Abstract
HIF prolyl 4-hydroxylases (PHD) are a family of enzymes that mediate key physiological responses to hypoxia by modulating the levels of hypoxia inducible factor 1-α (HIF1α). Certain benzimidazole-2-pyrazole carboxylates were discovered to be PHD2 inhibitors using ligand- and structure-based methods and found to be potent, orally efficacious stimulators of erythropoietin secretion in vivo.
Keywords: Prolyl 4-hydroxylase inhibitors, PHD inhibitors, hypoxia inducible factor, HIF, erythropoietin, epo, erythropoietin stimulating agents, anemia
HIF prolyl 4-hydroxylases (PHD1, PHD2, and PHD3) are a family of highly conserved, iron-containing, 2-oxoglutarate- (2OG) and dioxygen-dependent enzymes that play a crucial role in adaptation and survival during oxygen-deficient conditions.1−3 Under normoxia, the transcription factor hypoxia inducible factor 1-α (HIF1α) is constitutively expressed and rapidly degraded, with a half-life of less than 5 min.4 The degradation of HIF1α is regulated in part by PHD enzymes, which hydroxylate HIF1α proline residues 564 and 402, thereby enabling binding to the von Hippel-Lindau protein and subsequent proteasomic degradation.5 Under a sufficiently low partial pressure of oxygen, the rate of PHD-mediated hydroxylation of HIF1α is lessened such that the production of HIF1α outpaces its degradation. The accumulated HIF1α then translocates to the nucleus, leading to the formation of a HIF1α-HIF1β heterodimer and the activation of genes involved in hypoxic responses such as erythropoietin (epo) secretion and erythropoiesis, anaerobic glycolysis, and angiogenesis.6,7 Therefore, positive modulation of HIF transcriptional activity holds promise as a mode of treatment for a variety of debilitating ischemia-associated diseases, including anemia, myocardial infarction, stroke, and metabolic disorders, by conducting an orchestrated response to hypoxic challenge.8,9
Because of their key role in the regulation of HIF levels, inhibition of PHD enzymes is an attractive strategy by which to potentiate the transcriptional activity of HIF in a therapeutically relevant manner. Indeed, during the last several years, intense efforts to discover PHD inhibitors have been adumbrated primarily in the patent literature,10 and to a much lesser degree in the peer-reviewed literature,11−19 leading to four compounds entering clinical trials.20,21 In this letter, we report our efforts in this area, which led to the discovery of novel PHD inhibitors that are potent, orally active erythropoietin secretagogues.
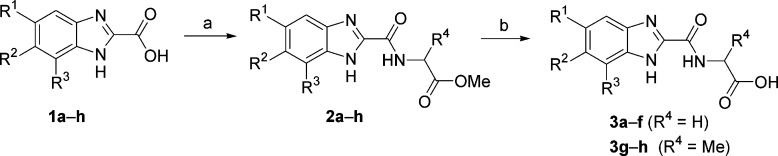
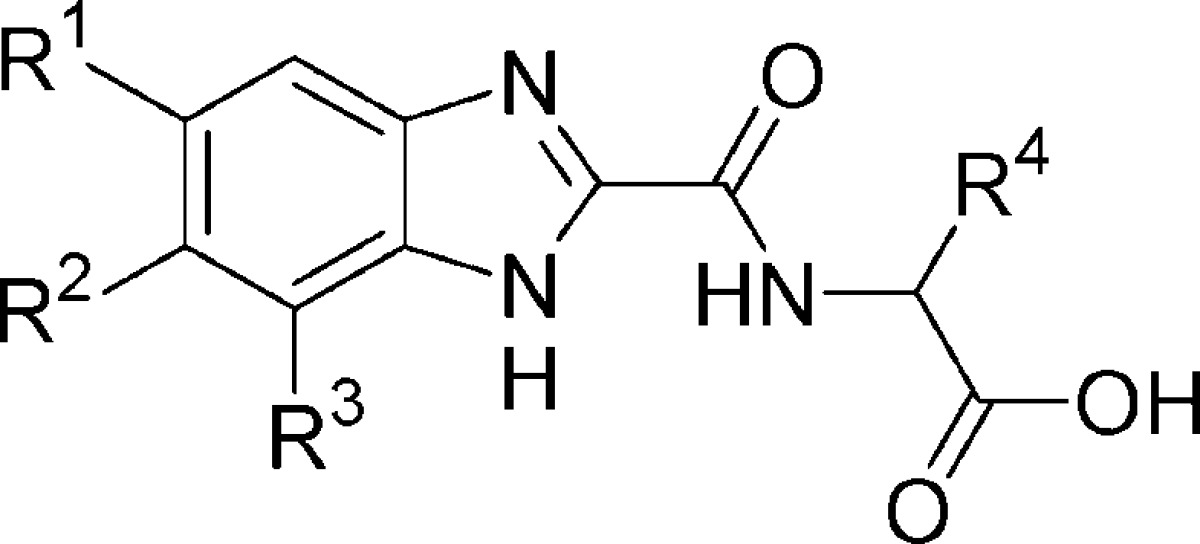
Screening of an internal compound library uncovered numerous strongly metal-ligating molecules that, upon closer investigation, appear to exert their inhibitory activity through iron sequestration and subsequent enzyme deactivation.22 As this mechanism is unlikely to translate into a viable therapeutic method, we undertook an alternative approach to lead generation. A number of benzimidazole-2-glycinamides emerged as targets for chemical synthesis and subsequent evaluation as inhibitors, using a PHD2 enzymatic assay previously reported from this laboratory.23 The synthesis of this initial set of potential inhibitors began with known or available benzimidazole-2-carboxylic acids 1a−h (Scheme 1).24 These acids were coupled to amino esters 2a−h, which were then saponified to provide the corresponding benzimidazole acids 3a−h.
Scheme 1. Synthesis of Benzimidazole Amides 3a−h.
Reagents and conditions: (a) HATU, amino acid methyl ester hydrochloride, DIEA, DMF, 23 °C, 16 h. (b) LiOH, THF/H2O, 23 °C, 2−16 h. For R1−3 groups, see Table 1.
Evaluation of compounds 3a−h showed several to possess measurable inhibitory activity in our PHD2 enzymatic assay (Table 1). Dichloride analogue 3b proved to be the most potent, with a pIC50 value of 4.9. Representative members of this set were found to have low iron affinity in solution relative to literature reference compounds, suggesting that the observed inhibitory activity does not result from metal center depletion.25 However, these particular benzimidazole inhibitors failed to significantly stimulate epo release from Hep3B cells at concentrations up to 100 μM, while efforts to optimize the biopharmaceutical properties of the benzimidazole-2-carboxamide compounds proved challenging. As part of a larger search for suitable isosteric glycine replacement groups, we prepared a number of benzimidazole-2-pyrazoles as shown in Scheme 2.26
Table 1. PHD2 Enzyme Inhibition and Epo Release in Hep3B Cells for Compounds 3a−h.
| compd | R1 | R2 | R3 | R4 | pIC50a | epo release (%)b |
|---|---|---|---|---|---|---|
| 3a | H | H | H | H | <4 | <20 |
| 3b | Cl | Cl | H | H | 4.9 | <20 |
| 3c | Br | H | H | H | 4.6 | <20 |
| 3d | F | H | H | H | <4 | <20 |
| 3e | OMe | H | H | H | 4.3 | <20 |
| 3f | CF3 | H | CF3 | H | 4.7 | <20 |
| 3g | CF3 | H | CF3 | (S)-Me | 4.3 | <20 |
| 3h | CF3 | H | CF3 | (R)-Me | 4.1 | <20 |
Negative logarithm of the concentration required to achieve 50% enzyme inhibition; the value is ±0.3 log units.
Percent increase in epo release from Hep3B cells at 100 μM concentration relative to positive control measured at 24 h. See the Supporting Information for assay details.
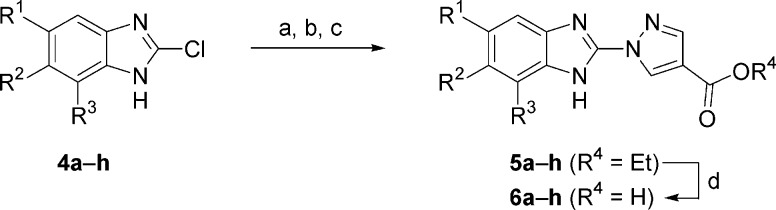
Scheme 2. Synthesis of Benzimidazole-2-pyrazoles 6a−h.
Reagents and conditions: (a) SEMCl or MEMCl, DIEA or NaH, THF. (b) Ethyl pyrazole-4-carboxylate, Cs2CO3, DMF, 80 °C, 1−5 h. (c) 4 M HCl/dioxane, EtOH, reflux, 1 h. (d) LiOH, THF/H2O. For R1−3 groups, see Table 2.
The synthesis of benzimidazole-2-pyrazoles 6a−h began from known or available 2-chlorobenzimidazoles 4a−h. Protection of the benzimidazole nitrogen atom as a MEM or SEM aminal facilitates displacement of the chloride atom by ethyl pyrazole-4-carboxylate in the presence of Cs2CO3. Removal of the protective group under acidic conditions provided ethyl esters 5a−h, which were saponified to afford the desired carboxylic acids 6a−h.
Benzimidazole-2-pyrazoles 6a−e proved to be potent inhibitors of PHD2 relative to the benzimidazole-2-carboxamides 3a−h (Table 2). For example, compounds 6a−c each show an increase in enzyme inhibitory activity of about 2 orders of magnitude relative to their direct comparators 3a, 3c, and 3e, while several other compounds display pIC50 values of 7.0 or more. As with the glycine amide compounds, selected members of this set also displayed low iron chelation ability in solution, indicating that the increased inhibitory activity was not a result of iron sequestration.25 Of greater significance, these compounds effected a robust increase of epo secretion from Hep3B cells measured after 24 h of incubation. To demonstrate that the observed increase in epo secretion was a consequence of PHD inhibition, 6a−e were tested in a HIF1α accumulation assay also using Hep3B cells, and compounds from this group were found to promote increases in HIF1α accumulation by as much as 92 ± 4% (e.g., 6g, Table 2).
Table 2. PHD2 Enzyme Inhibition, Epo Release, and HIF1α Accumulation for Compounds 6a−h.
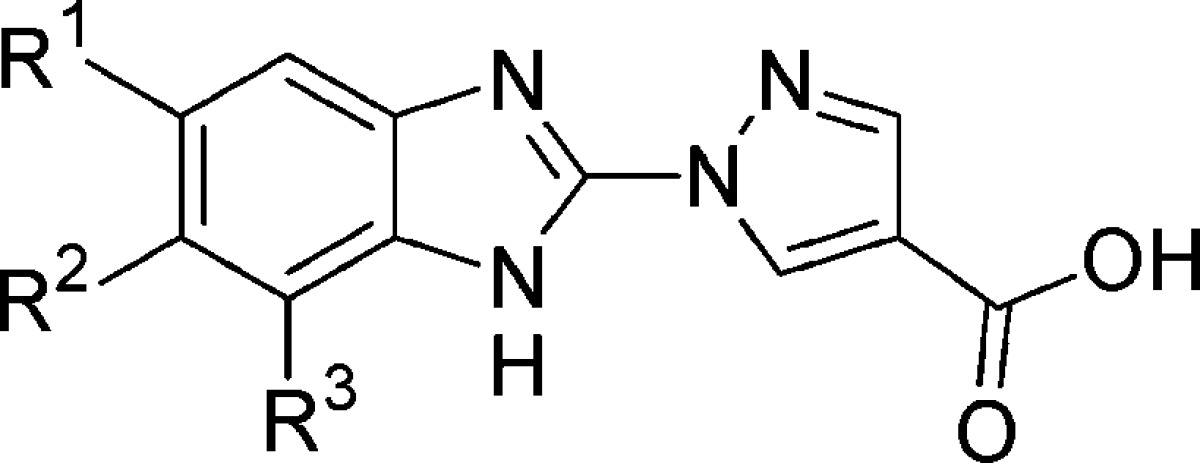
| compd | R1 | R2 | R3 | pIC50a | epo (%)b | HIF1α (%)c |
|---|---|---|---|---|---|---|
| 6a | H | H | H | 6.0 | <20 | <20 |
| 6b | Br | H | H | 6.4 | 105 ± 6 | 70 ± 3 |
| 6c | OMe | H | H | 6.4 | <20 | <20 |
| 6d | Cl | F | H | 7.1 | 129 ± 11 | 72 ± 7 |
| 6e | OCF3 | H | H | 6.5 | 142 ± 10 | 63 ± 2 |
| 6f | OCF3 | H | Br | 6.8 | 56 ± 4 | 21 ± 1 |
| 6g | CF3 | Cl | H | 7.1 | 176 ± 7 | 92 ± 4 |
| 6h | CF3 | F | H | 7.0 | 28 ± 9 | 92 ± 5 |
Negative logarithm of the concentration required to achieve 50% enzyme inhibition; the value is ±0.3 log units.
Percent increase in erythropoietin release from Hep3B cells at 100 μM concentration relative to positive control at 24 h.
Percent increase in HIF1α accumulation in Hep3B cells at 100 μM concentration relative to positive control measured at 24 h. See the Supporting Information for assay details.
Having discovered compounds that potently inhibit PHD2 with concomitant increases in both HIF1α accumulation and epo release in Hep3B cells, we evaluated 6g and 6h in an in vivo, orally dosed epo release model. Measured 6 h after a single oral dose of 100 μmol/kg, compounds 6g and 6h elicited increases in epo levels of 56- and 18-fold, respectively (Figure 1). This robust increase corresponds to plasma epo levels of 1410 ± 410 pg/mL versus the vehicle group value of 25 ± 8 pg/mL for the compound 6g cohort and 1270 ± 760 pg/mL versus the vehicle group value of 72 ± 20 pg/mL for the 6h cohort. For compound 6h, this pharmacodynamic response occurred with 1 h plasma concentrations of 14 μM and 6 h concentrations of 0.1 μM.
Figure 1.
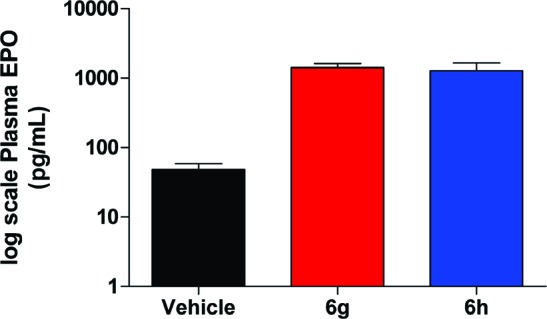
Epo levels vs vehicle for compounds 6g and 6h after a single oral dose of 100 μmol/kg in the mouse.
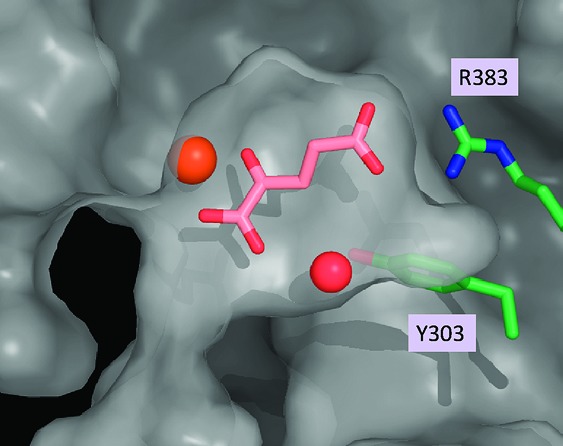
With these novel and structurally distinct PHD2 inhibitors in hand, we undertook structural studies to elucidate their mode of inhibition. Schofield, Syed, and co-workers, as well as Evodokimov and co-workers, have independently reported X-ray cocrystal structures of a truncated form of PHD2 with two structurally related isoquinoline inhibitors, both of which clearly mimic the endogenous cofactor 2OG in the active site of the enzyme.27,28 To supplement these data for our own studies, we obtained the X-ray cocrystal structure of PHD2 with its natural ligand, 2OG (Figure 2).29 In agreement with the previously published structures, the carboxylic acid group is clearly participating in a salt bridge interaction with R383, while the α-keto acid moiety is ligated to the iron atom in the active site. Furthermore, an ordered water cascade suggests the possibility that the α-ketocarboxylic acid moiety may be involved in a water-bridged interaction with Y303, analogous to the direct interaction observed between Y303 and the isoquinoline hydroxyl groups of the previously reported structures. Taken together, these X-ray crystal structures illustrate several key interactions between PHD2 and these known ligands.
Figure 2.
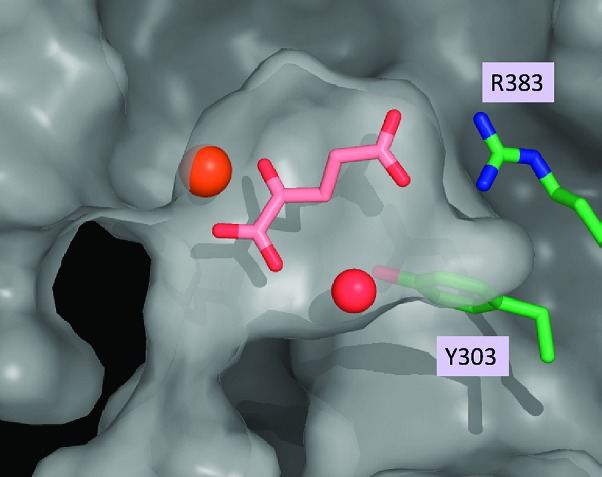
X-ray cocrystal structure of PHD2 and the endogenous ligand 2OG.
In the case of benzimidazole glycine amide 3b and pyrazole 6d, the iron atom is engaged by the two sp2 nitrogen atoms of the benzimidazole and pyrazole rings, respectively, while the carboxylic acid group appears well poised to interact with Arg383 (Figure 3).29 Once again, the benzimidazole NH group is proximal to an ordered water molecule, suggesting a possible water-bridged interaction with Y303. These structures clearly demonstrate that compounds 3b and 6d act as 2OG mimetics and that they adopt a similar binding mode to that of the endogenous ligand.
Figure 3.
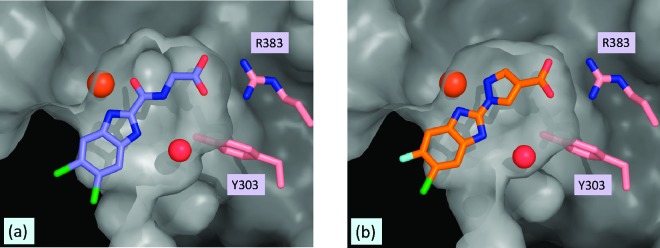
X-ray cocrystal structures of (a) PHD2/3b and (b) PHD2/6d.
In summary, we disclose the discovery and preliminary pharmacological profiling data for novel benzimidazole-based PHD2 inhibitors. The X-ray cocrystal structures of PHD2 with 2OG, 3b, and 6d are reported, and these structures elucidate key protein interactions and verify target engagement. Several compounds were identified that were efficacious in a series of assays, including biochemical enzyme inhibition, cell-based epo release and HIF1α accumulation, and in vivo epo release. This led to the discovery of compounds 6g and 6h, which were demonstrated to act as potent epo secretagogues when dosed orally in the mouse.
Acknowledgments
We thank Dr. Jiejun Wu and Heather McAllister for analytical assistance.
Supporting Information Available
Experimental details for the synthesis and characterization of 3a−h and 6a−h as well as assay details. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Ivan M.; Kondo K.; Yang H.; Kim W.; Valiando J.; Ohh M.; Salic A.; Asara J. M.; Lane W. S.; Kaelin W. G. Jr. HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 2001, 292, 464–468. [DOI] [PubMed] [Google Scholar]
- Berra E.; Benizri E.; Ginouves A.; Volmat V.; Roux D.; Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 2003, 22, 4082–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield C. J.; Ratcliffe P. J. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 2004, 40, 343–354. [DOI] [PubMed] [Google Scholar]
- Webb J. D.; Coleman M. L.; Pugh C. W. Hypoxia, hypoxia-inducible factors (HIF), HIF hydroxylases and oxygen sensing. Cell. Mol. Life Sci. 2009, 66, 3539–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salceda S.; Caro J. Hypoxia-inducible factor 1α (HIF-1α) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J. Biol. Chem. 1997, 272, 22642–22647. [DOI] [PubMed] [Google Scholar]
- Wenger R. H.; Stiehl D. P.; Camenisch G. Integration of oxygen signaling at the consensus HRE. Science Signal 2005, 306, re12. [DOI] [PubMed] [Google Scholar]
- Ivan M.; Harris A. L.; Martelli F.; Kulshreshtha R. Hypoxia response and microRNAs: no longer two separate worlds. J. Cell. Mol. Med. 2008, 12, 1426–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katschinski D. M. In vivo functions of the prolyl-4-hydroxylase domain oxygen sensors: direct route to the treatment of anaemia and the protection of ischaemic tissue. Acta Physiol. 2009, 195, 407–414. [DOI] [PubMed] [Google Scholar]
- Siddiq A.; Aminova L. R.; Ratan R. R. Hypoxia inducible factor prolyl 4-hydroxylase enzymes: Center stage in the battle against hypoxia, metabolic compromise, and oxidative stress. Neurochem. Res. 2007, 32, 931–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Review: Yan L.; Colandrea V. J.; Hale J. J. Prolyl hydroxylase domain-containing protein inhibitors as stabilizers of hypoxia-inducible factor: small molecule-based therapeutics for anemia. Expert Opin. Ther. Pat. 2010, 20, 1219–1245. [DOI] [PubMed] [Google Scholar]
- Rabinowitz M. H.; Barrett T. D.; Rosen M. D.; Venkatesan H.. Inhbitors of HIF Prolyl Hydroxylase. In Annual Reports in Medicinal Chemistry; Macor J. E., Robichaud A. J., Stamford A. W., Weinstein D., McAlpine S., Primeau J., Lowe J., Desai M. C., Eds.; Elsevier: London, 2010; Vol. 45, in press. [Google Scholar]
- Murray J. K.; Balan C.; Allgeier A. M.; Kasparian A.; Viswanadhan V.; Wilde C.; Allen J. R.; Yoder S. C.; Biddlecome G.; Hungate R. W.; Miranda L. P. Dipeptidyl-quinolone derivatives inhibit hypoxia inducible factor-1α prolyl hydroxylases-1, -2, and -3 with altered selectivity. J. Comb. Chem. 2010, 12, 676–686. [DOI] [PubMed] [Google Scholar]
- Mecinovic J.; Loenarz C.; Chowdhury R.; Schofield C. J. 2-Oxoglutarate analogue inhibitors of prolyl hydroxylase domain 2. Bioorg. Med. Chem. Lett. 2009, 19, 6192–6195. [DOI] [PubMed] [Google Scholar]
- Frohn M.; Viswanadhan V.; Pickrell A. J.; Golden J. E.; Muller K. M.; Burli R. W.; Biddlecome G.; Yoder S. C.; Rogers N.; Dao J. H.; Hungate R.; Allen J. R. Structure-guided design of substituted aza-benzimidazoles as potent hypoxia inducible factor-1α prolyl hydroxylase-2. Bioorg. Med. Chem. Lett. 2008, 18, 5023–5026. [DOI] [PubMed] [Google Scholar]
- Tegley C. M.; Viswanadhan V. N.; Biswas K.; Frohn M. J.; Peterkin T. A. N.; Chang C.; Buerli R. W.; Dao J. H.; Veith H.; Rogers N.; Yoder S. C.; Biddlecome G.; Tagari P.; Allen J. R.; Hungate R. W. Discovery of novel hydroxyl-thiazoles as HIF-α prolyl hydroxylase inhibitors: SAR, synthesis, and modeling evaluation. Bioorg. Med. Chem. Lett. 2008, 18, 3925–3928. [DOI] [PubMed] [Google Scholar]
- Warshakoon N. C.; Wu S.; Boyer A.; Kawamoto R.; Renock S.; Xu K.; Pokross M.; Evdokimov A. G.; Zhou S.; Winter C.; Walter R.; Mekel M. Design and synthesis of a series of novel pyrazolylpyridines as HIF 1-α prolyl hydroxylase inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 5687–5690. [DOI] [PubMed] [Google Scholar]
- Warshakoon N. C.; Wu S.; Boyer A.; Kawamoto R.; Sheville J.; Bhatt R. T.; Renock S.; Xu K.; Pokross M.; Zhou S.; Walter R.; Mekel M.; Evdokimov A. G.; East S. Design and synthesis of substituted pyridine derivatives as HIF-1α prolyl hydroxylase inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 5616–5620. [DOI] [PubMed] [Google Scholar]
- Warshakoon N. C.; Wu S.; Boyer A.; Kawamoto R.; Sheville J.; Renock S.; Xu K.; Pokross M.; Evdokimov A. G.; Walter R.; Mekel M. A novel series of imidazo[1,2-a]pyridine derivatives as HIF-1α prolyl hydroxylase inhibitors. Bioorg. Med. Chem. Lett. 2006, 16, 5598–5601. [DOI] [PubMed] [Google Scholar]
- Mole D. R.; Schlemminger I.; McNeill L. A.; Hewitson K. S.; Pugh C. W.; Ratcliffe P. J.; Schofield C. J. 2-Oxoglutarate analogue inhibitors of HIF prolyl hydroxylase. Bioorg. Med. Chem. Lett. 2003, 13, 2677–2680. [DOI] [PubMed] [Google Scholar]
- Melnikova I. Anaemia therapies. Nature Rev. Drug Discovery 2006, 5, 627–628. [DOI] [PubMed] [Google Scholar]
- Clinicaltrials.gov identifiers: NCT00750256, NCT00761657, NCT00840320, and NCT01083888.
- Wang J.; Buss J. L.; Chen G.; Ponka P.; Pantopoulos K. The prolyl 4-hydroxylase inhibitor ethyl-3,4-dihydroxybenzoate generates effective iron deficiency in cultured cells. FEBS Lett. 2002, 529, 309–312. [DOI] [PubMed] [Google Scholar]
- Kanelakis K. C.; Palomino H. L.; Li L.; Wu J.; Yan W.; Rosen M. D.; Rizzolio M. C.; Triveda M.; Morton M. F.; Yang Y.; Venkatesan H.; Rabinowitz M. H.; Shankley N. P.; Barrett T. B. Characterization of a robust enzymatic assay for inhibitors of 2-oxoglutarate-dependent hydroxylases. J. Biomol. Screening 2009, 14, 627–635. [DOI] [PubMed] [Google Scholar]
- Ennis B. C.; Holan G.; Samuel E. L. 2-(Trihalomethyl)benzazoles. III. Reactions of 2-(trichloromethyl)benzimidazole with nucleophiles. J. Chem. Soc. C 1967, 30–33. [Google Scholar]
- Breuer W.; Epsztejn S.; Millgram P.; Cabantchik I. Z. Transport of iron and other transition metals into cells as revealed by a fluorescent probe. Am. J. Physiol. Cell Physiol. 1995, 268, 1354–1361. [DOI] [PubMed] [Google Scholar]
- Zorc B. Peptidomimetics. Farm. Glas. 2008, 64, 113–121. [Google Scholar]
- McDonough M. A.; Li V.; Flashman E.; Rasheduzzaman C.; Mohr C.; Lienard B. M. R.; Zondlo J.; Oldham N. J.; Clifton I. J.; Lewis J.; McNeill L. A.; Kurzeja R. J. M.; Hewitson K. S.; Yang E.; Jordan S.; Syed R. S.; Schofield C. J. Cellular oxygen sensing: crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2). Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 9814–9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evodkimov A. G.; Walter R. L.; Mekel M.; Pokross M. E.; Kawamoto R.; Boyer A. RCSB Protein Data Bank ID: 2HBT, 2006.
- The coordinates have been deposited with the RSCB Protein Data Bank. PHD2/2OG, PDB ID 3OUJ; PHD2/3b, 3OUI; and PHD2/6d, 3OUH.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.