Abstract
The Fas death receptor-activated death-inducing signaling complex (DISC) regulates apoptosis in many normal and cancer cells. Qualitative biochemical experiments demonstrate that calmodulin (CaM) binds to the death domain of Fas. The interaction between CaM and Fas regulates Fas-mediated DISC formation. A quantitative understanding of the interaction between CaM and Fas is important for the optimal design of antagonists for CaM or Fas to regulate the CaM–Fas interaction, thus modulating Fas-mediated DISC formation and apoptosis. The V254N mutation of the Fas death domain (Fas DD) is analogous to an identified mutant allele of Fas in lpr-cg mice that have a deficiency in Fas-mediated apoptosis. In this study, the interactions of CaM with the Fas DD wild type (Fas DD WT) and with the Fas DD V254N mutant were characterized using isothermal titration calorimetry (ITC), circular dichroism spectroscopy (CD), and molecular dynamics (MD) simulations. ITC results reveal an endothermic binding characteristic and an entropy-driven interaction of CaM with Fas DD WT or with Fas DD V254N. The Fas DD V254N mutation decreased the association constant (Ka) for CaM–Fas DD binding from (1.79 ± 0.20) × 106 to (0.88 ± 0.14) × 106 M–1 and slightly increased a standard state Gibbs free energy (ΔG°) for CaM–Fas DD binding from −8.87 ± 0.07 to −8.43 ± 0.10 kcal/mol. CD secondary structure analysis and MD simulation results did not show significant secondary structural changes of the Fas DD caused by the V254N mutation. The conformational and dynamical motion analyses, the analyses of hydrogen bond formation within the CaM binding region, the contact numbers of each residue, and the electrostatic potential for the CaM binding region based on MD simulations demonstrated changes caused by the Fas DD V254N mutation. These changes caused by the Fas DD V254N mutation could affect the van der Waals interactions and electrostatic interactions between CaM and Fas DD, thereby affecting CaM–Fas DD interactions. Results from this study characterize CaM–Fas DD interactions in a quantitative way, providing structural and thermodynamic evidence of the role of the Fas DD V254N mutation in the CaM–Fas DD interaction. Furthermore, the results could help to identify novel strategies for regulating CaM–Fas DD interactions and Fas DD conformation and thus to modulate Fas-mediated DISC formation and thus Fas-mediated apoptosis.
The Fas death receptor is a member of the tumor necrosis factor family, and formation of the Fas death receptor-mediated death-inducing signaling complex (DISC) regulates cell death in a variety of normal and cancer cells.1,2 Binding of Fas ligand to the Fas receptor leads to oligomerization of the Fas receptor into homotrimers. Oligomerized Fas recruits the adaptor protein Fas-associated death domain protein (FADD) via its death domain (DD), which in turn recruits procaspase-8 to form the DISC.3 The intracellular protein calmodulin (CaM) functions as a mediator of Ca2+ signals, and an increased level of expression of CaM is associated with cancer.4 Immunoprecipitation experiments show CaM binds to Fas in cholangiocarcinoma,5 Jurkat,6 and osteoclast1 cells. CaM is recruited into the Fas-mediated DISC after Fas activation; furthermore, EGTA, a Ca2+ chelator, and trifluoperazine (TFP), a CaM antagonist, inhibit the recruitment of CaM into the Fas-mediated DISC.5 TFP was shown to decrease the level of Fas-mediated DISC-induced cleavage of caspase-8.5
Qualitative experiments demonstrate that CaM binds to the Fas death domain (Fas DD) in a calcium-dependent manner.6 A fragment containing helices 1–3 of the Fas DD associates with CaM.6 The structures of Ca2+-bound CaM [Protein Data Bank (PDB) entry 3cln]7 and the Fas death domain (PDB entry 1ddf) with the binding region for CaM colored blue6,8 are shown in Figure S1 of the Supporting Information. Dysregulation of the Fas receptor and mediators of its signaling pathways have been associated with diseases in humans and mice such as the autoimmune lymphoproliferative syndrome (ALPS) in humans.9−11 The expression of mutant Fas with impaired function has been observed in numerous types of cancers.12−14 Fas mutations that cause an alteration of the structure and function of Fas have been detected in many cancers.15,16 Lymphoproliferation complementing Gld (lpr-cg) mice have a mutation in Fas causing defective Fas signaling.17 The valine 254 to asparagine (V254N) mutation of the Fas DD is analogous to the mutant allele of Fas in lpr-cg mice.6 The lpr-cg mice Fas mutant reduces the level of binding to FADD, which could be due to an altered structure.8 Qualitative experiments show that the Fas DD V254N mutation reduced the level of CaM–Fas DD binding.6
Our lab has previously investigated the binding thermodynamics and conformation of CaM–Fas DD complexes using explicit solvent molecular dynamics simulations and implicit solvent binding free energy calculations, presenting structural evidence of CaM–Fas DD binding and the effect of the Fas DD V254N mutation on CaM–Fas DD interaction.18 Furthermore, our lab has shown, using molecular dynamics simulations, the structural and molecular mechanisms whereby the CaM antagonist, TFP, regulates CaM–Fas binding, indicating that the number of TFP molecules bound to CaM directly contributed to conformational changes in CaM and these changes affected the binding of CaM to Fas DD.19 These computational results provided insight into the structural and molecular mechanisms of the CaM–Fas DD interaction that regulate Fas-mediated DISC formation and the mechanism by which TFP affects the CaM–Fas interaction and modulates Fas-mediated DISC formation.
Understanding the protein interactions underlying Fas-mediated cell death pathways is potentially important for identifying novel targets and agents for the effective initiation of cancer cell death. Therefore, a quantitative characterization of the CaM–Fas interaction is important for designing optimized antagonists for CaM or Fas that regulate CaM–Fas interaction, thus modulating Fas-mediated DISC formation and apoptosis. In this study, the interactions of CaM with wild-type Fas DD (Fas DD WT) and the Fas DD V254N mutant were characterized using isothermal titration calorimetry (ITC), circular dichroism spectroscopy (CD), and molecular dynamics (MD) simulations. The results provide structural and thermodynamic evidence to understand the role of Fas DD V254N in CaM and Fas DD interaction.
Materials and Methods
We characterized the interactions of CaM with the Fas DD wild type (Fas DD WT) and the Fas DD V254N mutant using sedimentation velocity experiment, isothermal titration calorimetry (ITC), circular dichroism spectroscopy (CD) and molecular dynamics (MD) simulations. The sedimentation velocity experiment, ITC, and CD experiments were performed at 37 °C with the same buffer that consisted of 50 mM sodium acetate (pH 5), 150 mM NaCl, 20 mM CaCl2, and 0.5 mM tris(2-carboxyethyl)phosphine (TCEP).
Recombinant Protein Expression and Purification
Escherichia coli BL21(DE3) (pLysS) strain cells containing 6xHis-SUMO-calmodulin or 6xHis-SUMO-Fas(191–335) genes cloned into pET28 vectors were kindly provided by J. M. McDonald (Department of Pathology, University of Alabama at Birmingham). Fas DD V245N was created by site-directed mutagenesis of pET28 6xHis-SUMO-Fas(191–335) using the QuikChange II site-directed mutagenesis kit (Agilent Technologies). The procedure for the expression and purification of recombinant 6xHis-SUMO fusion proteins was performed as follows. Transformed E. coli cells were incubated in LB medium with 50 μg/mL kanamycin and 10 μg/mL chloramphenicol (Fisher Scientific) on a shaking incubator at 30 °C. E. coli cell population growth was monitored hourly by measuring the optical density of the E. coli culture at 600 nm. Recombinant protein expression was induced by the addition of 0.1 mM isopropyl β-d-thiogalactopyranoside (Promega). The culture was then incubated for an additional 4 h at 30 °C, and expression of the 6xHis-SUMO fusion protein was evaluated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). After protein expression, E. coli cultures were pelleted by centrifugation and stored at −80 °C until they were lysed. Cells were lysed using an EmulsiFlex-C3 French press (Avestin); lysates were centrifuged, and the supernatant was collected. The 6xHis-SUMO fusion proteins were purified by nickel ion affinity chromatography according to the manufacturer’s direction for the 6xHis-SUMO fusion protein purification system (Qiagen). The 6xHis-SUMO tag was cleaved off by overnight incubation with the SUMO tag protease (Ulp), and the 6xHis-SUMO tag and SUMO protease were removed by nickel ion affinity chromatography. Recombinant CaM, Fas DD WT, and Fas DD V254N were further purified by size exclusion chromatography using a HiLoad 16/600 Superdex 75 prepgrade column on an AKTA purifier (GE life sciences) with a final purity evaluated by SDS–PAGE. CaM, Fas DD WT, and Fas DD V254N concentrations were measured by UV absorbance at 280 nm using a Cary 300 UV–vis spectrophotometer (Varain Inc., Palo Alto, CA). The calculated extinction coefficients of 8480 M–1 cm–1 for Fas DD WT and Fas DD V254N and 2980 M–1 cm–1 for CaM were determined from their primary residue sequence using the edelhoch method,20 adjusted with the extinction coefficients determined by Pace et al.21 in Protpram (ExPASy SIB Bioinformatics Recourse Portal).
Sedimentation Velocity Experiments
Sedimentation velocity experiments were performed using a Beckman Proteome Lab XL-I analytical ultracentrifuge to assess the oligomeric state of the protein used for ITC and CD experiments. Sedimentation velocity experiments, using absorbance optics, were conducted by loading 10 μM Fas DD WT or 10 μM Fas DD V254N mutant samples (380 μL) and the protein dialysate (400 μL) into a double-sector Epon charcoal-filled centerpiece. Samples were subjected to an angular velocity of 50000 rpm at 37 °C. Absorbance scans as a function of radial position were collected by scanning the sample cells at a wavelength of 280 nm, as indicated in the text, per a distance of 0.003 cm. Scans were collected every 1 min. Sedimentation velocity boundaries were analyzed using SedFit,22 where sedimentation coefficient distributions [c(s)] and differential molar mass distribution [c(M)] analyses were applied by modeling the sedimentation boundaries as solutions of the Lamm equation for noninteracting species. All sedimentation coefficients reported in the text were corrected to 20 °C and water, i.e., s20,w. The partial specific volume of Fas DD WT and Fas DD V254N and the density and viscosity of the 50 mM sodium acetate (pH 5)/150 mM NaCl/20 mM CaCl2/0.5 mM tris(2-carboxyethyl) phosphine (TCEP) buffer at 37 °C were calculated using sednterp23 and subjected to c(s) analysis and c(M) analysis. The weighted average molecular weight was obtained by integrating the peak area of the c(M) distribution.
Isothermal Titration Calorimetry
Calorimetric titrations were performed using an Auto-ITC200 microcalorimeter from MicroCal (Northampton, MA). The acquisition and analysis of data were performed using the Origin software supplied by MicroCal. CaM, Fas DD WT, and Fas DD V254N were dialyzed into a 50 mM sodium acetate (pH 5)/150 mM NaCl/20 mM CaCl2/0.5 mM tris(2-carboxyethyl)phosphine (TCEP) buffer for isothermal titration calorimetry (ITC) experiments. A typical titration was performed by injections of 400 μM CaM into the calorimeter cell containing a 10 μM Fas DD WT or 10 μM Fas DD V254N solution. Successive injections were separated by 210 s intervals to allow the endothermic peak resulting from the reaction to return to the baseline. A constant stirring speed of 500 rpm and a temperature of 37 °C were maintained throughout the titration. Dilution heats of CaM were measured by titrating 400 μM CaM from the syringe into the cell containing only buffer. The resulting CaM dilution heat changes were subtracted from the measured heats of CaM–Fas DD binding.
Circular Dichroism Spectroscopy
Circular dichroism spectroscopy (CD) studies were performed using a JASCO J-810 spectropolorimeter (JASCO Corp.) fitted with a thermostatic cell holder and a thermostatic water bath. Fas DD WT and Fas DD V245N were dialyzed into the same buffer that was used for ITC experiments. The observed ellipticities were recorded as a function of wavelength every 1 nm using a 0.1 mm path length quartz cell at 37 °C, and each spectrum is reported as an average of four scans.
Molecular Dynamics Simulations
We performed 80 ns molecular dynamics (MD) simulations for Fas DD WT and Fas DD V254N using the AMBER 10 MD package.24 The conformational and structural changes of Fas DD and Fas DD V254N were analyzed to elucidate the structural basis and molecular mechanisms for the effect of the Fas DD V254N mutation on CaM–Fas DD binding as observed via ITC. The initial protein structure of the Fas death domain (PDB entry 1ddf)8 was obtained from the Protein Data Bank as shown in Figure S1 of the Supporting Information. The Fas DD V254N mutation was performed with the mutagenesis function of PyMol.25 The “parm99” parameters of the AMBER force field were assigned to the atoms in the simulated system, and a standard MD simulation protocol as used in our previous studies19,26−28 was performed for the simulated systems. The protein was first minimized and then solvated in a TIP3P29 water box with a physiological salt concentration of 150 mM NaCl. A buffer of 10 Å was set between the protein and the box boundaries to reduce potential artifacts arising from periodicity. Additional ions were added to each system to neutralize the charge of the protein. The solvent was then minimized and equilibrated for 10 ps, while the proteins and ions were constrained. The solvent was further equilibrated with the protein and ions restrained at a constant number–pressure–temperature (NpT) at 50 K and 1 atm for 20 ps. The simulated system was warmed via constant number–volume–temperature (NVT) MD simulations to 310 K by steps of 50 K lasting 10 ps each, with SHAKE constraints30 and a 2 fs time step. A production MD simulation of 80 ns at NpT of 310 K and 1 atm was performed for the simulated systems. The temperature of 310 K for MD simulations was the same as that used in ITC and CD experiments. The SHAKE constraints were applied to all hydrogen heavy bonds to permit a dynamics time step of 2 fs. Electrostatic interactions were calculated with the particle mesh Ewald method (PME).31 Both the direct space PME and Lennard-Jones cutoffs were set at 10 Å. The data for the MD simulations were collected every 2 ps. The simulations were performed on a local AMD Opteron cluster and on the AMD Opterons cluster in the Alabama Supercomputer Center.
Conformational and Structural Analyses
With the MD simulation trajectories after equilibration, we performed secondary structure analysis, conformational analysis, and dynamical motion analysis to better understand the conformational, structural, and motion characteristics of the Fas DD and its mutants. Root-mean-square deviations (rmsds) of protein backbone atoms were analyzed to determine the system’s equilibration tendencies and its convergence. We calculated the changes of the residues of α-helix occupancy for the Fas DD and Fas DD mutant to evaluate secondary structural changes of the protein caused by the Fas V254N mutation. An α-helix was defined to be at least five residues long in a coiled or spiral conformation. Whether an amino acid belonged to the α-helix was determined with the DSSP method by Kabsch and Sander32 implemented in the AMBER10 MD program. The occupancy of each residue in an α-helix was determined on the basis of the percentage of time that the residue existed in the α-helix over the simulation. The percentage helix for the Fas DD was calculated from the overall helicity divided by the total number of Fas residues. Root-mean-square fluctuations (rmsfs) of the proteins were calculated on a residue-by-residue basis and averaged over the production simulation trajectories for the observation of the conformational fluctuation of protein domains. Dynamical cross-correlation maps (normalized covariance matrices) between residues provided insight into which residues have general movements correlating with the motions of other residues and the degree of this correlation.19,26−28,33 These analyses were performed with the MD simulation trajectories obtained after the initial equilibration using the ptraj program of AMBER, and Matlab34 was used to generate the cross-correlation plots. We also analyzed hydrogen bond formation within the CaM binding region, the contact numbers of each residue, and the electrostatic potential for the CaM binding region for both the Fas DD and the Fas DD V254N mutant. A hydrogen bond was determined when the distance between the hydrogen and acceptor was <3.5 Å and the hydrogen–donor–acceptor angle was >150°. The number of hydrogen bonds between residues within the CaM binding region was calculated by summing all the hydrogen bonds within the regions and was averaged over the production simulations. A contact was identified if the atoms were within 5 Å of the specific residue. Then contact numbers for each residue were calculated by averaging each residue’s contact numbers over the production run. The electrostatic potential for the CaM binding region in the Fas DD was calculated by APBS.35 To calculate the electrostatic potential, dielectric constants of 1 and 78.54 were used separately for the protein and solvent, respectively. An ion concentration of 150 mM was used in the APBS calculation. The electrostatic potential was mapped onto the molecular surface of the Fas DD using VMD.36
Results and Discussion
Sedimentation Velocity Results for 10 μM Fas DD WT and 10 μM Fas DD V254N
The Fas DD WT protein tends to self-associate and is present predominately in a monomeric form in a low-pH buffer.8 Sedimentation velocity experiments were performed with 10 μM Fas DD WT and 10 μM Fas DD V254N in the same buffer that was used for the ITC and CD experiments at 37 °C to assess the oligomeric state of the protein. An example of raw sedimentation velocity data for Fas DD WT is shown in Figure S2 of the Supporting Information. Sedimentation coefficient distributions [c(s)] show that under the conditions used for upcoming ITC and CD experiments, the monomer is largely populated in these solution conditions for both 10 μM Fas DD WT and 10 μM Fas DD V254N (Figure 1A,B). From the c(s) analysis, the weighted average sedimentation coefficients (s20,w) for Fas DD WT and Fas DD V254N were determined to be 1.5 ± 0.3 and 1.6 ± 0.4 S, respectively, where the standard deviation describes the broadness of the peak. For both proteins, the weighted average sedimentation coefficient is consistent with a monomer. To further examine this, we examined the data by the molecular weight probability distributions [c(M)]. The molecular weight probability distributions [c(M)] of 10 μM Fas DD WT and 10 μM Fas DD V254N are shown in panels C and D of Figure 1, respectively, with weighted average molecular weights of 20 ± 5 and 18 ± 3 kDa, respectively. This result is consistent with a monomer molecular weight of 17 kDa for the Fas DD, indicating that under the experimental conditions used for the upcoming ITC and CD experiments, the monomers are largely populated for both 10 μM Fas DD WT and 10 μM Fas DD V254N.
Figure 1.
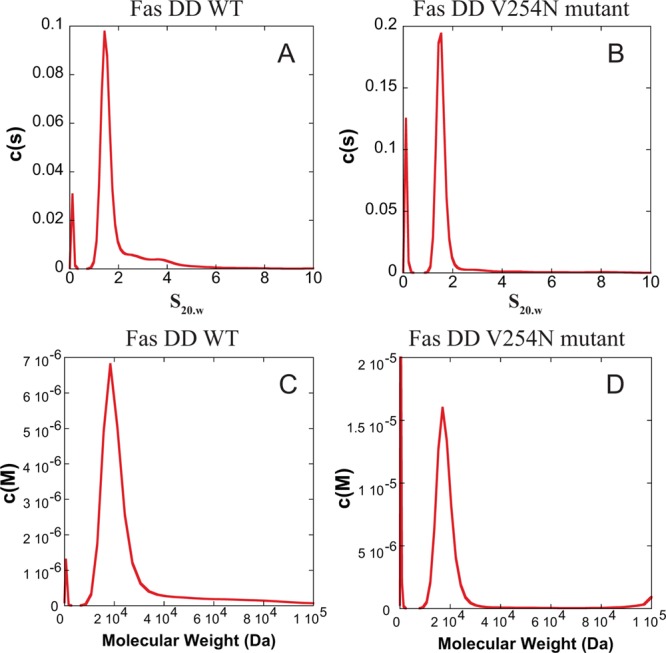
Sedimentation coefficient distribution [c(s)] as a function of s20,w for 10 μM Fas DD WT (A) and 10 μM Fas DD V254N (B). Molecular weight probability distribution [c(M)] as a function of molecular weight for 10 μM Fas DD WT (C) and 10 μM Fas DD V254N (D).
CaM with Fas DD Binding Assessed by Isothermal Titration Calorimetry
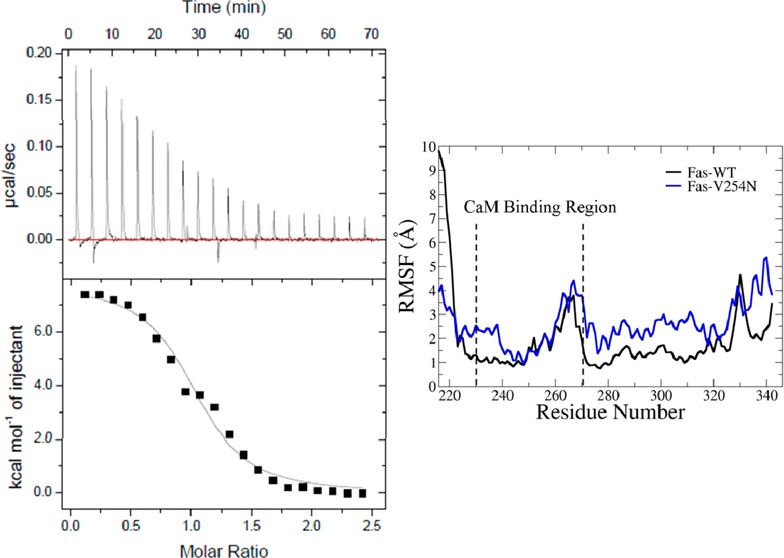
The thermodynamics of CaM and Fas DD WT binding was investigated using ITC. The calorimetric isotherm of binding of CaM to Fas DD WT illustrates an endothermic binding characteristic between CaM and Fas DD WT at 37 °C (Figure 2, top panel). The heat absorbed per injection of CaM into Fas DD WT was highest at low molar ratios of CaM to Fas DD WT. The heat absorbed decreased as the molar ratio of CaM to Fas DD WT increased until only the CaM dilution enthalpy was observed. The CaM dilution enthalpy was measured by titrating CaM from the syringe into the cell containing buffer. The CaM dilution enthalpy was subtracted from the CaM–Fas DD WT isotherm. The resulting CaM–Fas DD WT isotherms (Figure 2, bottom panel) were fit using the one-site model in Origin (MicroCal). The enthalpy change (ΔH°), association constant (Ka), and stoichiometry (n) and the standard error for each variable were derived on the basis of the one-site model fit of the CaM–Fas DD WT isotherm (Figure 2). The enthalpy change (ΔH°), association constant (Ka), and stoichiometry (n) listed in Table 1 are the error-weighted mean values and the standard error of the mean for each variable from three repeated experiments calculated as described in the previous study.37 The Gibbs free energy (ΔG°) was calculated on the basis of the Ka with eq 1.
| 1 |
where R is the gas constant (1.9872041 × 10–3 kcal mol–1 deg–1) and T is the absolute temperature of 310.15 K in this study. The entropy of binding (−TΔS°) was calculated using Ka and ΔH° via eq 2.
| 2 |
Figure 2.
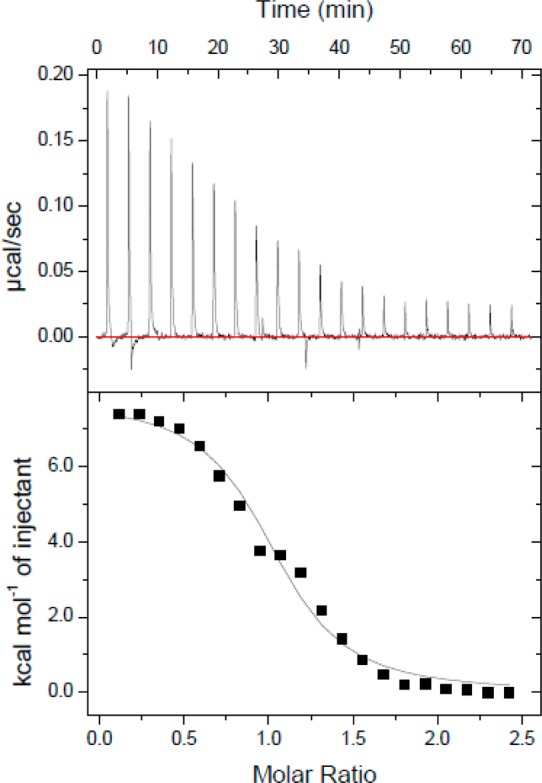
CaM–Fas DD WT isotherm. Thermogram of 400 μM CaM titrated into the calorimeter cell containing 10 μM Fas DD WT.
Table 1. Thermodynamic Parameters of CaM–Fas DD Binding Determined by ITC at 37 °C and pH 5.
| Ka (M–1)a | ΔG°a (kcal/mol) | ΔH°a (kcal/mol) | –TΔS°a (kcal/mol) | n | |
|---|---|---|---|---|---|
| CaM–Fas DD WT | (1.79 ± 0.20) × 106 | –8.87 ± 0.07 | 6.00 ± 0.09 | –14.87 ± 0.12 | 1.03 ± 0.01 |
| CaM–Fas DD V254N | (0.88 ± 0.14) × 106 | –8.43 ± 0.10 | 7.40 ± 0.27 | –15.84 ± 0.28 | 1.09 ± 0.02 |
Differences are statistically significant between CaM–Fas DD WT and CaM–Fas DD V254N mutant complexes (Student’s t test; p < 0.05).
The standard errors of the mean for ΔG° and −TΔS° in Table 1 were estimated on the basis of the standard error of the mean of Ka and ΔH° using the statistical error propagation method.38 With an enthalpy (ΔH°) of 6.00 ± 0.09 kcal/mol and an association constant (Ka) of (1.79 ± 0.20) × 106 M–1 for CaM–Fas DD binding, the entropy (−TΔS°) for CaM–Fas DD binding was calculated to be −14.87 ± 0.12 kcal/mol and the Gibbs free energy (ΔG°) for CaM–Fas DD binding to be −8.87 ± 0.07 kcal/mol.
The entropy-driven CaM–Fas DD WT binding could be due to the removal of aqueous solvent from the hydrophobic surface areas on Ca2+-bound CaM and Fas DD WT upon binding, resulting in a favorable −TΔS° being the dominant contribution to an overall favorable ΔG°. This phenomenon is the hydrophobic effect, which is the energetics involved in transferring nonpolar compounds from a liquid organic phase to an aqueous phase.39 When considering protein–protein interaction, the interactions of nonpolar residues in the interior of a protein or at a protein–protein interface, such as during binding, could be considered to be bsimilar to the interactions of compounds undergoing the organic to aqueous phase transfer.40 Binding of Ca2+ to CaM regulates CaM’s binding activity by modulating the amount of exposed apolar surface area available to interact with various CaM binding substrates.41 Binding of Ca2+ to CaM induces conformational changes that result in the exposure of the hydrophobic surface area to the aqueous solvent.42,43 During CaM–Fas DD WT binding, the burial of the apolar surface area of CaM to the apolar CaM binding site on Fas DD WT, seven nonpolar residues in the binding region,6 could result in a hydrophobic effect that would drive CaM–Fas DD WT binding and would explain the favorable −TΔS value of CaM–Fas DD WT binding at 37 °C observed via ITC.
The Fas DD V254N mutation is analogous to the mutant allele of Fas in lpr-cg mice.6 In lpr-cg mice, the mutant Fas has a reduced level of binding to Fas-associated death domain protein, which could be due to an altered structure and conformation.8 To evaluate the effect of the V254N mutation of the Fas DD on CaM–Fas DD binding, the interaction between CaM and Fas DD V254N was characterized using ITC experiments. Binding of CaM to Fas DD V254N resulted in an endothermic isotherm (Figure 3, top panel) similar to that for binding of CaM to Fas DD WT. The CaM dilution enthalpy was subtracted from the CaM–Fas DD V254N isotherms to obtain the resulting isotherms (Figure 3, bottom panel). The resulting CaM–Fas DD V254N isotherms were fit to a one-site model in Origin (MicroCal) to derive ΔH°, Ka, and n. Using the same method as described above to obtain the mean values and the standard errors of the mean from three repeated experiments for ΔH°, Ka, n, −TΔS°, and ΔG° for CaM–Fas DD WT binding, the thermodynamic parameters for CaM–Fas DD V254N binding were determined as shown in Table 1. The CaM–Fas DD V254N isotherm yielded the following binding parameters: an association constant (Ka) of (0.88 ± 0.14) × 106 M–1, an enthalpy (ΔH°) of 7.40 ± 0.27 kcal/mol, with a calculated entropy (−TΔS°) of −15.84 ± 0.28 kcal/mol, and a Gibbs free energy (ΔG°) of −8.43 ± 0.1 kcal/mol. Similar to CaM–Fas WT binding, CaM–Fas V254N binding was an entropy-driven interaction at 37 °C, which could be due to the interaction of hydrophobic residues at the Fas binding site.
Figure 3.
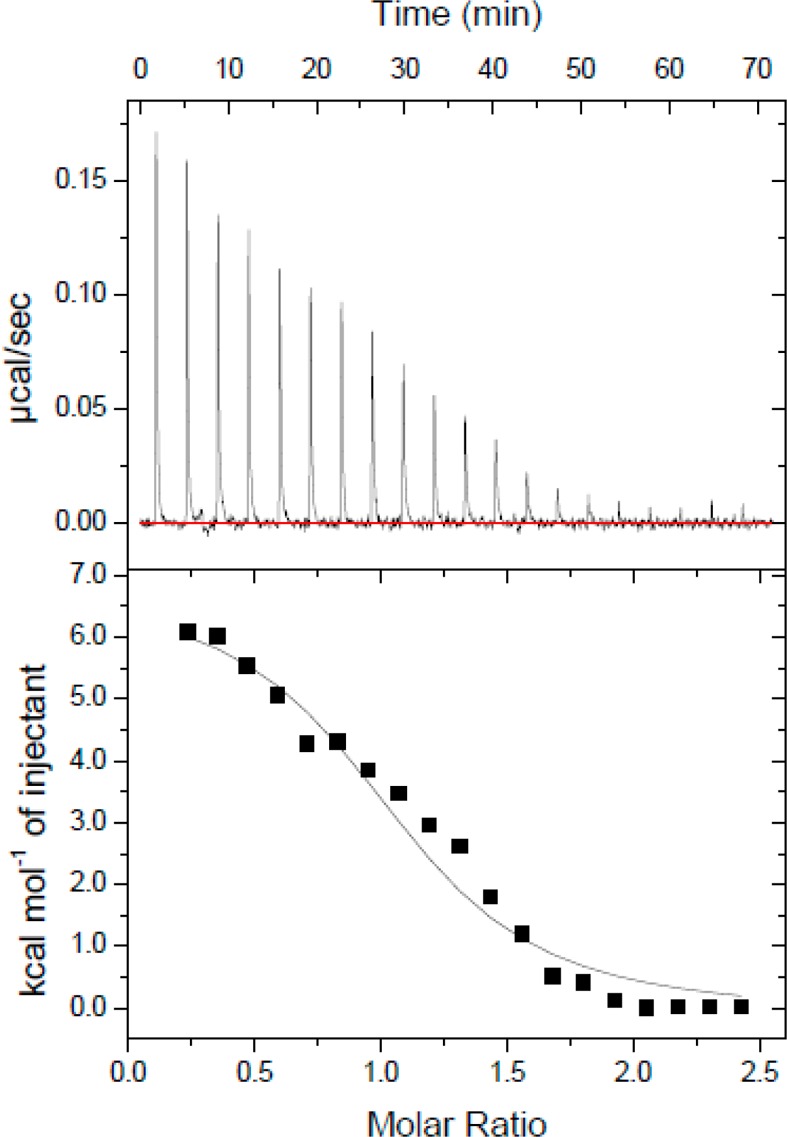
CaM–Fas DD V254N isotherm. Thermogram of 400 μM CaM titrated into the calorimeter cell containing 10 μM Fas DD V254N.
The Fas DD V254N mutation resulted in an association constant (Ka) for CaM–Fas DD binding that was decreased from (1.79 ± 0.20) × 106 to (0.88 ± 0.14) × 106 M–1 (Student’s t test; p < 0.05). The enthalpy (ΔH°) of CaM–Fas DD binding was increased from 6.00 ± 0.09 to 7.40 ± 0.27 kcal/mol (Student’s t test; p < 0.05). The standard state Gibbs free energy (ΔG°) for CaM–Fas DD binding was slightly increased from −8.87 ± 0.07 to −8.43 ± 0.10 kcal/mol, and the entropy (−TΔS°) for CaM–Fas DD binding was decreased from −14.87 ± 0.12 to −15.84 ± 0.28 kcal/mol (Student’s t test; p < 0.05).
Circular Dichroism Analysis of the Fas DD Secondary Structure
The V254N mutation of the Fas DD is analogous to the mutant allele of Fas in lpr-cg mice.6 The mutant Fas in lpr-cg mice may have an altered structure resulting in a reduced level of binding to Fas-associated death domain protein.8 To examine a possible structural basis for the decreased binding affinity of CaM for Fas DD V254N, we used far-UV CD to assess the secondary structure content of Fas DD WT and Fas DD V254N. Figure 4 shows the Fas CD spectra of Fas DD WT and Fas DD V254N. The far-UV CD spectrum of Fas DD WT shows a baseline signal at >245 nm and exhibits two negative peak minima at 222 and 208 nm and a positive CD signal trend toward 198 nm, which is consistent with a protein containing a majority α-helical content.44−46 The Fas DD V254N mutation is located in the CaM binding site in the Fas DD on the loop between helices α1 and α2.6 The V254N mutation could alter the proper formation of the structure of the turn or adjacent helices 2 and 3, resulting in a change in the α-helical secondary structure of Fas DD V254N. The far-UV CD spectrum of Fas DD V254N revealed the magnitude of the negative peak at 222 nm decreased by −2154 deg cm2 dmol–1, and there was a noticeable difference in the CD signal from 232 to 215 nm and from 205 to 198 nm for Fas DD V254N compared to that of Fas DD WT. The dynode voltage from far-UV CD spectra of Fas DD WT and Fas DD V254N shows that differences in Fas DD WT and Fas V254N CD spectra were not due to differences in Fas DD WT and Fas DD V254N concentrations (Figure S3 of the Supporting Information). The decrease in the magnitude of the negative peak at 222 nm and differences in the CD signal suggest a subtle decrease in α-helical content in the secondary structure of the Fas DD caused by the V254N mutation.47
Figure 4.
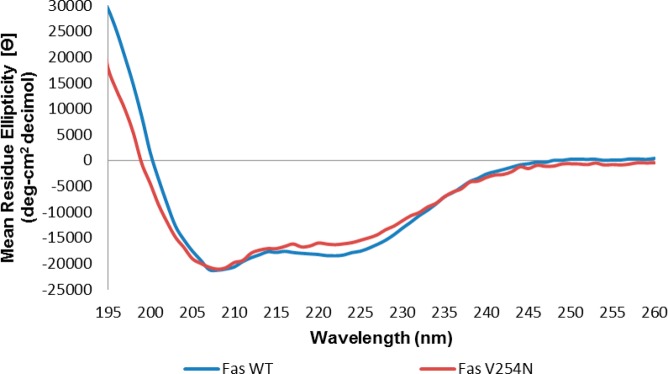
Far-UV CD spectra of Fas DD WT and Fas DD V254N at 37 °C and pH 5 (blue for Fas DD WT and red for Fas DD V254N).
Fas DD WT and Fas DD V254N secondary structure contents were estimated from their respective far-UV CD spectra using SELCON 3.48 SELCON 3 estimated Fas DD WT to have 62% α-helical, 0% β-sheet, 14% turn, and 24% random secondary structure content. SELCON 3 estimated Fas DD V254N to have 51% α-helical, 9% β-sheet, 21% turn, and 28% random secondary structure content. The Fas DD V254N mutation appears to produce a subtle decrease in α-helical content, but no significant changes in β-sheet, turn, or random secondary structure content estimated by SELCON 3 compared to Fas DD WT. The noticeable difference in the CD signal was not sufficiently significant to conclude that the Fas DD V254N mutation resulted in a significant change in the Fas DD secondary structure. The overall secondary structure of Fas DD V254N was similar to that of Fas DD WT; however, the orientation of secondary structure elements in the tertiary structure of the Fas DD could differ for Fas DD V254N compared to that of Fas DD WT, which could alter CaM–Fas DD interactions.
Conformational and Structural Changes in the Fas DD Caused by the Fas DD V254N Mutation from MD Simulations
We investigated the effect of the Fas DD V254N mutation on CaM–Fas DD binding from structural and conformational perspectives based on equilibrated MD simulation trajectories. The rmsds of Fas DD WT and Fas DD V254N over 80 ns MD simulations (Figure S4 of the Supporting Information) showed the systems reached equilibration after 40 ns. The last 40 ns MD simulation trajectories were used for the analyses of the structural and conformational changes of Fas DD WT and Fas DD V254N. We examined Fas DD secondary structural changes by analyzing the α-helix formation, evaluated the conformational stability of the Fas DD via rmsf analyses, and assessed the motion of the Fas DD with dynamical cross-correlation map analyses.
The structure of the Fas DD is primarily composed of six α-helices,8 and the CaM binding region is located within helices α1 and α2 of the Fas DD.6 To investigate the effect of the V254N mutation on the secondary structure change of Fas, α-helix formation analysis was performed and is shown in Figure 5. The helix percentages for Fas DD WT and Fas DD V254N were calculated from the overall helicity divided by the total number of Fas residues. The helix percentage was 41.3% for Fas DD WT and 40.0% for Fas DD V254N. The overall helicity of Fas DD WT was very similar to that of Fas DD V254N. The results were consistent with the observation from CD experiments that the overall secondary structure of Fas DD V254N is similar to that of Fas DD WT (Figure 4). The rmsf comparison of Fas DD WT and Fas DD V254N showed that the Fas DD V254N mutation resulted in the changes in the conformational flexibility of the Fas DD, including the conformational flexibility changes of the CaM binding site in the Fas DD (Figure 6).
Figure 5.
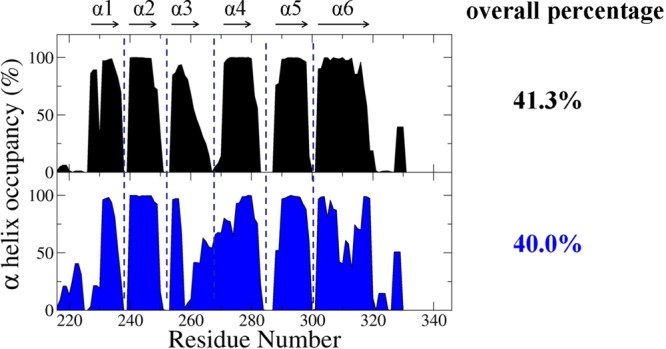
Residues of α-helix occupancy of Fas DD WT and Fas DD V254N. The overall percentage helix was calculated from the overall helicity divided by the total number of Fas residues (black for Fas DD WT and blue for Fas DD V254N).
Figure 6.
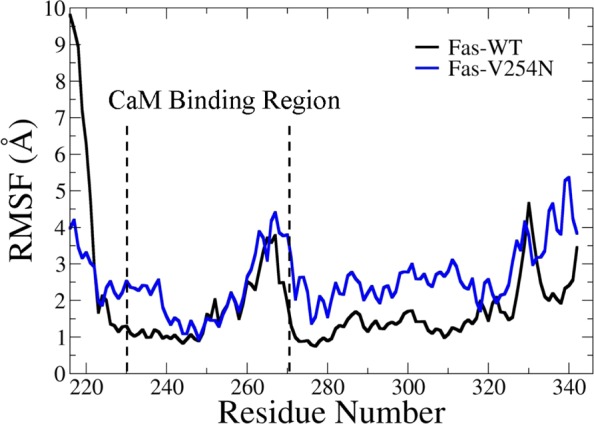
Root-mean-square fluctuation comparison of Fas DD WT and Fas DD V254N (black for Fas DD WT and blue for Fas DD V254N), calculated from the MD simulation trajectories after equilibration.
Dynamical cross-correlation maps that represent the degree of correlated motion between the residues in the protein provided further insight into the effect of the Fas DD V254N mutation on Fas DD conformational and structural changes (Figure 7). The bottom left half of Figure 7 is the cross-correlation map for Fas DD WT, while the top right half of Figure 7 is the cross-correlation map for Fas DD V254N. In Figure 7, the black boxes represent the changed regions of positive correlation, whereas the red boxes represent the changed regions of anticorrelation. Figure 7 shows that the V254N mutation of the Fas DD resulted in an increased degree of correlated motion (red color) between residues 227–240 (α1 and N-terminus of α2) and residues 276–316 (α4−α6) (region I) compared to those of Fas DD WT. The Fas DD V254N mutation also resulted in an increased degree of anticorrelated motion (blue color) between residues 227–240 (α1 and N-terminus of α2) and residues 320–340 (loop after α6) (region II), between residues 290–316 (α5 and α6) and residues 326–340 (loop after α6) (region III), between residues 256–270 (α3 and loop between α3 and α4) and residues 290–316 (α6) (region IV), and between residues 256–270 (α3 and loop between α3 and α4) and residues 220–240 (α1) (region V). In addition, Leu 242, Val 245, Gly 247, Phe 248, Val 249, Gly 253, and Val 254 of the Fas DD are seven nonpolar residues in the Fas DD binding region for CaM.6,8 The Fas DD V254N mutation changed the correlation motion patterns of the seven nonpolar residues relative to other residues (shown as the dotted boxes in Figure 7). The Fas DD V254N mutation increased the degree of correlated motion for these seven nonpolar residues in the CaM binding region relative to residues 290–316 (α5 and α6) (region VI) and the degree of anticorrelated motions relative to residues 256–280 (α3 and α4) (region VII). The degree of correlated motion change of these nonpolar residues relative to other residues in the Fas DD caused by the V254N mutation could cause the conformational change in the Fas DD binding region for CaM and further affect the van der Waals and electrostatic interactions between CaM and the Fas DD. In summary, the Fas DD V254N mutation resulted in a changed degree of correlated (red) and anticorrelated motion (blue) between residues in the Fas DD (boxed regions) and those in Fas DD WT, which could affect the binding of CaM to Fas DD.
Figure 7.
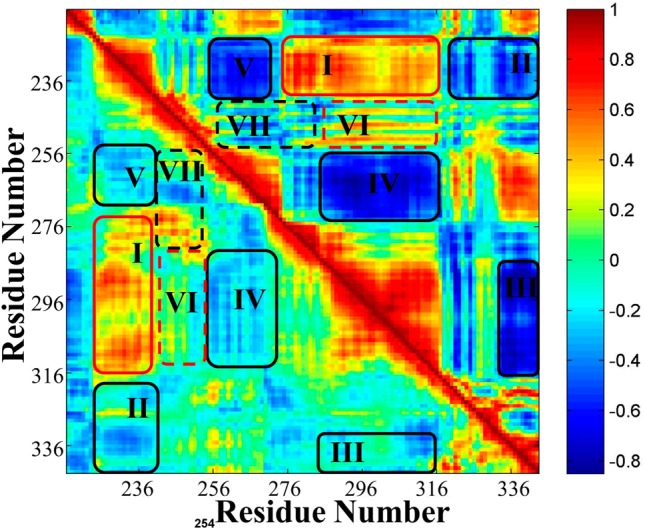
Dynamical cross-correlation maps for the comparison of the degree of correlated motion of the residues in Fas DD WT (bottom left) and Fas DD V254N (top right) (red for correlation between residues and blue for anticorrelation between residues). The black boxes represent the changed regions of positive correlation, whereas the red boxex represent the change regions of anticorrelation.
To investigate the potential effect of the Fas DD V254N mutation on the van der Waals and electrostatic interactions of the Fas DD with CaM, we analyzed hydrogen bond formation within the CaM binding region (Figure S5 of the Supporting Information), the contact numbers of each residue (Figure S6 of the Supporting Information), and the electrostatic potential for the CaM binding region (Figure S7 of the Supporting Information) for both the Fas DD and the Fas DD V254N mutant. The Fas DD V254N mutation resulted in changed hydrogen bond formation within the CaM binding region (Figure S5 of the Supporting Information) and the changed contact numbers of each residue, including the seven nonpolar residues in the binding site for CaM (Figure S6 of the Supporting Information). These changes could potentially affect van der Waals interactions between the Fas DD and CaM. The different electrostatic potential distribution for the CaM binding region in the Fas DD was observed between the Fas DD and the Fas DD V254N mutant (Figure S7 of the Supporting Information), which could affect the electrostatic interactions between the Fas DD and CaM.
Finally, the Fas DD V254N mutation did not significantly affect the Fas DD secondary structure as observed in CD experiments (Figure 4) and α-helix formation analyses (Figure 5). The altered conformational flexibility (Figure 6), the degree of correlated motion between residues in the Fas DD (Figure 7), hydrogen bond formation within the CaM binding region (Figure S5 of the Supporting Information), the contact number of each residue (Figure S6 of the Supporting Information), and the electrostatic potential for the CaM binding region (Figure S7 of the Supporting Information) caused by the Fas DD V254N mutation could influence the van der Waals interactions and electrostatic interactions between CaM and the Fas DD. Therefore, the Fas DD V254N mutation affected the CaM–Fas DD interactions and resulted in the decreased affinity of CaM for the Fas DD as observed in ITC experiments.
Conclusion
In this study, we characterize the interactions of CaM with Fas DD WT and Fas DD V254N using ITC, CD, and MD simulations. ITC results illustrate an endothermic binding characteristic and an entropy-driven interaction between CaM and the Fas DD or Fas DD V254N. ITC results showed that the Fas DD V254N mutation resulted in an association constant (Ka) for CaM–Fas DD binding that decreased from (1.79 ± 0.20) × 106 to (0.88 ± 0.14) × 106 M–1 and a standard state Gibbs free energy (ΔG°) for CaM–Fas DD binding that slightly increased from −8.87 ± 0.07 to −8.43 ± 0.10 kcal/mol. Both CD experiments and secondary structural analyses from MD simulations showed no significant secondary structural changes of the Fas DD by the V254N mutation. The conformational and dynamical motion analyses, hydrogen bond formation within the CaM binding region, the contact number of each residue, and the electrostatic potential for the CaM binding region based on MD simulations demonstrated the changes caused by the Fas DD V254N mutation. These changes caused by the Fas DD V254N mutation could affect the van der Waals interactions and electrostatic interactions between CaM and the Fas DD, thereby affecting CaM–Fas DD interactions and resulting in the decreased affinity of CaM for the Fas DD.
The results from the integrated computational and experimental studies quantitatively characterized CaM–Fas DD interactions and provided structural and thermodynamic evidence of the role of the V254N mutation of the Fas DD in altering CaM–Fas DD interaction. The results suggested that the biological effect of the Fas DD V254N mutation could be due to a change in the affinity of CaM for the Fas DD and the conformational changes in the Fas DD caused by the V254N mutation. Further, the results could help identify novel strategies for regulating CaM–Fas DD interactions and the Fas DD conformation to further modulate Fas-mediated DISC formation and apoptosis.
Acknowledgments
We acknowledge Dr. Donald Muccio of the Department of Chemistry for his assistance during CD experiments and CD data analysis, Dr. Christie G. Brouillette for her assistance during ITC experiments and ITC data analyses, and Dr. David Graves of the Department of Chemistry and Drs. Scott Wilson and David Sweatt of the Department of Neurobiology for allowing us access to their equipment for experiments. We also acknowledge the technical assistance received from Mr. Lei Ding, Mrs. Katherine Hayden, Mr. David McPherson, Dr. Michael Jablonsky, and Dr. Zhengrong Yang. Access to the Auto-ITC200 was provided by the Biocalorimetry Lab supported by National Institutes of Health Shared Instrumentation Grant 1S10RR026478 and the Shared Facility Program of the UAB Comprehensive Cancer Center (Grant 316851). Finally, we acknowledge the UAB VP Office for Equity and Diversity and ALSAMP Bridge to the Doctorate Fellowship Program.
Glossary
Abbreviations
- CaM
calmodulin
- Fas DD
Fas death domain
- DISC
death-inducing signaling complex
- WT
wild type
- CD
circular dichroism spectroscopy
- ITC
isothermal titration calorimetry
- MD
molecular dynamics
- V254N
substitution of valine 254 with asparagine.
Supporting Information Available
Structure of Ca2+-bound CaM and structure of the Fas DD (Figure S1), raw sedimentation velocity data for 10 μM Fas DD WT (Figure S2), a plot of dynode voltage from far-UV CD spectra of Fas DD WT and Fas DD V254N (Figure S3), rmsds for Fas DD WT, Fas DD V254N, and the Fas DD protein core (residues 225–318) over the 80 ns MD simulations (Figure S4), a comparison of the number of H-bonds formed between the CaM binding region in the Fas DD and the other part of the Fas DD (Figure S5), contact numbers for residues of the Fas DD (Figure S6), and an electrostatic potential map for the CaM binding site in Fas DD WT and Fas DD V254N (Figure S7). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This work was supported in part by a National Institutes of Health K25 Award (5K25CA140791) to Y.S. and a Veterans Administration Merit Award to J.M.M.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Wu X.; Ahn E.; McKenna M.; Yeo H.; McDonald J. (2005) Fas binding to calmodulin regulates apoptosis in osteoclasts. J. Biol. Chem. 280, 29964–29970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Xu J.; Jhala N.; Pawar P.; Zhu Z.; Ma L.; Byon C.; McDonald J. (2006) Fas-mediated apoptosis in cholangiocarcinoma cells is enhanced by 3,3′-diindolylmethane through inhibition of AKT signaling and FLICE-like inhibitory protein. Am. J. Pathol. 169, 1833–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kischkel F.; Hellbardt S.; Behrmann I.; Germer M.; Pawlita M.; Krammer P.; Peter M. (1995) Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 14, 5579–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hait W. N.; Lazo J. S. (1986) Calmodulin: A potential target for cancer chemotherapeutic agents. J. Clin. Oncol. 4, 994–1012. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Pawar P.; Pan G.; Ma L.; Liu H.; McDonald J. (2008) Calmodulin binding to the Fas-mediated death-inducing signaling complex in cholangiocarcinoma cells. J. Cell. Biochem. 103, 788–799. [DOI] [PubMed] [Google Scholar]
- Ahn E. Y.; Lim S. T.; Cook W. J.; McDonald J. M. (2004) Calmodulin binding to the Fas death domain. Regulation by Fas activation. J. Biol. Chem. 279, 5661–5666. [DOI] [PubMed] [Google Scholar]
- Babu Y. S.; Sack J. S.; Greenhough T. J.; Bugg C. E.; Means A. R.; Cook W. J. (1985) Three-dimensional structure of calmodulin. Nature 315, 37–40. [DOI] [PubMed] [Google Scholar]
- Huang B.; Eberstadt M.; Olejniczak E. T.; Meadows R. P.; Fesik S. W. (1996) NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature 384, 638–641. [DOI] [PubMed] [Google Scholar]
- Fisher G. H.; Rosenberg F. J.; Straus S. E.; Dale J. K.; Middelton L. A.; Lin A. Y.; Strober W.; Lenardo M. J.; Puck J. M. (1995) Dominant interfering fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell 81, 935–946. [DOI] [PubMed] [Google Scholar]
- Rieux-Laucat F.; Le Deist F.; Hivroz C.; Roberts I.; Debatin K.; Fischer A.; de Villartay J. (1995) Mutations in Fas associated with human lymphoproliferative syndrome and autoimmunity. Science 268, 1347–1349. [DOI] [PubMed] [Google Scholar]
- Bi L.; Pan G.; Atkinson T. P.; Zheng L.; Dale J.; Makris C.; Reddy V.; McDonald J.; Siegel R.; Puck J.; Lenardo M.; Straus S. (2007) Dominant inhibition of Fas ligand-mediated apoptosis due to a heterozygous mutation associated with autoimmune lymphoproliferative syndrome (ALPS) Type Ib. BMC Med. Genet. 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane M. M.; Ettenberg S. A.; Lowrey G. A.; Russell E. K.; Lipkowitz S. (1996) Fas expression and function in normal and malignant breast cell lines. Cancer Res. 56, 4791–4798. [PubMed] [Google Scholar]
- Maeda T.; Yamada Y.; Moriuchi R.; Sugahara K.; Tsuruda K.; Joh T.; Atogami S.; Tsukasaki K.; Tomonaga M.; Kamihira S. (1999) Fas gene mutation in the progression of adult T cell leukemia. J. Exp. Med. 189, 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G.; Vickers S.; Pickens A.; Phillips J.; Ying W.; Thompson J.; Siegal G.; McDonald J. (1999) Apoptosis and tumorigenesis in human cholangiocarcinoma cells. Involvement of Fas/APO-1 (CD95) and calmodulin. Am. J. Pathol. 155, 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H.; Shin M. S.; Lee H. S.; Bae J. H.; Lee H. K.; Kim H. S.; Kim S. Y.; Jang J. J.; Joo M.; Kang Y. K.; Park W. S.; Park J. Y.; Oh R. R.; Han S. Y.; Lee J. H.; Kim S. H.; Lee J. Y.; Yoo N. J. (2001) Expression of Fas and Fas-related molecules in human hepatocellular carcinoma. Hum. Pathol. 32, 250–256. [DOI] [PubMed] [Google Scholar]
- Takakuwa T.; Dong Z.; Takayama H.; Matsuzuka F.; Nagata S.; Aozasa K. (2001) Frequent mutations of Fas gene in thyroid lymphoma. Cancer Res. 61, 1382–1385. [PubMed] [Google Scholar]
- Adachi M.; Watanabe-Fukunaga R.; Nagata S. (1993) Aberrant transcription caused by the insertion of an early transposable element in an intron of the Fas antigen gene of lpr mice. Proc. Natl. Acad. Sci. U.S.A. 90, 1756–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suever J.; Chen Y.; McDonald J.; Song Y. (2008) Conformation and free energy analyses of the complex of calcium-bound calmodulin and the Fas death domain. Biophys. J. 95, 5913–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D.; Yan Q.; Chen Y.; McDonald J. M.; Song Y. (2011) Trifluoperazine regulation of calmodulin binding to Fas: A computational study. Proteins 79, 2543–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelhoch H. (1967) Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6, 1948–1954. [DOI] [PubMed] [Google Scholar]
- Pace C. N.; Vajdos F.; Fee L.; Grimsley G.; Gray T. (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. (1998) Sedimentation Analysis of Noninteracting and Self-Associating Solutes Using Numerical Solutions to the Lamm Equation. Biophys. J. 75, 1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laue T. M. (2001) Analytical Ultracentrifugation. In Current Protocols in Protein Science, John Wiley & Sons, Inc., New York. [DOI] [PubMed] [Google Scholar]
- Case D. A.; Cheatham T. E. III; Darden T.; Gohlke H.; Luo R.; Merz K. M. Jr.; Onufriev A.; Simmerling C.; Wang B.; Woods R. J. (2005) The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA. [Google Scholar]
- Yan Q.; Murphy-Ullrich J. E.; Song Y. H. (2010) Structural Insight into the Role of Thrombospondin-1 Binding to Calreticulin in Calreticulin-Induced Focal Adhesion Disassembly. Biochemistry 49, 3685–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D.; Song Y. H. (2010) Role of Altered Sialylation of the I-Like Domain of β1 Integrin in the Binding of Fibronectin to β1 Integrin: Thermodynamics and Conformational Analyses. Biophys. J. 99, 208–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suever J. D.; Chen Y.; McDonald J. M.; Song Y. (2008) Conformation and free energy analyses of the complex of calcium-bound calmodulin and the Fas death domain. Biophys. J. 95, 5913–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935. [Google Scholar]
- Ryckaert J.; Ciccotti G.; Berendsen H. J. C. (1977) Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341. [Google Scholar]
- Darden T.; York D.; Pedersen L. (1993) Particle Mesh Ewald: An N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 98, 10089–10092. [Google Scholar]
- Kabsch W.; Sander C. (1983) Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Li L.; Uversky V. N.; Dunker A. K.; Meroueh S. O. (2007) A computational investigation of allostery in the catabolite activator protein. J. Am. Chem. Soc. 129, 15668–15676. [DOI] [PubMed] [Google Scholar]
- MATLAB, MathWorks, Natick, MA: (2013). [Google Scholar]
- Baker N. A.; Sept D.; Joseph S.; Holst M. J.; McCammon J. A. (2001) Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. (1996) VMD: Visual molecular dynamics. J. Mol. Graphics 14, 27–38. [DOI] [PubMed] [Google Scholar]
- Demers J. P.; Mittermaier A. (2009) Binding mechanism of an SH3 domain studied by NMR and ITC. J. Am. Chem. Soc. 131, 4355–4367. [DOI] [PubMed] [Google Scholar]
- Tellinghuisen J. (2001) Statistical Error Propagation. J. Phys. Chem. A 105, 3917–3921. [Google Scholar]
- Kauzmann W. (1959) Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 14, 1–63. [DOI] [PubMed] [Google Scholar]
- Privalov P. L.; Gill S. J. (1988) Stability of protein structure and hydrophobic interaction. Adv. Protein Chem. 39, 191–234. [DOI] [PubMed] [Google Scholar]
- O’Neil K. T.; DeGrado W. F. (1990) How calmodulin binds its targets: Sequence independent recognition of amphiphilic α-helices. Trends Biochem. Sci. 15, 59–64. [DOI] [PubMed] [Google Scholar]
- Zhang M.; Tanaka T.; Ikura M. (1995) Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat. Struct. Biol. 2, 758–767. [DOI] [PubMed] [Google Scholar]
- Finn B. E.; Evenas J.; Drakenberg T.; Waltho J. P.; Thulin E.; Forsen S. (1995) Calcium-induced structural changes and domain autonomy in calmodulin. Nat. Struct. Biol. 2, 777–783. [DOI] [PubMed] [Google Scholar]
- Corrêa D. H. A.; Ramos C. H. I. (2009) The use of circular dichroism spectroscopy to study protein folding, form and function. Afr. J. Biochem. Res. 3, 164–173. [Google Scholar]
- Chang C. T.; Wu C. S.; Yang J. T. (1978) Circular dichroic analysis of protein conformation: Inclusion of the β-turns. Anal. Biochem. 91, 13–31. [DOI] [PubMed] [Google Scholar]
- Myer Y. P. (1968) Conformation of cytochromes. II. Comparative study of circular dichroism spectra, optical rotatory dispersion, and absorption spectra of horse heart cytochrome c. J. Biol. Chem. 243, 2115–2122. [PubMed] [Google Scholar]
- Sreerama N.; Woody R. W. (2004) Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 383, 318–351. [DOI] [PubMed] [Google Scholar]
- Sreerama N.; Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


