Abstract
Mice deficient small heterodimer partner (SHP) are protected from diet induced hepatic steatosis due to increased fatty acid oxidation and decreased lipogenesis. The decreased lipogenesis appears to be a direct consequence of very low expression of peroxisome proliferator activated receptor gamma 2 (PPARγ2), a potent lipogenic transcription factor, in the SHP−/− liver. The current study focuses on the identification of a SHP dependent regulatory cascade that controls PPARγ2 gene expression, thereby regulating hepatic fat accumulation. Illumina BeadChip array and real-time polymerase chain reaction were used to identify genes responsible for the linkage between SHP and PPARγ2 using hepatic RNAs isolated from SHP−/− and SHP-overexpressing mice. The initial efforts identify that hairy and enhancer of split 6 (Hes6), a novel transcriptional repressor, is an important mediator of the regulation of PPARγ2 transcription by SHP. The Hes6 promoter is specifically activated by the retinoic acid receptor (RAR) in response to its natural agonist ligand all-trans retinoic acid (atRA), and is repressed by SHP. Hes6 subsequently represses hepatocyte nuclear factor 4 alpha (HNF4α) activated-PPARγ2 gene expression via direct inhibition of the HNF4α transcriptional activity. Furthermore, we provide evidences that atRA treatment or adenovirus-mediated RARα overexpression significantly reduced hepatic fat accumulation in obese mouse models as observed in earlier studies and the beneficial effect is achieved via the proposed transcriptional cascade.
Conclusions
Our study describes a novel transcriptional regulatory cascade controlling hepatic lipid metabolism that identifies retinoic acid signaling as a new therapeutic approach to non-alcoholic fatty liver diseases.
Keywords: Lipid homeostasis, Nuclear Hormone Receptors, Fsp27, SHP, Hes6
SHP, an orphan nuclear hormone receptor, functions as a corepressor and antagonizes coactivator recruitment to target transcription factors with which it interacts (1, 2). Molecular and genetic studies have suggested that SHP also plays a role in obesity and diabetes (2-6). Supporting this, deletion or overexpression of SHP gene in mice disrupts lipid and glucose homeostasis (7-10). Congenic C57BL/6 SHP−/− mice show a more insulin resistant phenotype in response to a western diet (WestD), but are protected against development of hepatic steatosis and obesity (7, 11). The protection against steatosis was attributed to enhancement of hepatic β-oxidation and reduced expression of fatty acid synthetic genes. Gene expression profiling revealed that PPARγ was markedly down-regulated in SHP−/− mouse liver regardless of type of diets, suggesting that the effect of SHP on PPARγ expression is not a secondary complication of fat accumulation.
PPARγ expression is low in normal livers, but is strongly induced in models of nonalcoholic fatty liver diseases (NAFLD). Given its role in adipogenesis, this induction has been considered a key factor contributing to fat accumulation in the livers of animal models and human patients associated with obesity and diabetes (12-16). Indeed, adenoviral overexpression of PPARγ in the liver is sufficient to drive marked steatosis (17-19), and liver specific deletion of PPARγ strongly decreases steatosis in the ob/ob background (13) and high fat-fed mice (14, 15, 18). One proposed mechanism for PPARγ-dependent fat accumulation in ob/ob mice is direct induction of fat specific factor 27 (Fsp27), a lipid droplet binding protein promoting lipid accumulation in adipocytes (20). As observed in white adipose tissue, deletion of Fsp27 decreases the size of lipid droplets in liver and increases lipolysis and fatty acid oxidation due to increased accessibility of lipolytic enzymes and increased availability of non esterified free fatty acids (FFA) without affecting FFA uptake, triglyceride (TG) export, and de novo TG synthesis (20, 21). Thus, decreased PPARγ expression in SHP−/− liver (7, 11) may protect the mice from development of hepatic steatosis at least in part by decreasing Fsp27 expression.
Hes6 is a novel member of the family of mammalian homologues of Drosophila hairy and enhancer of split, and was initially identified as an inhibitor of its relative Hes1, a basic helix-loop-helix transcription factor regulating neuronal and muscle differentiation negatively (22). Like SHP, Hes6 alone cannot bind directly to DNA, but represses target gene transcription through protein-protein interaction. Intriguingly, Hes6 was recently identified as a repressor of hepatic PPARγ expression via inhibition of HNF4α transactivation and its expression is directly regulated by HNF4α, which is one of the direct targets of SHP-mediated repression (2, 23). These results suggest that Hes6 may function as a mediator in the regulation of fatty acid synthesis by SHP.
In the present study, we discovered a novel transcriptional cascade consisting of RAR, Hes6, HNF4α, and PPARγ in the regulation of hepatic fat mobilization. In the proposed transcriptional cascade, SHP and atRA coordinately regulates transcription of Hes6, and Hes6 subsequently suppresses Pparγ2 expression via repression of HNF4α transcriptional activity. We confirmed the physiologic effects of atRA and RAR on fat mobilization via the cascade using mouse models of fatty liver disease.
Materials and Methods
Mice and Treatment
The congenic SHP−/− mice (11, 24) were bred and housed with age-matched C57BL/6 mice (Harlan, Indianapolis, IN). Only male mice were used throughout experiments. The mice were fed a WestD (Harlan) or chow diet (CD) (Lab Diet, Brentwood, MO) as described (11). To overexpress SHP in mouse liver, AdSHP (1 × 109 pfu/mouse) (7) was delivered into 3 month-old C57BL/6 mice via tail vein. The mice were euthanized 7 days after injection and their livers were collected for RNA isolation. For atRA treatment, C57BL/6 mice were on WestD regimen for 4 months to induce hepatic steatosis and ob/ob mice were purchased from Harlan. Either ob/ob or the WestD-fed mice were challenged orally with corn oil containing atRA (15mg/kg/day) or vehicle (DMSO) for 7 days before euthanasia. For hepatic RARα overexpression, 4 month-old C57BL/6 mice fed WestD for 2 month were injected with adenovirus expressing mouse RARγ (1 × 109 pfu/mouse) and maintained for 7 days. Blood glucose was measured on tail vein samples using AlphaTRAK glucose monitoring system (Abbott Lab, Abbott Park, IL). Mice were maintained in the accredited pathogen-free facility at Northeast Ohio Medical University on a 12-hour light/dark cycle and fed a CD and water ad libitum. All animal experiments were performed in accord with the protocols approved by the Institutional Animal Care and Use Committee of the University.
Illumina Beadchip Array Analysis and Quantitative Real-time PCR
Livers of wild type C57BL/6 (WT) and SHP−/− mice fed either chow or WestD (n=4) for 22 weeks were collected after overnight fasting. Their RNAs were processed and analyzed on the MouseRef-8 v2.0 BeadChip (Illumina, San Diego, CA) array to assess gene expression at Lerner Research Institute in Cleveland Clinic (Cleveland, OH). The array results are deposited in Gene Expression Omnibus data bank at the National Cancer for Biotechnology Information (Accession number: GSE38013). Quantitative PCR analysis was performed using SYBR green (11).
Transient Transfection and Chromatin Immunoprecipitation
Luciferase reporter genes driven by mouse Hes6 promoters were constructed by insertion of polymerase chain reaction (PCR)-amplified promoters into pGL3 reporter plasmid (Promega, Madison, WI). All the plasmid and transfection conditions used in this study were described previously (2, 11). Mouse Hes6 cDNA in pCMV-SPORT6 was purchased from Open Biosystems (Lafayette, CO). HeLa cells were used for transfection assays. For ChIP assay, mouse liver nuclei were isolated and processed as described (11). Antibodies against SHP, RARα, Hes6, or HNF4α and IgG were purchased from Santa Cruz (Santa Cruz, CA). The immunocomplexes formed from overnight incubation were precipitated using protein A agarose (Millipore, Bedford, MA). Decrosslinked DNA fragments were amplified and quantified by PCR using primers designed for Hes6 promoter regions (Supplemental Table 1).
Adenovirus Expressing RARα
Mouse RARα in pCMV-SPORT6 was purchased from Open Biosystems and subcloned into pacAd5 CMV shuttle vector to generate AdRARα using RAPAd Expression System (Cell Biolabs, San Diego, CA). Viral titration was performed using Adeno-X Rapid Titer Kit (Clontech, Mountain View, CA).
Hepatic Lipid Extraction and Histology
Lipid extraction from liver was performed as described (11). Extracted TG and cholesterol contents were normalized to wet liver weight. For histology, livers collected from mice after overnight fasting were fixed in 10% formalin and embedded in paraffin. After sectioning, samples were stained with hematoxylin and eosin (H&E) for microscopic observation. For Oil red O staining, livers were fixed in 10% formalin for 1hr and incubated in 30% sucrose for 24hrs before cryosectioning and staining.
Results
Illumina BeadChip Array analysis
PPARγ is an imporant contributor to the development of hepatic steatosis, and its expression is markedly decreased in SHP−/− livers under both normal and high fat diet conditions. To explore the mechanism for this repression, we carried out gene expression profiling using Illumina BeadChip array analysis with total RNA isolated individual liver (n=4) from WT and SHP−/− mice fed chow or WestD. Since metabolic phenotypes and differential gene expression in SHP−/− mice had been evident upon WestD challenge, we searched genes differentially expressed between the two genotypes in the WestD-fed group and identified total 1010 using the screening criteria described in Table 1 (Supplemental table 2). Among these 1010 genes, 624 genes are downregulated and 386 genes are upregulated in SHP−/− mice compared to WT mice. We identified 7 transcription factors among the 386 upregulated genes (Table 1). As similarly observed earlier, none of the transcription factors are differentially expressed between the two genotypes in chow-fed condition. Major corepressors and chromatin remodeling proteins were eliminated by the screening criteria (Supplemental table 3), which additionally identified some genes involved in lipid and cholesterol metabolism (Supplemental table 4). Hes6 was particularly interesting among the 7 transcription factors because of its repression function and its suggested role in hepatic lipid homeostasis through inhibition of PPARγ gene transcription (23). The lipid droplet forming factor Fsp27, a direct target of PPARγ, was also significantly down-regulated in the livers of SHP−/− mice (Supplemental Table 4) (20). The array results suggested that Hes6 repression by SHP could directly increase PPARγ gene expression and promote hepatic lipid accumulation.
Table 1.
Transcription factors expressed higher in SHP−/− mice fed WestD than in wild type counterparts
| Gene Symbol | Name | Genbank | KOC/WTC | KOWD/WTWD | p-value |
|---|---|---|---|---|---|
| Foxa3 | Forkhead box A3, Hnf3gamma | NM_008260 | 1.06 | 1.54 | 0.06 |
| Hes6 | Hairy and enhancer of split 6 | NM_019479 | 0.96 | 1.97 | 0.2 |
| Hsf2 | Heat shock factor 2 | NM_008297 | 1.09 | 1.56 | 0.13 |
| Nr1i3 | Nuclear hormone receptor CAR | NM_009803 | 0.85 | 1.69 | 0.07 |
| Rorc | RAR-related orphan receptor gamma | NM_011281 | 0.98 | 2.54 | 0.07 |
| Srebf2 | Sterol regulatory element binding factor 2 | NM_033218 | 1.15 | 1.81 | 0.01 |
| Tfb1m | Mitochondrial transcription factor B1 | NM_146074 | 0.98 | 1.53 | 0.07 |
Gene expression profile results from livers of wild type and SHP−/− fed chow or WestD (n=4) using Illumina BeadChip array were screened for genes differentially regulated between the two genotypes when fed WestD. Group averages after quantile normalization were used to obtain p values and expression ratios of KO vs WT. The screening parameters are as follows. P values of WestD-fed group should be ≤ 0.2. Any group average should be higher than 40 if p values are between 0.05 and 0.2. Fold changes should be greater than 1.5. Among the 1010 screened genes (supplemental Table 2), well known transcription factor genes upregulated in SHP−/− mice compared to WT are presented.
A new molecular mechanism in SHP-regulated hepatic lipid accumulation: the SHP-Hes6-PPARγ2-Fsp27 cascade
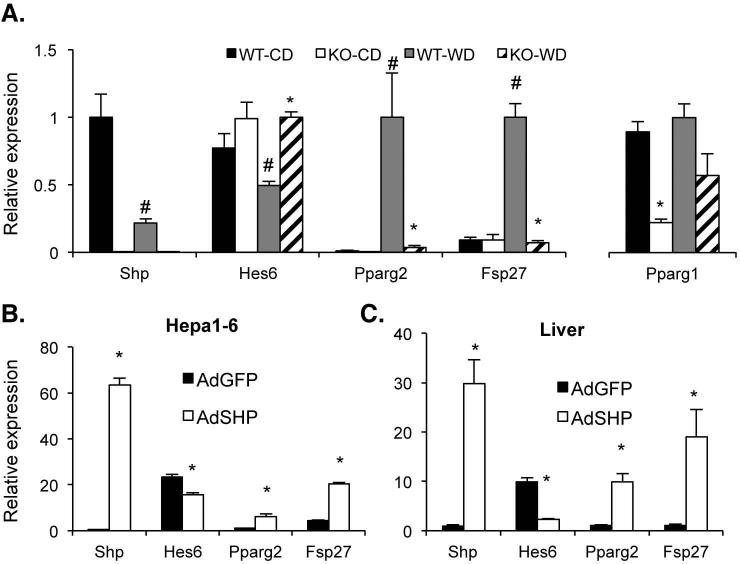
The proposed transcriptional regulation of Hes6 by SHP prompted us to examine the expression of their potential downstream target genes such as PPARγ and Fsp27 (20, 23) in the development of fatty liver using WT and SHP−/− mice fed WestD. As shown in Fig. 1A, WestD-challenged C57BL/6 WT mouse liver showed decreased Hes6 mRNA level and increased PPARγ2 and Fsp27 mRNA levels compared with those of chow-fed counterparts. However, expression of these genes was not changed in SHP−/− mice upon WestD challenge, with higher Hes6 expression and dramatically reduced expression of PPARγ2 and Fsp27. PPARγ1 expression was not significantly different between the two genotypes fed WestD as observed previously. These data strongly suggest that SHP is upstream of a Hes6-PPARγ2-Fsp27 transcriptional cascade to determine hepatic TG levels.
Fig. 1. mRNA levels of hepatic Hes6, PPARγ, and Fsp27 in the suggested transcriptional cascade upon various challenges.
(A) RNAs isolated from livers of SHP−/− and WT mice after 22 weeks WestD regimen were processed for qPCR to quantify the expression of the indicated genes. (B) Hepa1-6, a mouse hepatoma cell line, cells were infected with adenovirus expressing SHP (AdSHP) or GFP (AdGFP) for 3 days. Isolated RNAs were quantified for the indicated genes using qPCR. n = 3. (C) Hepatic RNAs from WT mice infected with AdSHP or AdGFP for 7 days were prepared and quantified for the indicated genes using qPCR. n = 3, *; p < 0.05 vs WT counterparts, #; p < 0.03 vs chow-fed control.
As an initial functional test of this suggested cascade, SHP was overexpressed in Hepa1-6, a mouse hepatoma cell line, and WT mouse livers using AdSHP infection. SHP overexpression results in a significant decrease in Hes6 expression and a strong induction of PPARγ2 and Fsp27 mRNA levels in both the Hepa1-6 cells and mouse livers (Fig. 1 B&C). As reported earlier, AdSHP infection increased fat accumulation almost 2 fold (22.8 ± 3.7 vs. 13.0 ± 0.5 mg/g of dried liver weight) compared to AdGFP control infection (8).
Regulation of Hes6 by SHP and all-trans-retinoic acid
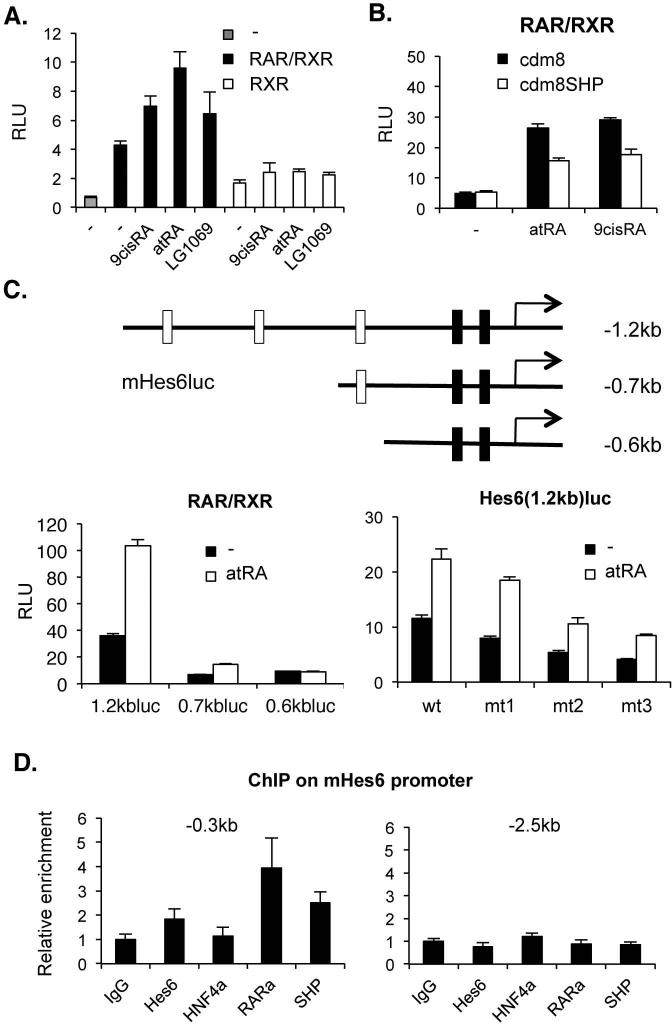
Based on the suggested regulation of Hes6 gene transcription by HNF-α (23) which is also a potential target of SHP (2), we examined the direct impact of SHP on Hes6 expression using Hes6 promoter driven luciferase reporters. In contrast to previous observations, the 2.7kb Hes6 promoter driven luciferase reporter was not transactivated by HNF-α or repressed by additional SHP transfection (Supplemental Fig. 1). Since we identified a number of potential nuclear receptor response elements in the 2.7kb promoter region, we screened several other nuclear hormone receptors for the promoter activation. This revealed a robust activation in response to RAR/RXR heterodimers and their ligands (Fig. 2A) including the synthetic RAR agonist TTNPB (data not shown) but not the RXR specific agonist LG1069. As expected from the strong interaction of SHP with RXR (2), SHP efficiently repressed Hes6 promoter activity induced by RAR-RXR heterodimers with either atRA or 9-cis RA (Fig. 2B). In order to locate the atRA response elements, we constructed a series of 5’ deletion promoter reporters. As shown in Fig. 2C, the -1.2kb promoter retained full atRA response (compared to -2.7kb promoter, data not shown), while the -0.7kb promoter had a reduced response, and deletion to -0.6kb abolished the response completely. These results are consistent with the presence of 3 potential RAR response elements (DR5s) between -1.2 and -0.6kb (Fig. 2C, top panel). It has been reported that atRA can also serve as a ligand for the nuclear hormone receptor PPARδ (25-28). However, GW0742, a PPARδ-specific agonist (29), did not activate the reporter gene activity (data not shown). Moreover, specific recruitment of RAR and SHP was detected on the Hes6 promoter region in the mouse liver (Fig. 2D). In accord with the transfection results, recruitment of HNF-α to the Hes6 promoter was not significant.
Fig. 2. Regulation of Hes6 promoter by SHP and atRA.
(A) Hes6(2.7kb)luc activities by cotransfection of RAR plus RXR or RXR alone with treatment of 1μM of the indicated ligands. (B) Repression of RAR-mediated Hes6(2.7kb)luc activity by SHP. (C) Top panel, mHes6luc reporter plasmids. open bars; potential RAR-RXR response elements (RARE), black bars; reported HNF4α binding sites (23). Bottom left, atRA responsiveness of each deletion reporter. Bottom right, The luciferase activities of Hes6(1.2kb)luc containing mutations on each RARE with cotransfection of RAR/RXR in the presence or absence of 1μM atRA. (E) ChIP analysis on Hes6 promoter region from 2 mouse livers using indicated antibodies. 0.3kb upstream covers HNF4α and RAR binding sites. 2.5kb upstream for negative control.
Effect of atRA in mouse models of NAFLD
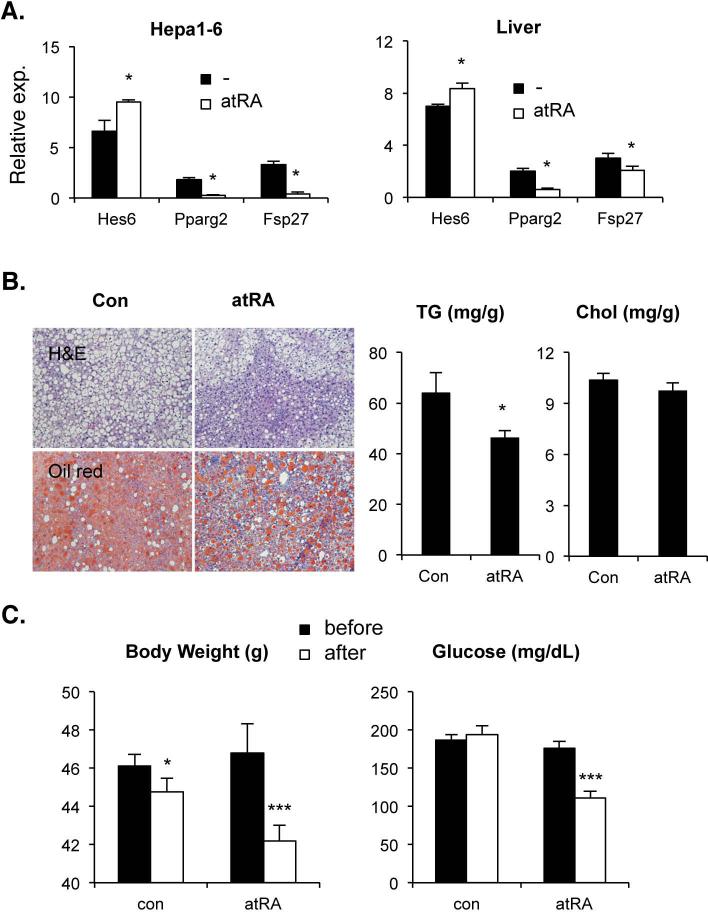
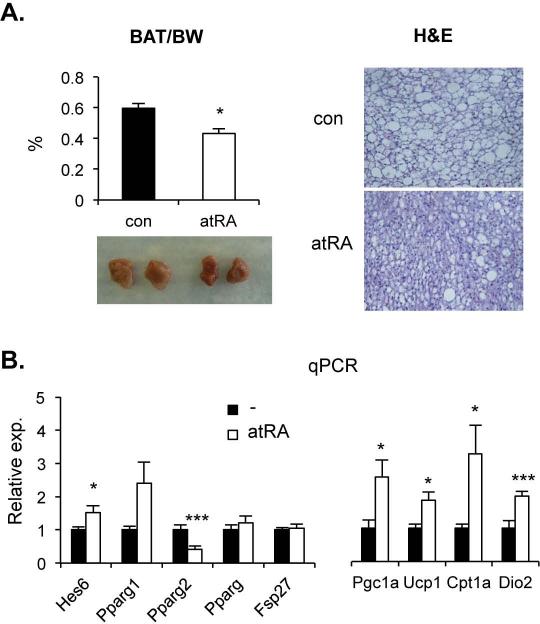
Berry et al. recently reported that long-term atRA treatment alleviated hepatic steatosis in obese mice fed a high fat diet (28). Our current results strongly suggest that the proposed transcriptional cascade could contribute to this response. To test this, mRNA levels of these genes were evaluated using Hepa1-6 and mouse livers after 24h atRA treatment (Fig. 3A). As predicted, Hes6 mRNA was induced, while PPARγ2 and Fsp27 transcripts were significantly reduced when compared to vehicle treated controls. Short-term atRA treatment (15mg/kg/day for 7 days) also effectively ameliorated the hepatic TG but not cholesterol accumulation in diet-induced obese and ob/ob mice (Fig. 3B and supplemental Fig. 2A). Along with hepatic TG reduction, the short-term atRA treatment significantly reduced body weight and serum glucose concentration (Fig. 3C and supplemental Fig. 2B). Though statistical significance was diminished in some genes, similar pattern of mRNA levels was observed in the 7-day-atRA-treated obese mice (Supplemental Fig. 3). Contrary to previous reports, however, our short-term treatment was not able to reduce white adipose tissue mass and cell size and serum TG content (Supplemental Fig. 2C, 4A, &5A). Instead we observed significant reduction of brown adipose tissue mass and fat content (Fig. 4A). Thus we examined the effect of atRA on the levels of mRNA of the cascade genes in these tissues. As shown in Fig. 4B and supplemental Fig. 4B, Hes6 and PPARγ2 were up- and down-regulated by atRA treatment, respectively, in both of the adipose tissues. However, the expression of Fsp27 was not significantly changed, which is consistent with the unaltered expression of total PPARγ mRNA due to higher expression of PPARγ1 in the atRA-treated adipocytes. Contrary to liver, the downregulated PPARγ2 expression by atRA in adipose tissues appears independent of HNF4α, since the expression of HNF4α mRNA was low in white adipose tissues but almost absent in brown adipose tissues (data not shown).
Fig. 3. Effect of atRA on hepatic lipid homeostasis.
(A) Hepa1-6 cells (left) or C57BL/6 mice (right) were treated with 1μM or 15mg/kg, respectively, of atRA or vehicle for 24hrs. mRNA levels of the indicated genes were isolated from cells or mouse liver and quantified using qPCR and plotted with SEM. n=3, *; p < 0.05 (B) A group of mice were challenged with WestD for 2 month and were daily gavaged with corn oil containing vehicle or atRA (15mg/kg) for 7 days while on the WestD regimen. Their livers were collected and processed for indicated stainings (left) or TG and total cholesterol measurement (right). (C) Their body weights and serum glucose levels were measured before and after atRA treatment, plotted and compared with those of vehicle treated control mice. Graphs are presented as means ± SEM (n=4). *; p < 0.05, ***; p < 0.005.
Fig. 4. Effect of atRA on brown adipocyte function.
(A) Brown adipose tissues were isolated from the atRA or vehicle-treated mice described in Fig. 3. Gross morphology and weight to body weight ratio are presented at left panel. Their fixed tissue sections were processed for hematoxilin and eosin staining. A representative image from each group is shown (right panel). (B) Total RNAs were isolated from brown adipose tissues and quantified for the expression level of indicated genes using real time PCR (n=4). *; p < 0.05, ***; p < 0.005.
Reduction of adiposity in brown fat by atRA treatment may directly link to the reduced body weight (30). Thus, we examined the expression of genes involved in thermogenesis. All the major genes tested were upregulated upon atRA treatment (Fig. 4B, right panel), suggesting that atRA may increase the brown adipocyte function probably via direct activation of UCP-1 (31).
Hepatic RARα overexpression ameliorates the progression of fatty liver in diet-induced obese mice
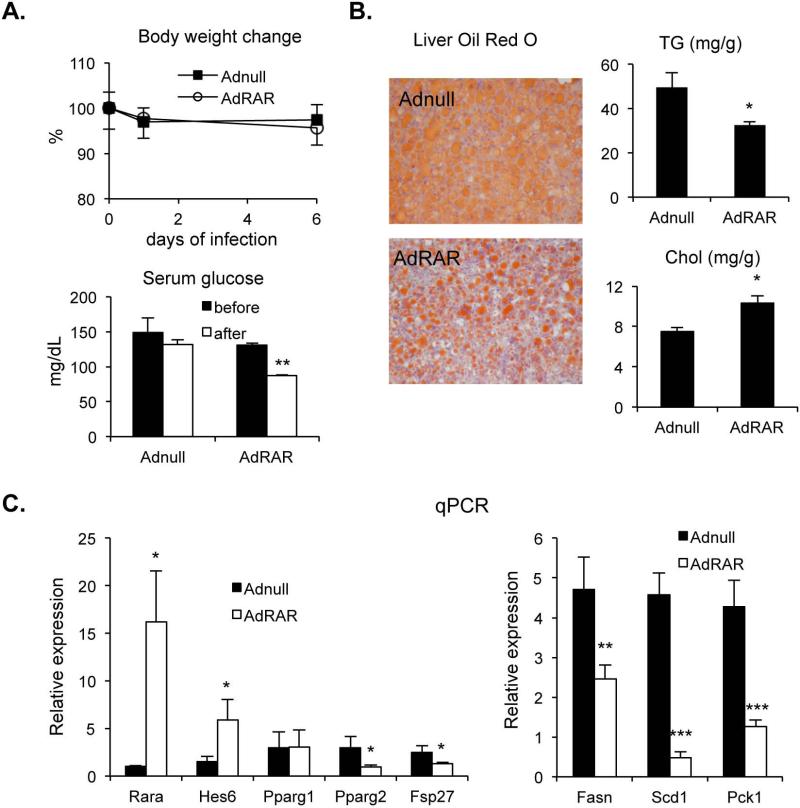
In order to test liver and RAR-specificity for the observed hepatic atRA effect, WT mice fed WestD were injected with an adenovirus expressing mouse RARα and hepatic gene expression and physiological parameters were examined. Seven days after injection the body weights of both Adnull and AdRAR-injected mice were decreased but were not significantly different between the two groups (Fig. 5A). The liver specific transduction was demonstrated by a lack of changes in the expression of key genes including RARα in brown and white adipocytes (Supplemental Fig. 6A, data not shown). Nonetheless serum glucose was significantly reduced after AdRAR injection as observed in atRA treated mice, suggesting suppression of hepatic gluconeogenesis contributes to the observed serum glucose reduction as evidenced by reduced gluconeogenic Pck1 gene expression (Fig. 5C). More importantly, hepatic overexpression of RARα alone is sufficient enough to reduce hepatic TG accumulation via the proposed transcriptional cascade and downregulation of the expression of numerous PPARγ target genes (Fig. 5B & 5C and supplemental Fig. 6B) but not serum TG and cholesterol (Supplemental Fig. 5B). These data suggest that atRA signals through RAR to inhibit PPARγ2 and its target gene expression via a novel transcriptional cascade thereby reducing hepatic TG accumulation (Fig. 6).
Fig. 5. Amelioration of hepatic lipid accumulation in diet-induced obese mice by RAR-α.
(A) WT mice were fed WestD for 2 months and infected with 1×109 pfu of Adnull or AdRAR for 7 days (4 mice per group). Their body weights and fasting serum glucose levels were measured before and after the viral injection and plotted with standard deviations. (B) Livers were collected for Oil Red O staining, TG and cholesterol contents. (C) Hepatic gene expression was examined using real time PCR with the indicated primers and plotted with SEM. *; p < 0.05, **; p < 0.01, ***; p < 0.005.
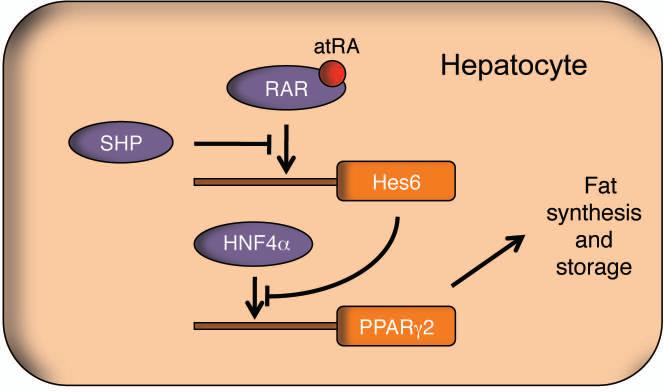
Fig. 6. Transcriptional regulatory network in atRA/SHP-controlled fat mobilization in hepatocyte.
In the suggested fat mobilization pathway, SHP represses Hes6 expression via RAR. Decreased level of Hes6 then derepresses HNF4α-regulated PPARγ2 expression. Eventually, the overexpressed PPARγ2 induces fat accumulation via turning on lipogenic programs in the liver. On the contrary, RAR ligand atRA results in fat utilization via shutting down PPARγ2 expression. Purple and orange represent protein and gene, respectively. Arrow indicates promoter region binding and activation.
Discussion
As a metabolic derivative of vitamin A, atRA functions as a natural ligand for the retinoic acid receptors (RARs), nuclear hormone receptors that regulate transcription of different sets of genes controlling cell differentiation, embryonic development, and physiological homeostasis (32, 33). Because of its specific activation of RARs, atRA is an effective chemotherapeutic agent for acute promyelocytic leukemia, which is associated with inactivation of RARα due to chromosomal translocation (34-36). Via unknown mechanisms, atRA also inhibits the growth of many other types of tumors (reviewed in (37)). atRA also exhibits inhibitory effects on adipocyte differentiation (38, 39). Interestingly, Xue et al. observed a marked reduction of PPARγ gene expression in adipocytes treated with atRA, suggesting that decreased PPARγ expression could account for its inhibitory effects on adipogenesis (40). In addition, recent in vivo studies revealed that atRA effectively reduced adiposity not only in fat but also in liver via increased TG hydrolysis and fat oxidation (28, 41, 42). Although changes in many downstream target genes have been reported in these studies, the exact molecular mechanisms behind the fat mobilization mediated by atRA and its metabolic effects outside adipose tissue have not been established.
Our current study discovered a novel transcriptional cascade, which underlies the aforementioned hepatic TG clearance mediated by atRA. The cascade is also shared by one of the pathways controlling TG accumulation mediated by orphan nuclear receptor SHP. One of the significant findings from current study includes a critical role of nuclear hormone receptor PPARγ2 in the development of hepatic steatosis. Recent studies also demonstrated that liver PPARγ2 is a major factor in the development of fatty liver in rodent models (12, 18, 43). Especially, its expression appears to be limited in hepatocyte, demonstrated by complete absence of PPARγ2 expression in the liver of hepatocyte specific PPARγ knockout mouse (14, 15). Hepatic overexpression of PPARγ2 using adenovirus was sufficient enough to increase liver TG along with an increase in SREBP-1c and other lipogenic gene mRNAs (18). In contrast, activation of atRA signals or deletion of SHP caused concomitant reduction of Srebp1c expression along with PPARγ2 (Supplemental Fig 3 & 6), which is in accord with earlier observations (9, 11, 42). Though it is not addressed whether the reduced Srebp1c expression is resulted from direct regulation by PPARγ2 or overall reduction in lipogenic program, the marked down-regulation of PPARγ2 appears to be a driving force against hepatic lipogenesis and TG accumulation observed in mouse models (11).
Another significant finding is an emergence of the transcriptional repressor Hes6 as a novel regulator of hepatic TG homeostasis. In accord with earlier observations (23), our data strongly suggest that Hes6 is an important upstream regulator of PPARγ2 expression. We suggest that Hes6 maintain low PPARγ2 expression in the normal liver to prevent excess lipid accumulation. However, some pathophysiological conditions such as obesity or diabetes may reduce the Hes6 expression as we observed (Fig. 1A) and unleash hepatic lipogenic program by activation of PPARγ2 expression. The linkage between Hes6 and PPARγ2 may complement the previously described CREB-mediated regulation of PPARγ1 expression via Hes1 in hepatic lipid homeostasis during fasting and feeding (44).
Interestingly, the cascade is responsive to atRA in all metabolic tissues tested and one of the studies also observed a PPARγ2 isoform-specific effect of atRA in adipose tissues (41). Lack of lipid-lowering effect in white adipose tissues of our atRA-treated mice is likely due to the overall constant PPARγ expression based on higher PPARγ1 isoform expression. While this result is contradictory to other atRA studies but it should be noted that administration method of atRA differs from these studies (oral gavage vs. subcutaneous injection). Moreover, complexity has been added in retinoid-mediated fat tissue biogenesis and differentiation by recent studies, in which retinal aldehydrogenase deficiency, increasing tissue retinal aldehyde content but decreasing retinoic acid content, leads to induction of brown fat biogenesis in white adipocytes (45, 46). It is possible that the clear reduction of white adipose tissues in other studies is mediated indirectly by enhanced TG handling and energy expenditure in liver and brown adipose tissues by long term atRA treatment or directly by enhanced thermogenic program via retinaldehyde, a precursor of atRA, -activated RARα (45, 46). Nonetheless, our observation may agree with the lack of inhibitory effect of atRA on adipogenesis in fully differentiated adipocytes (45).
atRA treatment also reduced serum glucose efficiently both in diet-induced obese and ob/ob mice. The result observed with ob/ob mice is contradictory to a recent report that the glucose lowering effect of atRA is absent in ob/ob mice, suggesting a leptin-dependent regulatory pathway (47), but is consistent with the effective amelioration of hyperglycemia in ob/ob mice by atRA observed in a different study (48). Our results from adenovirus-mediated RARα overexpression suggest that atRA may lower serum glucose through RAR and liver dependent manner. Similar observations from liver specific PPARγ KO mice additionally emphasize an importance of the proposed transcriptional cascade in hepatic glucose metabolism (13, 15). However, the glucose regulatory pathways appear to be dependent on SHP since SHP deficiency leads to hepatic insulin resistance with higher expression of genes responsible for glucose production (11).
Our current study identifies a novel transcriptional pathway linking RAR/SHP signals with PPARγ2 expression that determines fate of hepatic lipids along with expression of genes involved in fatty acid synthesis. Though there are some differences with earlier works (see references in (49)), our results are consistent with several previous studies linking retinoic acid signaling to beneficial metabolic effects in many aspects, and reveal specifically underlying molecular mechanisms in hepatic lipid metabolism regulated by retinoid signaling. Along with the finding of altered retinoid signals in human NAFLD patients (50), the current findings will provide additional insights into the fatty liver development and its therapeutic approaches.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by NEOMED startup fund and NIH grants R01DK093774 to YKL, DK068804 to DDM, DK44442 and DK58379 to JYLC.
Abbreviations
- SHP
small heterodimer partner
- PPARγ
peroxisome proliferator activated receptor gamma
- ChIP
chromatin immunoprecipitation
- RAR
retinoic acid receptor
- atRA
alltrans-retinoic acid
- HNF4α
hepatocyte nuclear factor 4 alpha
- WestD
western diet
- NAFLD
nonalcoholic fatty liver disease
- Fsp27
fat specific factor 27
- TG
triglyceride
- CD
chow diet
References
- 1.Johansson L, Thomsen JS, Damdimopoulos AE, Spyrou G, Gustafsson J, Treuter E. The orphan nuclear receptor SHP inhibits agonist-dependent transcriptional activity of estrogen receptors ERalpha and ERbeta. J Biol Chem. 1999;274:345–353. doi: 10.1074/jbc.274.1.345. [DOI] [PubMed] [Google Scholar]
- 2.Lee YK, Dell H, Dowhan DH, Hadzopoulou-Cladaras M, Moore DD. The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol Cell Biol. 2000;20:187–195. doi: 10.1128/mcb.20.1.187-195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shih DQ, Bussen M, Sehayek E, Ananthanarayanan M, Shneider BL, Suchy FJ, Shefer S, et al. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet. 2001;27:375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- 4.Nishigori H, Tomura H, Tonooka N, Kanamori M, Yamada S, Sho K, Inoue I, et al. Mutations in the small heterodimer partner gene are associated with mild obesity in Japanese subjects. Proc Natl Acad Sci U S A. 2001;98:575–580. doi: 10.1073/pnas.021544398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hung CC, Farooqi IS, Ong K, Luan J, Keogh JM, Pembrey M, Yeo GS, et al. Contribution of variants in the small heterodimer partner gene to birthweight, adiposity, and insulin levels: mutational analysis and association studies in multiple populations. Diabetes. 2003;52:1288–1291. doi: 10.2337/diabetes.52.5.1288. [DOI] [PubMed] [Google Scholar]
- 6.Enya M, Horikawa Y, Kuroda E, Yonemaru K, Tonooka N, Tomura H, Oda N, et al. Mutations in the small heterodimer partner gene increase morbidity risk in Japanese type 2 diabetes patients. Hum Mutat. 2008;29:E271–277. doi: 10.1002/humu.20865. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Liu J, Saha P, Huang J, Chan L, Spiegelman B, Moore DD. The orphan nuclear receptor SHP regulates PGC-1alpha expression and energy production in brown adipocytes. Cell Metab. 2005;2:227–238. doi: 10.1016/j.cmet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Boulias K, Katrakili N, Bamberg K, Underhill P, Greenfield A, Talianidis I. Regulation of hepatic metabolic pathways by the orphan nuclear receptor SHP. Embo J. 2005;24:2624–2633. doi: 10.1038/sj.emboj.7600728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J, Iqbal J, Saha PK, Liu J, Chan L, Hussain MM, Moore DD, et al. Molecular characterization of the role of orphan receptor small heterodimer partner in development of fatty liver. Hepatology. 2007;46:147–157. doi: 10.1002/hep.21632. [DOI] [PubMed] [Google Scholar]
- 10.Hartman HB, Lai K, Evans MJ. Loss of small heterodimer partner expression in the liver protects against dyslipidemia. J Lipid Res. 2009;50:193–203. doi: 10.1194/jlr.M800323-JLR200. [DOI] [PubMed] [Google Scholar]
- 11.Park YJ, Kim SC, Kim J, Anakk S, Lee JM, Tseng HT, Yechoor V, et al. Dissociation of diabetes and obesity in mice lacking orphan nuclear receptor small heterodimer partner. J Lipid Res. 2011;52:2234–2244. doi: 10.1194/jlr.M016048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edvardsson U, Bergstrom M, Alexandersson M, Bamberg K, Ljung B, Dahllof B. Rosiglitazone (BRL49653), a PPARgamma-selective agonist, causes peroxisome proliferator-like liver effects in obese mice. J Lipid Res. 1999;40:1177–1184. [PubMed] [Google Scholar]
- 13.Matsusue K, Haluzik M, Lambert G, Yim SH, Gavrilova O, Ward JM, Brewer B, Jr., et al. Liver-specific disruption of PPARgamma in leptin-deficient mice improves fatty liver but aggravates diabetic phenotypes. J Clin Invest. 2003;111:737–747. doi: 10.1172/JCI17223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavrilova O, Haluzik M, Matsusue K, Cutson JJ, Johnson L, Dietz KR, Nicol CJ, et al. Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J Biol Chem. 2003;278:34268–34276. doi: 10.1074/jbc.M300043200. [DOI] [PubMed] [Google Scholar]
- 15.Moran-Salvador E, Lopez-Parra M, Garcia-Alonso V, Titos E, Martinez-Clemente M, Gonzalez-Periz A, Lopez-Vicario C, et al. Role for PPARgamma in obesity-induced hepatic steatosis as determined by hepatocyte- and macrophage-specific conditional knockouts. Faseb J. 2011;25:2538–2550. doi: 10.1096/fj.10-173716. [DOI] [PubMed] [Google Scholar]
- 16.Pettinelli P, Videla LA. Up-regulation of PPAR-gamma mRNA expression in the liver of obese patients: an additional reinforcing lipogenic mechanism to SREBP-1c induction. J Clin Endocrinol Metab. 2011;96:1424–1430. doi: 10.1210/jc.2010-2129. [DOI] [PubMed] [Google Scholar]
- 17.Yu S, Matsusue K, Kashireddy P, Cao WQ, Yeldandi V, Yeldandi AV, Rao MS, et al. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor gamma1 (PPARgamma1) overexpression. J Biol Chem. 2003;278:498–505. doi: 10.1074/jbc.M210062200. [DOI] [PubMed] [Google Scholar]
- 18.Yamazaki T, Shiraishi S, Kishimoto K, Miura S, Ezaki O. An increase in liver PPARgamma2 is an initial event to induce fatty liver in response to a diet high in butter: PPARgamma2 knockdown improves fatty liver induced by high-saturated fat. J Nutr Biochem. 2011;22:543–553. doi: 10.1016/j.jnutbio.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Bai L, Jia Y, Viswakarma N, Huang J, Vluggens A, Wolins NE, Jafari N, et al. Transcription coactivator mediator subunit MED1 is required for the development of fatty liver in the mouse. Hepatology. 2011;53:1164–1174. doi: 10.1002/hep.24155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsusue K, Kusakabe T, Noguchi T, Takiguchi S, Suzuki T, Yamano S, Gonzalez FJ. Hepatic steatosis in leptin-deficient mice is promoted by the PPARgamma target gene Fsp27. Cell Metab. 2008;7:302–311. doi: 10.1016/j.cmet.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishino N, Tamori Y, Tateya S, Kawaguchi T, Shibakusa T, Mizunoya W, Inoue K, et al. FSP27 contributes to efficient energy storage in murine white adipocytes by promoting the formation of unilocular lipid droplets. J Clin Invest. 2008;118:2808–2821. doi: 10.1172/JCI34090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bae S, Bessho Y, Hojo M, Kageyama R. The bHLH gene Hes6, an inhibitor of Hes1, promotes neuronal differentiation. Development. 2000;127:2933–2943. doi: 10.1242/dev.127.13.2933. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Jimenez CP, Kyrmizi I, Cardot P, Gonzalez FJ, Talianidis I. Hepatocyte nuclear factor 4alpha coordinates a transcription factor network regulating hepatic fatty acid metabolism. Mol Cell Biol. 2010;30:565–577. doi: 10.1128/MCB.00927-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park YJ, Qatanani M, Chua SS, LaRey JL, Johnson SA, Watanabe M, Moore DD, et al. Loss of orphan receptor small heterodimer partner sensitizes mice to liver injury from obstructive cholestasis. Hepatology. 2008;47:1578–1586. doi: 10.1002/hep.22196. [DOI] [PubMed] [Google Scholar]
- 25.Shaw N, Elholm M, Noy N. Retinoic acid is a high affinity selective ligand for the peroxisome proliferator-activated receptor beta/delta. J Biol Chem. 2003;278:41589–41592. doi: 10.1074/jbc.C300368200. [DOI] [PubMed] [Google Scholar]
- 26.Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129:723–733. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schug TT, Berry DC, Toshkov IA, Cheng L, Nikitin AY, Noy N. Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARbeta/delta to RAR. Proc Natl Acad Sci U S A. 2008;105:7546–7551. doi: 10.1073/pnas.0709981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berry DC, Noy N. All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation-activated receptor beta/delta and retinoic acid receptor. Mol Cell Biol. 2009;29:3286–3296. doi: 10.1128/MCB.01742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sznaidman ML, Haffner CD, Maloney PR, Fivush A, Chao E, Goreham D, Sierra ML, et al. Novel selective small molecule agonists for peroxisome proliferator-activated receptor delta (PPARdelta)--synthesis and biological activity. Bioorg Med Chem Lett. 2003;13:1517–1521. doi: 10.1016/s0960-894x(03)00207-5. [DOI] [PubMed] [Google Scholar]
- 30.Wang YX, Lee CH, Tiep S, Yu RT, Ham J, Kang H, Evans RM. Peroxisomeproliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell. 2003;113:159–170. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 31.Puigserver P, Vazquez F, Bonet ML, Pico C, Palou A. In vitro and in vivo induction of brown adipocyte uncoupling protein (thermogenin) by retinoic acid. Biochem J. 1996;317(Pt 3):827–833. doi: 10.1042/bj3170827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petkovich M, Brand NJ, Krust A, Chambon P. A human retinoic acid receptor which belongs to the family of nuclear receptors. Nature. 1987;330:444–450. doi: 10.1038/330444a0. [DOI] [PubMed] [Google Scholar]
- 33.Giguere V, Ong ES, Segui P, Evans RM. Identification of a receptor for the morphogen retinoic acid. Nature. 1987;330:624–629. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- 34.Huang ME, Ye YC, Chen SR, Chai JR, Lu JX, Zhoa L, Gu LJ, et al. Use of all-trans retinoic acid in the treatment of acute promyelocytic leukemia. Blood. 1988;72:567–572. [PubMed] [Google Scholar]
- 35.Warrell RP, Jr., Frankel SR, Miller WH, Jr., Scheinberg DA, Itri LM, Hittelman WN, Vyas R, et al. Differentiation therapy of acute promyelocytic leukemia with tretinoin (all-transretinoic acid). N Engl J Med. 1991;324:1385–1393. doi: 10.1056/NEJM199105163242002. [DOI] [PubMed] [Google Scholar]
- 36.Lin RJ, Evans RM. Acquisition of oncogenic potential by RAR chimeras in acute promyelocytic leukemia through formation of homodimers. Mol Cell. 2000;5:821–830. doi: 10.1016/s1097-2765(00)80322-6. [DOI] [PubMed] [Google Scholar]
- 37.Soprano DR, Qin P, Soprano KJ. Retinoic acid receptors and cancers. Annu Rev Nutr. 2004;24:201–221. doi: 10.1146/annurev.nutr.24.012003.132407. [DOI] [PubMed] [Google Scholar]
- 38.Sato M, Hiragun A, Mitsui H. Preadipocytes possess cellular retinoid binding proteins and their differentiation is inhibited by retinoids. Biochem Biophys Res Commun. 1980;95:1839–1845. doi: 10.1016/s0006-291x(80)80113-6. [DOI] [PubMed] [Google Scholar]
- 39.Kuri-Harcuch W. Differentiation of 3T3-F442A cells into adipocytes is inhibited by retinoic acid. Differentiation. 1982;23:164–169. doi: 10.1111/j.1432-0436.1982.tb01279.x. [DOI] [PubMed] [Google Scholar]
- 40.Xue JC, Schwarz EJ, Chawla A, Lazar MA. Distinct stages in adipogenesis revealed by retinoid inhibition of differentiation after induction of PPARgamma. Mol Cell Biol. 1996;16:1567–1575. doi: 10.1128/mcb.16.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ribot J, Felipe F, Bonet ML, Palou A. Changes of adiposity in response to vitamin A status correlate with changes of PPAR gamma 2 expression. Obes Res. 2001;9:500–509. doi: 10.1038/oby.2001.65. [DOI] [PubMed] [Google Scholar]
- 42.Amengual J, Ribot J, Bonet ML, Palou A. Retinoic acid treatment enhances lipid oxidation and inhibits lipid biosynthesis capacities in the liver of mice. Cell Physiol Biochem. 2010;25:657–666. doi: 10.1159/000315085. [DOI] [PubMed] [Google Scholar]
- 43.Vidal-Puig A, Jimenez-Linan M, Lowell BB, Hamann A, Hu E, Spiegelman B, Flier JS, et al. Regulation of PPAR gamma gene expression by nutrition and obesity in rodents. J Clin Invest. 1996;97:2553–2561. doi: 10.1172/JCI118703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herzig S, Hedrick S, Morantte I, Koo SH, Galimi F, Montminy M. CREB controls hepatic lipid metabolism through nuclear hormone receptor PPAR-gamma. Nature. 2003;426:190–193. doi: 10.1038/nature02110. [DOI] [PubMed] [Google Scholar]
- 45.Ziouzenkova O, Orasanu G, Sharlach M, Akiyama TE, Berger JP, Viereck J, Hamilton JA, et al. Retinaldehyde represses adipogenesis and diet-induced obesity. Nat Med. 2007;13:695–702. doi: 10.1038/nm1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiefer FW, Vernochet C, O'Brien P, Spoerl S, Brown JD, Nallamshetty S, Zeyda M, et al. Retinaldehyde dehydrogenase 1 regulates a thermogenic program in white adipose tissue. Nat Med. 2012;18:918–925. doi: 10.1038/nm.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsuchiya H, Ikeda Y, Ebata Y, Kojima C, Katsuma R, Tsuruyama T, Sakabe T, et al. Retinoids ameliorate insulin resistance in a leptin-dependent manner in mice. Hepatology. 2012;56:1319–1330. doi: 10.1002/hep.25798. [DOI] [PubMed] [Google Scholar]
- 48.Manolescu DC, Sima A, Bhat PV. All-trans retinoic acid lowers serum retinol-binding protein 4 concentrations and increases insulin sensitivity in diabetic mice. J Nutr. 2010;140:311–316. doi: 10.3945/jn.109.115147. [DOI] [PubMed] [Google Scholar]
- 49.Bonet ML, Ribot J, Palou A. Lipid metabolism in mammalian tissues and its control by retinoic acid. Biochim Biophys Acta. 2012;1821:177–189. doi: 10.1016/j.bbalip.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Ashla AA, Hoshikawa Y, Tsuchiya H, Hashiguchi K, Enjoji M, Nakamuta M, Taketomi A, et al. Genetic analysis of expression profile involved in retinoid metabolism in non-alcoholic fatty liver disease. Hepatol Res. 2010;40:594–604. doi: 10.1111/j.1872-034X.2010.00646.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.