Abstract
We herein report recent advances in our understanding of transport protein evolution. The Drug-Metabolite Transporter (DMT) superfamily (TC# 2.A.7) arose from a 2TMS precursor to give 4TMS proteins which then added one and duplicated to give 10. The proposed pathway is 2 –> 4 –> 5 –> 10. This superfamily provides a rare example where all proposed topological intermediates in this evolutionary pathway have been identified in current protein databases. Another family, the Oligopeptide Transporter (OPT) family (TC# 2.A.67), also started with a 2 TMS peptide precursor, but it followed the pathway:
Only 16 and 17 TMS OPT family members have been identified in current databases. The TRIC family of K+ channels, characterized in animals, arose via the pathway:
where the seventh TMS was added c-terminally to the 6 TMS precursor that resulted from a 3 TMS duplication. Surprisingly, animal TRIC channels proved to have numerous 7 TMS homologues in prokaryotes, none of which had been identified previously. We found that two families of integral membrane proteins gave rise to multiple current topological types. Members of the SdpC killer factor immunity protein family, SdpI (TC# 9.A.32) probably arose via the pathway:
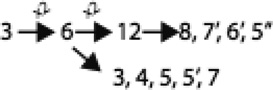 |
while members of the Heme Handling Protein (HHP) Family (TC# 9.B.14) arose via the pathway:
 |
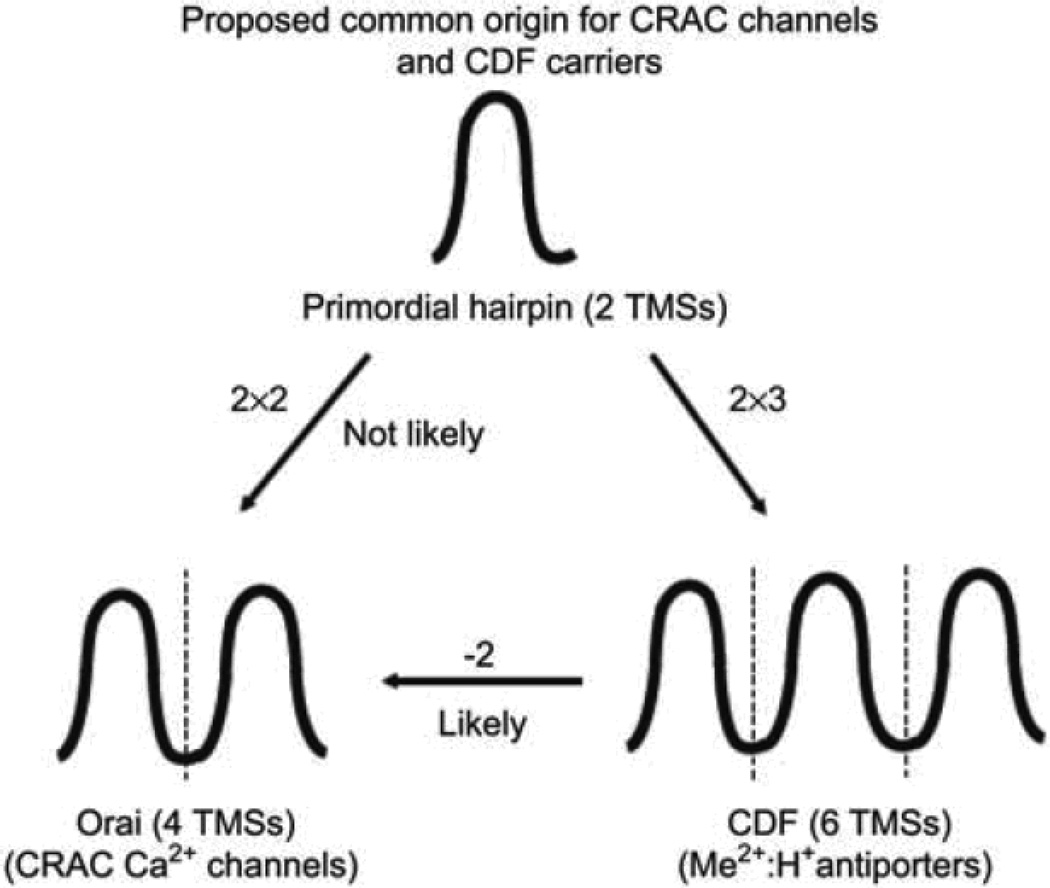
Predictions are also made for an evolutionary pathway giving rise to the seven topological types of P-type ATPases so far identified in the P-ATPase superfamily. Finally, the ubiquitous CDF carriers (TC# 1.A.4) of 6TMSs probably gave rise to CRAC channels of 4TMSs by loss of the first two TMSs an unusual example of retroevolution.
Keywords: Transport proteins, evolutionary pathways, intragenic duplication, repeat sequences
A. Introduction
Transport systems serve the cell in numerous capacities (Saier, 1999a, b, c; Saier, 2000b–e; Saier, 2007; Saier, 2009). First, they allow entry of all essential nutrients into the cytoplasmic compartment and subsequently into organelles. Second, they provide a means for the regulation of metabolite, drug and toxin concentrations by catalyzing the excretion of deleterious metabolites, xenobiotics and end products of metabolic pathways from organelles and cells, and even from the periplasmic spaces of Gram-negative bacteria. Third, they mediate uptake and efflux of ionic species that must be maintained at cytoplasmic concentrations that differ drastically from those in the external milieu, generating a membrane potential, bio-electricity and requisite ion concentration gradients, as well as appropriate cytoplasmic concentrations of all essential trace minerals that participate as cofactors in metabolic processes. Fourth, transporters participate in the secretion of macromolecules (proteins, complex carbohydrates, and lipids) into and beyond the cytoplasmic membrane. Fifth, transport systems allow the transfer of nucleic acids across cell membranes, allowing genetic exchange between organisms. Sixth, transporters facilitate the uptake and release of pheromones, alarmones, hormones, neurotransmitters, and a variety of other signaling molecules. Finally, transport proteins allow living organisms to conduct biological warfare, secreting, for example, biosynthesized toxins, antibiotics, antiviral agents, antifungal agents, etc., many of which are in use in medicine and agriculture. Some of these toxins are themselves channel-forming proteins or peptides that serve a cell-disruptive transport function. Thus, from a functional standpoint, the importance of molecular transport to all facets of life cannot be overestimated. It is not surprising that over 10% of all functionally characterized genes in living organisms encode transport proteins (Saier & Ren, 2006).
The capacity to deduce and extrapolate structural and mechanistic information illustrates the value of phylogenetic data. However, another benefit is that it allows an understanding of the mechanistic restrictions that were imposed upon an evolving family due to architectural constraints. Specific architectural features may allow one family to diversify in function with respect to substrate specificity, substrate affinity, velocity of transport, polarity of transport, and even mechanism of energy coupling. By contrast, the architectural constraints imposed on a second family may not allow functional diversification. Knowledge of the architectural constraints imposed on a permease family provides clues as to the reliability of functional predictions for uncharacterized but related gene products, revealed, for example, by genome sequencing. Conversely, the functional diversity of the members of a permease family must be assumed to reflect architectural constraints, and thus phylogenetic and functional analyses lead to architectural predictions (Hvorup et al., 2003; Saier, 2001; Saier & Ren, 2006).
Over the past two decades, our laboratory has provided information about the evolutionary histories of numerous permease families and these advances have been published and reviewed. (Saier, 1994; Saier, 1996; Saier, 1998; Saier, 2000a; Saier, 2003a, b; Saier et al., 2008). This work led to the formulation of a novel classification system superficially similar to that implemented years before for enzymes by the Enzyme Commission. The transporter classification (TC) system has been adopted by the International Union of Biochemistry and Molecular Biology (IUBMB). In contrast to the Enzyme Commission, which based its classification system solely on function, we have chosen to classify permeases on the basis of both function and phylogeny. Detailed descriptions of the TC system can be found in our database (http://www.tcdb.org) and several of our publications (Saier, Tran & Barabote, 2006; Saier et al., 2009). We continuously update TCDB as new relevant physiological, biochemical, genetic, biophysical, and sequence data become available. The system already provides user-friendly search tools so that the TC system can be readily accessed by keyword, TC number, gene name, protein name, and sequence motifs. Further advances in progress will render TCDB more comprehensive and increasingly accessible to the entire scientific community providing a guide for functional analyses and an essential tool for genome annotation purposes (Saier et al., 2009).
As noted above, our bioinformatics laboratory has identified many pathways by which many trans-membrane transport proteins have evolved from simple pore-forming peptides with 1, 2 or 3 transmembrane segments (TMSs) (Saier, 2003a; Saier, 2003b). Our recent work has identified novel evolutionary pathways and established previously unrecognized superfamily relationships based on statistical analyses of sequence similarity. In this summary article we will discuss our most recent research, much of which has not yet been published or reviewed previously. The Superfamilies discussed exemplify unique features of the evolutionary process.
B. Novel Evolutionary Pathways
1. The Drug/Metabolite Transporter Superfamily (TC# 2.A.7)
Members of the DMT superfamily possess 4, 5 or 10 TMSs, but in earlier work, we could not determine if a 4 or a 5 TMS protein predated the other (Jack et al., 2001). Our published pathway was therefore: Our published pathway was therefore:
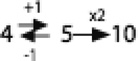 |
Recently we identified DMT family members with only two putative TMSs. Further investigation revealed that the two halves of the 4 TMS proteins possessed two 2 TMS repeat units (V. Lam & M.H. Saier, unpublished results). This observation allowed us to complete the evolutionary pathway as follows:
Surprisingly, in plants, a 30 TMS DMT family members was found that possessed 3 clear 10 TMS repeats, a probable consequence of a late intragenic triplication event.
2. The Oligopeptide Transporter (OPT) Family (TC# 2.A.67)
The Oligopeptide Transporter (OPT) family of peptide and iron-siderophore transporters includes members from both prokaryotes and eukaryotes but with restricted distribution in the latter domain. Eukaryotic members were found only in fungi and plants with a single slime mold homologue clustering with the fungal proteins. All functionally characterized peptide transporters of this family segregate from the known iron-siderophore transporters on a phylogenetic tree. Prokaryotic members derive from many different phyla, but they belong only to the iron-siderophore subdivision. These facts suggest that this family arose in prokaryotes, and that the peptide transporters arose from iron-siderophore transporters in eukaryotes.
OPT family proteins have 16, or occasionally 17 putative transmembrane spanning α-helical segments. We provided statistical evidence that the 16 TMS topology arose via three sequential duplication events followed by a gene fusion event for proteins with a seventeenth TMS (Gomolplitinant & Saier, 2010). The proposed pathway is: 2 TMSs → 4 TMSs → 8 TMSs → 16 TMSs → 17 TMSs (see Figure 1). The seventeenth C-terminal TMS, which probably arose just once, is found in a restricted phylogenetic group of these homologues. Analyses for orthology revealed that a few phylogenetic clusters consist exclusively of orthologues, but most have undergone intermixing, suggestive of horizontal transfer. It appears that in this family, horizontal gene transfer was frequent among prokaryotes, rare among eukaryotes and largely absent between prokaryotes and eukaryotes as well as between plants and fungi. These observations provide guides for future structural and functional analyses of OPT family members.
Figure 1.
Proposed pathway for the evolution of the OPT family of peptide and iron siderophore transporters.
3. The Heme Handling Protein (HHP) Family (TC# 9.B.14)
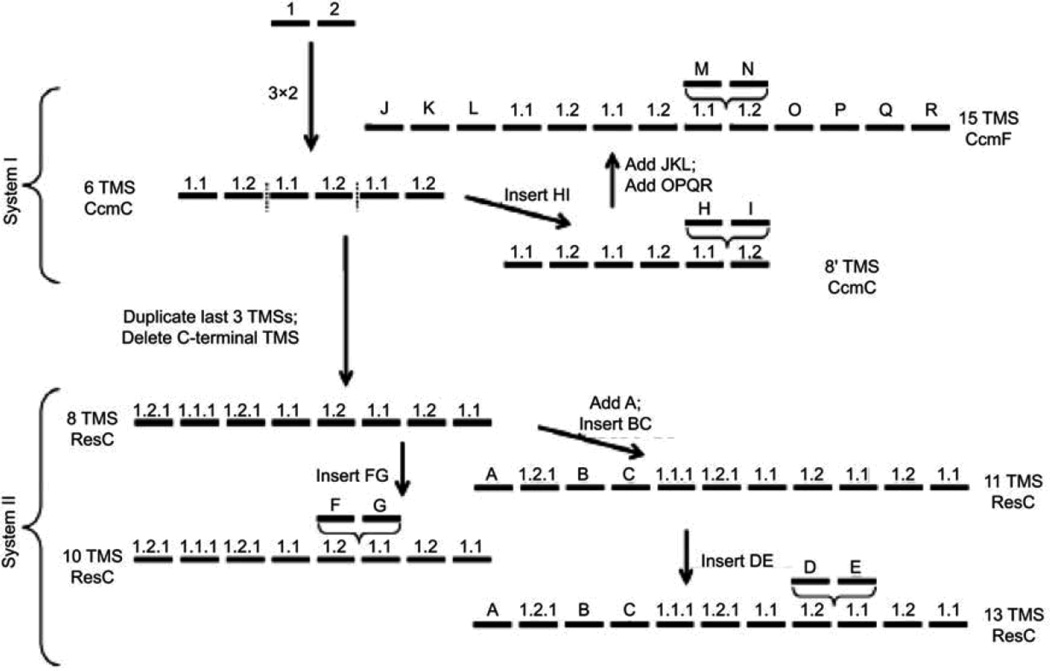
We have investigated the evolutionary origins of members of the Heme Handling Protein (HHP) family of integral membrane cytochrome c biogenesis proteins, which may be transporters for heme. Specifically, the relationships of homologues of the Escherichia coli CcmC protein were examined for probable topological features and evolutionary relationships. Bioinformatic evidence was obtained suggesting that the integral membrane proteins CcmC (an E. coli cytochrome c biogenesis system I protein), CcmF (another E. coli; cytochrome c biogenesis system I constituent) and ResC (a Bacillus subtilis; cytochrome c biogenesis system II component) are all related (Lee et al., 2007). Though the molecular functions of these proteins have not been fully described, they appear to be involved in the provision of heme to extracytoplasmic c-type cytochromes, and we have therefore named them the putative Heme Handling Protein (HHP) family (TC# 9.B.14).
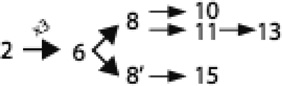
Members of this family exhibit 6, 8, 10, 11, 13 or 15 putative TMSs, thus exhibiting far more topological variation than observed for most other families of integral membrane proteins. We showed that intragenic triplication of a 2 TMS element gave rise to a protein with a 6 TMS topology, exemplified by CcmC (Lee et al., 2007). This basic 6 TMS unit then gave rise to two distinct types of proteins with 8 TMSs, exemplified by ResC and archaeal CcmC homologues, respectively. These underwent further fusional or insertional events yielding proteins with 10, 11 and 13 TMSs (ResC homologues) as well as 15 TMSs (CcmF homologues). Specific evolutionary pathways taken were proposed and are illustrated in Figure 2. This work provided the first evidence for a pathway of appearance of distantly related proteins required for post-translational maturation of c-type cytochromes in bacteria, plants, protozoa and archaea.
Figure 2.
Proposed pathway for the evolutionary appearance of the many topological types of the integral membrane proteins of the HHP family.
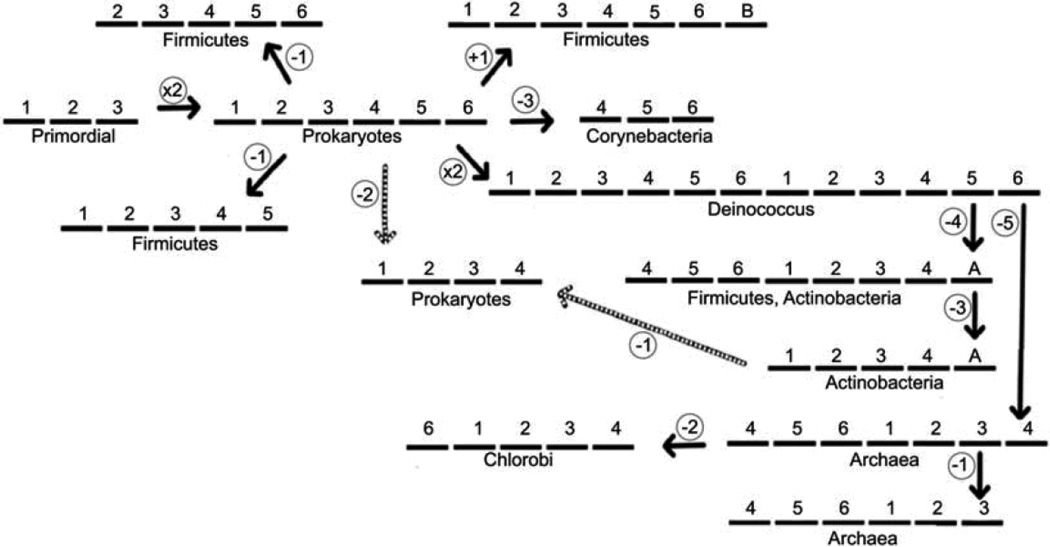
4. The SdpC (Peptide-Antibiotic Killer Factor) Immunity Protein, SdpI Family (TC# 9.A.32)
The SdpI protein of B. subtilis provides defense against cannibalism by its neighbors of the same species that produce a peptide toxin called SdpC. In 2006, Ellermeier et al. (2006) described a three-protein signal-transduction pathway that governs immunity to the SdpC toxin involved in “cannibalism” by this spore-forming bacterium. Cells of B. subtilis enter a regulatory progression leading to sporulation under conditions of nutrient limitation, but they delay becoming committed to spore formation by killing nonsporulating siblings and feeding on the dead cells. Killing is mediated by the exported toxic protein SdpC. Ellermeier et al. (2006) reported that extracellular SdpC induces the synthesis of an immunity protein, SdpI, that protects toxin-producing cells from being killed. SdpI, a polytopic membrane protein, is encoded by a two-gene operon under sporulation control that also contains the gene for an autorepressor, SdpR. The autorepressor binds to and blocks the promoter for the SdpIR operon. Evidence indicated that SdpI is also a signal-transduction protein that responds to the SdpC toxin by binding it on the outside and sequestering the SdpR autorepressor at the inner surface of the cytoplasmic membrane. Sequestration relieves repression and stimulates synthesis of the immunity protein.
Recognized SdpI family members are transmembrane proteins with 3, 4, 5, 6, 7, 8 or 12 putative transmembrane α-helical segments (TMSs). These varied topologies appear to be genuine rather than artifacts due to sequencing errors. The basic and most frequently occurring members of the SdpI family have 6 TMSs. In our studies of this family, homologues of all topological types were aligned to determine the homologous TMSs and loop regions, and the positive-inside rule was used to determine membrane sidedness. The two most conserved motifs were identified between TMSs 1 and 2 and TMSs 4 and 5 of the 6 TMS proteins (Povolotsky et al., 2010). These showed significant sequence similarity, leading to the suggestion that the primordial precursor of these proteins was a 3 TMS-encoding genetic element that underwent intragenic duplication to yield the standard 6 TMS proteins. Various deletional and fusional events that occurred during the evolution of this family, as well as intragenic duplication of the 6 TMS unit to 12 TMS proteins appear to have yielded SdpI homologues with topologies of varying numbers and positions of TMSs (see Figure 3). One 3 TMS element is believed to bind the toxin SdpC, on the outside, while the other binds the relevant transcriptional regulator (e.g. SdpR) on the inside (Ellermeier et al., 2006; see TCDB TC# 9.A.32). Binding of the toxin on the outside presumably transmits a signal, changing the affinity of SdpI for the regulator on the inside. Thus, the B. subtilis SdpI is a signal transduction protein with two independent protein binding functions associated with the two 3-TMS halves. Povolotsky et al. (2010) proposed the evolutionary pathway shown in Figure 3, which could have given rise to all of these distantly related putative bacterial immunity proteins. It was further shown that genes encoding SdpI homologues often appear in operons with genes for homologues of SdpR, SdpI’s autorepressor, although other types of transcriptional regulators may instead be present. These analyses allowed proposal of structure-function relationships that may be applicable to many, if not all family members (Povolotsky et al., 2010).
Figure 3.
Proposed pathway for the evolutionary appearance of the many topological types of the integral membrane proteins of the SdpI family.
5. Evolution of P-type ATPases (TC# 3.A.3)
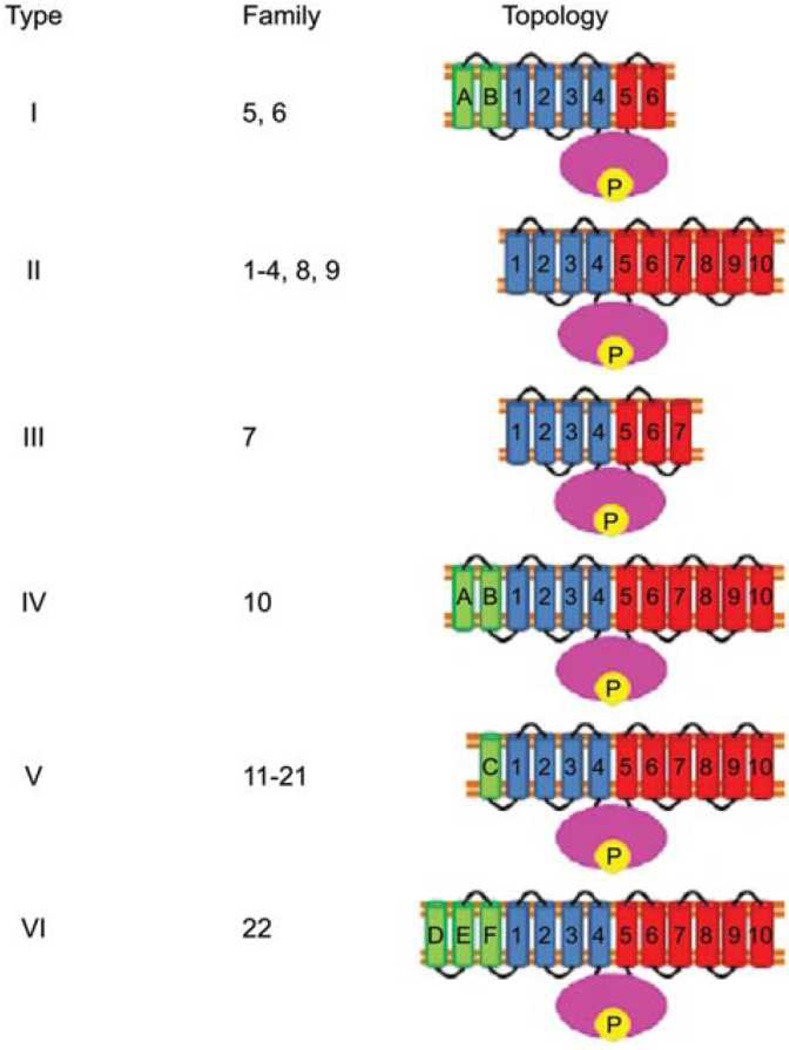
P-type ATPases play essential roles in numerous processes, which in humans include nerve impulse propagation, relaxation of muscle fibers, secretion and absorption in the kidney, acidification of the stomach and nutrient absorption in the intestine. Published evidence suggests that uncharacterized families of P-type ATPases with novel specificities and topologies exist. In a recent study, Thever and Saier (2009) examined the fully sequenced genomes of 26 eukaryotes, including animals, plants, fungi and several unicellular eukaryotes for P-type ATPases. The organismal distributions, phylogenetic relationships, probable topologies and conserved motifs of nine functionally characterized families and 13 uncharacterized families of these enzyme transporters were reported. These proteins were classified according to the conventions of the functional and phylogenetic T.C. system (www.tcdb.org, Saier et al., 2006; 2009). Knowledge of their topologies (Figure 4) suggested a pathway for their evolutionary appearance (see Figure 5). In later unpublished work, prokaryotic ATPases were similarly analyzed.
Figure 4.
Six putative topological types of P-type ATPases. Topologies I–III have been defined previously. Types IV–VI were identified by Thever and Saier (2009). In type IV, A’ and B’ are the two proposed TMSs preceding TMS 1, so designated because they exhibit sequence similarity to TMSs A and B in family 5 and 6 proteins (type I topology). TMS C in type V ATPases and TMSs D–F in type VI ATPases many in part share a common origin with A and B, but they lack significant sequence similarity with A and B and are designated C–F accordingly. The presence of odd numbers of TMSs preceding TMS 1 implies that, in contrast to all well-characterized P-type ATPases, the N-terminal sequences of these proteins are extracellular (Thever and Saier, 2009).
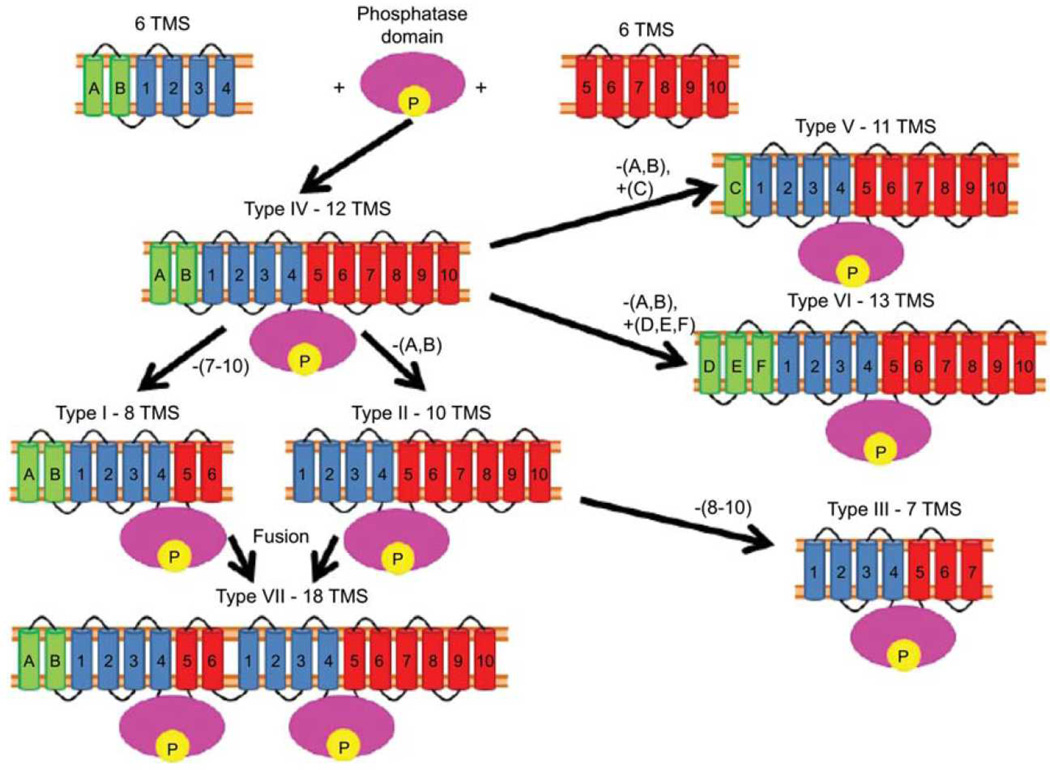
Figure 5.
Proposed pathway for the evolution of the different topological types of P-type ATPases established (I–III) or proposed (IV–VII). See figure 4 for a depiction of these topologies I–VI. Topological type VII represents a fusion of ATPase types I and II with loss of sequence motifs, clear topological features and ATPase activity for the N-terminal halves of these proteins.
Three standard P-type ATPase topologies are currently established and recognized by researchers in the field. They are numbered types I, II and III (see Figure 4). Based on hydropathy plots for individual proteins, and average hydropathy plots for the various families of P-type ATPases, an additional four topologies (types IV-VII) have been discovered. Type I ATPases have 8TMSs which occur in four pairs: A, B, 1,2, 3,4 and 5,6 (see Figure 4). Type II ATPases lack TMSs A and B, but possess TMSs 1–10 (Figure 4). Type III ATPases lack TMSs A and B as well as TMSs 8–10. Type III ATPases relegate their transport function to another subunit in the ATPase complex and thus serve only as a transporter energizer. For this reason, it is not surprising that in these type III Kdp K+ uptake porters, TMSs 5–7 show little or no sequence similarity with other P-type ATPases. This may reflect their lack of transport function since TMSs 3–6 are known to provide the cation transport pathway. Figure 5 shows our proposed pathway for the interconversion of the seven topological types. Strikingly, type IV ATPases have 12 TMSs (A, B and 1–10), all of the TMSs observed for both types I and II enzymes. We propose that type IV enzymes resulted from the fusion of two dissimilar 6 TMS proteins with a central phosphatase domain, “P” in figure 5, to give a present-day type IV ATPase. This then lost TMSs 7–10 to yield type I enzymes, lost TMSs A and B to give type II ATPases, and lost both TMSs A, B and TMSs 8–10 to give type III ATPases. We then proposed that topological types V and VI arose from Type IV by loss and gain of a single N-terminal TMS, respectively, and that type VII resulted from a fusion of a type II and a type I ATPase to yield an 18 TMS protein which retained its C-terminal ATPase function while losing the N-terminal ATPase activity. Presumably the enzymatically inactive N-terminal half became of use for another purpose such as a membrane anchor, a stabilizing element, a regulatory domain or a protein-protein-interaction domain, promoting for multi component complex formation (Charbit, Reizer & Saier, 1996).
6. Duplications that generate membrane proteins with odd numbers of TMSs
An examination of the numbers of TMSs in membrane transport systems have revealed that a minority of them have odd numbers of TMSs, yet duplication of a precursor peptide with either an odd or even number of TMSs should yield an even number of TMSs. Examination of families of such proteins has revealed that they either have the “extra” non-homologous TMS in the middle, between the repeat units, or at one end or the other. Thus for example, the microbial rhodopsins (TC# 3.E.1) consist of seven TMS proteins where TMSs 1–3 are homologous to TMSs 5–7. The extra one, TMS 4, is not demonstrably homologous to any of the others. It could have arisen by duplication of 3 with the generation of a new one in the middle, allowing for the 3 TMS repeat units to retain the same orientation in the membrane. However, it is just as likely that these proteins arose by duplication of a 4 TMS precursor followed by loss (by genetic deletion) of TMS 1 or TMS 8. Available methods cannot distinguish these possibilities.
On the other hand, analysis of recently identified Trimeric Cation Channel (TRIC) family (TC# 1.A.62) revealed that the seven TMSs of these proteins have a 3+3+1 arrangement where TMSs 1–3 are homologous to TMSs 4–6, and 7 is non homologous to the others. In this case a 3 TMS-encoding genetic element must have duplicated to give six with the two repeat units having opposite orientations in the membrane, and then the seventh TMS was added. According to Occam’s razor, this is the most probable pathway.
C. Retroevolution
Normally, the evolutionary process uses simple molecules and organisms and builds towards complexity. Thus, for example, according to the fossil record and numerous molecular biological inferences, simple bacteria were on Earth for hundreds of million years before complex eukaryotic cells arose and multicellular eukaryotes, animals, plants, and fungi, appeared much much later. In recent studies we have uncovered a case of “retroevolution” where a complex protein gave rise to a fundamentally and structurally more simple protein (Matias et al., 2010). The example cited here involved derivation of a cation-specific channel protein present in animals (Orai or CRAC; TC# 1.A.52) from a larger and fundamentally more complex carrier (CDF; TC# 2.A.4).
Antigen stimulation of immune cells triggers Ca2+ entry through Ca2+ release-activated Ca2+ (CRAC) channels, promoting an immune response to pathogens. Defects in a CRAC (Orai) channel in humans gives rise to the hereditary Sever Combined Immune Deficiency (SCID) syndrome. Matias et al. (2010) first reported results that defined the evolutionary relationship of the CRAC channel proteins of animals to the ubiquitous Cation Diffusion Facilitator (CDF) carrier proteins (see Table 1).
Table 1.
Comparisons of CDF carriers with CRAC channels.
| CDF (2.A.4) | CRAC-C (1.A.52) |
|---|---|
| Secondary carriers: catalyze Me2+H+ antiport | Channels: catalyze bidirectional Ca2+ flux |
| Ubiquitous; in plasma and intracellular membranes of eukaryotes | Present only in eukaryotes; at the plasma membrane/endoplasmic reticulum junctions |
| Six TMSs; N- and C-termini inside; dimeric | Four TMSs; N- and C-termini inside; tetrameric |
| Much size and sequence divergence | Little size and sequence divergence |
| Two aspartates are crucial for Me2+ binding | Two glutamates are crucial for Ca2+ binding |
| Me2+, divalent heavy metal ion. |
CDF antiporters derived from a primordial 2 TMS hairpin structure by intragenic triplication to yield 6 TMS proteins Matias et al. (2010) used four programs (IC/GAP, GGSEARCH, HMMER and SAM) to identify sequence similarity, thus establishing homology using statistical means, and these programs were also evaluated for their relative abilities to determine distant relationships. Overall, the order of sensitivity (similarity detection) was IC/GAP > GGSEARCH > HMMER > SAM, but the use of all four programs was superior to the use of any two or three of them. Regardless, all programs gave results consistently with the conclusion that the last four TMSs of members of the CDF family were homologous to the 4 TMSs of Orai channel proteins (Table 1).
Thus, the smaller CRAC channels probably derived from CDF carriers by loss of the first two TMSs of the latter. In other words TMSs 3–6 in CDF carriers are homologous to TMSs 1–4 in CRAC channels, and the former was the precursor of the latter. This is an unusual example of how a functionally and structurally more complex protein may have predated a simpler one.
D. Conclusions and Perspectives
As noted by Francois Jacob over 30 years ago, “Evolution is a tinkerer,” and as P.Z. Meyers elaborated, “Evolution is a tinkerer that cobbles together new functions from old ones, and the genome is a kind of parts bin of recyclable elements. When new features evolve, the parts in the bin are co-opted to operate in new roles. As a result, the same parts appear in anatomically and evolutionarily distinct structures because it is faster and easier to reuse an old gene network that almost does what is needed, than it is to spend another few million years evolving a distinct gene for the function.”
Thus, we have seen that complex carriers have originated from simple membrane embedded 2 TMS hairpin structures (e.g. DMT, CDF and OPT family members). Moreover, while some families of transport proteins exhibit topological constancy (e.g. MFS, TC# 2.A.1), others (e.g., HHP and SdpI) vary tremendously. Finally, while evolution at the molecular level tends towards complexity as illustrated here and in many of our earlier publications (Saier, 2003a, b), examples of the opposite, such as CRAC channels deriving from CDF carriers, have been identified. Generally, alteration of function may correlate with structural differences as, for example, is observed for P-type ATPases.
The study of evolution has yielded many surprising results, all of which illustrate the wisdom of Francois Jacob and P.Z. Meyers. Further studies are likely to reveal the unlimited, mindless “imaginations” of evolutionary processes. These far supersede those of the most intellectually advanced organisms on Earth. We can only observe and marvel.
Acknowledgments
We thank the NIH (GM 077 402) for financial support.
References
- Charbit A, Reizer J, Saier MH., Jr Function of the duplicated IIB domain and oligomeric structure of the fructose permease of Escherichia coli. J Biol Chem. 1996;271:9997–10003. doi: 10.1074/jbc.271.17.9997. [DOI] [PubMed] [Google Scholar]
- Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Hvorup RN, Winnen B, Chang AB, Jiang Y, Zhou XF, Saier MH., Jr The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur J Biochem. 2003;270:799–813. doi: 10.1046/j.1432-1033.2003.03418.x. [DOI] [PubMed] [Google Scholar]
- Lee JH, Harvat EM, Stevens JM, Ferguson SJ, Saier MH., Jr Evolutionary origins of members of a superfamily of integral membrane cytochrome c biogenesis proteins. Biochim Biophys Acta. 2007;1768:2164–2181. doi: 10.1016/j.bbamem.2007.04.022. [DOI] [PubMed] [Google Scholar]
- Matias MG, Gomolplitinant KM, Tamang DG, Saier MH., Jr Animal Ca2+ release-activated Ca2+ (CRAC) channels appear to be homologous to and derived from the ubiquitous cation diffusion facilitators. BMC Res Notes. 2010;3:158. doi: 10.1186/1756-0500-3-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povolotsky TL, Orlova E, Tamang DG, Saier MH., Jr Defense Against Cannibalism: The SdpI Family of Bacterial Immunity/Signal Transduction Proteins. J Membr Biol. 2010 doi: 10.1007/s00232-010-9260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58:71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH., Jr Phylogenetic approaches to the identification and characterization of protein families and superfamilies. Microb Comp Genomics. 1996;1:129–150. doi: 10.1089/mcg.1996.1.129. [DOI] [PubMed] [Google Scholar]
- Saier MH., Jr Molecular phylogeny as a basis for the classification of transport proteins from bacteria, archaea and eukarya. Adv Microb Physiol. 1998;40:81–136. doi: 10.1016/s0065-2911(08)60130-7. [DOI] [PubMed] [Google Scholar]
- Saier MH., Jr . Classification of transmembrane transport systems in living organisms. In: L VW, editor. Biomembrane Transport. San Diego: Academic Press; 1999a. pp. 265–276. [Google Scholar]
- Saier MH., Jr Eukaryotic transmembrane solute transport systems. Int Rev Cytol. 1999b;190:61–136. doi: 10.1016/s0074-7696(08)62146-4. [DOI] [PubMed] [Google Scholar]
- Saier MH., Jr Genome archeology leading to the characterization and classification of transport proteins. Curr Opin Microbiol. 1999c;2:555–561. doi: 10.1016/s1369-5274(99)00016-8. [DOI] [PubMed] [Google Scholar]
- Saier MH., Jr A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev. 2000a;64:354–411. doi: 10.1128/mmbr.64.2.354-411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH., Jr Families of proteins forming transmembrane channels. J Membr Biol. 2000b;175:165–180. doi: 10.1007/s00232001065. [DOI] [PubMed] [Google Scholar]
- Saier MH., Jr Families of transmembrane sugar transport proteins. Mol Microbiol. 2000c;35:699–710. doi: 10.1046/j.1365-2958.2000.01759.x. [DOI] [PubMed] [Google Scholar]
- Saier MH., Jr Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology. 2000d;146(Pt 8):1775–1795. doi: 10.1099/00221287-146-8-1775. [DOI] [PubMed] [Google Scholar]
- Saier MH., Jr Vectorial metabolism and the evolution of transport systems. J Bacteriol. 2000e;182:5029–5035. doi: 10.1128/jb.182.18.5029-5035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH., Jr Evolution of transport proteins. Genet Eng (N Y) 2001;23:1–10. doi: 10.1007/0-306-47572-3_1. [DOI] [PubMed] [Google Scholar]
- Saier MH., Jr Answering fundamental questions in biology with bioinformatics. ASM News. 2003a;69:175–181. [Google Scholar]
- Saier MH., Jr Tracing pathways of transport protein evolution. Mol Microbiol. 2003b;48:1145–1156. doi: 10.1046/j.1365-2958.2003.03499.x. [DOI] [PubMed] [Google Scholar]
- Saier MH., Jr Active transport in communication, protection and nutrition. J Mol Microbiol Biotechnol. 2007;12:161–164. doi: 10.1159/000099638. [DOI] [PubMed] [Google Scholar]
- Saier MH., Jr . Cell Membrane, Prokaryotic. In: Schaechter M, editor. Encyclopedia of Microbiology. Oxford: Elsevier; 2009. pp. 341–356. [Google Scholar]
- Saier MH, Jr, Ma CH, Rodgers L, Tamang DG, Yen MR. Protein secretion and membrane insertion systems in bacteria and eukaryotic organelles. Adv Appl Microbiol. 2008;65:141–197. doi: 10.1016/S0065-2164(08)00606-0. [DOI] [PubMed] [Google Scholar]
- Saier MH, Jr, Ren Q. The bioinformatic study of transmembrane molecular transport. J Mol Microbiol Biotechnol. 2006;11:289–290. doi: 10.1159/000095630. [DOI] [PubMed] [Google Scholar]
- Saier MH, Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The Transporter Classification Database: recent advances. Nucleic Acids Res. 2009;37:D274–D278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thever MD, Saier MH., Jr Bioinformatic Characterization of P-Type ATPases Encoded Within the Fully Sequenced Genomes of 26 Eukaryotes. J Membr Biol. 2009 doi: 10.1007/s00232-009-9176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]