primary |
7% |
0 |
575 |
92 |
256 |
8 |
266 |
15 |
189 |
6174 |
132 |
2% |
| BC#14 |
Primary |
8% |
654 |
745 |
45 |
413 |
0 |
279 |
2 |
253 |
3294 |
71 |
2% |
| BC#15 |
Metastasis |
11% |
4 |
912 |
50 |
461 |
1 |
311 |
0 |
286 |
3458 |
186 |
5% |
| BC#16 |
Primary |
2% |
0 |
199 |
188 |
94.6 |
4 |
73.6 |
2 |
69.7 |
610 |
100 |
14% |
| BC#17 |
Primary |
4% |
13 |
413 |
346 |
134 |
66 |
173 |
32 |
101 |
2131 |
197 |
8% |
| BC#18 |
Primary |
10% |
<1 |
1545 |
168 |
743 |
3 |
788 |
11 |
382 |
6550 |
167 |
2% |
| BC#19 |
Metastasis |
16% |
<1 |
2304 |
167 |
1188 |
5 |
1049 |
10 |
771 |
6004 |
280 |
4% |
| BC#20 |
Primary |
17% |
0 |
1967 |
340 |
959 |
1 |
709 |
20 |
634 |
11533 |
357 |
3% |
| BC#21 |
Primary |
9% |
0 |
730 |
334 |
332 |
2 |
476 |
91 |
202 |
8166 |
520 |
6% |
| BC#22 |
Primary |
6% |
0 |
598 |
451 |
222 |
10 |
334 |
90 |
124 |
5088 |
264 |
5% |
| BC#23 |
Primary |
4% |
0 |
377 |
331 |
179 |
2 |
195 |
19 |
83.3 |
1742 |
51 |
3% |
| BC#24 |
Primary |
22% |
1 |
1128 |
858 |
982 |
79 |
999 |
<1 |
495 |
21363 |
668 |
3% |
| BC#25 |
Primary |
10% |
0 |
1165 |
666 |
573 |
94 |
627 |
12 |
474 |
14542 |
244 |
2% |
| BC#26 |
Primary |
10% |
0 |
1446 |
429 |
572 |
0 |
685 |
230 |
510 |
5771 |
257 |
4% |
| BC#27 |
Primary |
14% |
2 |
880 |
4 |
452 |
94 |
415 |
<1 |
222 |
1505 |
299 |
17% |
| BC#28 |
Primary |
5% |
<1 |
182 |
91 |
113 |
7 |
119 |
6 |
80.3 |
1221 |
50 |
4% |
| BC#29 |
Primary |
9% |
<1 |
656 |
530 |
469 |
32 |
532 |
9 |
473 |
50360 |
255 |
1% |
| BC#30 |
Primary |
7% |
3 |
823 |
94 |
341 |
247 |
403 |
34 |
244 |
9097 |
373 |
4% |
| BC#31 |
Primary |
4% |
0 |
345 |
166 |
216 |
0 |
161 |
3 |
145 |
1085 |
79 |
7% |
| BC#32 |
Primary |
7% |
<1 |
629 |
13 |
276 |
4 |
237 |
19 |
194 |
1544 |
198 |
11% |
| BC#33 |
Primary |
9% |
<1 |
725 |
397 |
428 |
232 |
549 |
6 |
231 |
5414 |
144 |
3% |
| BC#34 |
Primary |
14% |
5 |
1061 |
245 |
557 |
0 |
457 |
0 |
308 |
3866 |
176 |
4% |
| BC#35 |
Primary |
5% |
2 |
484 |
103 |
358 |
3 |
506 |
50 |
185 |
23896 |
360 |
1% |
| BC#36 |
Primary |
13% |
0 |
1085 |
221 |
544 |
0 |
333 |
13 |
347 |
1153 |
231 |
17% |
| BC#37 |
Primary |
4% |
0 |
376 |
193 |
149 |
<1 |
144 |
13 |
120 |
1081 |
289 |
21% |
| BC#38 |
Primary |
6% |
2 |
776 |
90 |
415 |
8 |
326 |
12 |
281 |
4683 |
178 |
4% |
| BC#39 |
Primary |
14% |
0 |
961 |
5 |
731 |
83 |
691 |
4 |
606 |
4829 |
152 |
3% |
|
Median
|
|
7.9%
|
0.7
|
745
|
193
|
413
|
5
|
403
|
10
|
253
|
5088
|
244
|
4%
|
|
Cutaneous melanoma PDX |
|
|
|
|
|
|
|
|
|
|
|
|
|
| CM#1 |
Metastasis |
11% |
0 |
1725 |
2985 |
825 |
0 |
953 |
83 |
1026 |
10386 |
1401 |
12% |
| CM#2 |
Metastasis |
7% |
27 |
544 |
1391 |
426 |
768 |
360 |
0 |
184 |
10696 |
315 |
3% |
| CM#3 |
Metastasis |
4% |
1 |
282 |
257 |
180 |
0 |
122 |
1 |
155 |
599 |
102 |
14% |
| CM#4 |
Primary |
20% |
0 |
3306 |
784 |
1178 |
427 |
883 |
38 |
836 |
9188 |
718 |
7% |
| CM#5 |
Metastasis |
8% |
9 |
936 |
872 |
363 |
15 |
587 |
0 |
289 |
5590 |
201 |
3% |
| CM#6 |
Metastasis |
3% |
<1 |
342 |
648 |
196 |
6 |
188 |
5 |
236 |
960 |
23 |
2% |
| CM#7 |
Metastasis |
1% |
2 |
176 |
382 |
83.6 |
2 |
135 |
<1 |
84.3 |
5962 |
37 |
1% |
| CM#8 |
Primary |
10% |
9 |
4760 |
876 |
705 |
0 |
2230 |
5 |
841 |
16732 |
239 |
1% |
| CM#9 |
Metastasis |
1% |
0 |
118 |
284 |
61.6 |
20 |
125 |
14 |
73.6 |
3704 |
24 |
1% |
| CM#10 |
Primary |
10% |
0 |
876 |
756 |
300 |
0 |
248 |
279 |
285 |
387 |
126 |
25% |
| CM#11 |
Metastasis |
8% |
2 |
641 |
1102 |
427 |
0 |
355 |
1 |
309 |
8837 |
266 |
3% |
| CM#12 |
Primary |
5% |
0 |
530 |
112 |
186 |
0 |
440 |
0 |
423 |
683 |
26 |
4% |
| CM#13 |
Metastasis |
2% |
<1 |
243 |
145 |
101 |
<1 |
116 |
1 |
82.2 |
466 |
31 |
6% |
|
Median
|
|
7.1%
|
0.9
|
544
|
756
|
300
|
0.8
|
355
|
1
|
285
|
5590
|
126
|
3%
|
|
Renal cell carcinoma PDX |
|
|
|
|
|
|
|
|
|
|
|
|
|
| RCC#1 |
Primary |
17% |
0 |
1179 |
569 |
1002 |
0 |
680 |
2 |
473 |
19769 |
1513 |
7% |
| RCC#2 |
Primary |
12% |
0 |
3362 |
16 |
1929 |
0 |
1934 |
0 |
3043 |
27096 |
54 |
0% |
| RCC#3 |
Primary |
27% |
0 |
5431 |
411 |
2376 |
0 |
1866 |
0 |
1974 |
25792 |
211 |
1% |
| RCC#4 |
Metastasis |
11% |
120 |
2117 |
256 |
1430 |
4 |
1512 |
0 |
1337 |
13968 |
89 |
1% |
| RCC#5 |
Primary |
16% |
5 |
2906 |
33 |
1942 |
3 |
1624 |
0 |
2102 |
25817 |
87 |
0% |
| RCC#6 |
Primary |
1% |
1 |
341 |
2 |
125 |
2 |
157 |
47 |
119 |
609 |
13 |
2% |
| RCC#7 |
Primary |
21% |
0 |
768 |
549 |
1908 |
0 |
1292 |
0 |
1324 |
27232 |
157 |
1% |
| RCC#8 |
Metastasis |
17% |
1 |
842 |
410 |
778 |
0 |
466 |
0 |
286 |
1756 |
930 |
35% |
| RCC#9 |
Metastasis |
13% |
17 |
2024 |
230 |
1258 |
3 |
827 |
1 |
904 |
37839 |
55 |
0% |
| RCC#10 |
Primary |
11% |
0 |
2010 |
856 |
1359 |
0 |
1070 |
2 |
672 |
37217 |
83 |
0% |
| RCC#11 |
Primary |
5% |
2 |
597 |
907 |
350 |
0 |
487 |
0 |
253 |
5091 |
136 |
3% |
| RCC#12 |
Metastasis |
14% |
0 |
2546 |
257 |
1132 |
0 |
871 |
0 |
1040 |
16952 |
61 |
0% |
| RCC#13 |
Primary |
21% |
0 |
4963 |
38 |
3466 |
0 |
3281 |
0 |
3966 |
30645 |
155 |
1% |
| RCC#14 |
Primary |
6% |
330 |
1338 |
364 |
602 |
0 |
661 |
2 |
343 |
26952 |
52 |
0% |
| RCC#15 |
Primary |
6% |
77 |
565 |
1036 |
293 |
0 |
368 |
0 |
291 |
2210 |
59 |
3% |
|
Median
|
|
12.9%
|
1.2
|
2010
|
364
|
1258
|
0
|
871
|
0
|
904
|
25792
|
87
|
1%
|
|
Lung carcinoma PDX |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Small cell lung carcinoma
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| SCLC#1 |
Primary |
8% |
0 |
1030 |
3 |
387 |
0 |
250 |
0 |
196 |
419 |
43 |
9% |
| SCLC#2 |
Primary |
3% |
2 |
632 |
0 |
238 |
5 |
189 |
0 |
185 |
1300 |
52 |
4% |
| SCLC#3 |
Primary |
4% |
4 |
591 |
0 |
259 |
1 |
232 |
2 |
232 |
1117 |
49 |
4% |
| SCLC#4 |
Primary |
3% |
7 |
395 |
0 |
222 |
0 |
166 |
0 |
162 |
1498 |
46 |
3% |
| SCLC#5 |
Metastasis |
2% |
0 |
309 |
1 |
153 |
0 |
160 |
2 |
122 |
893 |
56 |
6% |
| SCLC#6 |
Primary |
7% |
2 |
670 |
9 |
221 |
471 |
208 |
72 |
192 |
954 |
86 |
8% |
|
Median
|
|
3.3%
|
1.7
|
612
|
0
|
230
|
1
|
198
|
1
|
189
|
1035
|
51
|
5%
|
|
Non small cell lung carcinoma
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| NSCLC#1 |
Primary |
28% |
3 |
1969 |
61 |
941 |
2 |
1145 |
14 |
637 |
18440 |
794 |
4% |
| NSCLC#2 |
Primary |
8% |
0 |
1270 |
0 |
611 |
0 |
511 |
335 |
639 |
5911 |
98 |
2% |
| NSCLC#3 |
Primary |
22% |
95 |
1590 |
31 |
1438 |
124 |
961 |
930 |
669 |
18346 |
429 |
2% |
| NSCLC#4 |
Primary |
5% |
2 |
686 |
5 |
339 |
4 |
212 |
59 |
221 |
875 |
85 |
9% |
| NSCLC#5 |
Primary |
20% |
3 |
1363 |
667 |
1387 |
184 |
896 |
3106 |
652 |
10612 |
688 |
6% |
| Median | 20% | 2.7 | 1363 | 31 | 941 | 4 | 896 | 335 | 639 | 10612 | 429 | 4% | |
Mouse cells encompass here a wide range of stromal cell types, including fibroblasts, inflammatory and immune cells, smooth muscle cells, and endothelial cells. We further focused on endothelial cells using expression of mouse Pecam1/Cd31 and Eng/Cd105 genes (hereinafter referred to as mCd31 and mCd105, respectively) to evaluate their proportion within xenografts. Vwf gene encoding von Willebrand factor was also preliminary selected but not kept because of a lower expression rate in the mouse and human controls (Ct > 30, data not shown).
As expected, all samples, collected from large xenografts without necrotic centre, expressed mCd31 and mCd105 genes. Nevertheless, mCd31 and mCd105 mRNA levels widely varied between the samples (Table 1), but remained highly correlated to each other (p < 10-7; Table 2). Noteworthy, mCd31 and mCd105 expression levels were highly correlated with the proportion of mouse cells (Table 2), suggesting that the relative amount of endothelial cells remains stable within diverse stromal cell populations, whatever the density of stroma component and the cancer type.
Table 2.
Relationships between mouse (m) and human (h) mRNA levels in the 150 human tumor xenografts
| hCD31 | mCd31 | hCD105 | mCd105 | hVEGFR1 | mVegfr1 | hVEGFR2 | mVegfr2 | hVEGFA | mVegfa | |
|---|---|---|---|---|---|---|---|---|---|---|
|
mCd31
|
0.0251 |
|
|
|
|
|
|
|
|
|
| |
0.762 |
|
|
|
|
|
|
|
|
|
|
hCD105
|
0.043 |
0.121 |
|
|
|
|
|
|
|
|
| |
0.60 |
0.14 |
|
|
|
|
|
|
|
|
|
mCd105
|
0.040 |
0.928 |
0.189 |
|
|
|
|
|
|
|
| |
0.63 |
<0.0000001 |
0.02 |
|
|
|
|
|
|
|
|
hVEGFR1
|
0.065 |
0.022 |
-0.076 |
0.004 |
|
|
|
|
|
|
| |
0.43 |
0.79 |
0.35 |
0.96 |
|
|
|
|
|
|
|
mVegfr1
|
0.076 |
0.851 |
0.305 |
0.877 |
0.006 |
|
|
|
|
|
| |
0.35 |
<0.0000001 |
<0.0002 |
<0.0000001 |
0.94 |
|
|
|
|
|
|
hVEGFR2
|
0.010 |
-0.029 |
0.232 |
-0.036 |
-0.036 |
0.070 |
|
|
|
|
| |
0.91 |
0.72 |
<0.005 |
0.66 |
0.66 |
0.40 |
|
|
|
|
|
mVegfr2
|
0.003 |
0.912 |
0.173 |
0.919 |
-0.017 |
0.858 |
-0.090 |
|
|
|
| |
0.98 |
<0.0000001 |
<0.05 |
<0.0000001 |
0.83 |
<0.0000001 |
0.27 |
|
|
|
|
hVEGFA
|
0.095 |
0.477 |
0.319 |
0.563 |
0.090 |
0.726 |
0.131 |
0.517 |
|
|
| |
0.25 |
<0.0000001 |
<0.0002 |
<0.0000001 |
0.27 |
<0.0000001 |
0.11 |
<0.0000001 |
|
|
|
mVegfa
|
0.031 |
0.505 |
0.194 |
0.524 |
0.304 |
0.514 |
0.062 |
0.413 |
0.328 |
|
| |
0.70 |
<0.0000001 |
<0.05 |
<0.0000001 |
<0.0002 |
<0.0000001 |
0.45 |
<0.0000001 |
<0.00005 |
|
|
% mouse cells |
-0.016 |
0.828 |
0.113 |
0.865 |
0.154 |
0.715 |
-0.145 |
0.797 |
0.364 |
0.666 |
| 0.84 | <0.0000001 | 0.17 | <0.0000001 | 0.06 | <0.0000001 | 0.08 | <0.0000001 | <0.000005 | <0.0000001 |
Results, expressed as N-fold differences in target gene expression relative to the mouse and human TBP genes (both the mouse and human TBP transcripts) and termed “Ntarget”, were determined as Ntarget = 2∆Ctsample , where the ∆Ct value of the sample was determined by subtracting the average Ct value of target gene (human or mouse) from the average Ct value of ‘Total-TBP’ gene). The Ntarget values of the tumor samples were subsequently normalized such that the value for mRNA level was 1 when Ct=35. Target mRNA levels that were total absence or very low (Ct > 38) in tumor samples were scored ‘0’ for non expressed. As for calculation of % of mouse cells, specific mouse Tbp gene expression and the expression of both the mouse and the human TBP genes were studied by real-time qRT-PCR using the mouse Tbp as target gene and the ‘Total-TBP’ as endogenous RNA control. Results, expressed as N-fold differences in specific mouse Tbp gene expression (using mouse Tbp primers) relative to the sum of the mouse and the human TBP gene expression (using ‘Total-TBP’ primers), termed NMm-TBP, are determined by theformula: NMm-TBP = 2DCtsample. The DCt value of the sample is determined by subtracting the Ct value of the mouse TBP gene from the Ct value of the Total TBP gene. The NMm-TBP values of the samples are subsequently normalized such that the median of NMm-TBP values of 4 mouse tissues was 100. As TBP is a ubiquitously expressed housekeeping gene, showing similar expression in our human and mouse tissues (Ct=27 for 5 ng cDNA), the final result (normalized NMm-TBP value) gives an estimate of the proportion of mouse cell content for a given xenograft. 1Spearman correlation coefficient, 2p value of Spearman rank correlation test, in bold when p is significant.
While numerous pro-angiogenic factors have been characterized, the VEGFA ligand has been identified as a predominant regulator of tumor angiogenesis and binds to VEGFR1 and VEGFR2 expressed on vascular endothelial cells. It mediates numerous changes within the tumor vasculature, including endothelial cell proliferation, migration, invasion, survival, chemotaxis of bone marrow-derived progenitor cells, vascular permeability and vasodilatation [1,2]. VEGFA expression by cancer cells is up-regulated by altered expression of oncogenes, a variety of growth factors and also hypoxia [2].
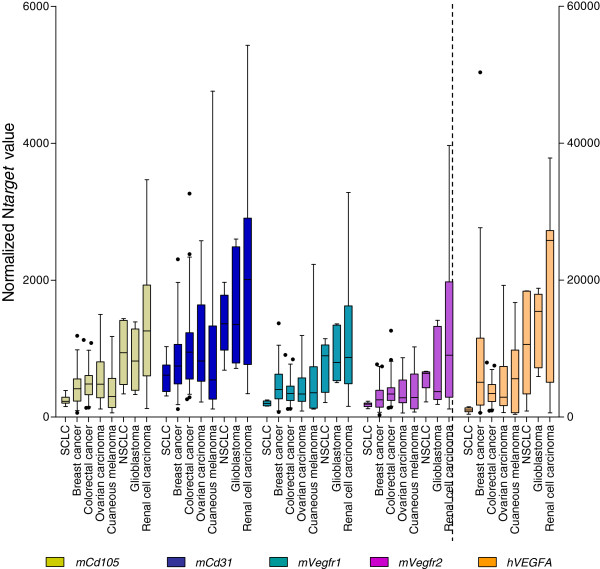
Unsurprisingly, we observed high levels of mouse Flt1/Vegfr1, mouse Kdr/Vegfr2 (hereby denominated mVegfr1 and mVegfr2) and human VEGFA (hVEGFA) transcripts, which correlated all with mCd31 and mCd105 RNA levels (Table 2). These strong positive correlations underline classical paracrine VEGFA-VEGFR1/2 signaling in tumorigenesis and crosstalk between the human ligand and mouse receptors. Expression of mVegfr1, mVegfr2 and hVEGFA however varied widely in the different tumor types. RCC, glioblastoma and NSCLC xenografts showed transcript level median of these three genes at least 2 times higher than in the 5 other tumor xenograft types (Table 1, Figure 1). According to the expression level of mCd105, mCd31, mVegfr1, mVegfr2 and hVEGFA (Figure 1), the most angiogenic PDXs are then renal cell carcinoma, glioblastoma, and NSCLs, tumor types well-known to be the most angiogenic tumors in patients [11], underlying the interest of PDX models to mimic patient tumors.
Figure 1.
Gene expression levels of mouse endothelial markers and hVEGFA in the 8 human tumor xenograft types. Box-and-whisker diagrams showing the expression of mouse endothelial marker genes (mCd31, mCd105, mVegfr1, mVegfr2), plot on left Y axis and hVEGFA gene plot on right Y axis. The box indicates the interquartile range, the centre horizontal line the median value and the black dots are outliers.
Surprisingly, we observed also marked level of mVegfa transcripts ranged from 50.7 (median in SCLC xenografts) to 429 (median in NSCLC xenografts). Individually, some xenografts showed more than 20% of the total VEGFA transcripts of mouse origin (Table 1). While VEGFA production by cancer cells is commonly reported, significant VEGFA expression has been also observed by fibroblasts and immune cells that surround and invade the tumor mass [12]. As reported by others [13], great attention has to be paid to mouse stromal VEGFA when anti-VEGF agents displaying specific human activity are tested in xenograft preclinical models.
Angiogenesis and vasculogenesis, mediated by angiogenic factors such as VEGFA are commonly accepted to support tumor vasculature. Vascular mimicry (ability of tumor cells to form functional vessel-like networks, devoid of endothelial cells) and cancer stem cell transdifferentiation into tumor endothelial cells are also two mechanisms recently reported in different tumors, including melanoma, breast, renal, ovarian cancer and glioblastoma [14-18] in which tumor cells directly participate in vascular channels. The presence of tumor-derived endothelial cells (TDECs) is usually investigated through the detection of CD31+ and CD105+ tumor cells [15-18]. TDEC cells are generally rare events and their identification needs highly sensitive methods (flow cytometry or confocal microscopy). Likewise, another approach to improving the detection of TDEC is to enhance the TDEC frequency by implanting into mice cancer stem cell enriched population. This prior enrichment could be done by culturing cells as tumor spheres [19,20] or by cell sorting for putative cancer stem cell markers [15,21]. Only one recent publication attempted to immunostain human CD31 directly in 3 human tumor xenografts, with no preliminary step of TDEC or CSC enrichment [22]. This study did not detect human CD31 and led the authors to conclude that endothelial cells in human hepatocellular carcinoma xenografts are of mouse rather than human origin, but did not allow them to absolutely exclude this possibility. Consequently, we apply in our PDX panel the real-time qRT-PCR method, known for its very high sensitivity, using human-specific PECAM1/CD31 (hCD31) and ENG/CD105 (hCD105) to gain more insight into TDECs.
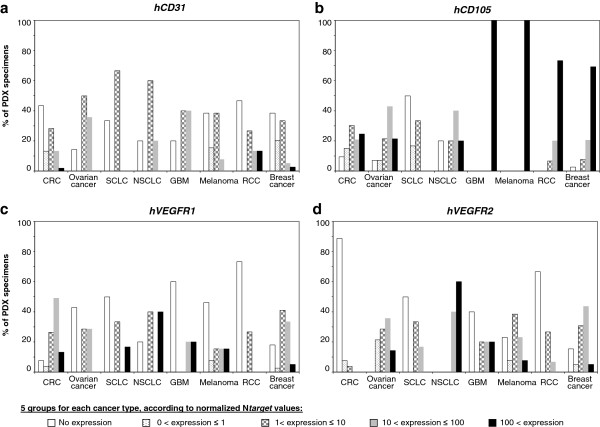
Surprisingly, we detected hCD31 and hCD105 transcripts in all types of PDXs, suggesting that TDECs can exist in virtually all types of cancer. The possibility of human endothelial marker signals due to very rare remaining human stroma cells can not be ignored, although the whole human stroma in tumor xenografts is reported to be eventually replaced by stroma of mouse origin [8,23,24]. But depending upon the types, the range of expression of hCD31 and hCD105 transcripts largely varied (Figure 2a-b). All tested samples of cutaneous melananoma and GBM highly expressed hCD105 gene (NHs-ENG >100). Literature indeed reports a large expression of CD105, a member of the transforming growth factor beta receptor family, on normal and neoplastic cells of the melanocytic lineage, including melanoma cell lines, and an up-regulation in gene signature of aggressive cutaneous melanoma in patients [14]. Likewise, CD105 is highly expressed in glioblastoma but essentially absent in normal brain [21]. RCC xenografts displayed a great proportion of samples expressed high levels of hCD31 or hCD105. These results fit with the literature that identified TDECs in patients mainly in glioblastoma and renal cancer [16,21]. By contrast, SCLCs show very low levels of both hCD31 and hCD105 mRNAs. A striking point is that hCD31 and hCD105 RNA levels did not correlate to each others (Table 2), even if their expression is analyzed for each cancer type (data not shown). It could be explained by different expression profiles for these 2 endothelial molecules: CD31 is considered as a pan-endothelial marker, whereas CD105 is a cell membrane glycoprotein predominantly expressed on cellular lineages within the vascular system, and over-expressed on proliferating endothelial cells [25]. These data underline that combination of markers is required to study the TDEC population.
Figure 2.
Variations of human hCD31 (a), hCD105 (b), hVEGFR1 (c) and hVEGFR2 (d) gene expression within the 8 human tumor xenograft types. Results are expressed for each cancer type as percent of PDX specimens showing normalized Ntarget values in the following categories: no expression, 0 to 1, 1 to 10, 10 to 100 or more than 100.
Initially, VEGFRs were thought to be expressed only on endothelial cells, but these receptors may also be expressed on tumor cells and play a role in tumor resistance to existing therapies [5-7]. The present species-specific real-time qRT-PCR assays combined with our series of 150 PDXs represents a powerful tool to obtain further insight into autocrine and paracrine VEGFA-VEGR1/2 signaling in tumorigenesis. We indeed observed human VEGFR expression in xenografts with a profile that varied widely according to tumor types (Table 1, Figure 2c-d): High levels of hVEGFR1 transcripts mainly observed in colon cancers and in NSCLCs; high levels of hVEGFR2 transcripts in NSCLCs. Individually, 2 out of 5 NSCLC xenografts (i.e.: NSCLC#3 and #5) showed more hVEGFR2 transcripts than mVegfr2 transcripts (Table 1). Conversely, SCLCs showed low levels of hVEGFR1 and hVEGFR2 transcripts and CRCs showed very low levels of hVEGFR2 transcripts (Absence in 89% of the 53 CRC xenografts). These results identified NSCLC as an attractive cancer type for anti-VEGFR2 treatment. Small-molecule inhibitors as Sunitinib and Sorafenib are oral multikinase inhibitors, including VEGFR2 among their targets. The development of antibodies that can selectively block VEGFR2 could potentially result in improved potency or tolerability [3].
Whereras mVegfr1 and mVegfr2 expressions were extremely correlated to mouse endothelial markers (p < 10-7), human VEGFR profiles did not correlate highly with neither hCD31 nor hCD105. Non exclusive hypotheses could explain this observation: i) human tumor cells expressing endothelial markers lead to VEGF- independent tumor vascularization with no expression of VEGFR1/2 [20]; ii) VEGFRs could be also expressed on carcinoma and participate to an essential autocrine/paracrine process for cancer cell proliferation and survival [1].
Collectively, VEGFA/VEGFR analyses suggest several autocrine and paracrine VEGFA-VEGFR1/2 signalings. In additional to the classical paracrine human tumoral VEGFA/mouse stromal VEGFR signalling, our data identified 3 others potential VEGFA-VEGFR signalings: a human cancer autocrine VEGFA/VEGFR signaling, an autocrine or paracrine mouse stromal VEGFA/VEGFR signaling, and a paracrine mouse stromal VEGFA/ human tumoral VEGFR signaling. It is noteworthy that the human cancer autocrine VEGFA/VEGFR signaling could occur intracellular, as well as by VEGFA secretion [6], limiting the quantity of extracellular VEGFA. Thus, VEGFR small-molecule inhibitors might be a more attractive therapy than VEGFA inhibitors which aim to sequestering free VEGFA.
To further investigate the potential value of species-specific PCR assays for in vivo evaluation of anti-angiogenesis therapy in PDX models, we analyzed in the same manner as described above, 2 NSCLC xenograft models after treatment with bevacizumab, a recombinant humanized monoclonal antibody to VEGF, approved for cancer therapy, including in NSCLC patients. These both models highly responded to one week-bevacizumab treatment in monotherapy: no tumor shrinkage but tumor stabilization throughout the experiment (Additional file 2: Figure S1).
As expected, the levels of mCd31, mCd105, mVegfr1 and mVegfr2 transcripts were significantly lower in the two bevacizumab-treated NSCLC xenografts as compared to matched non-treated xenografts (Table 3). Indeed, even if bevacizumab is able to bind and inhibit human VEGFA but unable to neutralize murine VEGFA, VEGFA in these 2 xenografts is produced by human cancer cells rather than by mouse stroma cells. It is noteworthy that one of the two xenografts (NSCLC#3) showed a significant upregulation of hVEGFA gene. More interestingly, the levels of hCD31, hCD105, hVEGFR1 and hVEGFR2 transcripts were not inferior in the two bevacizumab-treated NSCLC xenografts but on the contrary, hCD31 was upregulated by 3 times (p < 0.05 for NSCLC#3) in both bevacizumab-treated xenografts. These data suggest that the mouse endothelial cells are more sensitive to anti-VEGFA therapy than human cells. Indeed, cancer cells are able to take advantage of autocrine intracellular VEGFA/VEGFR signalling [6] while bevacizumab is directed against free fraction of VEGFA. Furthermore, transdifferentiation of tumor cells into endothelial cells has been reported to be VEGF-independent but induced by HIF-1α [20]. Finally, bevacizumab induces hypoxia through mouse endothelial cells destruction, which may lead in turn to TDEC expansion. These latter results are of interest to apprehend molecular mechanisms of bevacizumab resistance.
Table 3.
Target mRNA levels in 2 NSCLC xenografts after bevacizumab treatment
| |
NSCLC#3 |
NSCLC#5 |
|||||
|---|---|---|---|---|---|---|---|
| Control (n=5) | After bevacizumab reatment (n=5) | p-value 1 | Control (n=5) | After bevacizumab treatment (n=5) | p-value 1 | ||
|
PECAM1/CD31
mRNA |
Human
|
18.1 (7.34-43.1) |
57.6 (31.8-64.2) |
<0.05 |
2.38 (0.00-9.21) |
6.70 (2.41-17.1) NS |
|
| |
Mouse
|
863 (686-1790) |
578 (483-847) |
<0.05 |
2 334 (1 538-4 363) |
856 (699-980) |
<0.05 |
|
ENG/CD105
mRNA |
Human
|
29.1 (3.59-47.2) |
38.2 (15.1-71.4) |
NS |
57.64 (38.8-90.86) |
57.50 (47.2 - 84.4) |
NS |
| |
Mouse
|
619 (580-1098) |
414 (328-619) |
<0.05 |
1 519 (1120-1813) |
821 (610-860) |
<0.05 |
|
FLT1/VEGFR1
mRNA |
Human
|
59.6 (56.7-90.6) |
88.9 (62.3-118) |
NS |
3.84 (0.00-24.8) |
9.11 (3.87-20.3) |
NS |
| |
Mouse
|
589 (470-909) |
274 (212-362) |
<0.05 |
938 (633-1163) |
305 (216-344) |
<0.05 |
|
KDR/VEGFR2
mRNA |
Human
|
507 (361-622) |
545 (488-643) |
NS |
220 (140-274) |
574 (213-834) |
NS |
| |
Mouse
|
466 (386-800) |
204 (196-298) |
<0.05 |
1 175 (698-1 211) |
328 (316-349) |
<0.05 |
|
VEGFA
mRNA |
Human
|
20 503 (19162-24600) |
32 160 (30 331-35 680) |
<0.05 |
11 984 (5 368-13 961) |
12 235 (7 088-14 042) |
NS |
| Mouse | 160 (119-495) | 307 (184-614) | NS | 262 (170-680) | 267 (240-360) | NS | |
Results are expressed as normalized N-fold differences in target gene expression relative to the ‘Total-TBP’ expression. These Ntarget values of the tumor samples were normalized such that the value for the ’basal mRNA level‘ (Ct = 35) was 1Target mRNA levels that were total absence or very low (Ct > 38) in tumor samples were scored ‘0’ for non expressed.
Median and range in () are given for each gene in the different experimental conditions. 1Mann Whitney Test; NS, not significant; in bold, significant.
Conclusions
The screening of a large panel of xenografts established from various tumor types is appropriate to identify the human tumor types that are likely to benefit from a new targeted therapy, and next to identify predictive biomarkers for the response to this targeted therapy. Human tumor xenografted models, closely mimicking clinical situations in terms of biological features and response to treatment [8], will also provide the necessary experimental conditions to evaluate fundamental issues in cancer, including characteristics of metastasis, angiogenesis, and tumor-stroma interactions. The present approach combining species-specific real-time qRT-PCR assays with a large cohort of patient-derived xenografts identified tumor endothelial cells in the all 8 tumor types tested and also revealed a complex pattern of both stroma and tumoral and both autocrine and paracrine VEGFA-VEGFR1/2 signalings. These both findings should be taken into account when evaluating molecular mechanisms of resistance to tumor anti-angiogenic strategies.
Methods
Patient-derived xenografts
Tumor xenografts have been established directly from patient tumors and were routinely passaged by subcutaneous engraftment in Crl:NU(Ico)-Foxn1nu or CB17/Icr-Prkdcscid/IcrCrl [23,24,26-31] purchased from Charles River Laboratories (Les Arbresles, France), with protocol and animal housing in accordance with national regulation and international guidelines [32]. Xenografts were harvested here, after 5 to 12 passages into mice, when they reached around 2,000 mg in size.
Bevacizumab (Avastin, Roche) was given i.p. twice a week, one week, at 15 mg/kg in 0.9% NaCl. Omalizumab (Xolair, Novartis) is given as isotypic control. Lung carcinoma xenografts were transplanted into female 8-week-old Crl:NU(Ico)-Foxn1nu mice. Mice with tumors of 60–200 mm3 were randomly assigned to control or treated groups. Tumor growth was evaluated by measurement of two perpendicular tumor diameters with a caliper twice a week. Individual tumor volumes were calculated: V = a × b2/2, a being the largest diameter, b the smallest. Mice were ethically sacrificed when the tumor volume reached 2 500 mm3 for control groups or at D29 and D50 after first injection of bevacizumab for NSCLC#2 and NCSCLC#3, respectively.
Real-time RT-PCR
RNA extraction, cDNA synthesis and PCR conditions were previously described [33]. The precise amount and quality of total RNA in each reaction mix are both difficult to assess. Therefore, transcripts of the TBP gene encoding the TATA box-binding protein (a component of the DNA-binding protein complex TFIID) were quantified as an endogenous RNA control. The endogenous TBP control was selected due to the moderate prevalence of its transcripts and the absence of known TBP retropseudogenes (retropseudogenes lead to coamplification of contaminating genomic DNA and thus interfere with RT-PCR, despite the use of primers in separate exons) [9].
Quantitative values were obtained from the cycle number (Ct value) (Perkin-Elmer Applied Biosystems, Foster City, CA), according to the manufacturer’s manuals.
The gene primers (Additional file 1: Table S1) were chosen using the Oligo 6.0 program (National Biosciences, Plymouth, MN). The mouse and the human target genes primer pairs were selected to be unique when compared to the sequence of their respective orthologous gene. By contrast, a primer pair, referred as to ‘Total-TBP’ primer pair, was selected to amplify both the mouse and the human TBP genes. dbEST and nr databases were scanned to confirm the total gene specificity of the nucleotide sequences chosen for the primers and the absence of single nucleotide polymorphisms. To avoid amplification of contaminating genomic DNA, one of the two primers was always placed at the junction between two exons. Agarose gel electrophoresis was used to verify the specificity of PCR amplicons. For each human-specific primer pair validation, we performed no-template control (NTC), no-human-reverse-transcriptase control (human RT negative), mouse-reverse-transcriptase control (mouse RT positive from a pool of normal and tumoral mouse RNAs extracted from various tissues types) assays, which produced negligible signals (Ct >40), suggesting that primer–dimer formation, genomic DNA contamination and cross species contamination effects were negligible. Same controls were realized for each mouse-specific primer pair.
Statistical analysis
The distributions of mRNA levels were characterized by their median values and ranges. Relationships between mRNA levels of the different target genes were identified using nonparametric tests (GraphPad Prism 4.00, GraphPad Software, San Diego, CA).
Abbreviations
CRC: Colorectal cancer; CSC: Cancer stem cell; GBM: Glioblastoma; NSCLC: Non small cell lung carcinoma; PDX: Patient-derived tumor xenograft; RCC: Renal cell carcinoma; SCLC: Small cell lung carcinoma; TDEC: Tumor-derived endothelial cell; mCd31: Mouse Pecam1 gene encoding mouse CD31; mCd105: Mouse Eng gene encoding mouse CD105; mVegfr1: Mouse Flt1 gene encoding mouse VEGFR1; mVegfr2: Mouse Kdr gene encoding mouse VEGFR2; hCD31: Human PECAM1 gene encoding human CD31; hCD105: Human ENG gene encoding human CD105; hVEGFR1: Human FLT1 gene encoding human VEGFR1; hVEGFR2: Human KDR gene encoding human VEGFR2.
Competing interests
The authors declare no conflict of interest.
Authors’ contributions
IB and VDM initiated the project and its design, contributed with data analysis and co-drafted the manuscript. SV contributed to the molecular gene studies and performed the statistical analysis. DV participated in project development, SR in sample preparation. RH participated in the molecular gene study. LDM, AD, FN, EA produced the PDX tissues. EM, SRR, DD participated in revision of the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Sequences of oligonucleotides used.
Tumor growth curves of NSCLC#3 and NSCLC#5 xenografts as a function of time. Mice (at least 9 per group) were treated bevacizumab (•) at day 1 and 4; or not (o). Tumor volume was measured twice a week. Tumor growth was evaluated by plotting the mean of the RTV (relative tumor volume) ± SD per group over time after first treatment.
Contributor Information
Ivan Bieche, Email: ivan.bieche@curie.net.
Sophie Vacher, Email: sophie.vacher@curie.net.
David Vallerand, Email: david.vallerand@curie.fr.
Sophie Richon, Email: sophie.richon@parisdescartes.fr.
Rana Hatem, Email: rana.hatem@curie.fr.
Ludmilla De Plater, Email: Ludmilla.Deplater@curie.fr.
Ahmed Dahmani, Email: ahmed.dahmani@curie.fr.
Fariba Némati, Email: fariba.nemati@curie.fr.
Eric Angevin, Email: eric.angevin@igr.fr.
Elisabetta Marangoni, Email: elisabetta.marangoni@curie.fr.
Sergio Roman-Roman, Email: sergio.roman-roman@curie.fr.
Didier Decaudin, Email: didier.decaudin@curie.fr.
Virginie Dangles-Marie, Email: virginie.dangles-marie@curie.fr.
Acknowledgments
We thank Ludovic Bigot, Ludovic Lacroix, Franck Assayag and Dalila Labiod for the management of RNA, PDX tissues or PDX engrafted mice. We are grateful to Chantal Martin and Isabelle Grandjean for housing and care of mice in the animal facility of IMTCE and Institut Curie, respectively.
This work was supported by the Comité départemental des Hauts-de-Seine de la Ligue Nationale Contre le Cancer, the Conseil régional d'Ile-de-France, the Cancéropôle Ile-de-France and the Association pour la recherche en cancérologie de Saint-Cloud (ARCS), Genevieve and Jean-Paul Driot Transformative Research Grant, Philippe and Laurent Bloch Cancer Research Grant, Hassan Hachem Translational Medicine Grant and Sally Paget-Brown Translational Research Grant.
References
- Ellis LM, Hicklin DJ. VEGF-targeted therapy: mechanisms of anti-tumour activity. Nat Rev Cancer. 2008;8:579–591. doi: 10.1038/nrc2403. [DOI] [PubMed] [Google Scholar]
- Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23:1011–1027. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]
- Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signalling - in control of vascular function. Nat Rev Mol Cell Biol. 2006;7:359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- Ping YF, Bian XW. Consice review: Contribution of cancer stem cells to neovascularization. Stem Cells. 2011;29:888–894. doi: 10.1002/stem.650. [DOI] [PubMed] [Google Scholar]
- Knizetova P, Ehrmann J, Hlobilkova A, Vancova I, Kalita O, Kolar Z, Bartek J. Autocrine regulation of glioblastoma cell cycle progression, viability and radioresistance through the VEGF-VEGFR2 (KDR) interplay. Cell Cycle. 2008;7:2553–2561. doi: 10.4161/cc.7.16.6442. [DOI] [PubMed] [Google Scholar]
- Lichtenberger BM, Tan PK, Niederleithner H, Ferrara N, Petzelbauer P, Sibilia M. Autocrine VEGF signaling synergizes with EGFR in tumor cells to promote epithelial cancer development. Cell. 2010;140:268–279. doi: 10.1016/j.cell.2009.12.046. [DOI] [PubMed] [Google Scholar]
- Hamerlik P, Lathia JD, Rasmussen R, Wu Q, Bartkova J, Lee M, Moudry P, Bartek J Jr, Fischer W, Lukas J, Rich JN, Bartek J. Autocrine VEGF-VEGFR2-Neuropilin-1 signaling promotes glioma stem-like cell viability and tumor growth. J Exp Med. 2012;209:507–520. doi: 10.1084/jem.20111424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, Arcaroli JJ, Messersmith WA, Eckhardt SG. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9:338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieche I, Onody P, Laurendeau I, Olivi M, Vidaud D, Lidereau R, Vidaud M. Real-time reverse transcription-PCR assay for future management of ERBB2-based clinical applications. Clin Chem. 1999;45:1148–1156. [PubMed] [Google Scholar]
- Baird SM, Beattie GM, Lannom RA, Lipsick JS, Jensen FC, Kaplan NO. Induction of lymphoma in antigenically stimulated athymic mice. Cancer Res. 1982;42:198–206. [PubMed] [Google Scholar]
- Eberhard A, Kahlert S, Goede V, Hemmerlein B, Plate KH, Augustin HG. Heterogeneity of angiogenesis and blood vessel maturation in human tumors: implications for antiangiogenic tumor therapies. Cancer Res. 2000;60:1388–1393. [PubMed] [Google Scholar]
- Fukumura D, Xavier R, Sugiura T, Chen Y, Park EC, Lu N, Selig M, Nielsen G, Taksir T, Jain RK, Seed B. Tumor induction of VEGF promoter activity in stromal cells. Cell. 1998;94:715–725. doi: 10.1016/S0092-8674(00)81731-6. [DOI] [PubMed] [Google Scholar]
- Liang WC, Wu X, Peale FV, Lee CV, Meng YG, Gutierrez J, Fu L, Malik AK, Gerber HP, Ferrara N, Fuh G. Cross-species vascular endothelial growth factor (VEGF)-blocking antibodies completely inhibit the growth of human tumor xenografts and measure the contribution of stromal VEGF. J Biol Chem. 2006;281:951–961. doi: 10.1074/jbc.M508199200. [DOI] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Hess AR, Seftor RE. Vasculogenic mimicry and tumour-cell plasticity: lessons from melanoma. Nat Rev Cancer. 2003;3:411–421. doi: 10.1038/nrc1092. [DOI] [PubMed] [Google Scholar]
- Alvero AB, Fu HH, Holmberg J, Visintin I, Mor L, Marquina CC, Oidtman J, Silasi DA, Mor G. Stem-like ovarian cancer cells can serve as tumor vascular progenitors. Stem Cells. 2009;27:2405–2413. doi: 10.1002/stem.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussolati B, Bruno S, Grange C, Ferrando U, Camussi G. Identification of a tumor-initiating stem cell population in human renal carcinomas. Faseb J. 2008;22:3696–3705. doi: 10.1096/fj.08-102590. [DOI] [PubMed] [Google Scholar]
- Bussolati B, Grange C, Sapino A, Camussi G. Endothelial cell differentiation of human breast tumour stem/progenitor cells. J Cell Mol Med. 2009;13:309–319. doi: 10.1111/j.1582-4934.2008.00338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardee ME, Zagzag D. Mechanisms of Glioma-Associated Neovascularization. Am J Pathol. 2012;181:1126–1141. doi: 10.1016/j.ajpath.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T, Maira G, Parati EA, Stassi G, Larocca LM, De Maria R. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- Soda Y, Marumoto T, Friedmann-Morvinski D, Soda M, Liu F, Michiue H, Pastorino S, Yang M, Hoffman RM, Kesari S, Verma IM. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc Natl Acad Sci U S A. 2011;108:4274–4280. doi: 10.1073/pnas.1016030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Chadalavada K, Wilshire J, Kowalik U, Hovinga KE, Geber A, Fligelman B, Leversha M, Brennan C, Tabar V. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468:829–833. doi: 10.1038/nature09624. [DOI] [PubMed] [Google Scholar]
- Ghanekar A, Ahmed S, Chen K, Adeyi O. Endothelial cells do not arise from tumor-initiating cells in human hepatocellular carcinoma. BMC Cancer. 2013;13:485. doi: 10.1186/1471-2407-13-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien S, Merino-Trigo A, Lacroix L, Pocard M, Goere D, Mariani P, Landron S, Bigot L, Nemati F, Dartigues P, Weiswald LB, Lantuas D, Morgand L, Pham E, Gonin P, Dangles-Marie V, Job B, Dessen P, Bruno A, Pierre A, De Thé H, Soliman H, Nunes M, Lardier G, Calvet L, Demers B, Prevost G, Vrignaud P, Roman-Roman S, Duchamp O. et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin Cancer Res. 2012;18:5314–5328. doi: 10.1158/1078-0432.CCR-12-0372. [DOI] [PubMed] [Google Scholar]
- Reyal F, Guyader C, Decraene C, Lucchesi C, Auger N, Assayag F, De Plater L, Gentien D, Poupon MF, Cottu P, De Cremoux P, Gestraud P, Vincent-Salomon A, Fontaine JJ, Roman-Roman S, Delattre O, Decaudin D, Marangoni E. Molecular profiling of patient-derived breast cancer xenografts. Breast Cancer Res. 2012;14:R11. doi: 10.1186/bcr3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonsatti E, Altomonte M, Nicotra MR, Natali PG, Maio M. Endoglin (CD105): a powerful therapeutic target on tumor-associated angiogenetic blood vessels. Oncogene. 2003;22:6557–6563. doi: 10.1038/sj.onc.1206813. [DOI] [PubMed] [Google Scholar]
- Angevin E, Glukhova L, Pavon C, Chassevent A, Terrier-Lacombe MJ, Goguel AF, Bougaran J, Ardouin P, Court BH, Perrin JL, Vallancien G, Triebel F, Escudier B. Human renal cell carcinoma xenografts in SCID mice: tumorigenicity correlates with a poor clinical prognosis. Lab Invest. 1999;79:879–888. [PubMed] [Google Scholar]
- Cottu P, Marangoni E, Assayag F, de Cremoux P, Vincent-Salomon A, Guyader C, de Plater L, Elbaz C, Karboul N, Fontaine JJ, Chateau-Joubert S, Boudou-Rouquette P, Alran S, Dangles-Marie V, Gentien D, Poupon MF, Decaudin D. Modeling of response to endocrine therapy in a panel of human luminal breast cancer xenografts. Breast Cancer Res Treat. 2012;133:595–606. doi: 10.1007/s10549-011-1815-5. [DOI] [PubMed] [Google Scholar]
- de Plater L, Lauge A, Guyader C, Poupon MF, Assayag F, de Cremoux P, Vincent-Salomon A, Stoppa-Lyonnet D, Sigal-Zafrani B, Fontaine JJ, Brough R, Lord CJ, Ashworth A, Cottu P, Decaudin D, Marangoni E. Establishment and characterisation of a new breast cancer xenograft obtained from a woman carrying a germline BRCA2 mutation. Br J Cancer. 2010;103:1192–1200. doi: 10.1038/sj.bjc.6605900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuraud P, Taillandier L, Medioni J, Aguirre-Cruz L, Criniere E, Marie Y, Kujas M, Golmard JL, Duprez A, Delattre JY, Sanson M, Poupon MF. Distinct responses of xenografted gliomas to different alkylating agents are related to histology and genetic alterations. Cancer Res. 2004;64:4648–4653. doi: 10.1158/0008-5472.CAN-03-3429. [DOI] [PubMed] [Google Scholar]
- Nemati F, Bras-Goncalves R, Fontaine JJ, de Pinieux G, De Cremoux P, Chapelier A, Daniel C, Laurent-Puig P, Livartowski A, Judde JG, Bordier V, Poupon MF, Decaudin D. Preclinical assessment of cisplatin-based therapy versus docetaxel-based therapy on a panel of human non-small-cell lung cancer xenografts. Anticancer Drugs. 2009;20:932–940. doi: 10.1097/CAD.0b013e32833009cc. [DOI] [PubMed] [Google Scholar]
- Nemati F, Daniel C, Arvelo F, Legrier ME, Froget B, Livartowski A, Assayag F, Bourgeois Y, Poupon MF, Decaudin D. Clinical relevance of human cancer xenografts as a tool for preclinical assessment: example of in-vivo evaluation of topotecan-based chemotherapy in a panel of human small-cell lung cancer xenografts. Anticancer Drugs. 2010;21:25–32. doi: 10.1097/CAD.0b013e3283300a29. [DOI] [PubMed] [Google Scholar]
- Workman P, Aboagye EO, Balkwill F, Balmain A, Bruder G, Chaplin DJ, Double JA, Everitt J, Farningham DA, Glennie MJ, Kelland LR, Robinson V, Stratford IJ, Tozer GM, Watson S, Wedge SR, Eccles SA. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102:1555–1577. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieche I, Parfait B, Le Doussal V, Olivi M, Rio MC, Lidereau R, Vidaud M. Identification of CGA as a novel estrogen receptor-responsive gene in breast cancer: an outstanding candidate marker to predict the response to endocrine therapy. Cancer Res. 2001;61:1652–1658. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequences of oligonucleotides used.
Tumor growth curves of NSCLC#3 and NSCLC#5 xenografts as a function of time. Mice (at least 9 per group) were treated bevacizumab (•) at day 1 and 4; or not (o). Tumor volume was measured twice a week. Tumor growth was evaluated by plotting the mean of the RTV (relative tumor volume) ± SD per group over time after first treatment.