Abstract

Differential interactions between influenza A virus protein hemagglutinin (HA) and α2→3 (avian) or α2→6 (human) sialylated glycan receptors play an important role in governing host specificity and adaptation of the virus. Previous analysis of HA–glycan interactions with trisaccharides showed that, in addition to the terminal sialic acid linkage, the conformation and topology of the glycans, while they are bound to HA, are key factors in regulating these interactions. Here, the solution conformation and dynamics of two representative avian and human glycan pentasaccharide receptors [LSTa, Neu5Ac-α(2→3)-Gal-β(1→3)-GlcNAc-β(1→3)-Gal-β(1→4)-Glc; LSTc, (Neu5Ac-α(2→6)-Gal-β(1→4)-GlcNAc-β(1→3)-Gal-β(1→4)-Glc] have been explored using nuclear magnetic resonance and molecular dynamics simulation. Analyses demonstrate that, in solution, human and avian receptors sample distinct conformations, topologies, and dynamics. These unique features of avian and human receptors in solution could represent distinct molecular characteristics for recognition by HA, thereby providing the HA–glycan interaction specificity in influenza.
The interaction between the influenza A virus surface protein hemagglutinin (HA) and its cell surface glycan receptors is one of the critical steps governing virus selectivity. Several crystal structures of different HAs have been determined, both native and cocrystallized with glycan receptors.1–5 These studies have provided insight into the ability of HA to interact with receptor glycans. Human-adapted influenza viruses preferentially bind to sialylated glycan receptors terminated by N-acetylneuraminic acid (Neu5Ac) α2→6 linked to galactose (Gal) (human receptors), which are predominantly expressed on the apical surface of the upper respiratory epithelia in humans.6–8 In contrast, avian-adapted influenza viruses preferentially bind to sialylated glycans terminated by Neu5Ac α2→3 linked to Gal, which are expressed predominantly in the gut and respiratory tract of birds and, importantly, in the deep lung (alveolar region) of humans.7 Therefore, one of the characteristics of human adaptation of influenza A viruses is the change in glycan receptor binding preference of HA from avian to human receptors.9–14
Recent studies based on analysis of HA–glycan cocrystal structures15,16 have highlighted the importance of glycan conformation and topology in governing the receptor specificity of avian- and human-adapted HAs. In the case of avian receptors, the majority of contacts between HA and glycan involve the Neu5Ac residue at the nonreducing end. Chandrasekaran et al.16 proposed that, when the Neu5Ac is fixed in the glycan-binding pocket of HA, the other residues in this trisaccharide motif sample different torsional angles at their glycosidic linkages, such that the space occupied by the whole glycan resembles a cone with Neu5Ac at the vertex, characterized by a topological angle θ of >110°. The topological angle θ is defined by the C2, C1, and C1 atoms of the residues Neu5Ac, Gal, and GlcNAc (N-acetylglucosamine), respectively, in going from the nonreducing end to the reducing end. They also proposed for the case of human receptors that a longer glycan motif, comprising at least a tetrasaccharide Neu5Ac-α(2→6)-Gal-β(1→4)-GlcNAc-β(1→3)-Gal- at the nonreducing end, spans the glycan binding surface on HA. The additional C5–C6 bond of the Neu5Ac-α(2→6)-Gal glycosidic linkage provides added mobility to its glycosidic junction and the remaining backbone, allowing it to explore conformations similar to an umbrella progressing from a fully open form to a fully closed form (defined as having a θ of <110°).16 It is clear that the cone and umbrella topologies are generated by their motions while they are bound to the receptor, but when in their free states, they adopt conformations that are dependent on their respective nonreducing end connectivity, α(2→3) or α(2→6). The role of glycan shape and topology in regulating binding specificity for HA has also been investigated using molecular dynamics simulations,17 indicating that there are distinct differences in the conformational sampling of avian and human receptors from the different subtypes (H3, H5, and H9) upon binding to HA.
The previously mentioned studies highlighted differences in glycan receptor conformation and structural details when the receptor cocrystallized bound to HA, illustrating the structural specificity of HA–glycan interactions. Consequently, it is reasonable to suppose that receptor dynamic properties, strictly related to its structure, could play a role in the recognition and binding by the HA protein. To characterize human and avian sialylated glycan receptors in terms of conformation and topology in solution, detailed nuclear magnetic resonance (NMR) analyses, MD simulations, and subsequent numerical analysis of the data generated were performed on the two sialylated pentasaccharides: LSTc (Neu5Ac-α(2→6)-Gal-β(1→4)-GlcNAc-β(1→3)-Gal-β(1→4)-Glc), representing the human receptor, and LSTa (Neu5Ac-α(2→3)-Gal-β(1→3)-GlcNAc-β(1→3)-Gal-β(1→4)-Glc), representing the avian receptor (Figure 1 of the Supporting Information). We refer to Gal1 and Gal2 as labels for the first and second Gal residues encountered in the sequence of LSTc and LSTa, respectively, from the nonreducing to the reducing end. NOESY, 13C NMR relaxation measurements, and analysis of the glycan MD trajectories revealed that LSTa and LSTc have distinct conformational dynamics, topologies, and intramolecular hydrogen bond networks in solution. Specifically, LSTc preferentially adopts a bent shape in solution, which is consistent with the “umbrella”-like conformation, whereas LSTa adopts a bow shape in solution, which agrees with a “cone”-like structure. These observations suggest that the different species-adapted HAs could recognize specific shapes, conformations, and dynamic behaviors of glycan in solution.
EXPERIMENTAL PROCEDURES
NMR Chemical Shift Mapping
NMR measurements were taken using Bruker 500, 600, and 900 MHz AVANCE series NMR spectrometers (Bruker GmbH, Silberstreifen, Germany) with a high-sensitivity 5 mm TCI cryoprobe. Human milk lactosyl sialyl-N-tetraoses, namely, LSTa (PROzyme, Hayward, CA) [Neu5Ac-α(2→3)-Gal(Gal1)-β(1→3)-GlcNAc-β(1→3)-Gal(Gal2)-β(1→4)-Glc)] and LSTc (PROzyme) [Neu5Ac-α(2→6)-Gal(Gal1)-β(1→4)-GlcNAc-β(1→3)-Gal(Gal2)-β(1→4)-Glc], were dissolved in D2O (99.9%) or a buffered solution [150 mM sodium chloride, 100 mM sodium phosphate, and 10% D2O (pH 7.2)] according to the NMR experiment. Chemical shifts (1H and 13C) of the oligosaccharides were measured at 295 K in D2O (99.9%, 2 mg/0.2 mL) using a 3 mm NMR tube. Two-dimensional (2D) NMR experiments were conducted using edited HSQC, HSQC-TOCSY, HMBC, TOCSY, and NOESY (using different mixing times between 100 and 1000 ms), recorded for quadrature detection in the indirect dimension and acquired using 16–48 scans per series of 1024 × 320 data points, with zero filling in F1 (4096) prior to Fourier transformation. Figures of spectra were created using R18 and the libraries rNMR19 and lattice.20
Estimation of Intra- and Interproton Distances
Experimental distances were obtained by selective one-dimensional (1D) and 2D NOESY experiments conducted with mixing times between 100 and 800 ms (295 K) and used to construct the kinetic NOEs. Selected distances were estimated on the basis of the (two-spin model approximation) time-dependent NOE measurements extrapolated to short mixing times, considering the initial buildup enhancement up to 300 ms.21 The extrapolated NOEs signals were converted to proton distances considering the Neu5Ac distance (H3ax–H3eq) as a reference (1.78 Å).
13C NMR Relaxation Data
NMR relaxation times were obtained at 295 K using a 14.0 T magnetic field. 13C longitudinal relaxation (T1) and transverse relaxation (T2) were measured via 2D double INEPT-based inverse detection methods optimized to suppress the cross correlation between the chemical shift anisotropy (CSA) and the dipolar relaxation.22–24 The 2D spectra were recorded using 1024 × 192 data points, recording 24–32 scans at eight different relaxation delays arranged randomly to avoid systematic errors in every experiment. T1 relaxation delays were 0, 0.1, 0.2, 0.3, 0.5, 0.8, 1.0, and 1.4 s. T2 transverse relaxation times were obtained by the CPMG pulse sequences differing in spin echo delays (22.4, 44.8, 67.2, 89.6, 134.4, 179.2, 224.0, and 313.6 ms). The volumes of the cross-peaks were integrated using Topspin 2.1, and relaxation times were obtained by best nonlinear and linear fitting of normalized data of the evolution of the peak volumes as a function of the inversion recovery delays. All the values and estimated errors (2–10%) were calculated and plotted in Origin 8.0 (OriginLab Corp., Northampton, MA).
Determination of the nJC–H Heteronuclear Coupling Constants
Long-range JC–H couplings were measured using the J-HMBC pulse sequence (295 K), with a delta delay corresponding to a nJX–H of 2 Hz [LSTc (0.7 mg) and LSTa (1 mg) were dissolved in 220 μL of D2O]. Data matrices of 1024 × 320 points covering 2003.2 × 9054.9 Hz were expanded to 2048 × 1024 points using linear prediction and subsequently zero-filled to 8192 × 2048 points prior to echo/antiecho Fourier transformation. The traditional sequence was applied for LSTa, while the non uniform sampling (NUS) variant was used for LSTc (Bruker Topspin version 3.1).
Determination of Temperature Coefficients and Chemical Shift Deviations
Temperature coefficients were recorded for 0.5 mg/0.2 mL oligosaccharide solutions in buffered D2O or in a 90:10 H2O/D2O mixture at pH 7.2. All temperature gradients were measured by differences observed in spectra recorded at 3–9 K intervals between 278 and 313 K (1H). Temperature coefficients are expressed in units of parts per billion per kelvin, with a negative sign indicating an upfield shift upon warming; chemical shifts were measured using 1H and HSQC experiments.
Molecular Dynamics Simulations
The starting geometries of both oligosaccharides LSTa and LSTc were generated using the carbohydrate biomolecule builder;25 following the primary structure and the glycosidic constraints, both structures were minimized by the online energy minimizer.25 The energy description of both oligosaccharides included the amber/glycam06 force field.25,26 The atomic partial charges required by the oligosaccharide force field were calculated using the semiempirical bond-corrected AM1-BCC scheme.27 The MD simulations were conducted in explicit solvent using the TIP3P water model.28 Oligosaccharide partial charge management and topology files were constructed with AmberTools version 1.4.29 The MD trajectories were generated using the NAMD MD engine.30 Nonbonded interactions, i.e., electrostatic and van der Waals, are described using the cutoff approach set to 12.0 Å with a switching function beginning at 6.0 Å, and the time step for the integration algorithm is 1 fs. The simulation cell, including the oligosaccharide, was built enveloping the solute molecule in a layer of water 20.0 Å wide in each direction; the result is a cubic cell of approximately 60 Å × 67 Å × 59 Å. The oligosaccharide models that were compared to the experimental data were generated by the following procedure; the simulation cell was first optimized applying 10000 minimization steps of the NAMD minimizer algorithm, restraining each atom of the carbohydrate to the initial conformation by a harmonic restraint with a scaling constant of 500 kcal/mol. The next step was a MD simulation reproducing the constant-temperature and pressure statistical ensemble (NPT) applying a pressure on the cell walls of 1.01325 bar (atmospheric pressure). The simulation temperature was set at 295 K and was controlled by a Langevin thermostat as implemented in NAMD. The Nosé-Hoover Langevin piston algorithm controlled the pressure applied on the cell walls, with periodic boundary conditions being applied. The whole length of the simulation was 104 ns, where the first 4 ns was conducted restraining the carbohydrate atoms (solute) by a harmonic potential with a scaling constant of 50 kcal/mol to initially equilibrate the cell density without changing the solute conformation significantly, while the remaining simulation time was undertaken with the solute free to move. The cell densities calculated for LSTa and LSTc after equilibration (~4 ns) were in both cases 1.04 g/cm3. The MD trajectory consisted of the last 100 ns, with a single geometry being sampled every 10 ps. During the whole dynamic, the conformation of the residues remained in 2C5 for Neu5Ac and 4C1 for the others, as set from the beginning for both LSTa and LSTc, and in agreement with the three-bond proton–proton coupling constants (3JH–H).
The progression of the MD simulation was observed by determining the root-mean-square displacement (rmsd) of the trajectories, performed in R using the rmsd function from the Bio3D library,31 with the starting conformation as the reference. The rmsd distances were determined using eq 1:
| (1) |
where Drefi is the position of the ith atom of the reference structure (t = 0), Di(t) is the position of atom i at time t, and n is the number of atoms.
Glycosidic Torsion Angles
The torsional angles (ϕ and ψ) are defined in this paper by the following pairs of dihedral angles, from the nonreducing termini: ϕ1 and ψ1, ϕ2 and ψ2, ϕ3 and ψ3, and ϕ4 and ψ4. For LSTa, ϕ1 is the C1–C2–O3–C3 angle and ψ1 is the C2–O3–C3–H3 angle, while for LSTc, ϕ1 is the C1–C2–O6–C6 angle and ψ1 is the C2–O6–C6–C5 angle, in agreement with Xu et al.17 The successive pairs are defined as ϕi and ψi [H1–C1–O4′–C4′ and C1–O4′–C4′–H4′ (1→4 linkage) or H1–C1–O3′–C3′ and C1–O3′–C3′–H3′ (1→3 linkage)], as conventionally defined for sugars. Atoms labeled with a prime belong to the monosaccharide on the reducing side of the glycosidic linkage, while atoms without a prime are on the nonreducing side of the glycosidic linkage. To be consistent with the work of Chandrasekaran et al.16 and Xu et al.,17ϕ and ψ were illustrated in the range of –120° to 240°.
Topological θ Angle
The θ angle is defined by the C2, C1, and C1 atoms of residues Neu5Ac, Gal, and GlcNAc (N-acetylglucosamine), respectively, going from the nonreducing end to the reducing end
Comparison of the MD Results to the Experimental Data
The rmsdexp values (eq 2) were used to compare the MD trajectories with the experimental data, i.e., the estimated inter-residue proton–proton distances for both LSTc and LSTa using the two-spin model approximation, reported for LSTc (H5 Neu5Ac–CH3 GlcNAc, H1 GlcNAc–H3 Gal2, H1 GlcNAc–H4 Gal2, H1 Gal2–H4 Glc, and H1 Gal1–H4 GlcNAc) and LSTa (H3ax Neu5Ac–H3 Gal1, H3eq Neu5Ac–H3 Gal1, H1 Gal1–H3 GlcNAc, H1 Gal1–CH3 GlcNac, H1 GlcNAc–H3 Gal2, H1 GlcNAc–H4 Gal2, H1 Gal2–H4 Glc, and H1 Gal2–H5 Glc). In eq 2, Dexp,i is the experimental ith inter-residue proton–proton distance and DMD,i is the corresponding distance for a single MD-generated conformation. The number of the available inter-residue proton–proton distances is n.
| (2) |
2D NOESY Spectral Simulation (multispin model relaxation)
The 2D NOESY spectra were simulated using two different sets of 40 conformations each from the MD trajectory: 40 with the smallest rmsd (rmsdexp min) and highest (rmsdexp max). The 2D NOESY simulation was undertaken assuming isotropic motion with a single correlation time τc estimated to be 400 ps. The τc estimation was conducted on the smallest rmsd set of conformations (rmsdexp min) and simulating the intraresidue H1–H3 and H1–H5 2D NOE signals for the selected residues: GlcNAc for LSTc and Gal1, GlcNAc, and Gal2 for LSTa. The τc parameter was adjusted by trial and error until the NOE kinetic enhancement from 0 to 500 ms mixing time reproduced qualitatively the corresponding experimental values. The software for 2D NOESY simulation was NOEPROM.32
Principal Component Analysis of the MD Trajectory
The MD trajectories for LSTc and LSTa were exported as XYZ coordinates via VMD,33 allowing the data to be imported directly into R.18 Principal component analysis (PCA) was performed using the bio3d library, the pca.xyz function.31 The data were referred to the first conformer of the MD trajectory, removing translational and rotational effects from the analysis, and then mean centered before PCA was performed. The calculated eigenvalues (scree plot) provide a weighting for the importance of the eigenvectors (also known as loadings) (illustrated as porcupine plots), and the component scores (score plot) illustrate the relationship between the determined eigenvector (independent molecular motion) and the conformers that constitute the MD trajectory. Porcupine plots were produced by constructing a trajectory along the desired principal component, imposing the molecular motion extracted by that component onto the average geometry of the conformers.31 This was then visualized using VMD and the porcupine plot created using a VMD script.33,34
The convergence of the glycan PCA was observed by determining the root-mean-square internal product (rmsip) of the first 10 eigenvectors. If rmsip is equal to 1, then there is complete convergence between the sets of eigenvectors; a value of ≥0.70 shows that the eigenvectors are consistent with each other.35 In this circumstance, rmsip was used to see if the MD simulation was tending toward a stationary state. PCA was performed on the trajectory at different times, and this was compared to the eigenvectors derived for the trajectory at 100 ns. The rmsip was determined using R and the library Bio3d;31 the rmsip function was utilized. The rmsip is defined as
| (3) |
where ηi is the ith eigenvector of trajectory η and νi is the ith eigenvector of trajectory ν.
RESULTS
NMR Analyses of LSTa and LSTc
NMR characterization of sialyl-lacto-N-tetraoses LSTa and LSTc (Figure 1 of the Supporting Information) were conducted using homonuclear (COSY, TOCSY, and NOESY), heteronuclear (edited HSQC and HSQC-TOCSY), and heteronuclear multiple-bond correlation HMBC (1H/13C) experiments. This permitted the complete assignment of the hydrogen and carbon resonances for both LSTc and LSTa (Figure 1 and Table 1 of the Supporting Information). TOCSY analysis allowed the connectivity of the pyranosyl rings to be observed, with H–H axial correlations confirming the monosaccharide identities. The chemical shifts of the acetamide methyl groups of GlcNAc and Neu5Ac for both LSTa and LSTc were determined by 2D TOCSY (buffered system, 10% D2O), providing NH correlations with some of the hydrogens of each ring and with those from acetamide (Figure 2 of the Supporting Information). However, the strong peak superimposition did not permit the complete hydrogen assignment by homonuclear experiments; these were complemented by HSQC-TOCSY and by the COSY effect generated by homonuclear coupling in sensitivity-enhanced HSQC. These correlation peaks, usually considered as artifacts, are very useful in the assignment of signals in crowded spin systems when performing sensitivity-enhanced HSQC experiments36 (Figure 3 of the Supporting Information). Long-range correlations (1H/13C) of the quaternary C1 and C2 atoms from the sialyl residues were observed between the H3ax Neu5Ac atom at 1.782 ppm and the corresponding carbon signals at 176.7 and 102.3 ppm (C1 and C2, respectively). C2 correlated with H3 Gal1 at 4.082 and 78.2 ppm of LSTa, confirming the presence of the Neu5Ac α2→3 Gal linkage. Instead, LSTc correlated H3ax Neu5Ac at 1.711 ppm with carbon signals at 176.2 (C1) and 102.7 (C2) ppm, with the latter correlating with H6α/β Gal1 at 3.983α/3.544β/66.0 ppm, confirming that C2 Neu5Ac is O-linked to H6α/β Gal1. Anomeric proton correlations from LSTa were observed at 4.513/84.7 ppm (H1 Gal1/C3 GlcNAc), 4.739/84.5 ppm (H1 GlcNAc/C3 Gal2), and 4.446/81 ppm (H1 Gal2/C4 Glc). LSTc exhibited cross-peaks at 4.451/82.9 ppm (H1 Gal1/C4 GlcNAc), 4.73/84.6 ppm (H1 GlcNAc/C3 Gal2), and 4.445/80.9 ppm (H1 Gal2/C4 Glc). These measurements were confirmed by 1D selective and 2D NOESY experiments.
Three-bond proton–proton coupling constants, 3JH–H, were measured for both pentasaccharides, which indicated that all Hexp residues assumed the 4C1 conformation, because 3JH1–H2 values vary from 7.9 to 8.2 Hz and 3JH3–H4 values for Gal residues are ~3.5 Hz; Glc and GlcNAc had values of 9.0 and 9.8 Hz, respectively. The Neu5Ac residue had 3JH–H values compatible with those of a pyranosyl ring in the 2C5 conformation [3JH3–H4 = 4.8(eq) Hz/12.1(ax) Hz, and 3JH4–H5 = 10.1 Hz], thus suggesting no significant differences in the ring conformation between the pentasaccharides.
As in proteins, NH temperature coefficients can reflect the extent to which N-acetyl-amide protons are protected from exchange through hydrogen bonding. Evaluation of NH temperature coefficients showed different behavior between GlcNAc and Neu5Ac. The first had a value of –8.28 ppb/K for LSTc and –8.00 ppb/K for LSTa, while the NH group for Neu5Ac exhibited lower temperature coefficients with values of –6.77 and –6.60 ppb/K for LSTc and LSTa, respectively. This behavior suggested that the amide group from GlcNAc could be involved in an intramolecular hydrogen bond. This result is supported by a study of the half-height line widths (Δν1/2) extracted from each NH acetamide after a Lorentzian deconvolution and plotted at different temperatures. The NH data from the Neu5Ac N-acetyl group broadened before those of GlcNAc, suggesting that the NH group of GlcNAc is involved in intramolecular hydrogen bonds (Figure 4 of the Supporting Information).
13C NMR Relaxation Data
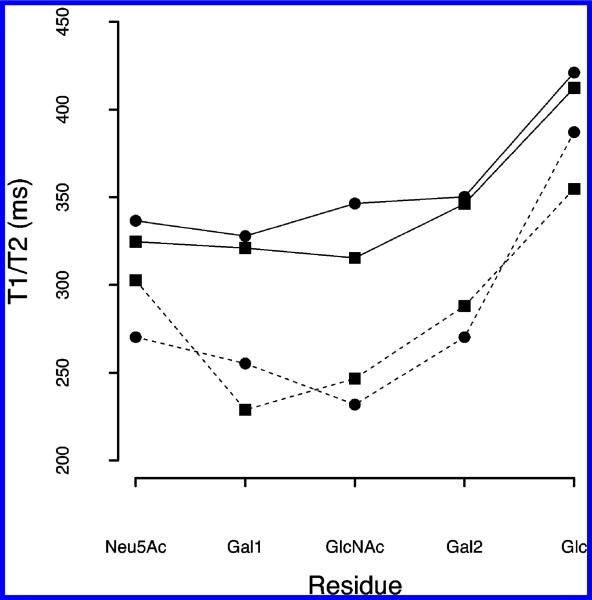
NMR relaxation studies have been utilized to interrogate the internal motion of the glycans, providing dynamic properties that can be qualitatively or quantitatively assessed.23,24,37,38T1 and T2 relaxation times and heteronuclear NOEs were measured at 14.0 T (Table 2 of the Supporting Information). T1 varied from 315 to 412 ms for LSTa and from 328 to 421 ms for LSTc. T2 varied from 229 to 355 ms and from 232 to 387 ms for LSTa and LSTc, respectively. To qualitatively observe the main differences in the motional properties between LSTc and LSTa, the average values of T1 and T2 of each CH ring were used (Figure 2). The numerical values of T1 for the different residues suggest an anisotropically tumbling molecule, because the Neu5Ac-Gal1-GlcNAc trisaccharide of both glycans gave rise to T1 values ranging from 315 to 346 ms, whereas values of the reducing Glc residue varied from 410 to 421 ms. This finding indicates a higher degree of motion for the reducing lactosyl disaccharide than for the nonreducing end of the oligosaccharide. The heteronuclear NOEs corroborate the T1 values, because larger heteronuclear NOEs are observed for the terminal units. Using the same approach, significantly lower T2 values were detected for both LSTa and LSTc, T2 values of the reducing Glc residue of both glycans (355 and 387 ms for LSTc and LSTa, respectively) confirmed the higher degree of motion of this residue than of the internal Gal1, and GlcNAc residues showed lower T2 values between 228 and 255 ms for both glycans. The main differences between LSTa and LSTc were the values observed for T2 of the Neu5Ac residues, being 303 and 270 ms, respectively; such a difference, larger than the experimental error, accounts for the significantly higher rigidity of the nonreducing end of LSTc. Contrary to that of the nonreducing side, the higher mobility of the reducing end residues has little biological relevance, because this part of the molecule in vivo is part of a greater structure.
Figure 2.
Average 13C relaxation times of CH group carbons for each monosaccharide unit from LSTc (●) and LSTa (■). The solid line represents the average T1 of each monosaccharide, while the dashed line represents the average T2 of each monosaccharide.
Inter- and Intra-NOESY Experimental Distances
The inter- and intraresidue proton–proton distances were measured by NOESY enhancement (see Experimental Procedures for details). Intraresidue distances (H1–H3 of Hexp units and H3–H5 of the Neu5Ac residues) were measured between 2.0 and 2.6 Å for all the residues (Table 3 of the Supporting Information). These values were in agreement with the expected 4C1 and 2C5 ring chair conformations, supporting the observed 3JH–H coupling constants. Inter-residue proton–proton distances were obtained mainly from O-glycosidic bonds (Table 1). LSTa exhibited NOE H3eq and H3ax Neu–H3 Gal1 contacts, providing distances of 4.7 and 2.8 Å, respectively, while LSTc exhibited an inter-residue NOE between H5 Neu5Ac and CH3 GlcNAc corresponding to an estimated distance of 4.2 Å (Figure 5 of the Supporting Information). This contact between GlcNAc and Neu5Ac residues, observed only in the NOESY spectra of LSTc, can be considered the experimental proof of a bent conformation that LSTc assumes compared to LSTa, which does not exhibit this signal at all.
Table 1.
Selected Interglycosidic Proton–Proton Distances (angstroms) for LSTa and LSTc Derived from NOE Measurements
| LSTc (Å) | LSTa (Å) | |
|---|---|---|
| H3ax Neu5Ac–H3 Gal1 | – | 2.76 |
| H3eq Neu5Ac–H3 Gal1 | – | 4.70 |
| H5 Neu5Ac–CH3 GlcNAc | 4.24 | – |
| H1 Gal1–H3 GlcNAc | – | 2.25 |
| H1 Gal1–CH3 GlcNAc | – | 4.00 |
| H1 GlcNAc–H3 Gal2 | 1.94 | 1.95 |
| H1 GlcNAc–H4 Gal2 | 3.05 | 2.84 |
| H1 Gal2–H4 Glc | 1.96 | 2.20 |
| H1 Gal2–H5 Glc | – | 2.80 |
Molecular Dynamics Simulations of LSTa and LSTc
To complement the NMR experiments, MD simulations were performed to compare the dynamics of the glycans in solution. The MD simulations were allowed to run for 100 ns, sampling conformers every 0.01 ns, therefore producing a comprehensive data set, with the simulation length considerably greater than the estimated correlation time (isotropic motion) of either glycan, of ~400 ps. After 70% of the simulation time had elapsed, several molecular parameters exhibited stationary behavior without significant drift; these include the end-to-end distance of the sugars, which is defined as the distance between C5 of Neu5Ac (nonreducing end) and C1 of Glc (reducing end). This property is one of the slowest-evolving degrees of freedom for an oligomer (Figures 7 and 8 of the Supporting Information). Stationary behavior is also observed for the θ angle of the glycans, which is characteristic for each glycan (Figures 9 and 10 of the Supporting Information) and the ω torsional angle of Gal1, which is defined as the H5–C5–C6–O6 angle (Figure 11 of the Supporting Information). Similar behavior was seen over the same time scale for the other four pairs of glycosidic torsional angles (ϕi and ψi) (see Experimental Procedures), the angles that define the backbone conformation of the two oligomers.
During the MD simulation, the residues comprising LSTc and LSTa adopted the 2C5 (Neu5Ac) and 4C1 (Gal, Glc, and GlcNAc) conformations, respectively, with only small deviations from the ideal chair of cyclohexane; the intraresidue proton–proton distances (H1–H3, H1–H5, and H3–H5) have values ranging between 2.5 and 2.7 Å for the five sugars, compared to the ideal value of 2.64 Å for cyclohexane. The same distances are estimated by NOE enhancement using the two-spin approximation (see Experimental Procedures); the values obtained ranged between 2.0 and 2.6 Å, in good agreement considering the strong coupling effects (Table 3 of the Supporting Information).
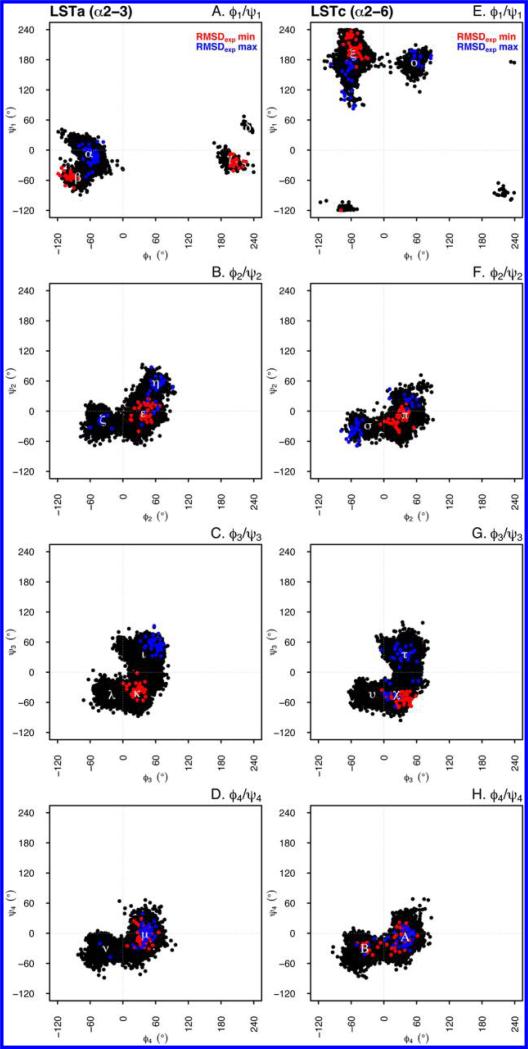
The intersugar unit conformations were defined using four pairs of inter-residue glycosidic dihedral angles, running from the nonreducing end to the reducing end of the glycans (ϕi and ψi) (see Experimental Procedures). The torsional angle values explored during the whole MD simulation are reported in Figure 3, and the torsional angles are displayed in the range of –120 to 240°. Visited torsional states are reported as cluster centroids with their relative populations in Table 2. The principal difference between LSTa and LSTc is located at the first glycosidic linkage (ϕ1 and ψ1), which is in agreement with those obtained by molecular modeling,39 and is related to the different connectivity at the nonreducing end [Neu5Ac-α(2→3)-Gal for LSTa and Neu5Ac-α(2→6)-Gal for LSTc]. For LSTa, four ϕ1 and ψ1 torsional states are observed, a principal state is located at –62° and –8° (89.8%), respectively, while the most significant member of the minor states is located at –83° and –55° (7.7%), respectively (Figure 3A, α and β, respectively). LSTc possesses two ϕ1 and ψ1 torsional states; the cluster with the greater population (96.2%) is located at –57° and 190°, respectively, and the minor population at 56° and 174°, respectively (Figure 3E, ξ and ο, respectively).
Figure 3.
Torsional states visited during the 100 ns simulation time for LSTa (left) and LSTc (right). For each glycan, the dihedral angle pairs [ϕ1 and ψ1 (A and E), ϕ2 and ψ2 (B and F), ϕ3 and ψ3 (C and G), and ϕ4 and ψ4 (D and H)] are shown. The red dots represent conformers that better fit the experimental NMR NOESY data (rmsdexp min set of conformations), while blue dots correspond to the conformers that exhibit poor agreement with the experimental constraints (rmsdexp max). The positions and populations of the torsional states are listed in Table 2.
Table 2.
Torsional States of LSTa and LSTc as Determined by Molecular Dynamics
| linkage | ϕ (deg) | ψ (deg) | no. | % | |
|---|---|---|---|---|---|
| LSTa (α2→3) Torsional angles | |||||
| 1 | α | –62 | –8 | 8982 | 89.8 |
| β | –83 | –55 | 770 | 7.7 | |
| γ | 196 | –19 | 227 | 2.3 | |
| δ | 230 | 47 | 21 | 0.2 | |
| 2 | ε | 37 | –5 | 6908 | 69.1 |
| ζ | –37 | –18 | 2402 | 24.0 | |
| η | 60 | 54 | 690 | 6.9 | |
| 3 | ι | 38 | 35 | 5534 | 55.3 |
| κ | 26 | –43 | 3134 | 31.3 | |
| λ | –21 | –44 | 1332 | 13.3 | |
| 4 | μ | 40 | –6 | 7732 | 77.3 |
| ν | –32 | –31 | 2268 | 22.7 | |
| LSTc (α2→6) Torsional angles | |||||
| 1 | ξ | –57 | 190 | 9622 | 96.2 |
| ο | 56 | 174 | 378 | 3.8 | |
| 2 | π | 40 | –9 | 9024 | 90.2 |
| σ | –30 | –30 | 976 | 9.8 | |
| 3 | τ | 39 | 34 | 4108 | 41.1 |
| χ | 23 | –46 | 4118 | 41.2 | |
| υ | –21 | –45 | 1774 | 17.7 | |
| 4 | A | 39 | –7 | 5840 | 58.4 |
| B | –36 | –30 | 4160 | 41.6 | |
To fully describe the first glycosidic linkage of LSTc [Neu5Ac-α(2→6)-Gal], the ω torsional angle (see Experimental Procedures for the definition) needed to be considered. This degree of freedom is absent for LSTa. LSTc has a single state characterized by an average ω value of –50°, which is in agreement with the prediction of Xu et al.17 (Figure 11 of the Supporting Information). The ω torsional angle of the Neu5Ac-α(2→6)-Gal-β disaccharide has previously been discussed by Sabesan et al.40 for similar di- or trisaccharides. They propose that two states are allowed for ω torsional angles, located approximately at –50° and 180°, corresponding to antiperiplanar (gt conformer) and synclinal (tg) for H5–H6(R) (Gal1), respectively. These states have characteristic JH5–H6(R) couplings, ~10 Hz for the former and a smaller value of ~3.5 Hz for the latter. We observed a coupling constant of 10 Hz for JH5–H6(R) in LSTc, which according to Sabesan et al.40 corresponds to the antiperiplanar H5–H6(R) Gal1 conformer, which supports the values determined by the MD simulation.
As we move along the glycans, from the nonreducing end to the reducing end, the glycosidic linkages exhibit progressively decreasing disparities between the glycosidic torsional state positions and populations (Figure 3). The ϕ2 and ψ2 distribution for LSTc and LSTa (Figure 3 and Table 2) qualitatively agree with those found previously by Sabesan et al.40 for disaccharides Gal-β(1→3)-GlcNAc and Gal-β(1→4)-GlcNAc, which are model compounds for LSTa and LSTc, respectively.
The torsional states determined for LSTa broadly agree with those described by Xu et al.,17 while the torsional angles determined for LSTc are less consistent with the data of the aforementioned work. Xu et al.17 found the following additional LSTc torsional states: ~180° and ~180° for ϕ1 and ψ1, ~60° and ~180° for ϕ2 and ψ2, ~30° and ~160° for ϕ3 and ψ3, and ~50° and ~180° for ϕ4 and ψ4, respectively. We predict a state in the first glycosidic linkage of LSTc located at 56° and 174°, which was not observed by Xu et al.,17 but this state is seen in the conformational map obtained at GlycoMapsDB for Neu5Ac-α(2→6)-Gal.41 This discrepancy is possibly related to the use of a different starting conformation for LSTc during the MD simulation, which prevents an ensemble of conformations from being visited as a consequence of the potential energy walls restricting exploration of the conformational space. To partially address this limitation, shorter MD simulations (20 ns) were performed at higher temperatures (395 and 495 K) using the same starting conformation at each temperature (Figures 12 and 13 of the Supporting Information). At these higher temperatures, the missing LSTc torsional states in linkages ϕ1 and ψ1 and ϕ4 and ψ4 started to be populated (Figure 12 of the Supporting Information). Interestingly, for both LSTc and LSTa, the first glycosidic torsional state (ϕ1 and ψ1) reproduces qualitatively in position and shape the torsional angle map of the Neu5Ac-α(2→6/3)-Gal glycosidic junction shown in GlycoMapsDB.41
A parameter used to monitor the evolution of a MD simulation is the root-mean-square displacement (rmsd) of the atoms in a molecule (determined using the conformation at time zero as a reference), which illustrates the different conformational states that the molecule assumes during the simulation, as compared to the starting state. Unsurprisingly, the conformational states assumed by LSTc and LSTa are related to the torsional angle states of their glycosidic linkages. During the simulation, LSTc moves between two conformational states, rmsd states located at ~2.10 and ~3.50 Å (Figure 14 of the Supporting Information), while LSTa assumes a single broad distribution of related conformational states (Figure 15 of the Supporting Information) centered at ~2.8 Å. The conformers that populate the different rmsd states observed for LSTc, located at rmsd values of ~2.10 and ~3.50 Å, have stark differences in the third glycosidic linkage (ϕ3 and ψ3), GlcNAc-β(1→3)-Gal-β (Figure 16 of the Supporting Information). This correlation is also true for LSTa; a subset of conformers located at rmsd values of 2.65 and 3.80 Å (±10 Å) also exhibit significant differences in the third glycosidic linkage (Figure 17 of the Supporting Information). This result is interesting for the case of LSTc, as it suggests that the third glycosidic linkage is the most conformationally mobile junction and not the first, extended linkage Neu5Ac-α(2→6)-Gal, as might be assumed. This could be due to the first residue forming hydrogen bonds with the rest of the molecule, the result being that the first linkage is static compared to the third.
NMR Data and MD Simulation Comparison
To compare the conformers derived by MD simulation and the experimental NMR data, an rmsdexp was derived from five experimentally estimated inter-residue proton–proton distances (see eq 2 and Table 1) and the corresponding MD predicted distances for each structure in the MD trajectories (see Experimental Procedures). The smaller the rmsdexp value, the more closely the conformer matches the NMR data.
The MD-generated conformers of LSTc have two distinct rmsdexp states. The first state located at 0.64 Å is very narrow, and these conformers match the NMR data well; this population ends at 1.05 Å, and thereafter, there is a very broad declining population of conformers with increasing disparity between the NMR-estimated inter-residue proton–proton distances and MD-predicted distances (Figure 18 of the Supporting Information). The conformers that populate these states have distinct glycosidic torsional states, with the conformers that more closely match the NMR data, the population with the smaller rmsdexp values (<1.05 Å), belonging to the following glycosidic torsional angle states: –57° and 190° for ϕ1 and ψ1 and 39° and –9° for ϕ2 and ψ2, respectively (clusters ξ and π, respectively, Figure 3E,F and Figure 19 of the Supporting Information). LSTa also has two rmsdexp populations, located at 0.75 and 0.97 Å (Figure 20 of the Supporting Information); again the conformers that comprise these different populations belong to differing torsional angle states. In this circumstance, the differentiating linkage is closer to the reducing end of the glycan, ϕ3 and ψ3; the MD-generated conformers that are in closer agreement to the NMR data are members of the torsional angle states located at –21° and –45° and 23° and –46° for ϕ3 and ψ3, respectively (clusters λ and κ, respectively, Figure 3C and Figure 21 of the Supporting Information). It is interesting to note that on the whole the MD conformers of LSTa more closely match the NMR data, with the whole population falling within the rmsdexp range of 0–1.5 Å, whereas the LSTc conformers have rmsdexp values spanning the range of 0–5 Å.
To further compare the experimental NMR data with the MD-determined structures, two subsets of conformers were chosen for each glycan, a subset containing the conformers with the 40 smallest rmsdexp values (rmsdexp min) (0.24–0.33 Å for LSTc and 0.12–0.43 Å for LSTa) and a subset comprising the structures with the 40 largest rmsdexp values (rmsdexp max) (1.26–1.41 Å for LSTa and 3.83–4.40 Å for LSTc). The 2D NOESY spectra were then simulated for these selected structures (see Experimental Procedures), with the calculated intra- and inter-residue proton–proton NOE signals then being compared (Table 4 of the Supporting Information). The H1–H3, H1–H5, and H3–H5 intraresidue NOEs signals are qualitatively reproduced for LSTc residues Neu5Ac, Gal1, GlcNAc, and Gal2, while for LSTa, the signals are reproduced for only Gal1, GlcNAc, and Gal2. Interestingly, the agreement between the simulated and experimental intraresidue 2D NOE signals is similar for both rmsdexp min and rmsdexp max conformer subsets, because the rmsdexp values were determined using inter-residue distances and not intraresidue distances. In fact, the H1 Gal1–H4 GlcNAc, H1 GlcNAc–H3 Gal2, H1 Gal2–H4 Glc, and CH3 GlcNAc–H5 Neu5Ac inter-residue distances compare well for the LSTc rmsdexp min conformer subset, while the rmsdexp max conformers reproduce only the H1 Gal2–H4 Glc distance. This trend is also true for LSTa, with the rmsdexp min set of structures reproducing the H3 Gal1–H3ax Neu5Ac, H1 Gal1–H3 GlcNAc, H1 GlcNAc–H3 Gal2, and H1 Gal2–H4 Glc inter-residue distances well and the rmsdexp max conformers replicating adequately only the experimentally determined H1 Gal2–H4 Glc distance (Table 4 of the Supporting Information). This illustrates the ability of the rmsdexp minimization procedure to select conformers from the MD simulation trajectory, providing a means of determining that quality of the MD simulation and allowing the structures that more closely match the experimental data to be extracted. To make a broader comparison of the modeled and measured data, with MD conformers not being chosen by experimental constraints, the last 60 geometries of the MD simulation for LSTa and LSTc were used to generate NOE magnitudes (Table 4 of the Supporting Information). For both glycans, the agreement between the NOE magnitudes derived from MD and experimental data is qualitatively good, but the NOE magnitudes for the rmsdexp min MD conformer subset more closely match the experimental data than the NOE magnitudes derived from the MD structures, which were chosen without experimental constraints. It appears that the first glycosidic linkage, Neu5Ac-α(2→6/3)-Gal, for both glycans is the least well described by the MD simulation, while the remaining interglycosidic NOEs are better modeled in LSTa. This could be due to the isotropic motion approximation introduced by NOEPROM to generate the theoretical NOE magnitudes, with LSTc exhibiting stronger motional anisotropy than LSTa, which is observed in the NMR relaxation measurements.
The torsional angles for the rmsdexp min and rmsdexp max conformer subsets are shown in Figure 3 for LSTa and LSTc, respectively (Table 5 of the Supporting Information). rmsdexp min structures are colored red, while the rmsdexp max conformers are colored blue. Both of the conformer subsets for LSTc and LSTa show significant differences in their torsional angle states; the ϕ4 and ψ4 linkage for both glycans is the only linkage where there is little divergence. Interestingly, the LSTa rmsdexp min conformers have two ϕ1 and ψ1 states, the linkage between Neu5Ac and Gal1, which are located at –100° and –53° and 207° and –24°, corresponding to clusters β and γ, respectively. The conformers in the state centered at –100° and –53° are in better agreement with the NOE values for the H3ax Neu5Ac–H3 Gal1 distance (Table 4 of the Supporting Information). Sabesan et al. also observed a similar NOE, between H3ax Neu5Ac and H3 Gal1, for the di- and trisaccharide structures.40 The glycosidic torsional angles of the conformers that more closely fit the NMR data are as follows: –55° and 197° for ϕ1 and ψ1, 28° and –15° for ϕ2 and ψ2, 34° and 54° for ϕ3 and ψ3, and 11° and 15° for ϕ4 and ψ4, respectively, for LSTc and 207° and –24° for ϕ1 and ψ1, 41° and –1° for ϕ2 and ψ2, 27° and 34° for ϕ3 and ψ3, and 38° and –5° for ϕ4 and ψ4, respectively, for LSTa. To corroborate this result, selected long-range coupling constants, 3JCH, across the glycosidic linkages (Jϕ for H1–C1–Ox–Cx and Jψ for C1–Ox–Cx–Hx) were measured; this allowed an indirect estimation of the glycosidic torsional angles for both glycans. The measured coupling constants (in hertz) starting from the first torsion (ϕ1 and ψ1) are as follows: Jψ1 = 4.4, Jϕ2 = 2.6, Jψ2 = 5.4, Jϕ3 = 4.2, Jψ3 = 4.5, and Jϕ4 = 4.1 for LSTa and Jψ1 = 2.6, Jϕ2 = 3.6, Jϕ3 = 4.1, Jψ3 = 4.4, and Jϕ4 = 3.4 for LSTc, respectively. Calculated pairs of couplings (Jϕ and Jψ), determined using the rmsdexp min subset of conformers, in accordance to the Karplus-type equation,42 are reported in Table 7 of the Supporting Information. The only measured long-range coupling that does not agree with the rmsdexp min selected subset of conformers is the ϕ1 and ψ1 coupling for LSTa; the measured coupling (Jexpψ1 = 4.4 Hz) is in better agreement with the glycosidic torsional angles ϕ1 and ψ1 of –99° and –53°, respectively, and not ϕ1 and ψ1 values of 207° and –24°, respectively, which were determined by the NMR-selected MD conformers. J-HMBC spectra of LSTa and LSTc, overlaid with HSQC spectra, can be found in Figure 6 of the Supporting Information.
Analyses of the Shape and Topology of LSTc and LSTa
The global shape of the two oligosaccharides is described by the topological θ angle, as defined by Xu et al.17 (see Experimental Procedures). Chandrasekaran et al. previously proposed that the θ angle of LSTc and LSTa defines their conformation, with LSTa adopting a linear form, coined a cone, which is defined as having a θ angle >110°, while LSTc, with its extra degree of freedom at the nonreducing end, Neu5Ac-α(2→6)-Gal-β(1→4)-, forms a bent conformation, termed the umbrella form, defined as having a θ angle of <110°.16
The LSTc conformers determined by MD have two θ angle populations; the much larger population is located at 87°, and the second smaller population starts above 109° (Figure 9 of the Supporting Information). These differences in θ values also correspond to differences in the torsional angle states for the linkage between Gal1 and GlcNAc; the subset of conformers with a θ angle centered at 87° correlates with the torsional angle state with values for ϕ2 and ψ2 of 39° and –9°, respectively, while the structures with a θ angle of >88° populate torsional states with values for ϕ2 and ψ2 of 39° and –9° and –30° and –30°, respectively (Figure 22 of the Supporting Information). This indicates that the second glycosidic linkage is important in the “opening and closing” of the umbrella conformation proposed for LSTc and not the first glycosidic linkage, which might be assumed because of the α2→6 linkage between Neu5Ac and Gal1.
LSTa also has two θ angle populations, with the more populated state centered at 153° and the less populated state located at 119° (Figure 10 of the Supporting Information). As for LSTc, the LSTa conformers that belong to the different θ angle populations correlate strongly with the torsional states of the second glycosidic linkage, ϕ2 and ψ2 (Figure 23 of the Supporting Information). The θ angle population centered at 153° correlates with the torsional state with values for ϕ2 and ψ2 of 37° and –5°, respectively, while the population located at 119° correlates with the torsional state with values for ϕ2 and ψ2 of –37° and 18°, respectively. These results highlight that the conformational and subsequent dynamic properties of both LSTc and LSTa are linked strongly to the second glycosidic linkage.
The LSTc conformers that more closely fit the NMR data (rmsdexp min structures) have an average θ angle of 87° (standard deviation of 4°), while the corresponding LSTa conformers have an average θ angle of 159° (standard deviation of 5°). These values, for the NMR-selected MD-generated conformers, agree with the results published by Xu et al.17 and the hypothesis proposed by Chandrasekaran et al.16
Principal Component Analysis of the LSTa and LSTc MD Trajectories
Principal component analysis (PCA) has been applied on the MD-simulated trajectory to extract the independent mode of motion of both oligosaccharides, allowing an easier correlation with its structural properties. PCA has been a powerful tool for the analysis of protein35,43 and glycan44 molecular dynamics. The convergence of the distinct ensembles of motion was observed by determining the rmsip of the MD trajectory eigenvectors, comparing the eigenvectors generated by PCA of the whole MD tracjectory (100 ns) to the PCA of the trajectory at different time points (increasing steps of 1 ns). The rmsip provides a measure of the similarity of the “essential modes” extracted; if the value is zero, then they are dissimilar, and if the value is one, the subspaces have converged. The rmsip for both LSTc and LSTa is ≥0.9 for at least 50% of the trajectory; Amadei et al.35 opined that values of >0.7 provide evidence of good convergence (Figure 24 of the Supporting Information).
Here, only the first four components generated by PCA for LSTa and LSTc are considered. They explain 73.8 and 66.2% of the total variance, respectively. The PCA scree, loading, and normalized eigenvector plots can be found in Figures 25–31 of the Supporting Information. The normalized eigenvector plots indicate the contribution each atom makes to the extracted components, i.e., whether an atom, or a group of atoms, is involved in the independent dynamic motions isolated by PCA.
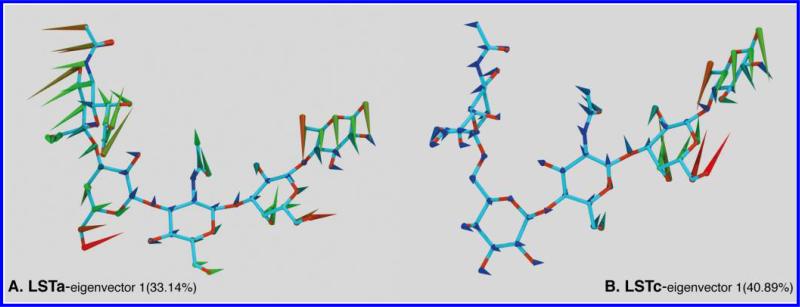
The independent modes of motion described by component one for LSTc have the nonreducing end of the glycan close to stationary compared to the other residues, while the reducing end (Gal2-Glc) rotates around their glycoside linkages, moving as a single unit. This is in agreement with the rmsd analysis of the LSTc MD trajectory. Figure 4 provides an illustration of this mode of motion, “porcupine” representations of the first eigenvector for LSTc and LSTa [supplementary data contain graphical representations of all four components (Figures 30 and 31 of the Supporting Information)]. The three other distinct modes of motion extracted for LSTc show both the nonreducing and reducing ends of the molecule are in motion, with the reducing end disaccharide (Gal2-Glc) moving as one unit and the sialyl group of Neu5Ac moving toward the N-acetyl group of GlcNAc. The least mobile residue is Gal1, which exhibits the smallest variation of the four components examined, which is illustrated in the normalized eigvector figure for LSTc (Figure 28 of the Supporting Information). These results are consistent with a small θ angle and a hydrogen bonding network between GlcNAc and Neu5Ac of LSTc, while the fact that the reducing end of the molecule moves in unison is indicative of a cellobiose-like hydrogen bonding network between Glc and Gal2. This dynamic description agrees with the results of the T1 and T2 relaxation measurements.
Figure 4.
Porcupine plot representations of the first eigenvectors for LSTa (A) and LSTc (B). The direction, size, and color of the spines are proportional to the direction and magnitude of the motion of the atoms. The illustration shows LSTa moving in a bowlike motion while LSTc is moving in two independent sections: Neu5Ac synchronized with the GlcNAc and the reducing-end disaccharide as a single unit. In LSTc, Gal1 is the least mobile residue.
The independent modes of motion extracted for LSTc correlate with structural parameters that have been previously discussed; the motion described by component 1 correlates with the rmsd of the conformers and is therefore related to the different torsional states in the third glycosidic linkage, GlcNAc-β(1→3)-Gal-β (Figures 16 and 32 of the Supporting Information). The molecular motion illustrated by the second component correlates with the structures that belong to the different θ angle populations, which are strongly tied to differences in the second glycosidic linkage (Gal-β(1→4)-GlcNAc) (Figures 22 and 34 of the Supporting Information). Finally, the dynamic that distinguishes whether a conformer agrees well with the experimental NMR data, rmsdexp, correlates with component 3, which is related to the different torsional states at glycosidic linkages 1 and 2 (Figures 19 and 36 of the Supporting Information).
The distinct modes of motion isolated for LSTa are very different from those of LSTc. LSTa has “bowlike” dynamics for the four components examined, the whole molecule moving in unison; this is particularly true of the motion isolated for component 1 (Figure 29 of the Supporting Information). Again, the modes of motion isolated by PCA correlate with previously discussed structural parameters. A mixture of components distinguishes the conformers with different rmsd values, for example, the mixture of components 1 and 2 and also components 1 and 4. These dynamics, which are described by the mixtures of components, are generated by specific states in the second and third glycosidic linkages (Figures 17 and 33 of the Supporting Information). Component 1 correlates strongly with the θ angle of the conformers, which in turn is related to the torsional state of ϕ2 and ψ2 (Figures 23 and 35 of the Supporting Information). Component 2 is the independent motion that discriminates the structures that fit well with the experimental NMR data from those that do not; high or low rmsdexp values are associated with differences in the torsional states of ϕ3 and ψ3 (Figures 20 and 37 of the Supporting Information).
Inter-Residue Hydrogen Bond Analysis
To further understand the consequences and driving forces behind the different conformations assumed by LSTc and LSTa, the possible inter-residue hydrogen bonds were determined for the rmsdexp min and rmsdexp max conformer subsets. The analysis employed distance and angular restrictions; the distance between the donated hydrogen and the acceptor was ≤2.6 Å, and the X–H···Y (X is the donor and Y the acceptor) angle was greater than 110°, tending toward 180°. Here we will discuss the proposed hydrogen bonds for the rmsdexp min subset of conformers, the structures that more accurately represent the experimental data; the hydrogen bond analysis for the rmsdexp max conformers, for comparison, is shown in Table 6 and Figure 38 of the Supporting Information.
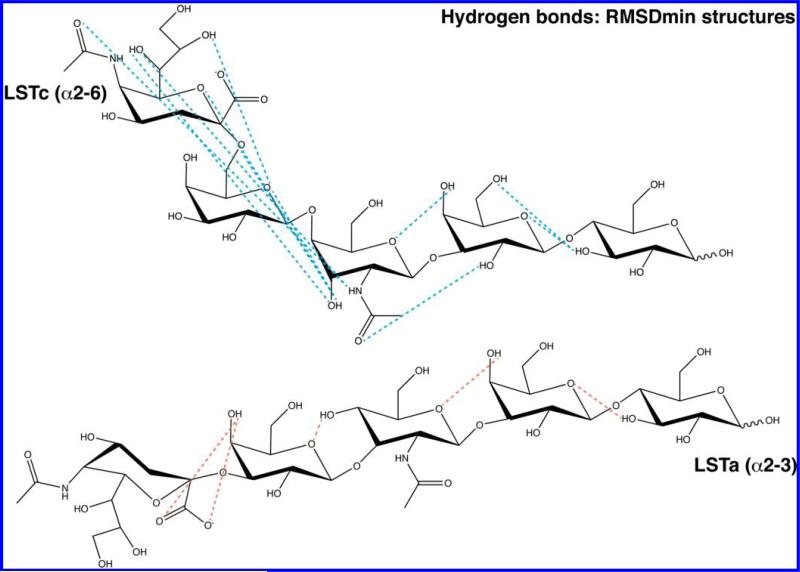
For both glycans, several possible hydrogen bonds were determined for the rmsdexp min conformers, 5 for LSTa and 11 for LSTc, illustrated in Figure 5, while the populations of each are reported in Table 6 of the Supporting Information. Both LSTc and LSTa have potential hydrogen bonds that offer an explanation for the tandem motion of the reducing end residues Glc and Gal2 isolated by PCA: H3O Glc–O5 Gal2 (percentage of conformers, 62.5% for LSTc and 80% for LSTa) (cellobiose-like), H4O Gal2–O5 GlcNAc (82.5% for LSTc and 22.5% for LSTa), H2O Gal2–O2N GlcNAc (42.5% for LSTc), and H3O Glc–O6 Gal2 (12.5% for LSTc). The cellobiose-like hydrogen bond between H3O of Glc and O5 of Gal2 has previously been observed in a structurally related molecule, Neu5Ac-α(2→6)-Gal-β(1→4)-Glc, sialyl-α(2→6)-lactose (S6L).45
Figure 5.
Proposed hydrogen bond network for LSTc and LSTa rmsdexp min conformers. The hydrogen bond network at the nonreducing end of each glycan is strongly affected by the different linkage between Neu5Ac and Gal1 [α(2→3) for LSTa and α(2→6) for LSTc]. The acceptor and donor atoms lie within 2.6 Å of each other, and the X–H···Y (X is the donor and Y the acceptor) angle was greater than 110°, tending toward 180°. Predicted hydrogen bonds for both LSTa and LSTc appear in Table 6 of the Supporting Information.
The hydrogen bond networks present at the nonreducing end of either glycan are a direct consequence of their θ angles. The nonreducing end hydrogen bond network for LSTc, which has a bent form in solution (θ = 86° for the selected conformers), originates from GlcNAc, with two predicted bonds to Gal1 [H3O GlcNAc–O5 Gal1 (100%) and H3O GlcNAc–O6 Gal1 (25.0%)] and five others spanning Gal1 to Neu5Ac [O3 GlcNAc–H7O Neu5Ac (55.0%), O5 Gal1–H8O Neu5Ac (20.0%), H2N GlcNAc–O5N Neu5Ac (15.0%), H3O GlcNAc–O6 Neu5Ac (12.5%), and H2N GlcNAc–O7 Neu (12.5%)] (Figure 5 and Table 6 of the Supporting Information). A hydrogen bond similar to the O3 GlcNAc–H7O Neu5Ac hydrogen bond is observed in S6L, between O3 of Glc and H7O of Neu5Ac.45 Because of the bowlike backbone conformation of LSTa, its hydrogen bonds can be formed only between directly adjacent residues: H4O GlcNAc–O5 Gal1 (92.5%), H4O Gal1–O1A/O1B (acid group) Neu5Ac (52.5%), H4O Gal2–GlcNAc (22.5%), and H3O Glc–O5 Gal2 (80.0%) (Figure 5 and Table 6 of the Supporting Information). All of the proposed hydrogen bonds are listed in Table 6 of the Supporting Information, while Figure 5 is a schematic showing the major proposed inter-residue hydrogen bonds.
DISCUSSION
This study has identified the distinct conformational, topological, and dynamic properties of the glycan receptors [two pentasaccharides, LSTc (Neu5Ac-α(2→6)-Gal-β(1→4)-GlcNAc-β(1→3)-Gal-β(1→4)-Glc) and LSTa (Neu5Ac-α(2→3)-Gal-β(1→3)-GlcNAc-β(1→3)-Gal-β(1→4)-Glc] of influenza A viruses in solution, i.e., without its receptor. The study involved utilizing extensive NMR measurements, including the complete 1H and 13C assignment for each glycan, MD simulations, and the numerical analysis of the data generated, to provide not only information concerning the conformation in solution but also, for the first time, details of its overall dynamic properties that could be related to the receptor binding ability.
Conformers from the MD simulations were selected using a rmsdexp function, comparing NOE enhancement-derived inter-residue proton–proton distances and the equivalent predicted distances. Using this approach, two subsets of conformers were selected, in good (rmsdexp min) or poor (rmsdexp max) agreement with the experimental data. In particular, the conformers that were in better agreement with the experimental restraints, rmsdexp min, generated 2D NOESY spectra, using the multispin approximation (NOEPROM), in good agreement with the values measured at 600 and 900 MHz. These results were subsequently corroborated by experimental nJC–H heteronuclear coupling constants, which were measured across the glycosidic linkages for LSTc and LSTa; these results supported the concept of using NMR experimental constraints to select conformers from MD simulation trajectories. LSTc exhibits a bent shape characterized by a topological angle θ of 87°, whereas LSTa has a more linear form and therefore a greater θ angle of 159°, a structural difference in agreement with the previous work of Chandrasekaran16 and Xu et al.17 that concurs with the glycan ligand cocrystallized with HA.15
As a consequence of these structural differences, the nonreducing end terminal Neu5Ac of LSTc could form an intricate hydrogen bond network with GlcNAc, which is not the case for its isomer, LSTa, because hydrogen bond analysis revealed interactions with only directly adjacent residues. PCA of the glycan trajectories extracted the independent modes of motion that differentiate the structures, with LSTa moving like a flexing bow and the reducing and nonreducing disaccharides moving around the GlcNAc fulcrum, whereas the dynamics of LSTc are different from those of LSTa, the sialyl group of Neu5Ac moving in tandem with the N-acetyl group of GlcNAc, which moves in a manner that is independent of the reducing end disaccharide.
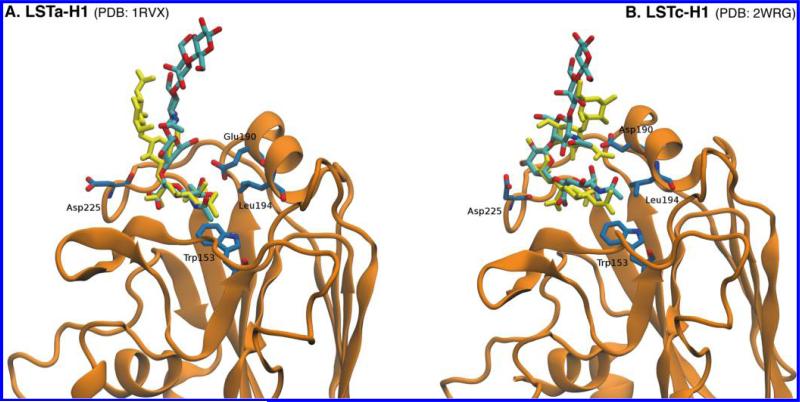
In Figure 6, the structures of LSTc and LSTa extracted from the respective rmsdexp min sets are superimposed on the Neu5Ac and Gal1 residues in the cocrystallized glycan–HA complexes [PDB entries 2WRG46 (panel B) and 1RVX2 (panel A)]. The rmsd values between the superimposed structures and the cocrystallized ligand, considering the three nonreducing end residues (i.e., Neu5Ac, Gal1, and GlcNAc), are 6.5 and 6.2 Å for LSTa and LSTc, respectively. These results illustrate how the NMR-selected conformation for LSTc and LSTa in solution, i.e., determined without HA, matches qualitatively the cocrystallized ligand bound to HA and also suggests that the respective sugar conformation does not significantly change upon interaction with HA. This observation supports the hypothesis that HA recognizes the solution conformation of the glycan, instead of the ligand and receptor undergoing a “bind and fit” process. This study contributes to an improved understanding of the structural basis for the glycan receptor specificity of HA in influenza.
Figure 6.
Comparison of the MD -generated LSTa (A) and LSTc (B) conformers (structures with the smallest rmsdexp value, rmsdexp min) and crystal structures of LSTa and LSTc bound to hemagglutinin from H1. The conformers generated via MD simulations are colored cyan (C atoms) and red (O atoms), while the conformers determined by X-ray crystallography are colored yellow. The rmsd values between the MD simulation-generated structures and the cocrystallized glycans are 6.2 and 6.5 Å for LSTa and LSTc, respectively.
ASSOCIATED CONTENT
Supplementary Material
Figure 1.
1H–13C HSQC NMR spectra of LSTc (A and B) and LSTa (C and D).
ACKNOWLEDGMENTS
We are very grateful for access to the NMR spectrometer (900 MHz) at the Utrecht NMR Facility and the assistance of Dr. Hans Wienk.
Funding
We thank Finlombardia SPA ‘Fondo per la promozione di Accordi Istituzionali’ and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for their financial support. This work was also funded in part by the National Institutes of Health (R37 GM057073-13) and the National Research Foundation supported Interdisciplinary Research group in Infectious Diseases of SMART (Singapore MIT Alliance for Research and Technology). The 900 MHz spectra were recorded at the SONNMR Large Scale Facility in Utrecht, by the financial support of the Access to Research Infrastructures activity in the seventh Framework Programme of the EC (contract no. 261863, EU-NMR).
ABBREVIATIONS
- HA
hemagglutinin
- LSTa
Neu5Ac-α(2→3)-Gal-β(1→3)-GlcNAc-β(1→3)-Gal-β(1→4)-Glc
- LSTc
Neu5Ac-α(2→6)-Gal-β(1→4)-GlcNAc-β(1→3)-Gal-β(1→4)-Glc
- MD
molecular dynamics
- Gal
galactose
- Neu5Ac
N-acetylneuraminic acid
- GlcNAc
N-acetylglucosamine
- Glc
glucose
- rmsip
root-mean-square internal product
- rmsd
root-mean-square displacement
- rmsdexp
comparison of root-mean-square displacements between experimental and theoretical data
- PCA
principal component analysis
Footnotes
Supporting Information
Supporting NMR data and analyses of the MD simulations. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Eisen MB, Sabesan S, Skehel JJ, Wiley DC. Binding of the influenza A virus to cell-surface receptors: Structures of five hemagglutinin-sialyloligosaccharide complexes determined by X-ray crystallography. Virology. 1997;232:19–31. doi: 10.1006/viro.1997.8526. [DOI] [PubMed] [Google Scholar]
- 2.Gamblin SJ, Haire LF, Russell RJ, Stevens DJ, Xiao B, Ha Y, Vasisht N, Steinhauer DA, Daniels RS, Elliot A, Wiley DC, Skehel JJ. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 3.Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc. Natl. Acad. Sci. U.S.A. 2001;98:11181–11186. doi: 10.1073/pnas.201401198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 5.Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 6.Ibricevic A, Pekosz A, Walter MJ, Newby C, Battaile JT, Brown EG, Holtzman MJ, Brody SL. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J. Virol. 2006;80:7469–7480. doi: 10.1128/JVI.02677-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: Influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 8.van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am. J. Pathol. 2007;171:1215–1223. doi: 10.2353/ajpath.2007.070248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jayaraman A, Pappas C, Raman R, Belser JA, Viswanathan K, Shriver Z, Tumpey TM, Sasisekharan R. A single base-pair change in 2009 H1N1 hemagglutinin increases human receptor affinity and leads to efficient airborne viral transmission in ferrets. PLoS One. 2011;6:e17616. doi: 10.1371/journal.pone.0017616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumari K, Gulati S, Smith DF, Gulati U, Cummings RD, Air GM. Receptor binding specificity of recent human H3N2 influenza viruses. Virol. J. 2007;4:42. doi: 10.1186/1743-422X-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4620–4624. doi: 10.1073/pnas.0308001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 13.Russell RJ, Stevens DJ, Haire LF, Gamblin SJ, Skehel JJ. Avian and human receptor binding by hemagglutinins of influenza A viruses. Glycoconjugate J. 2006;23:85–92. doi: 10.1007/s10719-006-5440-1. [DOI] [PubMed] [Google Scholar]
- 14.Shriver Z, Raman R, Viswanathan K, Sasisekharan R. Context-specific target definition in influenza A virus hemagglutinin-glycan receptor interactions. Chem. Biol. 2009;16:803–814. doi: 10.1016/j.chembiol.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bewley CA. Illuminating the switch in influenza viruses. Nat. Biotechnol. 2008;26:60–62. doi: 10.1038/nbt0108-60. [DOI] [PubMed] [Google Scholar]
- 16.Chandrasekaran A, Srinivasan A, Raman R, Viswanathan K, Raguram S, Tumpey TM, Sasisekharan V, Sasisekharan R. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat. Biotechnol. 2008;26:107–113. doi: 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- 17.Xu D, Newhouse EI, Amaro RE, Pao HC, Cheng LS, Markwick PR, McCammon JA, Li WW, Arzberger PW. Distinct glycan topology for avian and human sialopentasaccharide receptor analogues upon binding different hemagglutinins: A molecular dynamics perspective. J. Mol. Biol. 2009;387:465–491. doi: 10.1016/j.jmb.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2012. [Google Scholar]
- 19.Lewis IA, Schommer SC, Markley JL. rNMR: Open source software for identifying and quantifying metabolites in NMR spectra. Magn. Reson. Chem. 2009;47(Suppl. 1):S123–S126. doi: 10.1002/mrc.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkar D. Lattice: Multivariate Data Visualization with R. Springer; New York: 2008. [Google Scholar]
- 21.Neuhaus D, Williamson M. The Nuclear Overhauser Effect in Structural and Conformational Analysis. VHC Publishers Inc.; New York: 1989. [Google Scholar]
- 22.Angulo J, Hricovini M, Gairi M, Guerrini M, de Paz JL, Ojeda R, Martin-Lomas M, Nieto PM. Dynamic properties of biologically active synthetic heparin-like hexasaccharides. Glycobiology. 2005;15:1008–1015. doi: 10.1093/glycob/cwi091. [DOI] [PubMed] [Google Scholar]
- 23.Hricovini M, Torri G. Dynamics in aqueous solutions of the pentasaccharide corresponding to the binding site of heparin for antithrombin III studied by NMR relaxation measurements. Carbohydr. Res. 1995;268:159–175. doi: 10.1016/0008-6215(94)00334-c. [DOI] [PubMed] [Google Scholar]
- 24.Poveda A, Martin-Pastor M, Bernabe M, Leal JA, Jimenez-Barbero J. Solution conformation and dynamics of a fungal cell wall polysaccharide isolated from Microsporum gypseum. Glycoconjugate J. 1998;15:309–321. doi: 10.1023/a:1006909515343. [DOI] [PubMed] [Google Scholar]
- 25.Woods Group GLYCAM Web. 2005−2012.
- 26.Kirschner KN, Yongye AB, Tschampel SM, Gonzalez-Outeirino J, Daniels CR, Foley BL, Woods RJ. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J. Comput. Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 28.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 29.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts BP, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvai I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye J, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER 11. University of California; San Francisco: 2010. [Google Scholar]
- 30.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grant BJ, Rodrigues AP, ElSawy KM, McCammon JA, Caves LS. Bio3d: An R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 32.Martin-Pastor M. NOEPROM. 2005.
- 33.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graphics. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 34.Schulz R. 2009 http://www.ks.uiuc.edu/Research/vmd/mailing_list/vmd-l/att-15365/PorcupinePlot.tcl.
- 35.Amadei A, Linssen AB, Berendsen HJ. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 36.Turner CJ, Connolly PJ, Stern AS. Artifacts in sensitivity-enhanced HSQC. J. Magn. Reson. 1999;137:281–284. doi: 10.1006/jmre.1998.1692. [DOI] [PubMed] [Google Scholar]
- 37.Poppe L, Van Halbeek H. The rigidity of sucrose: Just an illusion? J. Am. Chem. Soc. 1992;114:1092–1094. [Google Scholar]
- 38.Poppe L, van Halbeek H, Acquotti D, Sonnino S. Carbohydrate dynamics at a micellar surface: GD1a headgroup transformations revealed by NMR spectroscopy. Biophys. J. 1994;66:1642–1652. doi: 10.1016/S0006-3495(94)80956-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raab M, Tvaroska I. The binding properties of the H5N1 influenza virus neuraminidase as inferred from molecular modeling. J. Mol. Model. 2011;17:1445–1456. doi: 10.1007/s00894-010-0852-z. [DOI] [PubMed] [Google Scholar]
- 40.Sabesan S, Bock K, Paulson JC. Conformational analysis of sialyloligosaccharides. Carbohydr. Res. 1991;218:27–54. doi: 10.1016/0008-6215(91)84084-r. [DOI] [PubMed] [Google Scholar]
- 41.Frank M, Lutteke T, von der Lieth CW. GlycoMapsDB: A database of the accessible conformational space of glycosidic linkages. Nucleic Acids Res. 2007;35:287–290. doi: 10.1093/nar/gkl907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tvaroška I, Hricovíni M, Petráková E. An attempt to derive a new Karplus-type equation of vicinal proton-carbon coupling constants for C-O-C-H segments of bonded atoms. Carbohydr. Res. 1989;189:359–362. [Google Scholar]
- 43.Kitao A, Go N. Investigating protein dynamics in collective coordinate space. Curr. Opin. Struct. Biol. 1999;9:164–169. doi: 10.1016/S0959-440X(99)80023-2. [DOI] [PubMed] [Google Scholar]
- 44.Gotsev MG, Ivanov PM. Molecular dynamics of large-ring cyclodextrins: Principal component analysis of the conformational interconversions. J. Phys. Chem. B. 2009;113:5752–5759. doi: 10.1021/jp805446w. [DOI] [PubMed] [Google Scholar]
- 45.Poppe L, Stuike-Prill R, Meyer B, van Halbeek H. The solution conformation of sialyl-α (2−6)-lactose studied by modern NMR techniques and Monte Carlo simulations. J. Biomol. NMR. 1992;2:109–136. doi: 10.1007/BF01875524. [DOI] [PubMed] [Google Scholar]
- 46.Liu J, Stevens DJ, Haire LF, Walker PA, Coombs PJ, Russell RJ, Gamblin SJ, Skehel JJ. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17175–17180. doi: 10.1073/pnas.0906849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.