ABSTRACT
Phosphorylation-dependent protein ubiquitylation and degradation provides an irreversible mechanism to terminate protein kinase signaling. Here, we report that mammary epithelial cells require cullin-5–RING–E3-ubiquitin-ligase complexes (Cul5-CRLs) to prevent transformation by a Src–Cas signaling pathway. Removal of Cul5 stimulates growth-factor-independent growth and migration, membrane dynamics and colony dysmorphogenesis, which are all dependent on the endogenous tyrosine kinase Src. Src is activated in Cul5-deficient cells, but Src activation alone is not sufficient to cause transformation. We found that Cul5 and Src together stimulate degradation of the Src substrate p130Cas (Crk-associated substrate). Phosphorylation stimulates Cas binding to the Cul5-CRL adaptor protein SOCS6 and consequent proteasome-dependent degradation. Cas is necessary for the transformation of Cul5-deficient cells. Either knockdown of SOCS6 or use of a degradation-resistant Cas mutant stimulates membrane ruffling, but not other aspects of transformation. Our results show that endogenous Cul5 suppresses epithelial cell transformation by several pathways, including inhibition of Src–Cas-induced ruffling through SOCS6.
KEY WORDS: Migration, Transformation, Ubiquitin, Cullin 5, Cul5, Src, Cas
INTRODUCTION
Cullin–RING–ligase (CRL) complexes constitute the largest known class of E3 ubiquitin ligases (Deshaies and Joazeiro, 2009; Petroski and Deshaies, 2005). Each CRL contains a cullin protein backbone that forms a bridge between a RING protein and one of a large number of alternative substrate-specific adaptors. Substrate binding to a CRL stimulates mono- or polyubiquitylation that, in most cases, targets the substrate to the 26S proteasome for degradation. CRL substrates include oncogenic and anti-oncogenic proteins, so CRLs can inhibit or stimulate transformation depending on their substrate specificity. Consequently, genes encoding CRL polypeptides are often altered in cancer cells (Lee and Zhou, 2010). For example, cancer cells often have loss-of-function mutations in Fbw7, VHL and Keap1, which target oncoproteins to Cul1, Cul2 and Cul3 respectively, whereas Cul4A, which ubiquitylates several tumor suppressors, is commonly amplified or overexpressed. Adenoviruses and Kaposi's-sarcoma-associated herpesvirus encode Cul5 adaptors that bind to the p53 tumor suppressor, thereby inducing degradation of p53 (Lee and Zhou, 2010). Cul5 thus mediates oncogenesis by these viruses. However, it is not known whether Cul5 targets oncogenic proteins or tumor suppressors in the absence of virus infection.
Cul5 resembles other cullin proteins in the large number of alternative adaptor proteins it uses to select substrates (Kile et al., 2002). Most Cul5 adaptors contain a sequence called a SOCS box that binds a linker protein, elongin C (EloC, Tceb1) and the N-terminus of Cul5 (Kile et al., 2002). EloC also binds EloB (Tceb2), and Cul5 binds RING protein Rbx2 (Rnf7), resulting in adaptor–EloBC–Cul5–Rbx2 CRLs (Huang et al., 2009; Kamura et al., 2004). The mammalian genome encodes approximately 40 SOCS box proteins, which can be classified according to their substrate-interaction domains, such as SH2 domains, WD40 motifs, SPRY domains, Ankyrin repeats and Tubby-like domains (Okumura et al., 2012). These substrate-interaction domains, in turn, bind a variety of substrates. This complexity has hindered the discovery of specific substrates and understanding their biological significance.
One family of SOCS box proteins contains SH2 domains and can thus target substrates containing phosphotyrosine (pY) degrons to Cul5-CRL for polyubiquitylation (Okumura et al., 2012; Pawson et al., 2001). There are eight SOCS-SH2 family members, named SOCS1–SOCS7 and CisH. Of these, SOCS1–SOCS3 and CisH have been intensively studied in leukocytes where they are induced by cytokines and chemokines. They inhibit JAK family tyrosine kinase signaling by Cul5-independent as well as Cul5-dependent mechanisms. The remaining family members, SOCS4–SOCS7, are widely expressed in many tissues and inhibit signaling by cytokines and peptide growth factors including insulin, Steel factor and EGF. The full complexity of SOCS–Cul5 CRL substrates is unknown.
Expression of CUL5 and several genes encoding SOCS-SH2 proteins is decreased in some types of human cancer, suggesting that SOCS–Cul5 CRL substrates include oncogenic proteins (Elliott et al., 2008; Fay et al., 2003; Hampton et al., 1994; Lai et al., 2010; Sasi et al., 2010; Wang et al., 2006). Tyrosine kinases tend to be oncogenic, and are frequently activated in human cancers (Hunter, 2009). Loss-of-function mutations, loss of heterozygosity and genetic silencing of CUL5 and SOCS genes in cancer cells might therefore be selected because their loss inhibits turnover of pY proteins and stimulates oncogenic signaling.
Src, the protein encoded by the SRC proto-oncogene, is a tyrosine kinase that is strongly implicated in human cancers (Ishizawar and Parsons, 2004; Krishnan et al., 2012). Src is negatively regulated by the ubiquitin-proteasome pathway: inactive Src is stable but active Src is polyubiquitylated and degraded (Hakak and Martin, 1999; Harris et al., 1999; Imamoto and Soriano, 1993; Nada et al., 1993). It has been unclear which ubiquitin ligase is involved. We and others have found that knockdown of Cul5 in mouse fibroblasts stabilizes active Src, suggesting that Cul5-CRLs are required for Src turnover in these cells (Laszlo and Cooper, 2009; Pan et al., 2011). Moreover, Cul5 knockdown induces transformation of fibroblasts in which Src is also genetically activated, either by SRC gene mutation or by deletion of the CSK gene, which encodes a Src-inhibitory kinase (Laszlo and Cooper, 2009). The transformation of Cul5-deficient, SRC mutant cells is not due simply to the increased activity of Src, suggesting that additional Cul5 substrates are also crucial. However, these substrates have not been identified. Two important questions remain unanswered. (1) Are Cul5-deficient cells only transformed if Src is also activated? (2) Which Cul5 substrates drive transformation when Cul5 is absent?
We now show that inhibition of CUL5 expression in human mammary epithelial cells induces transformation. Transformation does not require genetic activation of Src, but endogenous Src is enzymatically activated and required for transformation. However, ectopic Src does not induce transformation when Cul5 is present, suggesting that other Cul5 substrates are involved. We found that removal of Cul5 stabilizes p130Cas (also known as breast cancer anti-estrogen resistance 1, BCAR1). Cas is a substrate for Src and other tyrosine kinases. Cas interacts with focal adhesion proteins and becomes tyrosine phosphorylated in response to cytoskeletal tension, and thus binds to adaptors that regulate small GTPases (Bouton et al., 2001; Matsui et al., 2012). Cas is important for the motility and proliferation of cancer cells (Cabodi et al., 2006; Tornillo et al., 2011; van der Flier et al., 2000). It is required in Cul5-deficient cells for growth-factor-independent proliferation and increased migration. The Cul5 adaptor SOCS6 binds Cas when Cas is phosphorylated at specific tyrosine residues and thus stimulates turnover of Cas. Removal of SOCS6 or expression of degradation-resistant Cas stimulates membrane ruffling but not other aspects of the Cul5-deficient phenotype. The results suggest that Cul5 suppresses the transformation of epithelial cells by targeting phosphorylated Cas and other unidentified Src substrates for degradation.
RESULTS
Inhibition of Cul5 expression transforms epithelial cells in vitro
Two lines of evidence suggest that Cul5 might be a tumor suppressor: first, Cul5 expression is reduced in several carcinomas and second, expression of Cul5 (VACM1) in some cancer cell lines inhibits proliferation or survival (Fay et al., 2003; Johnson et al., 2007; Lai et al., 2010). To further test whether endogenous Cul5 inhibits transformation, we used RNA interference to knockdown Cul5 in the non-tumorigenic, spontaneously immortalized human mammary cell line MCF10A, which has been used extensively for transformation studies (Debnath and Brugge, 2005; Muthuswamy et al., 2001). Cells were infected with retroviruses that express a puromycin-resistance gene and short-hairpin RNA (shRNA) targeting Cul5 or, as a control, shRNA against Cul2. We selected puromycin-resistant, polyclonal cell lines and confirmed that Cul5 expression was inhibited (Fig. 1A). We then tested whether Cul5-deficient cells were transformed.
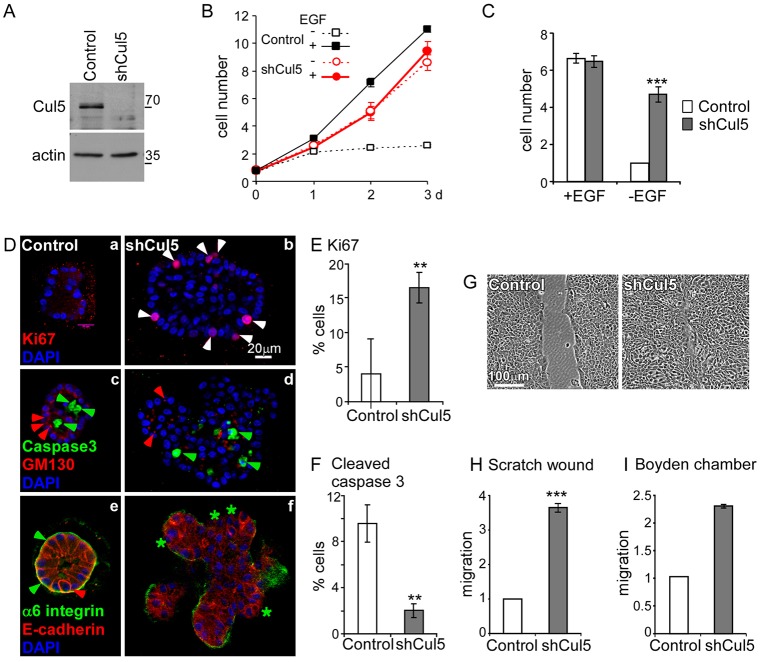
Fig. 1.
Cullin 5 suppresses EGF-independent proliferation, migration and formation of transformed acini. (A) Efficiency of Cul5 depletion. Western blot of control (pMXpuroII vector) and Cul5-deficient (pMXpuroII-shCul5) MCF10A cell lysates probed with antibodies against Cul5 and an internal control protein (actin). (B,C) Control and Cul5-deficient MCF10A cells were grown in the presence and absence of EGF, stained with Crystal Violet on days 1, 2 or 3 (B) or day 3 only (C) and intensity was measured. Cell counts were normalized to control cells grown in the absence of EGF. Mean and s.e.; n = 3. (D) Growth in Matrigel culture. Control and Cul5-deficient MCF10A cells cultured in growth factor-depleted Matrigel with 5 ng/ml EGF for 14 days. Colonies were fixed, permeabilized and stained for dividing cells (a,b, Ki67, white arrowheads), apoptotic cells (c,d, cleaved caspase 3, green arrowheads), Golgi complex (c,d, GM130, red arrowheads), basal membrane (e,f, α6 integrin, green arrowheads indicate presence; green stars indicate absence) and lateral membrane (e,f, E-cadherin, red arrowhead). Optical sections through the centers of representative colonies are shown. (E,F) Quantification of proliferation and apoptosis. Serial optical sections were used to determine the percentage of stained cells in five colonies of each type. (G,H) Scratch-wound healing assay in the absence of EGF. (G) Phase-contrast images of monolayers 24 hours after wounding. (H) Migration distance. Mean and s.e.; n = 3. (I) Migration in a Boyden chamber assay. Cells migrated through a collagen-coated filter in response to 5% horse serum and 20 ng/ml EGF. *P<0.05; **P<0.01 and ***P<0.001 by Student's t-test.
Cul5-deficient cells grew at a similar rate to control cells in normal medium, which contains epidermal growth factor (EGF), but grew significantly better than control cells in the absence of EGF (Fig. 1B,C). Stable knockdown of Rbx2, the RING protein needed by Cul5 but not other cullin proteins, also allowed EGF-independent growth, whereas knockdown of Cul2 did not (supplementary material Fig. S1A) (Huang et al., 2009; Kamura et al., 2004). To control for possible off-target effects of the Cul5 shRNA, we performed transient knockdown of Cul5 with siRNA targeting different sequences in CUL5 mRNA. Again, EGF-independent growth was significantly stimulated (supplementary material Fig. S1B). This suggests that endogenous Cul5 specifically inhibits EGF-independent cell proliferation.
Transformation of MCF10A cells can be assayed by colony formation in Matrigel (Debnath et al., 2003). Normal cells form hollow colonies comprising dead and dying inner cells and an outer, quiescent, polarized epithelium. By contrast, cells transformed by activated viral Src (vSrc), the combination of cyclin D1 and Bcl2, or activated tyrosine kinase receptors, form solid colonies containing cells that continue to proliferate, have decreased apoptosis and are poorly polarized (Debnath et al., 2002; Reginato et al., 2005; Wrobel et al., 2004). Like other transformed MCF10A cells, Cul5-deficient cells formed dysmorphic colonies that were larger than the control colonies and showed increased proliferation (Fig. 1Da,b, Ki67, red) and reduced apoptosis (Fig. 1Dc,d, caspase 3, green). The Golgi was not oriented consistently to the interior (Fig. 1Dc,d, GM130, red) and some cells lacked basal α6 integrin (Fig. 1De,f, green). E-cadherin was more diffuse than in control colonies (Fig. 1De,f, red).
To investigate whether Cul5 regulates cell motility, we performed migration assays using scratch wounds and Boyden chambers. Confluent monolayers of control and Cul5-deficient cells were transferred to EGF-deficient medium, wounded, and monitored by phase-contrast microscopy. Cul5-deficient cells migrated more rapidly than control cells (Fig. 1G,H) but remained as a sheet and did not undergo the epithelial–mesenchymal transition (Wrobel et al., 2004). Single-cell migration towards EGF in a Boyden chamber was also increased when Cul5 was absent (Fig. 1I). Migration was also stimulated when Cul5 was transiently knocked down using siRNAs targeting different regions of CUL5 mRNA (see below), reducing the chance of off-target effects.
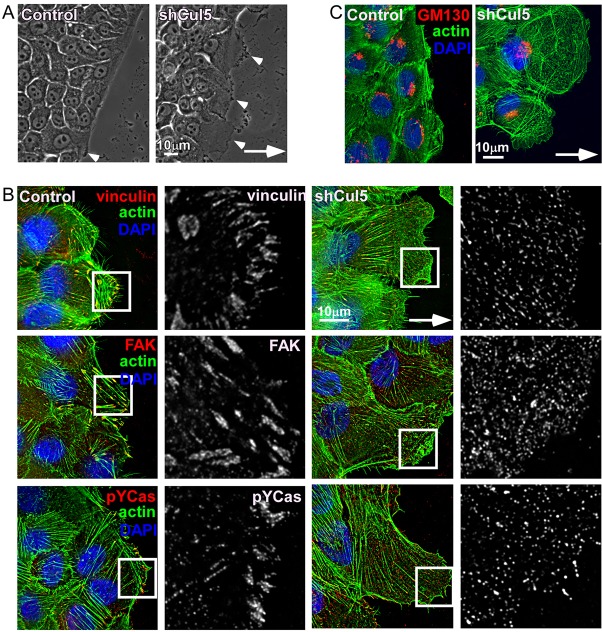
Cul5-deficient cells were morphologically altered when migrating or sub-confluent (Fig. 2; supplementary material Fig. S1C,D). Characteristically, they exhibited an elongated leading lamellipodium and phase-dark regions, indicating membrane ruffling (Fig. 2A). Increased ruffling was confirmed by time-lapse micrography of migrating cells (data not shown). Enlarged lamellipodia and ruffling were also evident when Cul5-deficient cells were plated in EGF-deficient medium at low density (supplementary material Fig. S1C,D). Cul5-deficient cells contained many tiny contact sites, detected with antibodies against vinculin, FAK and pY-Cas, in contrast to the prominent focal adhesions of normal cells (Fig. 2B). Stress fibers were also decreased (Fig. 2B,C). However, the Golgi complex was oriented in front of the nucleus, suggesting that Cul5-deficient cells polarize normally during migration (Fig. 2C).
Fig. 2.
Cullin-5-deficient cells show increased membrane ruffling, larger leading lamella and smaller adhesions. Confluent monolayers of control and Cul5-deficient MCF10A cells were transferred to EGF-deficient medium, scratched and cells at the leading edge imaged 6 hours later. Arrows indicate direction of migration. (A) Cul5-deficient cells have an elongated leading lamellipodium and increased membrane ruffling (arrowheads). (B,C) Cells were fixed, permeabilized and stained with anti-vinculin, anti-FAK and anti-pY-Cas to visualize focal adhesions (B, single 0.2 µm slices at the bottom of the cells) and anti-GM130, a Golgi marker (C, 3 µm flattened Z-projections). Phalloidin was used to stain actin fibers. The white boxes indicate the regions enlarged in the insets.
Taken together, these results suggest that endogenous Cul5 suppresses several hallmarks of transformation, including EGF-independent proliferation, colony dysmorphogenesis, EGF-independent migration, disruption of focal adhesions, and the formation of an extended, ruffling lamellipodium.
Src is required for transformation of Cul5-deficient cells
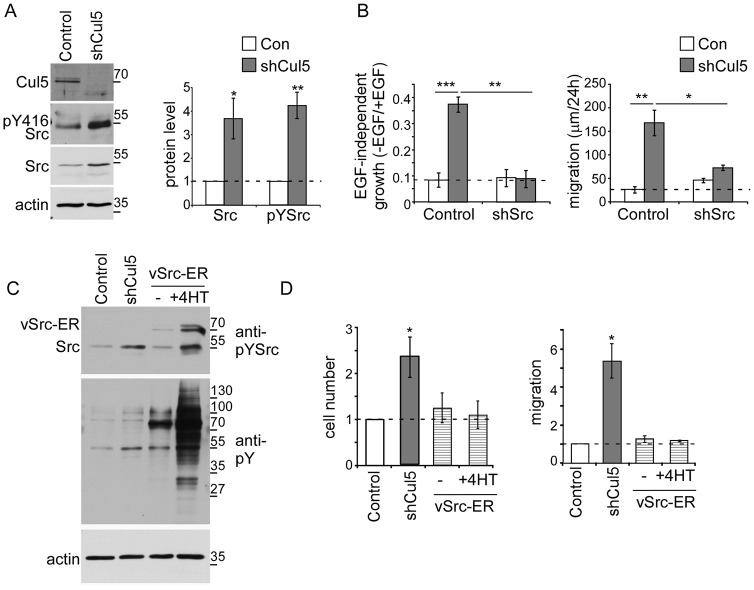
Cul5 is required for the degradation of activated but not inhibited Src in fibroblasts (Laszlo and Cooper, 2009; Pan et al., 2011). We tested whether endogenous Cul5 also regulates Src in MCF10A cells. Western blotting showed that Src protein levels increased in Cul5-deficient MCF10A cells, even though mRNA levels, measured by RT-PCR, were unaltered (Fig. 3A; supplementary material Fig. S2A). The increase in Src protein was quite variable between experiments, but Src activity, detected using a phosphoepitope antibody to autophosphorylation site Tyr416, was consistently increased (Fig. 3A). The results suggest that Cul5 represses Src activity, perhaps partly by regulating proteins that activate Src as well as by regulating Src expression or stability.
Fig. 3.
Src is necessary, but not sufficient, for EGF-independent proliferation and migration of Cul5-deficient cells. (A) Western blot analysis of MCF10A cell lysates. Cullin 5 deficiency increases the levels of Src and phospho-Src. Actin protein levels remained constant. Quantification of western blots is shown on the right; mean and s.e.; n = 3. (B) The transformed phenotypes of Cul5-deficient cells require Src. Proliferation: single and double knockdown MCF10A cells were cultured in the presence and absence of EGF and counted after 5–7 days. Ratios of cell numbers in the absence and presence of EGF were averaged across n = 3 experiments. Migration: scratch-wound healing assay in the absence of EGF. Distance measured 24 hours after wounding. Mean and s.e.; n = 4. (C,D) Active Src is not sufficient to induce proliferation or migration. (C) Western blot analysis of lysates of control and Cul5-deficient cells and cells expressing a vSrc–ER fusion protein. Note the phosphorylation of the vSrc–ER protein and increased tyrosine phosphorylation of cell proteins in vSrc–ER cells in the absence of 4-hydroxy-tamoxifen (4HT) compared with normal or Cul5-deficient cells. Phosphorylation was increased further with 0.25 µM 4HT. (D) Proliferation: cell counts were normalized to control cells grown in the absence of EGF; mean and s.e.; n = 3. Migration: scratch-wound healing assay in the absence of EGF. Distance measured 24 hours after wounding. Mean and s.e.; n = 3. *P<0.05, **P<0.01 and ***P<0.001 by Student’s t-test.
Some oncogenes require endogenous Src to transform MCF10A cells (Wrobel et al., 2004). We tested whether Src is required for transformation of Cul5-deficient MCF10A cells. Hygromycin- and puromycin-resistant retrovirus vectors encoding shRNAs against Cul5 and Src were introduced into MCF10A cells in various combinations and polyclonal populations were selected. Src shRNA inhibited Src expression and suppressed the EGF-independent proliferation and migration induced by Cul5 knockdown (Fig. 3B; supplementary material Fig. S2B). Similar results were obtained whether Cul5 shRNA was expressed from a puromycin-resistant vector and Src shRNA from a hygromycin-resistant vector or vice versa (data not shown). To control for off-target effects, cells were transiently transfected with siRNAs targeting Cul5 and Src, and migration was assayed. Src was required for the increased migration of Cul5-deficient cells (supplementary material Fig. S2C,D). The pharmacological Src inhibitor, SU6656, restored normal acini morphogenesis to Cul5-deficient cells (supplementary material Fig. S2E). These results indicate that the increased migration and dysmorphic colonies of Cul5-deficient cells require Src kinase activity.
To test whether Src activation is sufficient to stimulate MCF10A cell proliferation and migration, we made use of an MCF10A cell line expressing a 4-hydroxytamoxifen (4HT)-regulated vSrc–ER (estrogen receptor) fusion protein (Reginato et al., 2005). These cells are not transformed in normal medium, but become transformed in the presence of 4HT (Reginato et al., 2005). We found that vSrc–ER was partially active even in the absence of 4HT, and stimulated further by a low dose of 4HT, as judged by blotting for pY in total proteins or pY-Cas (Fig. 3C; supplementary material Fig. S2F). Despite having greater Src activity than Cul5-deficient cells, the vSrc–ER cells did not proliferate or migrate faster than control cells (Fig. 3D). Although it is possible that different substrates are phosphorylated by vSrc–ER and activated endogenous Src, it seems likely that Cul5 suppresses transformation by inhibiting other proteins in addition to Src.
Cas is required for transformation of Cul5-deficient cells
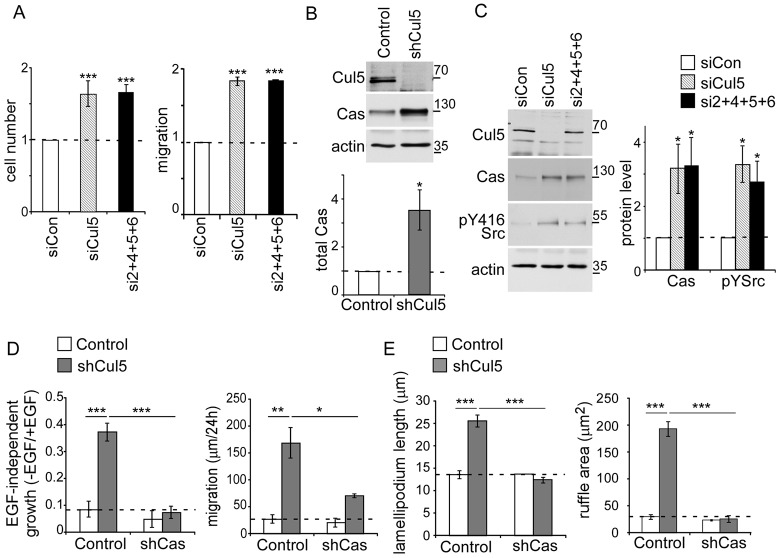
We searched for other proteins that might be Cul5 substrates and contribute to the transformation of Cul5-deficient cells. Cul5 has the potential to ubiquitylate many substrates, depending on which substrate adaptor is bound. We speculated that other Cul5 substrates that contribute to transformation could be Src substrates and contain pY. Such pY Cul5 substrates are likely to be recognized by the SH2 domain-containing SOCS adaptor proteins. RT-PCR analysis indicated that SOCS2, SOCS4, SOCS5 and SOCS6 are more abundant at the RNA level than other SH2-SOCS genes in MCF10A cells (supplementary material Fig. S3A). We then tested whether combined knockdown of SOCS2, SOCS4, SOCS5 and SOCS6 with siRNA would recapitulate the effects of Cul5 siRNA. Indeed, combined siRNA against SOCS2, SOCS4, SOCS5 and SOCS6 stimulated EGF-independent proliferation and migration to a similar extent as Cul5 siRNA (Fig. 4A). This suggests that Cul5 suppresses proliferation and migration by inhibiting pY proteins through SH2-SOCS adaptors.
Fig. 4.
Cas is necessary for leading edge abnormalities of Cul5-deficient cells and is regulated by Cul5. (A) Removal of SOCS2, SOCS4, SOCS5 and SOCS6 phenocopies knockdown of Cul5. MCF10A cells were incubated with siRNA against control, Cul5 or SOCS proteins, and analyzed 4 days later. Proliferation: Cell counts were normalized to control cells grown in the absence of EGF. Migration: scratch-wound healing assay in the absence of EGF. Distance measured 24 hours after wounding. Mean and s.e.; n = 3. (B,C) Western blot analysis of MCF10A cell lysates. (B) Cullin 5 deficiency increases the levels of Cas. As a control, actin protein levels remained constant. Quantification of western blots. Mean and s.e.; n = 5. (C) Transient knockdown of Cul5 or SOCS2, SOCS4, SOCS5 and SOCS6 increases the levels of Cas and pY-Src. Actin protein levels remained constant. Quantification of western blots. Mean and s.e.; n = 8. (D) The transformed phenotypes of Cul5-deficient cells require Cas. Proliferation: single- and double-knockdown MCF10A cells were cultured in the absence and presence of EGF and counted after 5–7 days. Ratios of cell numbers in the absence and presence of EGF were averaged across three independent experiments. Migration: scratch-wound healing assay in the absence of EGF. Distance measured 24 hours after wounding. Mean and s.e., n = 4. The control data are the same as those shown in Fig. 3B. (E) Cas removal from Cul5-deficient MCF10A cells rescued lamellipodium length and ruffle area. Confluent monolayers were transferred to EGF-deficient medium and then scratched. Phase-contrast images of cells at the leading edge were captured 6 hours after scratching. For each cell, the lamellipodia length (distance from the nucleus to the leading edge) and area of phase-dark membrane ruffles were measured. Graphs show the mean and s.e. for ∼50 cells/condition from n = 3 experiments. *P<0.05, **P<0.01 and ***P<0.001 by Student’s t-test.
We screened for pY-containing proteins that might be targets for SH2-SOCS–Cul5 CRLs. Briefly, pY peptides were recovered from control, Cul5- and Cul2-deficient MCF10A cells and identified by mass spectrometry (Zhang et al., 2005). Phosphopeptides corresponding to pY128 and pY249 of p130Cas were identified a total of 18 times in the sample from Cul5-deficient cells but were not detected in control or Cul2-deficient cells (supplementary material Fig. S3B). Western blotting confirmed that Cas levels were increased in Cul5-deficient cells and when Cul5 or SOCS2, 4, 5 and 6 were transiently knocked down with siRNA (Fig. 4B,C). Levels of pY-Src were also increased when Cul5 or SOCS2, SOCS4, SOCS5 and SOCS6 were transiently knocked down (Fig. 4C). Cas mRNA levels did not increase detectably (supplementary material Fig. S2A). These results suggest that Cul5 inhibits Cas protein expression post-transcriptionally.
Cas is a Src substrate implicated in transformation (Matsui et al., 2012). Cas-knockout mouse fibroblasts resist transformation by active mutant Src, and Cas knockdown in breast cancer cells suppresses transformation. Cas overexpression induces membrane ruffling and stimulates invasion by Src-transformed fibroblasts and cancer cells (Bouton et al., 2001; Cabodi et al., 2006; Matsui et al., 2012; Tornillo et al., 2011; van der Flier et al., 2000). We therefore tested whether Cas is required for transformation of Cul5-deficient MCF10A cells. Retroviruses were used to knock down Cas in control and Cul5-deficient cells (supplementary material Fig. S3C). Cas knockdown inhibited the EGF-independent proliferation and migration of Cul5-deficient cells (Fig. 4D). In addition, Cas knockdown restored the normal lamellipodium length and ruffle area to Cul5-deficient cells (Fig. 4E). To control for off-target effects, transient knockdown of Cas with siRNA also restored normal migration (supplementary material Fig. S3D,E). Therefore, Cas, like Src, is required for the increased proliferation and migration of Cul5-deficient cells, suggesting that Src and Cas work in the same signaling pathway.
Cul5 regulates Cas protein stability and binds Cas through SOCS6
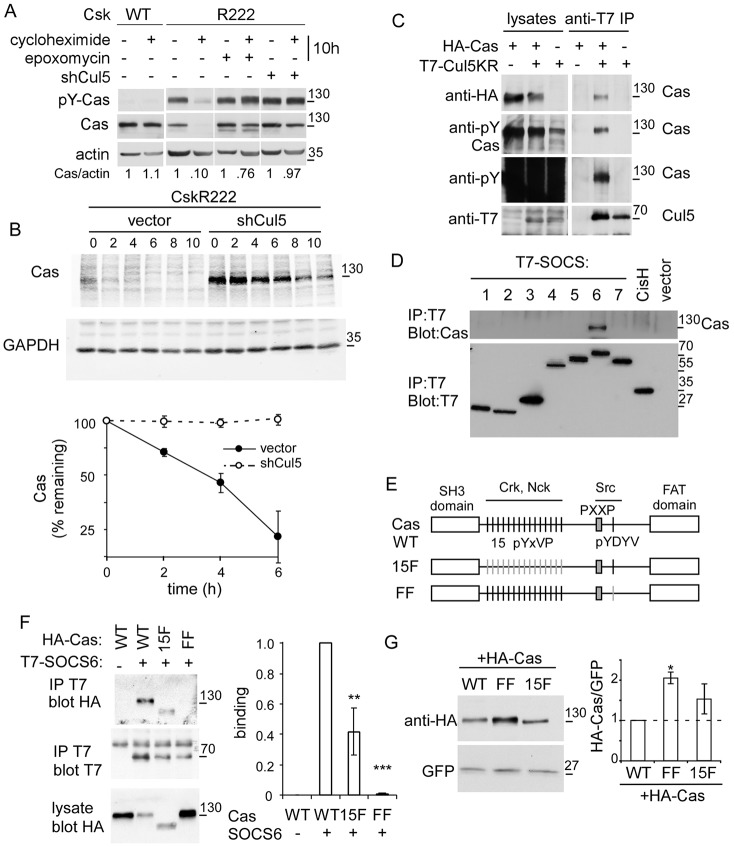
To test whether Cas stability is regulated by Cul5 when Src is active, we used matched mouse fibroblast lines derived from a Csk mutant embryo. Either kinase-defective Csk (R222) or wild-type Csk (Csk+) was re-expressed, to de-repress and repress Src, respectively (Hakak and Martin, 1999; Howell and Cooper, 1994). Cas phosphorylation, assayed with anti-pTyr165 antibodies, was greater in CskR222 cells than Csk+ cells, as expected (Fig. 5A, lanes 1 and 3). We generated lines of CskR222 cells expressing vector or shRNA against Cul5. When protein synthesis was inhibited with cycloheximide, Cas was rapidly degraded, dependent on Cul5 (Fig. 5A, lanes 3, 4, 7, 8). Cas half-life was ∼2–3 hours in CskR222 cells but greater than 10 hours when Cul5 was absent or Src was repressed (Fig. 5B; supplementary material Fig. S4A). Cas turnover was inhibited by epoxomycin, a proteasome inhibitor (Fig. 5A, lanes 5, 6). Cas was stable in Csk+ cells (Fig. 5A; supplementary material Fig. S4A). Although it was not technically feasible to show Cul5-dependent Cas ubiquitylation, the results are consistent with the hypothesis that Src promotes Cul5- and proteasome-dependent degradation of Cas. Physical interaction between Cas and Cul5 was assayed by co-expressing HA-tagged Cas with T7-tagged, inactivated Cul5 mutant (Cul5 K799R) in Cul5-deficient CskR222 cells. HA-tagged, pY-Cas co-precipitated with T7-tagged Cul5 (Fig. 5C).
Fig. 5.
Cul5 regulates Cas protein stability and binds Cas through SOCS6. (A) Cul5-dependent Cas turnover requires the proteasome. Western blots of lysates of control (vector) and Cul5-deficient (shCul5) CskR222 (active Src) and Csk+ (repressed Src) MEFs, incubated in the presence or absence of cycloheximide and epoxomycin for 10 hours. (B) Cas degradation. Western blots of Cas in lysates of vector and Cul5-deficient CskR222 MEFs, treated for various times with cycloheximide to inhibit new protein synthesis. GAPDH was used as a control. Half-life quantification is shown in bottom panel. (C) Cas associates with Cul5. HA-Cas and neddylation mutant T7-Cul5K799R were cotransfected into Cul5-deficient, CskR222 MEFs, in which endogenous Src is active. Cells were treated with pervanadate to stimulate tyrosine phosphorylation, lysates were prepared, immunoprecipitated with antibodies against T7 and western blots probed for HA-Cas, pY-Cas and total pY proteins. Note that many pY proteins were present in the cell lysate, but Cas was the only one immunoprecipitated with T7-Cul5K799R. (D) Cas associates with SOCS6. HeLa cells were transfected with control vector, T7-tagged SOCS1–SOCS7 or T7-tagged CisH, treated with pervanadate and lysed. Cell lysates were immunoprecipitated with anti-T7 antibody and immunoblotted with anti-Cas. (E) Scheme of Cas WT, 15F and FF mutants. (F) Cas tyrosine phosphorylation sites are required for SOCS6 binding. Wild-type or mutant HA-tagged Cas and T7-tagged SOCS6 were co-expressed in Cul5-deficient CskR222 MEFs. Cells were treated with pervanadate, lysed and immunoprecipitated with T7 antibodies. Quantification of co-immunoprecipitation is shown on right; mean and s.e.; n = 4. (G) CasFF was expressed at significantly higher level than the wild type or Cas15F. HA-tagged Cas WT, FF or 15F were transiently transfected into HeLa cells with active Src and GFP as a control. Quantification of western blots is shown on right; mean and s.e.; n = 3. *P<0.05, **P<0.01 and ***P<0.001 by Student’s t-test.
We tested which SH2-SOCS proteins could bind to pY-Cas. T7-tagged SH2-SOCS proteins were transiently overexpressed and, after brief treatment with pervanadate to inhibit pY phosphatases and increase pY-Cas, SOCS proteins were immunoprecipitated and western blotted for endogenous Cas. SOCS6 was the only SOCS protein that co-immunoprecipitated with Cas (Fig. 5D). Similarly, co-transfection of T7-SOCS proteins with HA-Cas into Cul5-deficient, CskR222 MEFs revealed that Cas binds to SOCS6 but not other SOCS proteins (supplementary material Fig. S4B). More Cas–SOC6 binding was detected when Src was activated or phosphorylation was increased with pervanadate, whereas binding decreased when Src was inhibited with PP2 (supplementary material Fig. S4C). These results are consistent with Src stimulating the binding of Cas to SOCS6. Cas is a scaffold protein with an N-terminal SH3 domain, central region and C-terminal focal adhesion targeting (FAT) domain (Fig. 5E) (Matsui et al., 2012). The central region contains 15 repeated tyrosine sequences YxVP, followed by a PxxP binding site for the Src SH3 domain and the sequence YDYV, which when phosphorylated, binds the Src SH2 domain. After Src binding, increased phosphorylation of the pYxVP repeats induces binding to a variety of signaling proteins, including Crk and Nck, which link to multiple downstream pathways. We tested whether the 15 YxVP tyrosines or the YDYV tyrosines were required for binding to SOCS6. HA-tagged Cas wild type, 15F (all YxVP sites mutated to FxVP) and FF (YDYV mutated to FDFV) mutants were co-transfected with T7-SOCS6 into CskR222, Cul5-deficient MEFs and binding was measured by co-immunoprecipitation. The FF mutation completely inhibited and the 15F mutation partly inhibited Cas binding to SOCS6 (Fig. 5F). CasFF was also expressed at a significantly higher level than the wild type or Cas15F when transiently transfected into cells with Src, consistent with reduced degradation (Fig. 5G).
Taken together, the results suggest that Src-catalyzed phosphorylation of Cas at the YDYV sequence stimulates binding to SOCS6 and Cul5-dependent Cas degradation.
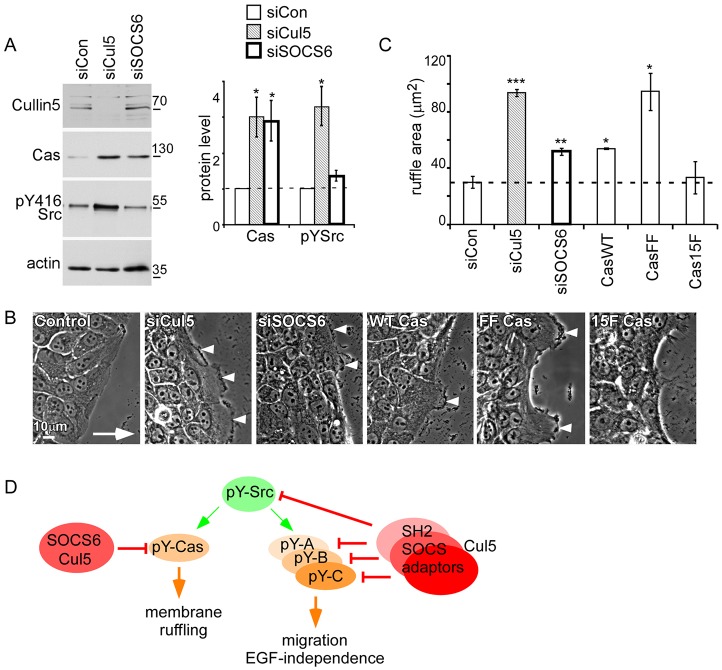
SOCS6 regulates membrane ruffling by targeting Cas for degradation
Our results so far suggested that Cas is necessary for the increased migration of Cul5-deficient cells. To test whether Cas is sufficient for the increase in migration, we stabilized Cas and other SOCS6–Cul5 CRL substrates by knocking down SOCS6. We first confirmed that SOCS6 siRNA increased the steady state level of Cas but not Src, suggesting that Src is a target for other SOCS–Cul5 CRLs (Fig. 6A). The increase in Cas is unlikely to be due to off-target effects because each of five individual SOCS6 siRNAs increased Cas and inhibited expression of SOCS6 mRNA but not other SOCS mRNAs (supplementary material Fig. S4D and data not shown). We then tested whether the stabilization of Cas by knockdown of SOCS6 was sufficient to stimulate migration. Although SOCS6 knockdown did not stimulate migration (supplementary material Fig. S4E), membrane ruffling was significantly increased, suggesting that stabilization of Cas or another SOCS6 substrate is sufficient for ruffling (Fig. 6B,C). To test whether stabilization of Cas was sufficient to stimulate ruffling, we established Cas-deficient MCF10A cell lines re-expressing CasWT, FF or 15F, fused to YFP. CasWT was expressed at similar level to endogenous Cas in Cul5-deficient MCF10A cells (supplementary material Fig. S4F). Degradation-resistant CasFF was expressed at a higher level, consistent with increased stability. Cas (re)-expression did not stimulate migration (supplementary material Fig. S4E) but did stimulate ruffling (Fig. 6B,C). Ruffling was stimulated most by degradation-resistant CasFF, less by CasWT, and least by signaling-defective Cas15F. Whether CasFF stimulates ruffling more strongly than CasWT because its expression is increased or its turnover is decreased is unclear.
Fig. 6.
SOCS6 targets active Cas for degradation and regulates membrane ruffling. (A) Transient knockdown of SOCS6 increases the level of Cas protein but not Src. Actin protein levels remained constant. Quantification of western blots is shown on right; mean and s.e.; n = 3. (B,C) Transient knockdown of SOCS6 or overexpression of Cas stimulates membrane ruffling at the leading edge. Confluent monolayers were transferred to EGF-deficient medium and then scratched. (B) Phase-contrast images of cells migrating into the wounded area show membrane ruffling in cells lacking Cul5, SOCS6 or overexpressing Cas (arrowheads). The arrow indicates direction of migration. (C) The area of phase-dark membrane ruffles was measured 6 hours after scratching. Mean and s.e. for ∼30 cells/condition from n = 3 experiments. (D) Model for the roles of Cul5 and Src in epithelial cells. Cul5 represses Src-dependent transformed phenotypes, including membrane ruffling, increased migration and EGF-independence. We propose that Cul5 targets a subset of Src substrates (pY-A, pY-B, pY-C) for downregulation by the proteasome. Various SOCS–Cul5 CRL complexes suppress different molecular pathways contributing to the transformed phenotype. One substrate for the SOCS6–Cul5 CRL is pY-Cas, whose upregulation when SOCS6 or Cul5 is absent is sufficient to stimulate membrane ruffling. Cas and Src are also required for other transformed phenotypes of Cul5-deficient cells. *P<0.05, **P<0.01 and ***P<0.001 by Student’s t-test.
The results suggest that SOCS6–Cul5 inhibits membrane ruffling at the leading edge of migrating cells by stimulating the downregulation of pY-Cas, and that increased migration and other phenotypes of Cul5-deficient cells involve other SOCS proteins and other substrates (Fig. 6D).
DISCUSSION
The evidence suggests that SOCS–Cul5 CRLs have vital functions in normal epithelial cells by restraining tyrosine kinase signaling and inhibiting Src-dependent transformation. Our data suggest a model in which different SOCS adaptors target phosphorylated Src substrates for downregulation by the ubiquitin/proteasome system (Fig. 6D). When Cul5 expression is reduced, these phosphoproteins are degraded less rapidly. The net accumulation of each substrate is small because only the phosphorylated subpopulation is downregulated, but the increases in several pY proteins combine to cause transformation. One key Cul5 substrate is Cas, which binds to SOCS6 contingent on Src-dependent phosphorylation at a pYDYV Src-SH2 binding site involved in Cas activation. The dependence of activation and degradation on the same phosphorylation site means that Cas molecules are only degraded after signaling, and the pool of unphosphorylated Cas is unaffected until it too becomes phosphorylated. This delayed negative-feedback system may provide temporal and spatial control of Cas activity that would not be possible if Cas was inactivated solely by pY phosphatases. In the absence of SOCS6 or Cul5, increased Cas activity stimulates dynamic membrane ruffling at the leading edge of migrating cells. Other SOCS proteins and their substrates presumably contribute to other aspects of transformation. By binding to proteins that contain pY, SH2-SOCS–Cul5 CRLs are exquisitely adapted to inhibit the transforming actions of deregulated tyrosine kinases.
Suppression of Src-dependent transformation by SOCS–Cul5 CRLs
Unlike other Cullins, Cul5 is restricted to multicellular animals (Hampton et al., 1994). SOCS genes also arose in metazoans, with four ancestral SOCS genes present in the sea anenome Nematostella and four to eight members in Bilateria (Liongue et al., 2012). The tyrosine kinase and phosphatase families also burgeoned in metazoans (King et al., 2008), raising the possibility that Cul5 evolved to counteract tyrosine kinase signaling. Indeed, we found here that Cul5 suppresses epithelial cell transformation. Cul5-deficient cells formed dysmorphic acini, with decreased polarity, reduced apoptosis and increased proliferation, when cultured in semi-solid medium. Their migration and proliferation were increased in the absence of EGF, and their morphology was altered, including a greatly extended lamellipodium, highly dynamic membrane ruffling and reduced focal adhesions and stress fibers. These phenotypes can be compared with those of MCF10A cells transformed by other mechanisms. Single genetic manipulations regulate proliferation, invasion, apoptosis and cell polarity to different degrees. For example, overexpression of cyclin D1 induces excess cell proliferation but not protection from apoptosis, whereas Bcl2 protects from apoptosis without increasing proliferation. Combined expression of cyclin D1 and Bcl2 generates filled multi-acinar structures similar to those created by activated mutant HER2, autocrine-activated CSF1R (Debnath and Brugge, 2005) or, as we show here, knockdown of Cul5. Like transformation by HER2 or CSF1R, transformation of Cul5-deficient cells requires endogenous Src. This suggests that Src activation could be sufficient for transformation. Indeed, highly active mutant Src causes similar changes in acinar morphogenesis (Reginato et al., 2005) and Src is a direct substrate for PTPN23, a pY phosphatase that inhibits Src and inhibits invasion by MCF10A cells (Lin et al., 2011). However, the increased migration and dysmorphic colonies of Cul5-deficient cells cannot be explained simply by the modest increase in Src activity. Moreover, highly activated mutant Src causes cell rounding, whereas Cul5-deficient cells remain spread. Therefore, some but not all of the abnormal phenotypes of Cul5-deficient cells might be attributed to increased Src activity.
Suppression of transformation by Cul5 in vitro is consistent with previous suggestions that SOCS–Cul5 CRLs suppress cancer in humans. Expression of CUL5 and SOCS genes is frequently decreased in poor-prognosis breast and gastric cancer (Elliott et al., 2008; Fay et al., 2003; Lai et al., 2010; Sasi et al., 2010). Ectopic expression of Cul5 inhibits proliferation of gastric and mammary cancer cells in culture (Johnson et al., 2007; Lai et al., 2010). The CUL5 gene maps to a region of frequent loss of heterozygosity in breast cancer and neuroblastoma (Hampton et al., 1994; Wang et al., 2006). In addition, an allele of SOCS6 is frequently lost and the gene is underexpressed in a large percentage of carcinomas of lung, colorectum and stomach (Lai et al., 2010; Sriram et al., 2012) (http://www.oncomine.org, accessed October, 2012). Other SOCS genes are also underexpressed in specific cancer types: expression of SOCS2 decreases with worsening grade of breast cancer and is hypermethylated in ovarian cancers (Farabegoli et al., 2005; Sutherland et al., 2004). Furthermore, ectopic SOCS2 and SOCS5 suppress transformation in vitro (Herranz et al., 2012; Sutherland et al., 2004).
The importance of Cas for transformation of Cul5-deficient cells is also consistent with data from human tumor cells. Cas phosphorylation by Src stimulates cancer cell invasion and also stimulates Akt, the anti-apoptotic pathway, growth factor-independence and, in breast cancer, induces resistance to anti-estrogens (Bouton et al., 2001; Cabodi et al., 2006; Schrecengost et al., 2007; Stupack et al., 2000; Tornillo et al., 2011; van der Flier et al., 2000). Because Src is highly active in many human tumors but is only weakly oncogenic in mice (Kline et al., 2008; Summy and Gallick, 2003), decreased expression of CUL5 and SOCS genes might contribute to some aspects of cancer progression by activating tyrosine phosphorylation of proteins such as Cas.
Cytoskeletal regulation by SOCS6–Cul5 and Cas
Cul5 profoundly affects cell motility. Knockdown of Cul5 stimulated migration, creating an oversized, dynamic leading lamellipodium, tiny focal adhesions and decreased stress fibers. Cell–cell contacts were maintained and the cells migrated as a sheet. Combined knockdown of SOCS2, SOCS4, SOCS5 and SOCS6 had the same effect. Because these SOCS proteins all contain SH2 domains, and because the increased migration of Cul5-deficient cells requires Src, we infer that many or all of the Cul5 substrates that limit migration in normal cells could be pY proteins. When Cul5 is absent, increased levels or decreased turnover of these substrates increases the speed of migration. However, no single SOCS protein had a significant effect on migration speed. This suggests that SOCS2, SOCS4, SOCS5 and SOCS6 are non-redundant, and target different pY proteins that collectively regulate cytoskeletal, membrane and adhesion dynamics and ensure normal migratory behavior.
Although no one SOCS protein is limiting for normal migration, SOCS6 has a special role in leading-edge ruffling. We observed that either inhibiting SOCS6 or increasing the expression or stability of Cas induced leading-edge ruffling, consistent with the hypothesis that the SOCS6–Cul5 CRL stimulates the turnover of Cas and possibly other phosphoproteins involved in actin dynamics at the leading edge. The importance of Src and Cas for leading-edge extension and ruffling is well established and might occur by the following mechanism (Cheresh et al., 1999; Klemke et al., 1998; Sawada et al., 2006; Schrecengost et al., 2007; Sharma and Mayer, 2008; Stupack et al., 2000). Cas could be stretched by tension in the cytoskeleton, revealing sites for Src-family-kinase-dependent phosphorylation. Src-dependent phosphorylation recruits a Crk–DOCK180–ELMO complex that activates Rac. Rac then stimulates dendritic actin networks that drive membrane ruffling and forward movement. Although the Src–Cas–Crk–Rac pathway is specifically associated with ruffling, it might also be involved, although not sufficient, to regulate the tiny focal adhesions and lack of stress fibers at the front of Cul5-deficient cells. One possible mechanism is that active Rac inhibits Rho-dependent tension in the actin cytoskeleton, with predictable defects in focal adhesion growth and formation of stress fibers. However, other Src and Cul5 targets are likely to be involved.
Inactivation of Cas by phosphorylation-dependent ubiquitylation and proteolysis provides a radically different alternative to inactivation by dephosphorylation. Cas turnover requires multiple steps, presumably including pY-Cas association with SOCS6 (and possibly EloB/C), interaction with Cul5–Rbx2, assembly of polyubiquitin chains and association with the proteasome. Presumably Cas loses activity at some stage of this process prior to complete degradation. If we allow for an estimated ∼10% phosphorylation stoichiometry and overall Cas half life of 2–3 hours in fibroblasts with active Src, individual phospho-Cas molecules might be active for ∼12–18 minutes before degradation. As pY-Cas leaves focal adhesions, new Cas molecules might enter, become phosphorylated, signal downstream and associate with SOCS6. Globally, at steady state, the rate of Cas turnover by pY-dependent and -independent mechanisms must match the rate of synthesis. Locally, increased Cas phosphorylation in focal adhesions will stimulate turnover, new Cas will be incorporated more rapidly, and the local concentration of Cas available for new assembly will decrease. Thus inactivation by ubiquitylation and proteolysis allows for negative feedback with a time delay. In the absence of Cul5 or SOCS6, there is more active Cas and it signals for a longer time. By contrast, dephosphorylation is typically rapid and reversible, and could occur while Cas is still in focal adhesions. Dephosphorylation does not have an inherent time delay and does not alter the local pool of free Cas molecules. For these reasons, the SOCS6–Cul5 CRL potentially plays a unique role in cytoskeletal regulation. An important task for the future will be identifying other SOCS adaptors and their substrates that regulate cell movements, and how the localization or activity of individual SOCS–Cul5 CRLs are regulated as cells move.
MATERIALS AND METHODS
Antibodies, DNA constructs and chemicals
Antibodies were obtained from the following sources: goat anti-actin, rabbit anti-Cul5, rabbit anti-Cas, rabbit anti-GAPDH and rabbit anti-FAK (Santa Cruz Biotechnology, Inc., Santa Cruz, CA); rabbit anti-Ki67 (Leica Biosystems, Buffalo Grove, IL); mouse anti-GM130 and mouse anti-extracellular signal-regulated kinase-2 (ERK2, BD Transduction, San Jose, CA); mouse anti-vinculin and rat anti-E-cadherin (Sigma-Aldrich, St Louis, MO); rabbit anti-phospho-p130 Cas (pTyr165); rabbit anti-phospho-Src (pTyr416) and rabbit anti-cleaved Caspase-3 (Asp175) (Cell Signaling Technology, Beverly, MA); mouse anti-RhoGAP (Upstate Biotechnology, Inc., Lake Placid, NY); mouse anti-pTyr (4G10) and rabbit anti-GFP (Life Technologies, Carlsbad, CA); mouse anti-GFP (Roche, Indianapolis, IN); rabbit anti-HA (Bethyl Laboratories, Inc., Montgomery, TX); mouse anti-T7 (EMD Millipore, Billerica, MA); 327 anti-Src mouse monoclonal antibody (Joan Brugge, Harvard Medical School, Boston, MA); and P5G10, mouse anti-integrin α6 IgG1 (Elizabeth Wayner, Fred Hutchinson Cancer Research Center).
pMXpuroII containing the ubiquitin promoter and shRNAs against the Cul5 target sequence GCTGCAGACTGAATTAGTAG (shCul5), the Cul2 target sequence GAGCTAGCATTGGATATGTGG (shCul2), the Rbx2 target sequence GGGTCCAGGTGATGGATGCCT (shRbx2-1) and the empty vector control plasmid pMXpuroII, were previously described (Kamura et al., 2004). The pMXpuroII-shSrc and pMXpuroII-shCas shRNA plasmids were made by PCR with pMXpuroII-shCul5 plasmid as template, a common primer 5′-GTCGACCACTGTGCTGGC-3′ and a unique primer containing sequence homologous to the U6 promoter in pMXpuroII followed by the sense target sequence, a loop sequence, the antisense target sequence, an XhoI site, and six bases of random sequence. The target sequences used were: 5′-ACATGAGCAAGGGGAGTT-3′ for Src and 5′-GGTCGACAGTGGTGTGTA-3′ for Cas. The PCR products were cut with NotI-XhoI and cloned into NotI-XhoI-cut pMXpuroII-shCul5. Clones were sequenced. pLXSH-shCul5, pLXSH-shSrc and pLXSH-shCas shRNA plasmids were made by ligation of NotI-XhoI fragments from corresponding pMXpuroII plasmids into NotI-XhoI-cut pLXSH shCul5′ as previously described (Laszlo and Cooper, 2009).
Mouse SOCS and Cul5 gene cDNA clones were obtained by RT-PCR from mouse brain mRNA and sequenced. They were inserted into pCAG-GFP, which had been modified by inserting a T7 epitope tag at the N-terminus and deleting GFP. Residue Lys799 was mutated to Arg to inhibit neddylation using the oligonucleotide ATCATACAAATAATGCGAATGAGAAAGAAAATT.
Mouse wild-type and 15F mutant Cas cDNA clones (Shin et al., 2004) were obtained from Steve Hanks (Vanderbilt University, Nashville, TN) and the FF mutant (double mutation of Y667 and Y669 in the sequence YDYV, corresponding to Y762 and Y764 in rat Cas) was made by Susumu Antoku. BamHI-NotI fragments were cloned into the pCAG2-nHA vector for transient expression with an N-terminal HA tag. The same fragments were cloned into pMSCV_puro_EYFPC1 (pMSCV with an N-terminal EYFP tag and modified polylinker; S. Antoku, unpublished) for retrovirus production. Clones were confirmed by sequencing. MG132, epoxomycin and mitomycin C were from Sigma-Aldrich (St Louis, MO), SU6656 from Sugen (San Francisco, CA) and 4-hydroxy-tamoxifen from Sigma-Aldrich.
Cell culture and retroviral infection
MCF10A cells were maintained in DMEM/F12 (Life Technologies) with 5% horse serum (Life Technologies), 20 ng/ml EGF (Life Technologies), 0.5 µg/ml hydrocortisone (Sigma-Aldrich), 0.1 µg/ml cholera toxin (EMD Millipore), 10 µg/ml insulin, and penicillin-streptomycin, both at 100 U/ml (Life Technologies). Recombinant retroviruses containing pLXSH empty vector, pMXpuroII empty vector, shCul5, shSrc or shCas were packaged using HEK 293T cells and infection of MCF10A cells was carried out by standard protocols (Miller and Rosman, 1989). Following initial selection of stable cell lines in 0.25–0.5 µg/ml puromycin or 50–100 µg/ml hygromycin for 10–15 days, lines (selected as pools, not clones) were maintained in the absence of selective agent. Cas-deficient MCF10A cells stably expressing EYFP-Cas from the MSCV promoter were prepared by retrovirus infection with pMSCV_puro_EYFPCas and selection with puromycin. MCF10A cells expressing vSrc-ER were a kind gift from Senthil Muthuswamy (University of Toronto, Toronto, Canada) (Reginato et al., 2005). 4-hydroxy-tamoxifen (0.25 µM, Sigma-Aldrich) was added 24 hours before scratch-wound assays to activate the vSrc-ER fusion protein.
SV40 large-T antigen immortalized Csk−/−, Csk and Csk R222 cell lines, infected with recombinant retroviruses containing pLXSH empty vector/pMXpuroII empty vector or shCul5, were described before (Howell and Cooper, 1994; Laszlo and Cooper, 2009). HeLa cells were cultured in DME supplemented with 10% FBS and penicillin/streptomycin both at 100 U/ml.
siRNA transfection
2×104 MCF10A cells were resuspended in 800 µl of growth medium (described above) and added directly to wells of a 12-well plate containing 50 pmol of a pool of four siRNA oligonucleotides specific for Cul5 (SMART pool siGENOME, Thermo Fisher Scientific, Waltham, MA), SOCS1–SOCS7, CisH, Src or Cas (Qiagen, Germantown, MD), and 1.25 µl Lipofectamine2000 (Life Technologies). Control siRNA (Qiagen) targeted the sequence AATTCTCCGAACGTGTCACGT, which lacks homology to any known mammalian gene. Assays were performed 96 hours after transfection.
DNA transfection
HeLa cells were grown in six-well plates to near confluence and transfected with HA-tagged Cas (WT, FF or 15F), GFP and active Src using Lipofectamine2000 (Life Technologies). Cells were lysed 24 hours later and analyzed by western blotting.
Proliferation
Cells were plated in growth medium containing 2% horse serum and 0 or 20 ng/ml EGF. For Fig. 1B,C; supplementary material Fig. S1B, Fig. S3D and Fig. S4A, triplicate wells were stained with Crystal Violet the next day (day 0) and on day 3 and the fold increase in cell number was measured. For supplementary material Fig. S1A, Fig. S3B and Fig. S4D, cell number was counted on day 8.
Matrigel 3D growth and imaging
Three-dimensional culture of MCF10A cells on reconstituted basement membrane (growth-factor-depleted Matrigel) using the overlay method, with EGF at 5 ng/ml (Debnath et al., 2003). Immunofluorescence staining of acini has been described (Debnath et al., 2003). Sequential optical sections were taken using a Zeiss LSM510 laser-scanning confocal microscope fitted with a Zeiss Plan Apochromat 20× /0.75 objective.
Scratch wound
Scratch-wound migration assays were performed as previously described (Dow et al., 2007). Briefly, confluent MCF10A monolayers were EGF-starved overnight, wounded by scratching the surface with a P200 micropipette tip, and the medium replaced with fresh medium lacking EGF and containing 1 ng/ml Mitomycin C. Phase-contrast micrographs of triplicate wounds were taken shortly after wounding and at intervals thereafter. The difference in distance between wound edges at the start and end of the experiment was converted to microns.
Boyden chamber assay
Migration assays were performed in a 48-well micro chemotaxis chamber (Neuroprobe, Gaithersburg, MD). The lower wells of the chamber were loaded with DMEM/F12 supplemented with 5% horse serum and 20 ng/ml EGF. An 8 µm pore size polycarbonate filter (VWR, West Chester, PA) coated with 2 µg/ml collagen IV separated the upper and lower wells. MCF10A cells were detached using a solution of 0.25% trypsin, 2 mM EDTA and PBS for 20 minutes at 37°C. After inactivating trypsin with 10% FBS, cells were resuspended in DMEM/F12 at 250,000 cells/ml and added to the top wells. The chamber was incubated in a humidified atmosphere of 5% CO2 at 37°C for 12 hours. The cells on the top side of the membrane were removed, and the migrated cells on the bottom side were stained with 0.1% Crystal Violet in 20% ethanol. Each condition was analyzed in triplicate.
Immunofluorescence of migrating cells
Cells were grown on coverslips. After scratch wounding and migration for 6 hours, cells were fixed and permeabilized with 0.1% Triton X-100 in PBS for 5 minutes at 25°C. Cells were blocked in a solution of 2% BSA, 5% normal goat serum and PBS for 30 minutes at 25°C. Primary antibodies were diluted in blocking solution and added either for 3–4 hours at 25°C. Coverslips were rinsed three times in PBS before the addition of Alexa-Fluor-labeled secondary antibodies (Life Technologies), diluted 1∶1000, for 1 hour at 25°C. Alexa-Fluor-tagged phalloidin (Life Technologies) was used to visualize actin. After several PBS rinses, coverslips were mounted in ProLong Gold solution (Life Technologies).
Imaging
Cells were visualized using 40×, 1.3 NA or 60×, 1.42 NA oil objectives on a DeltaVision microscope (IX71, Olympus). Images were recorded using fixed camera settings (Photometrics HQ2 CCD camera; Olympus). Images were acquired and deconvolved using SoftWorx (Applied Precision), and all exposure times and image scaling were equal within an experiment.
Movies of membrane ruffles were captured on a Nikon LiveScan (Prairie Technologies Swept Field) system, equipped with a high-resolution Photometrics Coolsnap HQ2 scientific CCD camera. A 40× phase objective was used. Images were recorded every minute for 60 minutes. The movies were used to confirm that phase dense areas were dynamically ruffling, then one frame of the movie was analyzed using ImageJ (National Institutes of Health) to measure ruffle area for each cell along the wound edge. Average ruffle area per cell was calculated for ∼50 cells along three wounds. The entire experiment was repeated to obtain biological replicates. Figures were assembled using Photoshop (Adobe) and Canvas (Deneba) software.
Protein analysis
Cells were washed twice in phosphate-buffered saline (PBS) and lysed in RIPA buffer containing 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate, 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1 mM phenylmethylsulfonyl fluoride, 20 µg/ml leupeptin, 20 µg/ml aprotinin, and 1 mM sodium orthovanadate. Protein concentration was assessed using Bradford assay. Samples were adjusted to 1× SDS sample buffer, boiled and resolved by 10% SDS-PAGE using 30∶1 or 75∶1 acrylamide:bisacrylamide ratio. Proteins were transferred onto nitrocellulose (Osmonics, Minnetonka, MN), blocked in 5% non-fat dry milk (or BSA for pY) in Tris-buffered saline with 0.1% Tween, probed first with the indicated primary antibody, followed by either goat anti-mouse- or goat anti-rabbit-horseradish peroxidase-conjugated secondary antibody and visualized using Amersham ECL Plus Western Blotting Detection System (GE Healthcare Biosciences, Piscataway, NJ).
Immunoprecipitation
For immunoprecipitation from HeLa cells, T7-tagged SOCS proteins were transfected into HeLa cells using Lipofectamine2000 according to the manufacturer's protocols. After 48 hours, cells were incubated with 1 mM sodium pervanadate for 30 minutes then lysed at 4°C in buffer containing 150 mM NaCl, 10 mM HEPES, pH 7.4, 2 mM EDTA, 50 mM NaF, 1% Triton-X-100 and protease inhibitors followed by centrifugation for 15 minutes at 14,000 g. Lysate was rotated with mouse anti-T7 antibody for 3 hours at 4°C. Protein A+G beads were added to the lysate and antibody mix for 1 hour at 4°C. After centrifugation, the protein–antibody–bead complex was washed three times in lysis buffer, resuspended in Laemmli buffer, then resolved by SDS-PAGE. Immunoprecipitation from Cul5-deficient CskR222 or Csk+ MEFs used a similar protocol, except that 10 µM Src inhibitor PP2 or 1 mM sodium pervanadate were added before lysis and lysates were prepared using RIPA buffer (Howell and Cooper, 1994).
Cycloheximide chase
Cells were grown to near confluence, then treated with 25 µg/ml cycloheximide or vehicle (1% ethanol final concentration) for various times. Control untreated cells received vehicle for 10 hours. In some cases, cells were pre-treated for 1 hour with 10 µM epoxomycin or vehicle (0.1% DMSO final concentration) followed with 25 µg/ml cycloheximide for various times.
RT-PCR
RNA was extracted and cDNA synthesized as described previously (Laszlo and Cooper, 2009). The abundance of mRNA encoding Cas, Src and β-actin was measured by RT-PCR as previously described (Laszlo and Cooper, 2009). Samples were removed after 21 and 24 PCR cycles and analyzed by 2% agarose gel electrophoresis and ethidium bromide stain. Amplification was in the linear range. For SOCS genes, qPCR was performed using QuantiTect SYBR green PCR kit (Qiagen), the 7900HT Real Time PCR System and SDS software (Applied Biosystems). Primers are listed in supplementary material Table S1.
Phosphoproteomics
MCF10A vector control and shCul5 cells were grown to 90% confluency in normal growth medium on three 15 cm plates. Cell lysis and protein digestion was performed as previously described (Zhang et al., 2005), except that a 20∶1 substrate∶trypsin ratio was used for digestion. Lyophilized peptides were resuspended in 0.5 ml of 100 mM Tris-HCl, pH 7.4, with 100 mM NaCl, 0.1% NP-40 and immunoprecipitated overnight at 4°C with 2.5 µg of anti-pY antibody-conjugated beads (PY-99, Santa Cruz). Beads were then rinsed three times with 0.5 ml of 100 mM Tris-HCl, pH 7.4, 100 mM NaCl. Phosphotyrosine-containing peptides were eluted using 50 µl of 100 mM glycine pH 2.5, and samples were desalted using microC18 Ziptips (Millipore, Billerca, CA) following the manufacturer's instructions. Dried samples were resuspended in 5 µl of 0.1% formic acid and analyzed with an LTQ-FT mass spectrometer in a configuration as described (Yi et al., 2003), collecting one MS scan followed by five MS/MS scans of the five most abundant ions in the MS scan. Peptide identifications were performed with X!Tandem using a variable modification of 79.9663 Da on tyrosine to search the IPI mouse (v3.44) protein database.
Supplementary Material
Acknowledgments
We are very grateful to Denis Smirnov, Natalie Toida, Moham Ansari and Melanie Rivera for technical assistance during the course of the research. The work would not have been possible without valuable reagents and suggestions from Susumu Antoku, Steve Hanks, Takumi Kamura, Bruce Clurman, Markus Welcker, Roland Walter, Elizabeth Wayner, Bill Carter and Senthil Muthuswamy.
Footnotes
Competing interests
The authors declare no competing financial interests.
Author contributions
All authors designed, performed and interpreted experiments. A.T., G.S.L. and J.A.C. wrote the paper.
Funding
This work was supported the National Institutes of Health [grant numbers R01-CA41072, R01-NS080194]; a pilot grant from the Hutchinson Center Synergy Fund; the Breast Cancer Research Program Pilot Project Fund at FHCRC; and the Safeway Foundation, postdoctoral fellowship from the American Cancer Society [grant number PF-07-287-01-MGO to G.L.]; and a National Science Foundation Graduate Research Fellowship to C.P. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.127829/-/DC1
References
- Bouton A. H., Riggins R. B., Bruce-Staskal P. J. (2001). Functions of the adapter protein Cas: signal convergence and the determination of cellular responses. Oncogene 20, 6448–6458 10.1038/sj.onc.1204785 [DOI] [PubMed] [Google Scholar]
- Cabodi S., Tinnirello A., Di Stefano P., Bisarò B., Ambrosino E., Castellano I., Sapino A., Arisio R., Cavallo F., Forni G. et al. (2006). p130Cas as a new regulator of mammary epithelial cell proliferation, survival, and HER2-neu oncogene-dependent breast tumorigenesis. Cancer Res. 66, 4672–4680 10.1158/0008-5472.CAN-05-2909 [DOI] [PubMed] [Google Scholar]
- Cheresh D. A., Leng J., Klemke R. L. (1999). Regulation of cell contraction and membrane ruffling by distinct signals in migratory cells. J. Cell Biol. 146, 1107–1116 10.1083/jcb.146.5.1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J., Brugge J. S. (2005). Modelling glandular epithelial cancers in three-dimensional cultures. Nat. Rev. Cancer 5, 675–688 10.1038/nrc1695 [DOI] [PubMed] [Google Scholar]
- Debnath J., Mills K. R., Collins N. L., Reginato M. J., Muthuswamy S. K., Brugge J. S. (2002). The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell 111, 29–40 10.1016/S0092-8674(02)01001-2 [DOI] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S. K., Brugge J. S. (2003). Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 10.1016/S1046-2023(03)00032-X [DOI] [PubMed] [Google Scholar]
- Deshaies R. J., Joazeiro C. A. (2009). RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- Dow L. E., Kauffman J. S., Caddy J., Zarbalis K., Peterson A. S., Jane S. M., Russell S. M., Humbert P. O. (2007). The tumour-suppressor Scribble dictates cell polarity during directed epithelial migration: regulation of Rho GTPase recruitment to the leading edge. Oncogene 26, 2272–2282 10.1038/sj.onc.1210016 [DOI] [PubMed] [Google Scholar]
- Elliott J., Hookham M. B., Johnston J. A. (2008). The suppressors of cytokine signalling E3 ligases behave as tumour suppressors. Biochem. Soc. Trans. 36, 464–468 10.1042/BST0360464 [DOI] [PubMed] [Google Scholar]
- Farabegoli F., Ceccarelli C., Santini D., Taffurelli M. (2005). Suppressor of cytokine signalling 2 (SOCS-2) expression in breast carcinoma. J. Clin. Pathol. 58, 1046–1050 10.1136/jcp.2004.024919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay M. J., Longo K. A., Karathanasis G. A., Shope D. M., Mandernach C. J., Leong J. R., Hicks A., Pherson K., Husain A. (2003). Analysis of CUL-5 expression in breast epithelial cells, breast cancer cell lines, normal tissues and tumor tissues. Mol. Cancer 2, 40 10.1186/1476-4598-2-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakak Y., Martin G. S. (1999). Ubiquitin-dependent degradation of active Src. Curr. Biol. 9, 1039 10.1016/S0960-9822(99)80453-9 [DOI] [PubMed] [Google Scholar]
- Hampton G. M., Mannermaa A., Winqvist R., Alavaikko M., Blanco G., Taskinen P. J., Kiviniemi H., Newsham I., Cavenee W. K., Evans G. A. (1994). Loss of heterozygosity in sporadic human breast carcinoma: a common region between 11q22 and 11q23.3. Cancer Res. 54, 4586–4589 [PubMed] [Google Scholar]
- Harris K. F., Shoji I., Cooper E. M., Kumar S., Oda H., Howley P. M. (1999). Ubiquitin-mediated degradation of active Src tyrosine kinase. Proc. Natl. Acad. Sci. USA 96, 13738–13743 10.1073/pnas.96.24.13738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz H., Hong X., Hung N. T., Voorhoeve P. M., Cohen S. M. (2012). Oncogenic cooperation between SOCS family proteins and EGFR identified using a Drosophila epithelial transformation model. Genes Dev. 26, 1602–1611 10.1101/gad.192021.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. W., Cooper J. A. (1994). Csk suppression of Src involves movement of Csk to sites of Src activity. Mol. Cell. Biol. 14, 5402–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. T., Ayrault O., Hunt H. W., Taherbhoy A. M., Duda D. M., Scott D. C., Borg L. A., Neale G., Murray P. J., Roussel M. F. et al. (2009). E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell 33, 483–495 10.1016/j.molcel.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. (2009). Tyrosine phosphorylation: thirty years and counting. Curr. Opin. Cell Biol. 21, 140–146 10.1016/j.ceb.2009.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto A., Soriano P. (1993). Disruption of the csk gene, encoding a negative regulator of Src family tyrosine kinases, leads to neural tube defects and embryonic lethality in mice. Cell 73, 1117–1124 10.1016/0092-8674(93)90641-3 [DOI] [PubMed] [Google Scholar]
- Ishizawar R., Parsons S. J. (2004). c-Src and cooperating partners in human cancer. Cancer Cell 6, 209–214 10.1016/j.ccr.2004.09.001 [DOI] [PubMed] [Google Scholar]
- Johnson A. E., Le I. P., Buchwalter A., Burnatowska-Hledin M. A. (2007). Estrogen-dependent growth and estrogen receptor (ER)-alpha concentration in T47D breast cancer cells are inhibited by VACM-1, a cul 5 gene. Mol. Cell. Biochem. 301, 13–20 10.1007/s11010-006-9392-3 [DOI] [PubMed] [Google Scholar]
- Kamura T., Maenaka K., Kotoshiba S., Matsumoto M., Kohda D., Conaway R. C., Conaway J. W., Nakayama K. I. (2004). VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 18, 3055–3065 10.1101/gad.1252404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile B. T., Schulman B. A., Alexander W. S., Nicola N. A., Martin H. M., Hilton D. J. (2002). The SOCS box: a tale of destruction and degradation. Trends Biochem. Sci. 27, 235–241 10.1016/S0968-0004(02)02085-6 [DOI] [PubMed] [Google Scholar]
- King N., Westbrook M. J., Young S. L., Kuo A., Abedin M., Chapman J., Fairclough S., Hellsten U., Isogai Y., Letunic I. et al. (2008). The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451, 783–788 10.1038/nature06617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemke R. L., Leng J., Molander R., Brooks P. C., Vuori K., Cheresh D. A. (1998). CAS/Crk coupling serves as a “molecular switch” for induction of cell migration. J. Cell Biol. 140, 961–972 10.1083/jcb.140.4.961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline C. L., Jackson R., Engelman R., Pledger W. J., Yeatman T. J., Irby R. B. (2008). Src kinase induces tumor formation in the c-SRC C57BL/6 mouse. Int. J. Cancer 122, 2665–2673 10.1002/ijc.23445 [DOI] [PubMed] [Google Scholar]
- Krishnan H., Miller W. T., Goldberg G. S. (2012). SRC points the way to biomarkers and chemotherapeutic targets. Genes Cancer 3, 426–435 10.1177/1947601912458583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai R. H., Hsiao Y. W., Wang M. J., Lin H. Y., Wu C. W., Chi C. W., Li A. F., Jou Y. S., Chen J. Y. (2010). SOCS6, down-regulated in gastric cancer, inhibits cell proliferation and colony formation. Cancer Lett. 288, 75–85 10.1016/j.canlet.2009.06.025 [DOI] [PubMed] [Google Scholar]
- Laszlo G. S., Cooper J. A. (2009). Restriction of Src activity by Cullin-5. Curr. Biol. 19, 157–162 10.1016/j.cub.2008.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Zhou P. (2010). Cullins and cancer. Genes Cancer 1, 690–699 10.1177/1947601910382899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G., Aranda V., Muthuswamy S. K., Tonks N. K. (2011). Identification of PTPN23 as a novel regulator of cell invasion in mammary epithelial cells from a loss-of-function screen of the ‘PTP-ome’. Genes Dev. 25, 1412–1425 10.1101/gad.2018911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liongue C., O'Sullivan L. A., Trengove M. C., Ward A. C. (2012). Evolution of JAK-STAT pathway components: mechanisms and role in immune system development. PLoS ONE 7, e32777 10.1371/journal.pone.0032777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H., Harada I., Sawada Y. (2012). Src, p130Cas, and Mechanotransduction in Cancer Cells. Genes Cancer 3, 394–401 10.1177/1947601912461443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. D., Rosman G. J. (1989). Improved retroviral vectors for gene transfer and expression. Biotechniques 7, 980–982– 984,–986– 989–990 [PMC free article] [PubMed] [Google Scholar]
- Muthuswamy S. K., Li D., Lelievre S., Bissell M. J., Brugge J. S. (2001). ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nat. Cell Biol. 3, 785–792 10.1038/ncb0901-785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nada S., Yagi T., Takeda H., Tokunaga T., Nakagawa H., Ikawa Y., Okada M., Aizawa S. (1993). Constitutive activation of Src family kinases in mouse embryos that lack Csk. Cell 73, 1125–1135 10.1016/0092-8674(93)90642-4 [DOI] [PubMed] [Google Scholar]
- Okumura F., Matsuzaki M., Nakatsukasa K., Kamura T. (2012). The Role of Elongin BC-Containing Ubiquitin Ligases. Front Oncol. 2, 10 10.3389/fonc.2012.00010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Qiao F., Gao C., Norman B., Optican L., Zelenka P. S. (2011). Cdk5 targets active Src for ubiquitin-dependent degradation by phosphorylating Src(S75). Cell. Mol. Life Sci. 68, 3425–3436 10.1007/s00018-011-0638-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T., Gish G. D., Nash P. (2001). SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 11, 504–511 10.1016/S0962-8924(01)02154-7 [DOI] [PubMed] [Google Scholar]
- Petroski M. D., Deshaies R. J. (2005). Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- Reginato M. J., Mills K. R., Becker E. B., Lynch D. K., Bonni A., Muthuswamy S. K., Brugge J. S. (2005). Bim regulation of lumen formation in cultured mammary epithelial acini is targeted by oncogenes. Mol. Cell. Biol. 25, 4591–4601 10.1128/MCB.25.11.4591-4601.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasi W., Jiang W. G., Sharma A., Mokbel K. (2010). Higher expression levels of SOCS 1,3,4,7 are associated with earlier tumour stage and better clinical outcome in human breast cancer. BMC Cancer 10, 178 10.1186/1471-2407-10-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y., Tamada M., Dubin-Thaler B. J., Cherniavskaya O., Sakai R., Tanaka S., Sheetz M. P. (2006). Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell 127, 1015–1026 10.1016/j.cell.2006.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrecengost R. S., Riggins R. B., Thomas K. S., Guerrero M. S., Bouton A. H. (2007). Breast cancer antiestrogen resistance-3 expression regulates breast cancer cell migration through promotion of p130Cas membrane localization and membrane ruffling. Cancer Res. 67, 6174–6182 10.1158/0008-5472.CAN-06-3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Mayer B. J. (2008). Phosphorylation of p130Cas initiates Rac activation and membrane ruffling. BMC Cell Biol. 9, 50 10.1186/1471-2121-9-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N. Y., Dise R. S., Schneider-Mergener J., Ritchie M. D., Kilkenny D. M., Hanks S. K. (2004). Subsets of the major tyrosine phosphorylation sites in Crk-associated substrate (CAS) are sufficient to promote cell migration. J. Biol. Chem. 279, 38331–38337 10.1074/jbc.M404675200 [DOI] [PubMed] [Google Scholar]
- Sriram K. B., Larsen J. E., Savarimuthu Francis S. M., Wright C. M., Clarke B. E., Duhig E. E., Brown K. M., Hayward N. K., Yang I. A., Bowman R. V. et al. (2012). Array-comparative genomic hybridization reveals loss of SOCS6 is associated with poor prognosis in primary lung squamous cell carcinoma. PLoS ONE 7, e30398 10.1371/journal.pone.0030398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupack D. G., Cho S. Y., Klemke R. L. (2000). Molecular signaling mechanisms of cell migration and invasion. Immunol. Res. 21, 83–88 10.1385/IR:21:2-3:83 [DOI] [PubMed] [Google Scholar]
- Summy J. M., Gallick G. E. (2003). Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev. 22, 337–358 10.1023/A:1023772912750 [DOI] [PubMed] [Google Scholar]
- Sutherland K. D., Lindeman G. J., Choong D. Y., Wittlin S., Brentzell L., Phillips W., Campbell I. G., Visvader J. E. (2004). Differential hypermethylation of SOCS genes in ovarian and breast carcinomas. Oncogene 23, 7726–7733 10.1038/sj.onc.1207787 [DOI] [PubMed] [Google Scholar]
- Tornillo G., Bisaro B., Camacho-Leal M. P., Galiè M., Provero P., Di Stefano P., Turco E., Defilippi P., Cabodi S. (2011). p130Cas promotes invasiveness of three-dimensional ErbB2-transformed mammary acinar structures by enhanced activation of mTOR/p70S6K and Rac1. Eur. J. Cell Biol. 90, 237–248 10.1016/j.ejcb.2010.09.002 [DOI] [PubMed] [Google Scholar]
- van der Flier S., Brinkman A., Look M. P., Kok E. M., Meijer-van Gelder M. E., Klijn J. G., Dorssers L. C., Foekens J. A. (2000). Bcar1/p130Cas protein and primary breast cancer: prognosis and response to tamoxifen treatment. J. Natl. Cancer Inst. 92, 120–127 10.1093/jnci/92.2.120 [DOI] [PubMed] [Google Scholar]
- Wang Q., Diskin S., Rappaport E., Attiyeh E., Mosse Y., Shue D., Seiser E., Jagannathan J., Shusterman S., Bansal M. et al. (2006). Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 66, 6050–6062 10.1158/0008-5472.CAN-05-4618 [DOI] [PubMed] [Google Scholar]
- Wrobel C. N., Debnath J., Lin E., Beausoleil S., Roussel M. F., Brugge J. S. (2004). Autocrine CSF-1R activation promotes Src-dependent disruption of mammary epithelial architecture. J. Cell Biol. 165, 263–273 10.1083/jcb.200309102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi E. C., Lee H., Aebersold R., Goodlett D. R. (2003). A microcapillary trap cartridge-microcapillary high-performance liquid chromatography electrospray ionization emitter device capable of peptide tandem mass spectrometry at the attomole level on an ion trap mass spectrometer with automated routine operation. Rapid Commun. Mass Spectrom. 17, 2093–2098 10.1002/rcm.1150 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wolf-Yadlin A., Ross P. L., Pappin D. J., Rush J., Lauffenburger D. A., White F. M. (2005). Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol. Cell. Proteomics 4, 1240–1250 10.1074/mcp.M500089-MCP200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.