SUMMARY
V(D)J recombination-associated DNA double-strand breaks (DSBs) are normally repaired by the high fidelity, classical non-homologous end joining (cNHEJ) machinery. Previous studies implicated the RAG/DNA post-cleavage complex (PCC) in regulating pathway choice by preventing access to inappropriate repair mechanisms such as homologous recombination (HR) and alternative NHEJ (aNHEJ). Here we report that RAG2’s “acidic hinge”, previously of unknown function, is critical for several key steps. Mutations that reduce the hinge’s negative charge destabilize the PCC, disrupt pathway choice, permit repair of RAG-mediated DSBs by the translocation-prone aNHEJ machinery, and reduce genomic stability in developing lymphocytes. Structural predictions and experimental results support our hypothesis that reduced flexibility of the hinge underlies these outcomes. Furthermore, sequence variants present in the human population reduce the hinge’s negative charge, permit aNHEJ, and diminish genomic integrity.
INTRODUCTION
DNA double-strand breaks (DSBs) must be detected promptly and repaired accurately to ensure genomic stability. While two major DSB repair pathways, classical NHEJ (cNHEJ) and HR, have been extensively studied (Weterings and Chen, 2008), additional error-prone joining mechanisms (aNHEJ), have been increasingly recognized (Corneo et al., 2007; Helmink and Sleckman, 2012; Kabotyanski et al., 1998; Simsek and Jasin, 2010; Zhu et al., 2002). aNHEJ causes chromosome translocations (Mladenov and Iliakis, 2011; Simsek et al., 2011; Simsek and Jasin, 2010; Yan et al., 2007; Zhu et al., 2002) and is implicated in oncogenesis (Wang et al., 2008; Zhu et al., 2002). aNHEJ’s ability to join chromosomal DSBs (Bogue et al., 1998; Bogue et al., 1997; Malynn et al., 1988; Soulas-Sprauel et al., 2007; Yan et al., 2007) suggests that pathway choice control could be an important safeguard of genomic integrity. Scant evidence, however, links defects in pathway choice control with genomic instability.
V(D)J recombination employs DSBs to generate antigen receptors (Helmink and Sleckman, 2012), providing a tractable, physiologically relevant model to investigate pathway choice. The RAG1/2 complex assembles on a pair of recombination signal sequences (RSSs), forms a synaptic complex (Hiom and Gellert, 1998; Schatz and Swanson, 2011), generates a pair of DSBs, and remains associated with the four broken DNA ends in a post-cleavage complex (PCC) (Agrawal and Schatz, 1997; Jones and Gellert, 2001), which is essential for pathway choice control (Arnal et al., 2010; Corneo et al., 2007; Lee et al., 2004). One model posits that the PCC forms a scaffold, retaining the broken DNA ends to facilitate proper repair by cNHEJ and discourage repair by HR and aNHEJ (Corneo et al., 2007; Lee et al., 2004; Tsai et al., 2002), both of which could lead to inappropriate and dangerous outcomes.
To investigate the role of pathway choice control in maintaining genomic integrity during V(D)J recombination, we focused on RAG2, which lacks known DNA binding or catalytic activity, but is essential for recombination and has several regulatory roles. Thymocytes from mice bearing a truncated RAG2 mutant (RAG2core /core) display genomic instability and, in the absence of p53, rapidly succumb to thymic lymphomas with complex chromosomal aberrations involving the Igh and Tcrα/δ loci (Deriano et al., 2011). This truncation removes the C-terminus and its regulatory domains (Figure 1A), including a plant homeodomain (PHD) finger that interacts with active chromatin (Callebaut and Mornon, 1998; Ji et al., 2010; Jones and Simkus, 2009; Matthews et al., 2007), and a CDK phosphorylation site (T490) that targets RAG2 for destruction at the G1-to-S transition (Figure 1A)(Li et al., 1996; Zhang et al., 2011). Mutating this residue alone (T490A) stabilizes RAG2 protein levels throughout the cell cycle, and T490A knock-in lymphocytes contain elevated levels of DSBs (Zhang et al., 2011). These persistent breaks are pathogenic: in a p53−/− background, these T490A mice develop thymic lymphomas with antigen receptor translocations. RAG2core /corep53−/− mice develop lymphomas much more rapidly and with substantially higher penetrance than RAG2T490A/T490A p53−/− mice, suggesting that additional, unidentified regulatory elements in RAG2’s C-terminus safeguard the genome.
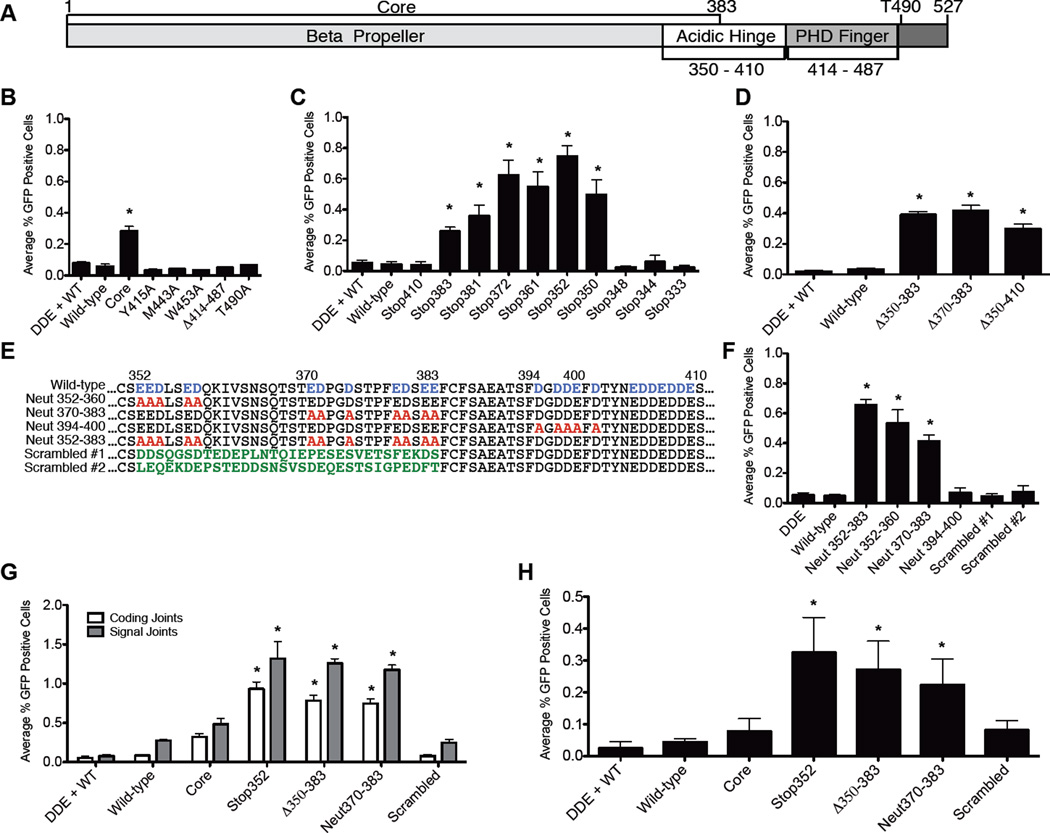
Figure 1. Acidic residues in the hinge region of RAG2 regulate repair pathway choice.
(A) RAG2 schematic. Quantification of FACS data from three separate experiments for aNHEJ in CHO-K1 cells measuring mutations to known regulatory regions (B), truncation mutations (C), internal deletions (D), and neutralizing mutations (E,F). (G) XRCC4-deficient CHO cells were tested the ability to bypass cNHEJ defects using CJGFP and SJGFP (H) Recombination on the chromosomal substrate pMX-INV in SCID-MEFs with the indicated RAG2 mutations. Values significantly different than wild-type RAG1 and RAG2 are marked with an asterisk (Student’s t-test, unpaired, two tailed, equal variance). Means from three independent experiments are plotted; error bars reflect the standard error of the mean. See also Figure S1.
The C-terminus also contains a conserved hinge region (Jones and Simkus, 2009), which lies after a predicted β-propeller (Callebaut and Mornon, 1998) and has a high density of acidic amino acids (Oettinger et al., 1990). A small region of the hinge (residues 402–407) interacts with core histones (West et al., 2005), but the significance of this interaction remains unclear. Nothing further has been reported as to the hinge’s function (Jones and Simkus, 2009).
Through a systematic analysis of RAG2’s C-terminus, we show that the hinge is critical for PCC stability, pathway choice control, and maintenance of genomic integrity. Based on our data and structural predictions, we hypothesize that the acidic hinge is an intrinsically disordered domain (IDD) that regulates joining by modulating the conformation of the RAG2 C-terminus. Furthermore, sequence variants (SVs) in the acidic hinge identified by human exome sequencing (Tennessen et al., 2012) impair pathway choice control and reduce genomic integrity. Together, our data reveal a novel functional role for RAG2's acidic hinge and suggest that RAG2 SVs may undermine genomic stability in humans.
RESULTS AND DISCUSSION
The acidic hinge restricts repair pathway choice
We employed established assays (Arnal et al., 2010; Corneo et al., 2007; Deriano et al., 2009) that measure coding and signal joint formation by cNHEJ, and coding joints formed by aNHEJ (CJGFP, SJGFP, and AltGFP respectively, Figure S1A). The AltGFP substrate only scores coding joints that have suffered large deletions and employ a specific microhomology (both characteristics of aNHEJ). As a negative control, we used an inactive RAG1 mutant (DDE) (Landree et al., 1999). As expected (Corneo et al., 2007), both wild-type and core RAG2 produced robust levels of coding and signal joints (Figure S1B), but only core RAG2 allowed aNHEJ (Figure 1B).
We first tested two candidate motifs: the PHD finger and the protein degradation domain. Previously described point mutations in the PHD finger that selectively disrupt H3K4me3-recognition (Y415A, M443A, and W453A) (Matthews et al., 2007) diminished recombination (Figure S1B), consistent with the reported decrease in protein stability and nuclear localization (Couedel et al., 2010), but did not allow aNHEJ (Figure 1B). Interestingly, a mutant lacking the entire PHD finger (Δ414–487) formed normal levels of coding and signal joints (Figure S1B), like core RAG2, which also lacks the PHD finger. These results, contrary to the PHD finger point mutations (Matthews et al., 2007), demonstrate that the PHD finger is not critical for recombination activity. Unlike core RAG2, the Δ414–487 mutant suppressed aNHEJ (Figure 1B) indicating that the PHD finger does not play a key role in pathway choice.
We next tested the cell cycle regulated degradation motif using the well-characterized T490A mutant (Li et al., 1996; Zhang et al., 2011). As expected, T490A formed normal levels of coding and signal joints (Figure S1B) and reached higher steady state levels than its wild-type counterpart (Figure S2A). We did not, however, observe aNHEJ (Figure 1B), indicating that the T490A mutant maintains proper pathway choice control.
We next examined a series of RAG2 C-terminal truncations (Figure S1C). Mutants truncated at (or C-terminal to) residue 350 were active for recombination, but activity dropped abruptly upon removal of two additional amino acids (Figure S1C), coinciding with the end of the predicted β-propeller (Callebaut and Mornon, 1998). Thus, the minimal core of RAG2 sufficient to support recombination includes only residues 1–350, 33 amino acids shorter than previously described (Cuomo and Oettinger, 1994; Sadofsky et al., 1994). Truncations that terminate RAG2 between residue 350 and 410 permit aNHEJ, identifying the region as critical for the pathway choice control (Figure 1C).
The Δ350–410, Δ350–383, and Δ370–383 internal deletion mutants were active for recombination and allowed aNHEJ (Figures S1D and 1D), narrowing the candidate region to amino acids 350–383. This sequence is highly acidic, an evolutionarily conserved feature noted upon the discovery of RAG2 (Oettinger et al., 1990)(Figure S2B). We next neutralized stretches of acidic residues in this region (Figure 1E). All “neutralized” mutants were active for recombination (Figure S1E). Neutralizations within the critical region (Neut352–360, Neut370–383, and Neut352–383) allowed substantial aNHEJ (Figure 1F), while Neut394–400 did not. These data identify acidic amino acids (352–383) important for pathway choice control. Furthermore, these mutants allow aNHEJ without causing protein accumulation (Figure S2A) indicating that they ablate pathway choice control without impairing other functions of the C-terminus.
We hypothesized that the neutralized mutants affect pathway choice control owning to decreased density of negative charge. To test this, we constructed two “scrambled” mutants (Figure 1E), which preserve the charge but scramble the hinge’s amino acid sequence. These mutants, fully active for recombination (Figure S1E), did not allow aNHEJ (Figure 1F), indicating the negative charge of this region, rather than its specific amino acid sequence, is important for pathway choice control.
As a definitive test, we asked whether RAG mutants that allow aNHEJ could rescue joining in cells deficient for cNHEJ. We used XRCC4-deficient cells (Li et al., 1995), which are unable to form coding and signal joints in the presence of wild-type RAG proteins unless complemented with wild-type XRCC4 (Figures 1G, Supplementary Figure S1F) (Corneo et al., 2007). RAG2 mutants that allow aNHEJ in cNHEJ-proficient cells (Stop352, Δ350–383, and Neut370–383) formed substantial levels of coding and signal joints (Figure 1G), whereas the scrambled mutant did not. We also tested a chromosomal substrate, pMX-INV, (Bredemeyer et al., 2006; Deriano et al., 2009) in an embryonic fibroblast cell line from Prkdcscid/scid mice, which, due to a crippled DNAdependent protein kinase, are cNHEJ-deficient (Blunt et al., 1995). In agreement with our results in XRCC4-deficient cells, Stop352, Δ350–383, and Neut370–383 supported cNHEJ-deficient recombination, but neither wild-type RAG2 nor a scrambled mutant was able to bypass the NHEJ defect (Figure 1H). Together, these results demonstrate that disrupting the hinge or reducing its negative charge allows RAG-generated DSBs to access aNHEJ.
Acidic hinge mutants destabilize the RAG post-cleavage complex
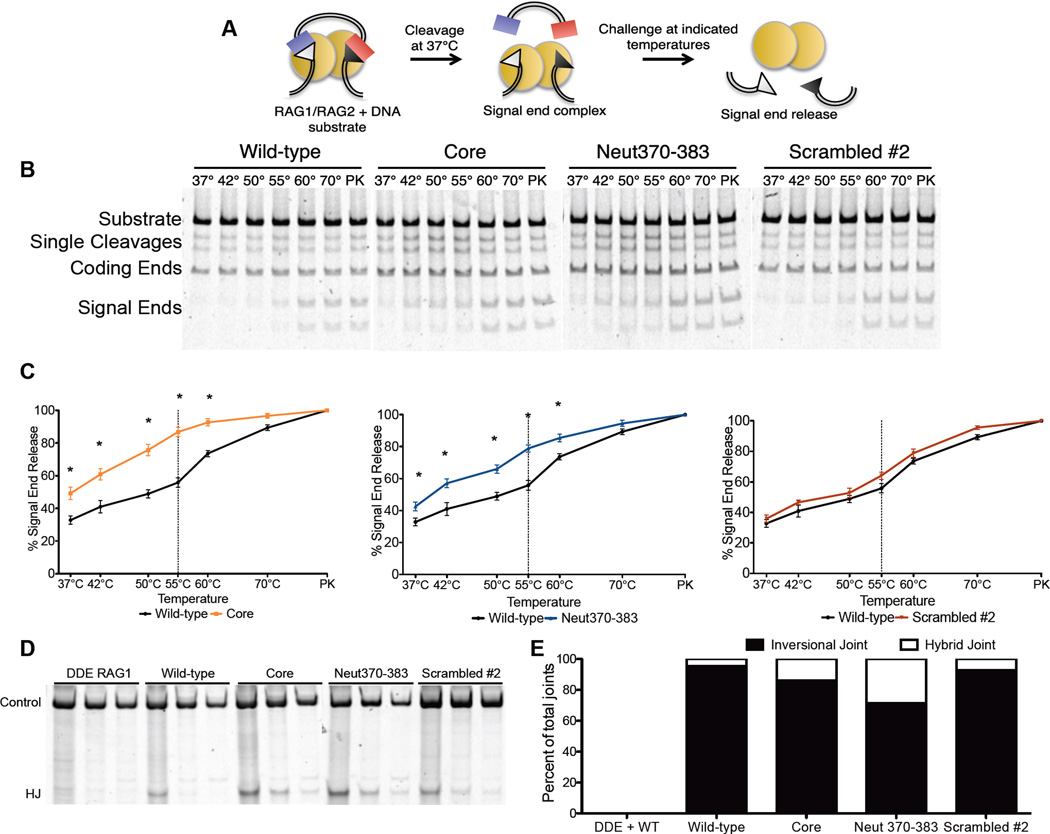
We hypothesized that the mechanism by which the RAG2’s acidic region affects pathway choice might involve the PCC (since, by retaining DNA ends, the PCC influences their fate after cleavage (Agrawal and Schatz, 1997; Jones and Gellert, 2001; Lee et al., 2004)). To test this hypothesis, we employed purified RAG proteins to measure the stability of the post-cleavage signal end complex (SEC) (Figure 2A). As expected (Deriano et al., 2011), SECs containing core RAG2 showed greater signal end release than complexes containing the wild-type protein (Figures 2B and 2C). SECs containing the Neut370–383 RAG2 mutant showed a similar increase in end release, while those containing a scrambled RAG2 mutant did not. These data implicate the acidic hinge in forming or maintaining the proper architecture of the SEC.
Figure 2. Charged residues in the acidic region stabilize the RAG-DNA post-cleavage complex.
(A) Biochemical end-release assay; (B) Gels for end-release assays, with the challenge temperatures indicated. PK samples were treated with proteinase K and SDS. (C) Quantification of signal end release (*P<0.05, Student’s ttest). (D) Hybrid joint formation measured by semi-quantitative PCR (D) and sequence analysis (E).
The influence of the hinge on the structure of the PCC can also be probed in cells by measuring inversional recombination. RAG mutants that destabilize the PCC in vitro also increase the formation of alternative, nonfunctional products in which signal and coding ends are incorrectly joined (Deriano et al., 2011; Deriano et al., 2009), indicating that post-cleavage processing has been altered. We observed increased hybrid joint formation with core RAG2 and Neut370–383 (Figures 2D and 2E), indicating that altering the acidic character of the hinge region favors formation of abnormal PCCs that fail to coordinate inversional joining, which requires coordination of all four ends. Thus, in every case for which we observe a breach in pathway choice control, we have also found evidence for an altered PCC.
RAG2’s acidic hinge is important for genomic stability
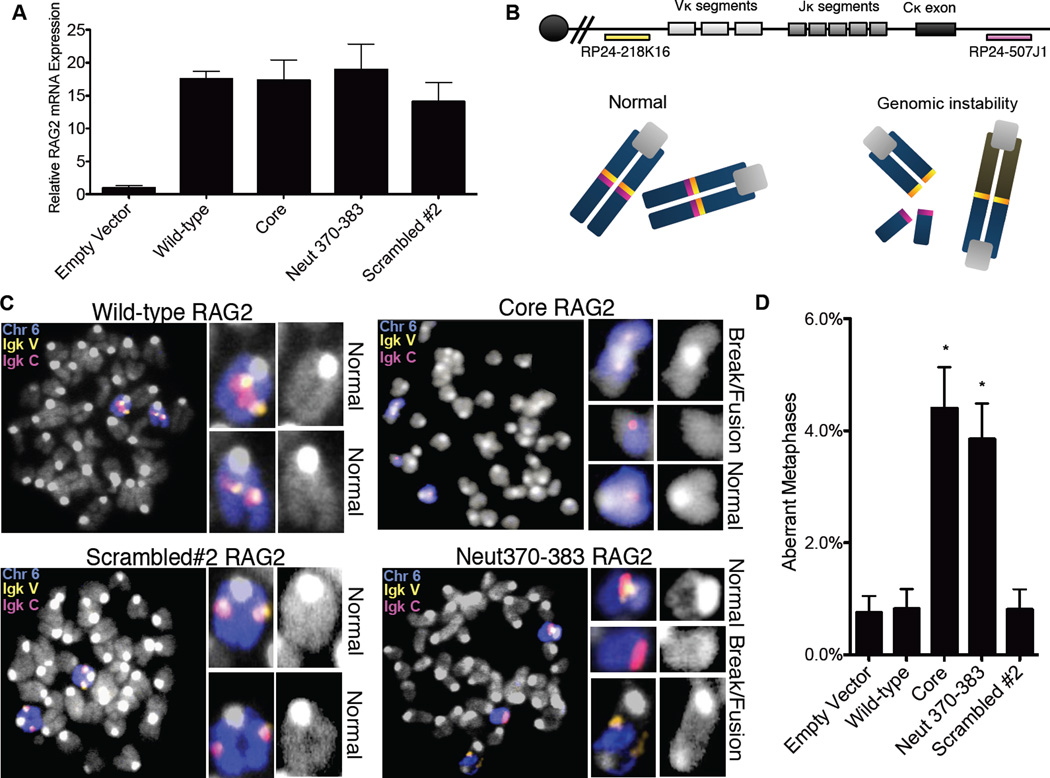
Using fluorescence in situ hybridization (FISH) on the Igκ locus, we tested the effects of RAG2 mutants on genomic stability in complemented RAG2−/− pre-B cells (Bredemeyer et al., 2006) (Figures 3A and 3B). As expected (Yin et al., 2009), we observed only background levels of aberrant events in cells complemented with wild-type RAG2 (0.8%, similar to levels observed in RAG2-deficient cells infected with an empty vector) (Figures 3C and 3D). In contrast, core RAG2-expressing cells displayed substantial levels (4.4%) of aberrant events, signifying genomic instability and in agreement with our previous analysis of RAG2core/core mouse thymocytes (Deriano et al., 2011). Cells expressing Neut370–383 also exhibited increased levels of aberrant metaphases (3.9%), whereas a scrambled mutant that retains the negative charge did not (0.8%). Aberrant metaphases, including chromosomal breakage and fusion events (Figure 3C), were quantified and the data from three separate experiments are shown in Figure S3A. Together, these results demonstrate that Neut370–383 RAG2, which bears only seven alanine substitutions and, unlike RAG2T490A and other truncation mutants, does not alter protein levels (Figure S2A), nevertheless promotes substantial genomic instability at an endogenous locus.
Figure 3. The acidic region in RAG2 promotes genomic stability.
RAG2 mRNA expression in complemented RAG2−/− pre-B cells(A). (B) Top; Igκ locus with BAC probes for DNA FISH. Bottom; Schematic of normal and aberrant metaphases. (C) Metaphases from complemented RAG2−/− preB cells with the BAC probes indicated in A. and a FITCchr6 paint; (D) Percentages of aberrant metaphases identified at the Igκ locus. Results from three separate experiments (Figure S3) were collected and combined. Statistical analysis was conducted using a one-tailed Fisher’s exact test for comparison to the wild-type complemented aberrant metaphases. Error bars reflect the standard error of the mean.
RAG2 neutralization mutations diminish the hinge flexibility
The acidic hinge is thought to be an unstructured stretch of amino acids linking the globular β-propeller and PHD finger (Callebaut and Mornon, 1998). We hypothesized that hinge-like flexibility might be critical for proper end processing. To investigate this, we employed software that predicts the inherent flexibility of a protein sequence based on defined parameters (sequence, sequence profiles, predicted secondary structure, solvent accessibility, backbone dihedral torsion angles, residue flexibility and B-factors) (Ferron et al., 2006; Mizianty et al., 2010). Three different algorithms predicted low flexibility within the proposed β-propeller region and the PHD finger, while the acidic hinge was far more flexible (Figure S3B). The Neut352–383 mutation, which destabilizes the PCC and ablates pathway choice restriction, substantially decreased predicted flexibility in this region, whereas a scrambled mutation, which retains the charge, did not. Together, these data indicate that the high concentration of charged residues in this region, which contribute to the flexibility of the acidic hinge, is necessary to enforce the pathway choice control.
Human RAG2 SVs decrease hinge flexibility, permit aNHEJ, and decrease genomic integrity
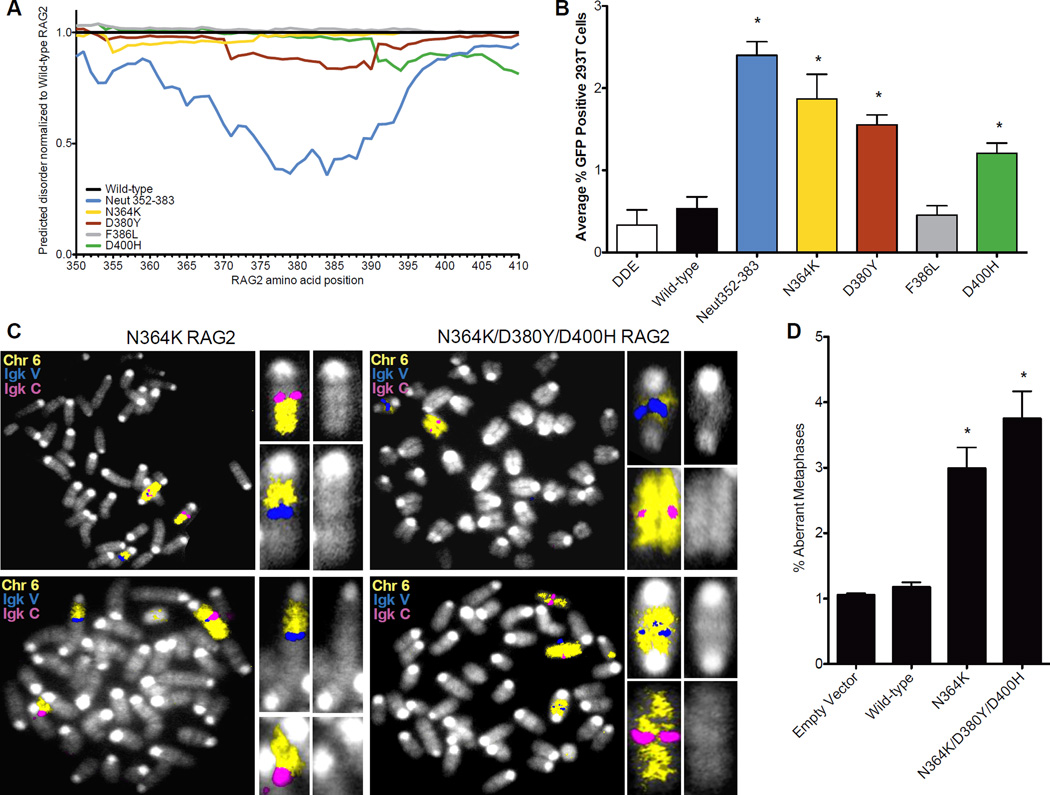
A search of the human exome project database (Tennessen et al., 2012) revealed four SVs in the hinge, three of which (N364K, D380Y, D400H) decrease negative charge and predicted hinge flexibility (Figure 4A). These three variants permit joining by aNHEJ (Figure 4B), while a fourth variant that does not alter the charge (F386L) or the predicted flexibility (Figure 4A) is not permissive for aNHEJ. RAG2−/− pre- B cells complemented with the indicated human SVs displayed increased aberrant metaphases with breaks at the endogenous Igk locus (Figure 4C). These data provide additional support for the importance of negative charge and flexibility of the acidic hinge, and suggest the intriguing possibility that RAG2 SVs present in the human population predispose to genomic instability.
Figure 4. Human SVs permit aNHEJ and impair genomic stability.
(A) Structurally predictive software to measure the inherent disorder and flexibility of the acidic hinge was used to compare the identified human RAG2 mutations with the wild-type sequence. (B) Human RAG2 acidic region SVs were tested for their ability to permit aNHEJ. (C) Metaphases from cells expressing the indicated RAG2 mutants with the Igk locus on chromosome 6 indicated. (D) Quantification of aberrant metaphases observed in C. Results from separate experiments (Figure S4) were collected and combined. Statistical analysis was conducted using a one-tailed Fisher’s exact test for comparison to the wild-type complemented aberrant metaphases. Error bars reflect the standard error of the mean.
The steps in which the broken DNA is moved from the PCC to the NHEJ pathway remain murky; however, our results show that DNA ends are not relinquished haphazardly. While the potential for joining broken DNA ends by aNHEJ is present in most of our experiments, unless we tamper with the acidic hinge, aNHEJ is excluded. Mutation of the acidic hinge creates correlated changes in the structure of the PCC, as probed in cells (by hybrid joint formation) and biochemically (by signal end release). At a more macroscopic level, we observe coincident increased genomic instability at the rearranging Igk locus. Interestingly, fusion events involving the Igk locus were only observed following recombination with charged-disrupted RAG2 mutants (Figure S4). Regardless of the cause and effect relationships between these observations (which are suggestive), we can safely conclude that the acidic hinge is a necessary element for directing the proper outcome of V(D)J recombination and that it is a key component of the overall genome guardian functions performed by RAG2.
We hypothesize that these post-cleavage effects require the hinge-like mobility of the acidic region. Although even a single neutralizing point mutation affects pathway choice, 33 amino acids within the acidic hinge can be shuffled without resulting in defects in post-cleavage function. This suggests that the hinge functions in the PCC without assuming a specific folded configuration. While this is compatible with the hinge serving as a simple linker, intrinsically disordered domains (IDDs) in other DNA binding and catalytic protein complexes are starting to be appreciated as exceptional components in protein structure (Dyson, 2011; Fuxreiter, 2012; Fuxreiter et al., 2011; Vuzman and Levy, 2012). IDDs can tune the binding affinities of a protein or complex, providing a flexible interaction surface for diverse protein partners, or regulating catalysis by shielding or unveiling catalytic sites.
IDDs seem well suited to the challenges of V(D)J recombination, for as in other site-directed DNA transactions (Li et al., 2006; Rice and Baker, 2001), it appears that protein:DNA complexes are restructured throughout the reaction. For V(D)J recombination, engagement of target sequences occurs stepwise, culminating in a paired synaptic complex. RAG2 influences the quality and degree of binding in the complex (Jones and Simkus, 2009; Schatz and Swanson, 2011) and is important in discriminating between proper and improper recombination targets (Grundy et al., 2010; Shimazaki et al., 2009). Conformational changes ensue at the point of full cleavage, based upon the demonstration that the PCC formed of core RAG proteins and signal ends is much more stable than the pre-cleavage (synaptic) complex (Jones and Gellert, 2001). Tight product binding, a strategy for imposing directionality toward cleavage (Li et al., 2006; Rice and Baker, 2001), may engender further remodeling of the complex to provide access to the cNHEJ machinery. All of the foregoing involves tuning binding interactions, something for which IDDs are well adapted (Dyson, 2011; Fuxreiter, 2012; Fuxreiter et al., 2011; Vuzman and Levy, 2012).
The notion that the PCC is a central hub for transmitting and responding to signals that ensure proper V(D)J joining is reinforced in light of the fact that the PCC recruits general DNA damage repair/damage sensors (ATM and the Mre11 complex). Both of these factors affect the stability of the PCC in vivo (Bredemeyer et al., 2006; Deriano et al., 2009). ATM−/− mice are unable to trigger ATM/p53-dependent DNA damage checkpoints (Zha et al., 2010). Interestingly, RAG2core/core p53−/− mice phenocopy the ATM−/− mice and may also be unable to correctly respond to the checkpoint (Deriano et al., 2011). We speculate that this response is a mediated by a conformational change involving the acidic hinge.
Interestingly, human SVs that reduce the acidic charge in the hinge region permit repair by the translocation-prone aNHEJ machinery and decrease genomic integrity. Currently there is no evidence to suggest that these rare SVs are pathogenic, but it is conceivable that, under the right circumstances, they might lead to diminished fidelity of V(D)J recombination. Given the enormous number of developing lymphocytes in humans (9×1011 each day, (Saada et al., 2007)), even a slight decrease in fidelity might have significant consequences.
EXPERIMENTAL PROCEDURES
Mutagenesis was carried out using QuikChange II Site-Directed Mutagenesis Kit (Stratagene) or overlapping PCR, confirmed by sequence analysis. GST-tagged RAG proteins were isolated from CHO cells and assayed for complex stability. To measure protein levels, HA-tagged RAG2 protein from transfected cells was detected using an antibody to the HA tag (Roche). NHEJ-deficient chromosomal recombination was measured in Prkdcscid/scid MEFs containing the pMX-INV substrate. Hybrid joining was measured on recovered pJH299 following transient transfection with RAG1 and RAG2. Instability at the Igκ locus was assayed by complementing V-abl-transformed RAG2−/− pre-B cells with a RAG2 retroviral construct (pMIT-RAG2-IRES-CD90.1), treating with STI571 (Novartis), and fixing. BAC RP24–507J1 and BAC RP24–218K16 were labeled with Alexa Fluor 594 and Cy3, respectively (Molecular Probes). Mouse FITC chromosome 6 paint was prepared following supplier’s instructions (Cambio). Metaphases were analyzed using a MetaSystems Metafer and Isis Fluorescence Imaging. A number of samples were independently re-scored by an outside investigator with fully consistent results. For disorder and flexibility predictions, RAG2 sequences were processed through the mFDP, IUPREDL, and IUPREDS algorithms. A detailed description of the materials and methods is provided in the Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
The acidic hinge in RAG2 prevents aberrant repair of RAG-mediated DNA breaks
Acidic residues strengthen RAG/DNA post-cleavage complex and enhance flexibility
Acidic residues preserve genomic stability in lymphocytes during VDJ recombination
Experiments with identified sequence variants suggest similar mechanism in humans
ACKNOWLEDGMENTS
We thank S. Hewitt and J. Skok (Department of Pathology, New York University Langone Medical Center) for the Igκ BAC probes and assisting with the M-FISH experiments, S. Lewis (Department of Pathology and Laboratory Medicine, Perelman School of Medicine at the University of Pennsylvania) for manuscript revisions, C. McPhee and E. Hernando-Monge (Department of Pathology, New York University Langone Medical Center) for generous support.
FUNDING
D.B.R. was supported by a National Institutes of Health [grant # CA104588] and the Irene Diamond Fund. L.D. is a Fellow of The Leukemia and Lymphoma Society. Funding for open access charge: National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
- Agrawal A, Schatz DG. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- Arnal SM, Holub AJ, Salus SS, Roth DB. Non-consensus heptamer sequences destabilize the RAG post-cleavage complex, making ends available to alternative DNA repair pathways. Nucleic Acids Res. 2010;38:2944–2954. doi: 10.1093/nar/gkp1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blunt T, Finnie NJ, Taccioli GE, Smith GC, Demengeot J, Gottlieb TM, Mizuta R, Varghese AJ, Alt FW, Jeggo PA, et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- Bogue MA, Jhappan C, Roth DB. Analysis of variable (diversity) joining recombination in DNAdependent protein kinase (DNA-PK)-deficient mice reveals DNA-PK-independent pathways for both signal and coding joint formation. Proc Natl Acad Sci U S A. 1998;95:15559–15564. doi: 10.1073/pnas.95.26.15559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogue MA, Wang C, Zhu C, Roth DB. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity. 1997;7:37–47. doi: 10.1016/s1074-7613(00)80508-7. [DOI] [PubMed] [Google Scholar]
- Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP. The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell Mol Life Sci. 1998;54:880–891. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- Couedel C, Roman C, Jones A, Vezzoni P, Villa A, Cortes P. Analysis of mutations from SCID and Omenn syndrome patients reveals the central role of the Rag2 PHD domain in regulating V(D)J recombination. J Clin Invest. 2010;120:1337–1344. doi: 10.1172/JCI41305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo CA, Oettinger MA. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 1994;22:1810–1814. doi: 10.1093/nar/22.10.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L, Chaumeil J, Coussens M, Multani A, Chou Y, Alekseyenko AV, Chang S, Skok JA, Roth DB. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature. 2011;471:119–123. doi: 10.1038/nature09755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ. Expanding the proteome: disordered and alternatively folded proteins. Q Rev Biophys. 2011;44:467–518. doi: 10.1017/S0033583511000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F, Longhi S, Canard B, Karlin D. A practical overview of protein disorder prediction methods. Proteins. 2006;65:1–14. doi: 10.1002/prot.21075. [DOI] [PubMed] [Google Scholar]
- Fuxreiter M. Fuzziness: linking regulation to protein dynamics. Mol Biosyst. 2012;8:168–177. doi: 10.1039/c1mb05234a. [DOI] [PubMed] [Google Scholar]
- Fuxreiter M, Simon I, Bondos S. Dynamic protein-DNA recognition: beyond what can be seen. Trends BiochemSci. 2011;36:415–423. doi: 10.1016/j.tibs.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Grundy GJ, Yang W, Gellert M. Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination. Proc Natl Acad Sci U S A. 2010;107:22487–22492. doi: 10.1073/pnas.1014958107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmink BA, Sleckman BP. The Response to and Repair of RAG-Mediated DNA Double-Strand Breaks. Annu Rev Immunol. 2012;30:175–202. doi: 10.1146/annurev-immunol-030409-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JM, Gellert M. Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc Natl Acad Sci U S A. 2001;98:12926–12931. doi: 10.1073/pnas.221471198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JM, Simkus C. The roles of the RAG1 and RAG2 "non-core" regions in V(D)J recombination and lymphocyte development. Arch Immunol Ther Exp (Warsz) 2009;57:105–116. doi: 10.1007/s00005-009-0011-3. [DOI] [PubMed] [Google Scholar]
- Kabotyanski EB, Gomelsky L, Han JO, Stamato TD, Roth DB. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 1998;26:5333–5342. doi: 10.1093/nar/26.23.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- Li M, Mizuuchi M, Burke TR, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Dordai DI, Lee J, Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity. 1996;5:575–589. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- Li Z, Otevrel T, Gao Y, Cheng HL, Seed B, Stamato TD, Taccioli GE, Alt FW. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- Malynn BA, Blackwell TK, Fulop GM, Rathbun GA, Furley AJ, Ferrier P, Heinke LB, Phillips RA, Yancopoulos GD, Alt FW. The scid defect affects the final step of the immunoglobulin VDJ recombinase mechanism. Cell. 1988;54:453–460. doi: 10.1016/0092-8674(88)90066-9. [DOI] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizianty MJ, Stach W, Chen K, Kedarisetti KD, Disfani FM, Kurgan L. Improved sequence-based prediction of disordered regions with multilayer fusion of multiple information sources. Bioinformatics. 2010;26:i489–i496. doi: 10.1093/bioinformatics/btq373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res. 2011;711:61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- Rice PA, Baker TA. Comparative architecture of transposase and integrase complexes. Nat Struct Biol. 2001;8:302–307. doi: 10.1038/86166. [DOI] [PubMed] [Google Scholar]
- Saada R, Weinberger M, Shahaf G, Mehr R. Models for antigen receptor gene rearrangement: CDR3 length. Immunol Cell Biol. 2007;85:323–332. doi: 10.1038/sj.icb.7100055. [DOI] [PubMed] [Google Scholar]
- Sadofsky MJ, Hesse JE, Gellert M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 1994;22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz DG, Swanson PC. V(D)J Recombination: Mechanisms of Initiation. Annu Rev Genet. 2011 doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- Shimazaki N, Tsai AG, Lieber MR. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocation. Mol Cell. 2009;34:535–544. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat Struct Mol Biol. 2010;17:410–416. doi: 10.1038/nsmb.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, Abramowski V, Olivier-Martin C, Goujet-Zalc C, Charneau P, de Villartay JP. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J Exp Med. 2007;204:1717–1727. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O'Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, et al. Evolution and Functional Impact of Rare Coding Variation from Deep Sequencing of Human Exomes. Science. 2012 doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CL, Drejer AH, Schatz DG. Evidence of a critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes Dev. 2002;16:1934–1949. doi: 10.1101/gad.984502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuzman D, Levy Y. Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Mol Biosyst. 2012;8:47–57. doi: 10.1039/c1mb05273j. [DOI] [PubMed] [Google Scholar]
- Wang JH, Alt FW, Gostissa M, Datta A, Murphy M, Alimzhanov MB, Coakley KM, Rajewsky K, Manis JP, Yan CT. Oncogenic transformation in the absence of Xrcc4 targets peripheral B cells that have undergone editing and switching. J Exp Med. 2008;205:3079–3090. doi: 10.1084/jem.20082271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West KL, Singha NC, De Ioannes P, Lacomis L, Erdjument-Bromage H, Tempst P, Cortes P. A direct interaction between the RAG2 C terminus and the core histones is required for efficient V(D)J recombination. Immunity. 2005;23:203–212. doi: 10.1016/j.immuni.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- Yin B, Savic V, Juntilla MM, Bredemeyer AL, Yang-Iott KS, Helmink BA, Koretzky GA, Sleckman BP, Bassing CH. Histone H2AX stabilizes broken DNA strands to suppress chromosome breaks and translocations during V(D)J recombination. J Exp Med. 2009;206:2625–2639. doi: 10.1084/jem.20091320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha S, Bassing CH, Sanda T, Brush JW, Patel H, Goff PH, Murphy MM, Tepsuporn S, Gatti RA, Look AT, et al. ATM-deficient thymic lymphoma is associated with aberrant tcrd rearrangement and gene amplification. J Exp Med. 2010;207:1369–1380. doi: 10.1084/jem.20100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Reynolds TL, Shan X, Desiderio S. Coupling of V(D)J recombination to the cell cycle suppresses genomic instability and lymphoid tumorigenesis. Immunity. 2011;34:163–174. doi: 10.1016/j.immuni.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.