Abstract
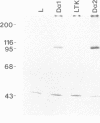
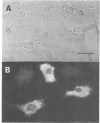
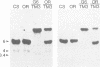
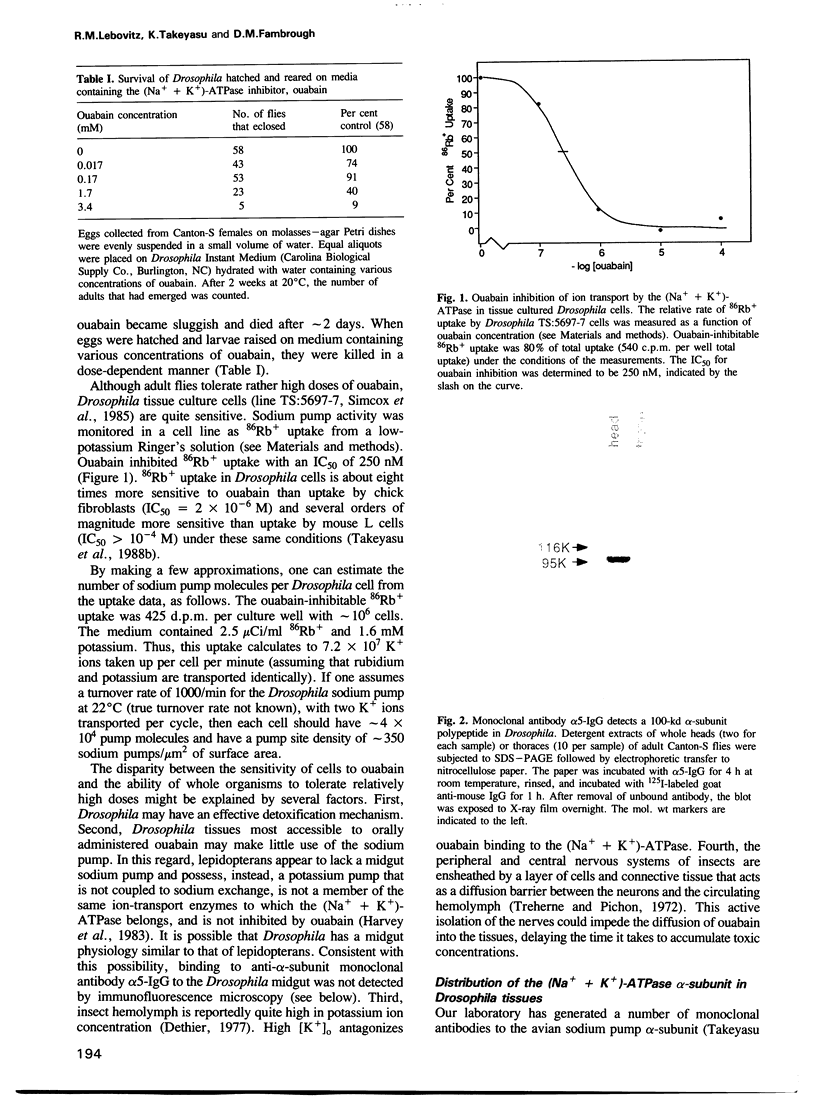
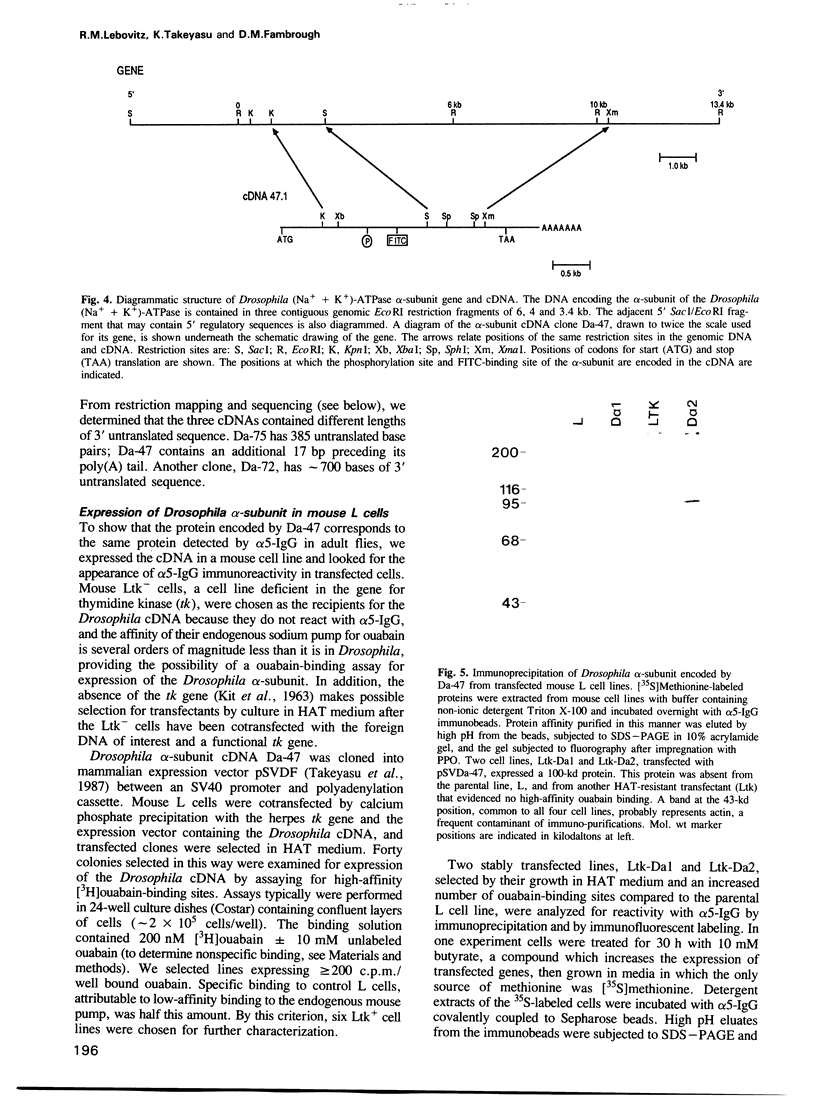
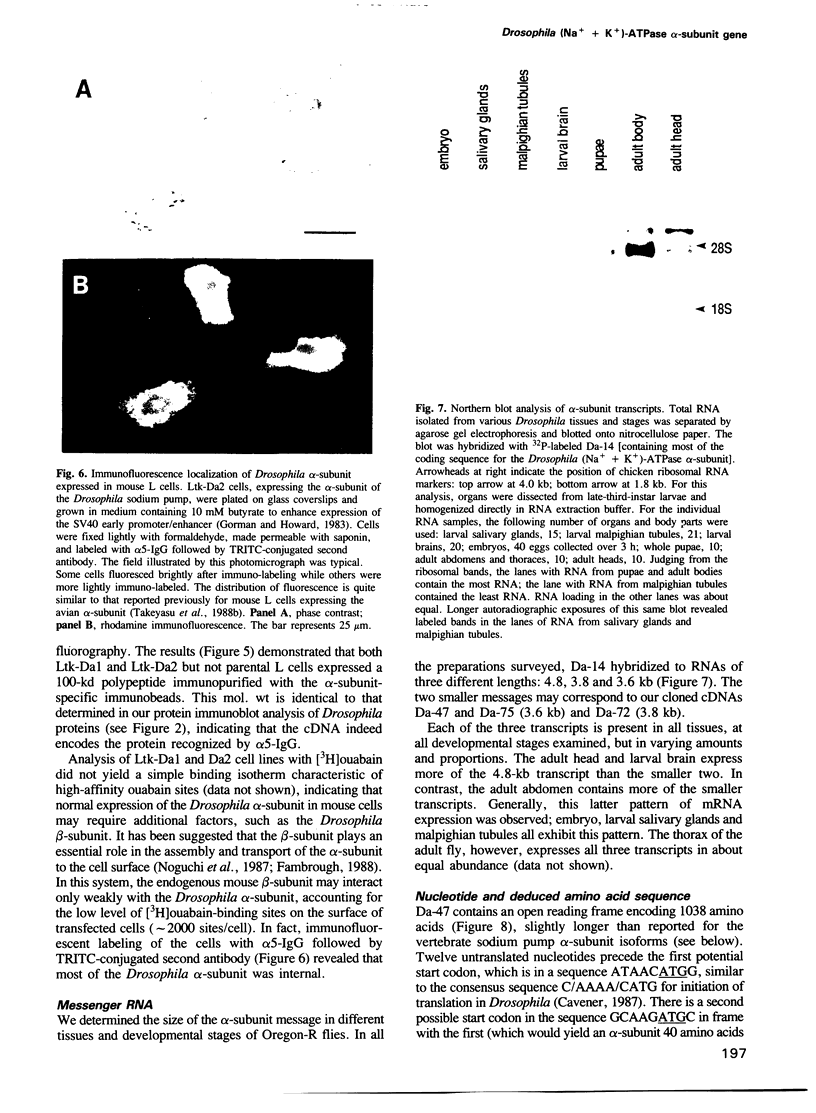
The (Na+ + K+)-ATPase (sodium pump) is an ouabain-sensitive, electrogenic ion pump responsible for maintaining the balance of sodium and potassium ions in almost all animal cells. Robust, ouabain-sensitive rubidium uptake, indicative of the sodium pump, was found in tissue-cultured Drosophila cells, and both larvae and adults die when fed a diet containing ouabain. A monoclonal antibody to the avian sodium pump alpha-subunit was found to cross-react with the Drosophila sodium pump alpha-subunit. Immunofluorescence microscopy was used to obtain a semi-quantitative view of the expression of the sodium pump in Drosophila tissues: high levels of the sodium pump were detected in malpighian tubules, indirect flight muscles and tubular muscles, and throughout the nervous system. The cDNA encoding this sodium pump alpha-subunit in Drosophila melanogaster was cloned, sequenced and expressed in mouse L cells. At the amino acid level, its deduced sequence of 1038 residues (the first such sequence for an invertebrate) is approximately 80% similar to alpha-subunit sequences reported for vertebrates. Only one gene was found in Drosophila, located on the third chromosome at position 93B. A restriction site polymorphism has been found, and several mutations exist that may involve the alpha-subunit gene.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biggs J., Searles L. L., Greenleaf A. L. Structure of the eukaryotic transcription apparatus: features of the gene for the largest subunit of Drosophila RNA polymerase II. Cell. 1985 Sep;42(2):611–621. doi: 10.1016/0092-8674(85)90118-7. [DOI] [PubMed] [Google Scholar]
- Cavener D. R. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987 Feb 25;15(4):1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethier V. G. The taste of salt. Am Sci. 1977 Nov-Dec;65(6):744–751. [PubMed] [Google Scholar]
- Donelson J. E., Wu R. Nucleotide sequence analysis of deoxyribonucleic acid. VII. Characterization of Escherichia coli exonuclease 3 activity for possible use in terminal nucleotide sequence analysis of duplex deoxyribonucleic acid. J Biol Chem. 1972 Jul 25;247(14):4661–4668. [PubMed] [Google Scholar]
- Fambrough D. M., Bayne E. K. Multiple forms of (Na+ + K+)-ATPase in the chicken. Selective detection of the major nerve, skeletal muscle, and kidney form by a monoclonal antibody. J Biol Chem. 1983 Mar 25;258(6):3926–3935. [PubMed] [Google Scholar]
- Fambrough D. M. The sodium pump becomes a family. Trends Neurosci. 1988 Jul;11(7):325–328. doi: 10.1016/0166-2236(88)90096-3. [DOI] [PubMed] [Google Scholar]
- Farley R. A., Tran C. M., Carilli C. T., Hawke D., Shively J. E. The amino acid sequence of a fluorescein-labeled peptide from the active site of (Na,K)-ATPase. J Biol Chem. 1984 Aug 10;259(15):9532–9535. [PubMed] [Google Scholar]
- Ganetzky B., Wu C. F. Indirect Suppression Involving Behavioral Mutants with Altered Nerve Excitability in DROSOPHILA MELANOGASTER. Genetics. 1982 Apr;100(4):597–614. doi: 10.1093/genetics/100.4.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Howard B. H., Reeves R. Expression of recombinant plasmids in mammalian cells is enhanced by sodium butyrate. Nucleic Acids Res. 1983 Nov 11;11(21):7631–7648. doi: 10.1093/nar/11.21.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HODGKIN A. L., KEYNES R. D. Active transport of cations in giant axons from Sepia and Loligo. J Physiol. 1955 Apr 28;128(1):28–60. doi: 10.1113/jphysiol.1955.sp005290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W. R., Cioffi M., Wolfersberger M. G. Chemiosmotic potassium ion pump of insect epithelia. Am J Physiol. 1983 Feb;244(2):R163–R175. doi: 10.1152/ajpregu.1983.244.2.R163. [DOI] [PubMed] [Google Scholar]
- Jan Y. N., Jan L. Y. Genetic dissection of short-term and long-term facilitation at the Drosophila neuromuscular junction. Proc Natl Acad Sci U S A. 1978 Jan;75(1):515–519. doi: 10.1073/pnas.75.1.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIT S., DUBBS D. R., PIEKARSKI L. J., HSU T. C. DELETION OF THYMIDINE KINASE ACTIVITY FROM L CELLS RESISTANT TO BROMODEOXYURIDINE. Exp Cell Res. 1963 Aug;31:297–312. doi: 10.1016/0014-4827(63)90007-7. [DOI] [PubMed] [Google Scholar]
- Kawakami K., Noguchi S., Noda M., Takahashi H., Ohta T., Kawamura M., Nojima H., Nagano K., Hirose T., Inayama S. Primary structure of the alpha-subunit of Torpedo californica (Na+ + K+)ATPase deduced from cDNA sequence. Nature. 1985 Aug 22;316(6030):733–736. doi: 10.1038/316733a0. [DOI] [PubMed] [Google Scholar]
- Kent R. B., Fallows D. A., Geissler E., Glaser T., Emanuel J. R., Lalley P. A., Levenson R., Housman D. E. Genes encoding alpha and beta subunits of Na,K-ATPase are located on three different chromosomes in the mouse. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5369–5373. doi: 10.1073/pnas.84.15.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Freund R., Schweber M., Wensink P. C., Meselson M. Sequence organization and transcription at two heat shock loci in Drosophila. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5613–5617. doi: 10.1073/pnas.75.11.5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L. The nucleotide sequence and transcript map of the herpes simplex virus thymidine kinase gene. Nucleic Acids Res. 1980 Dec 20;8(24):5949–5964. doi: 10.1093/nar/8.24.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi M., Kawamura M. Solubilization and purification of Artemia salina (Na,K)-activated ATPase and NH2-terminal amino acid sequence of its larger subunit. J Biol Chem. 1984 Dec 10;259(23):14928–14934. [PubMed] [Google Scholar]
- Noguchi S., Mishina M., Kawamura M., Numa S. Expression of functional (Na+ + K+)-ATPase from cloned cDNAs. FEBS Lett. 1987 Dec 10;225(1-2):27–32. doi: 10.1016/0014-5793(87)81125-0. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov YuA, Dzhandzhugazyan K. N., Lutsenko S. V., Mustayev A. A., Modyanov N. N., Dzhandzugazyan KN=Dzhadzhugazyan K. N. Affinity modification of E1-form of Na+, K+-ATPase revealed Asp-710 in the catalytic site. FEBS Lett. 1987 Jun 8;217(1):111–116. doi: 10.1016/0014-5793(87)81253-x. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov YuA, Modyanov N. N., Broude N. E., Petrukhin K. E., Grishin A. V., Arzamazova N. M., Aldanova N. A., Monastyrskaya G. S., Sverdlov E. D. Pig kidney Na+,K+-ATPase. Primary structure and spatial organization. FEBS Lett. 1986 Jun 9;201(2):237–245. doi: 10.1016/0014-5793(86)80616-0. [DOI] [PubMed] [Google Scholar]
- Robinson D., McGee R., Jr Agonist-induced regulation of the neuronal nicotinic acetylcholine receptor of PC12 cells. Mol Pharmacol. 1985 Apr;27(4):409–417. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull G. E., Greeb J., Lingrel J. B. Molecular cloning of three distinct forms of the Na+,K+-ATPase alpha-subunit from rat brain. Biochemistry. 1986 Dec 16;25(25):8125–8132. doi: 10.1021/bi00373a001. [DOI] [PubMed] [Google Scholar]
- Shull G. E., Schwartz A., Lingrel J. B. Amino-acid sequence of the catalytic subunit of the (Na+ + K+)ATPase deduced from a complementary DNA. Nature. 1985 Aug 22;316(6030):691–695. doi: 10.1038/316691a0. [DOI] [PubMed] [Google Scholar]
- Shull M. M., Lingrel J. B. Multiple genes encode the human Na+,K+-ATPase catalytic subunit. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4039–4043. doi: 10.1073/pnas.84.12.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simcox A. A., Sobeih M. M., Shearn A. Establishment and characterization of continuous cell lines derived from temperature-sensitive mutants of Drosophila melanogaster. Somat Cell Mol Genet. 1985 Jan;11(1):63–70. doi: 10.1007/BF01534735. [DOI] [PubMed] [Google Scholar]
- Sverdlov E. D., Monastyrskaya G. S., Broude N. E., Ushkaryov YuA, Allikmets R. L., Melkov A. M., Smirnov YuV, Malyshev I. V., Dulobova I. E., Petrukhin K. E. The family of human Na+,K+-ATPase genes. No less than five genes and/or pseudogenes related to the alpha-subunit. FEBS Lett. 1987 Jun 15;217(2):275–278. doi: 10.1016/0014-5793(87)80677-4. [DOI] [PubMed] [Google Scholar]
- Takeyasu K., Tamkun M. M., Renaud K. J., Fambrough D. M. Ouabain-sensitive (Na+ + K+)-ATPase activity expressed in mouse L cells by transfection with DNA encoding the alpha-subunit of an avian sodium pump. J Biol Chem. 1988 Mar 25;263(9):4347–4354. [PubMed] [Google Scholar]
- Takeyasu K., Tamkun M. M., Siegel N. R., Fambrough D. M. Expression of hybrid (Na+ + K+)-ATPase molecules after transfection of mouse Ltk-cells with DNA encoding the beta-subunit of an avian brain sodium pump. J Biol Chem. 1987 Aug 5;262(22):10733–10740. [PubMed] [Google Scholar]