Abstract
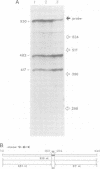
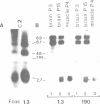
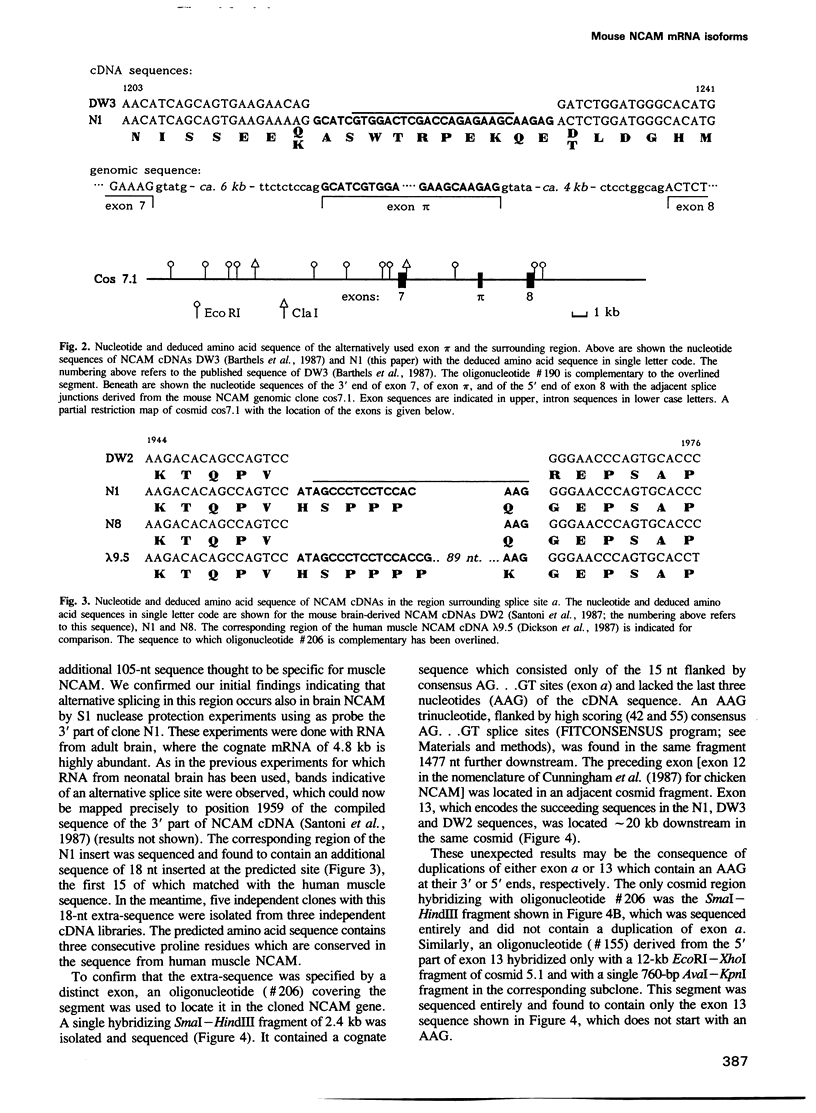
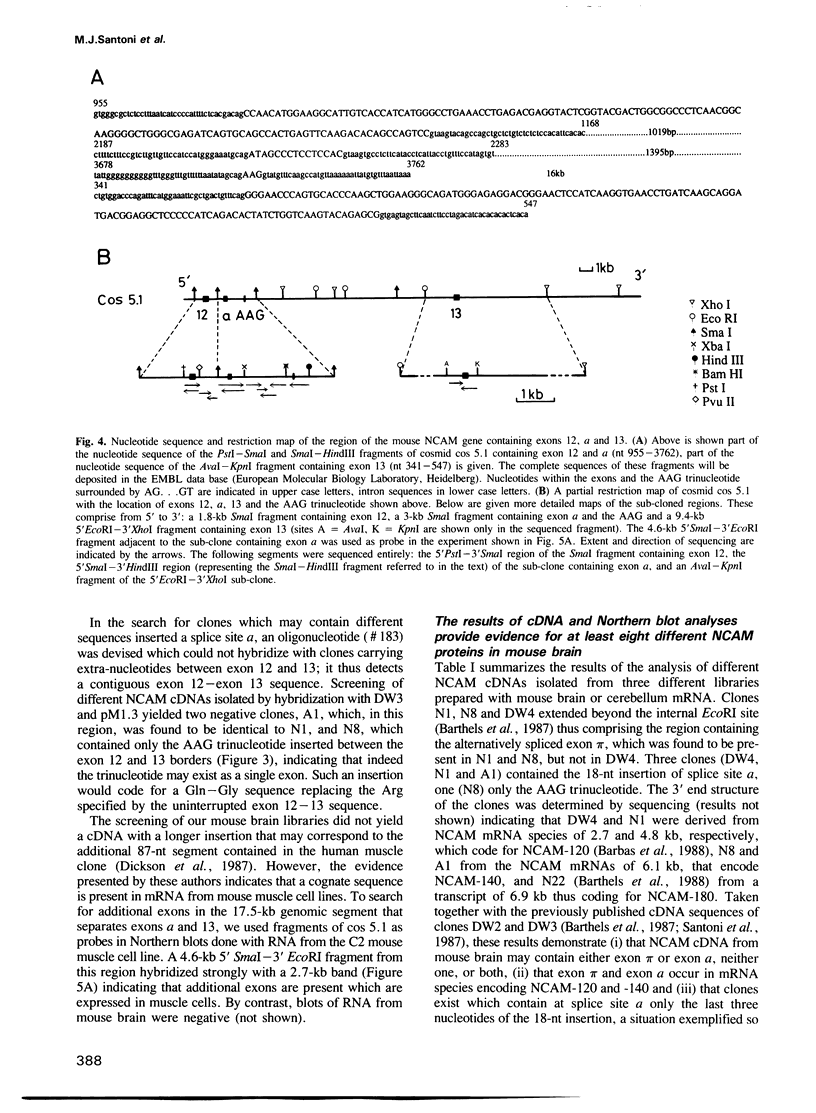
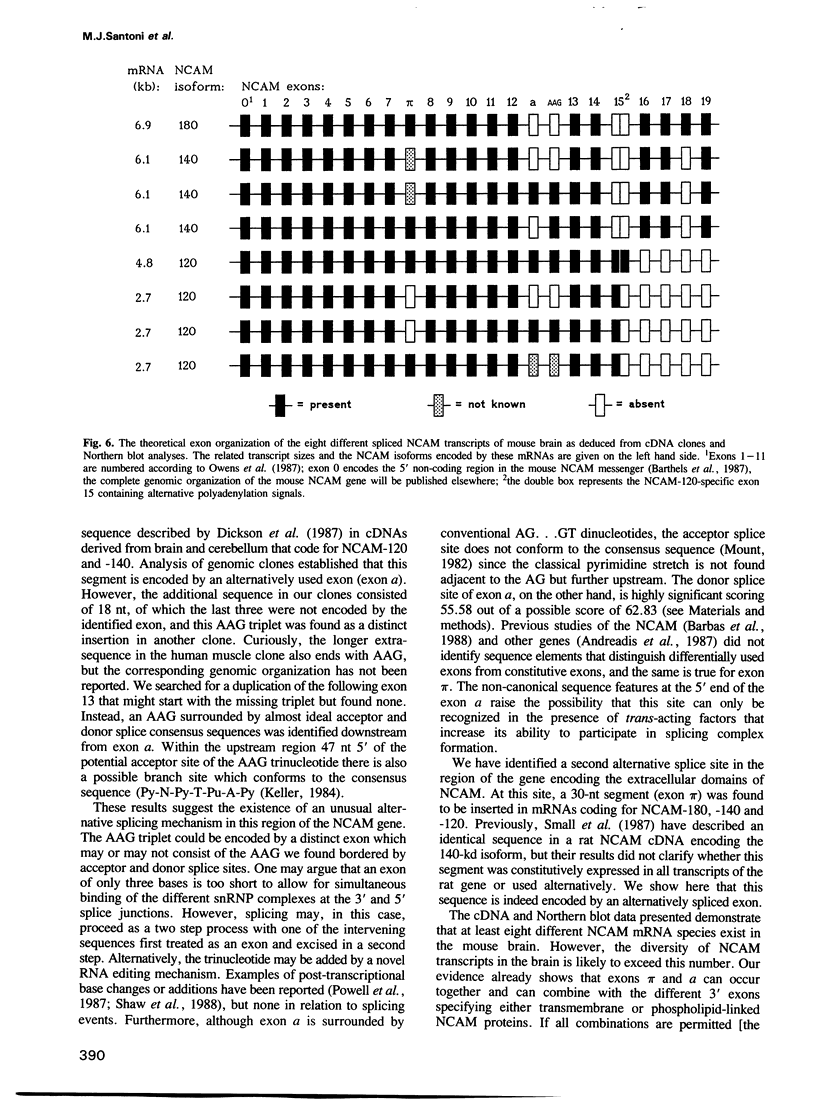
The murine neural cell adhesion molecule (NCAM) is known to exist in three isoforms of different size, NCAM-180, -140 and -120 coded for by four transcripts of 6.9, 6.1, 4.8 and 2.7 kb in length. Since the differences between these isoforms are due to alternative splicing in the coding region for the transmembrane and cytoplasmic domains, the extracellular, N-terminal portion of NCAM seemed to be shared by all three protein forms. Here we report that the coding region for N-terminal domains of NCAM also contains at least two sites of alternative splicing, termed alpha and pi. Short additional sequences of 3, 18 and 30 nt in length can be introduced at these sites, which are located in the membrane-proximal 'stem' between the Ig-like domains and the membrane attachment site and within the Ig-like domain IV, respectively. Proof for at least eight different mRNAs has been found by sequencing and S1 nuclease protection assays of selected independent cDNA clones, and Northern blot analyses. If most combination of the splice patterns identified so far in mouse brain occurred, 24 different mRNAs could be generated coding for 18 different proteins. The shortest extra-sequence found inserted at splice site alpha consisted only of the trinucleotide AAG, raising questions about the mechanism of this particular insertion.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreadis A., Gallego M. E., Nadal-Ginard B. Generation of protein isoform diversity by alternative splicing: mechanistic and biological implications. Annu Rev Cell Biol. 1987;3:207–242. doi: 10.1146/annurev.cb.03.110187.001231. [DOI] [PubMed] [Google Scholar]
- Barbas J. A., Chaix J. C., Steinmetz M., Goridis C. Differential splicing and alternative polyadenylation generates distinct NCAM transcripts and proteins in the mouse. EMBO J. 1988 Mar;7(3):625–632. doi: 10.1002/j.1460-2075.1988.tb02856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthels D., Santoni M. J., Wille W., Ruppert C., Chaix J. C., Hirsch M. R., Fontecilla-Camps J. C., Goridis C. Isolation and nucleotide sequence of mouse NCAM cDNA that codes for a Mr 79,000 polypeptide without a membrane-spanning region. EMBO J. 1987 Apr;6(4):907–914. doi: 10.1002/j.1460-2075.1987.tb04837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthels D., Vopper G., Wille W. NCAM-180, the large isoform of the neural cell adhesion molecule of the mouse, is encoded by an alternatively spliced transcript. Nucleic Acids Res. 1988 May 25;16(10):4217–4225. doi: 10.1093/nar/16.10.4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart R. E., Nadal-Ginard B. Complete nucleotide sequence of the fast skeletal troponin T gene. Alternatively spliced exons exhibit unusual interspecies divergence. J Mol Biol. 1986 Apr 5;188(3):313–324. doi: 10.1016/0022-2836(86)90157-9. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Nguyen H. T., Medford R. M., Destree A. T., Mahdavi V., Nadal-Ginard B. Intricate combinatorial patterns of exon splicing generate multiple regulated troponin T isoforms from a single gene. Cell. 1985 May;41(1):67–82. doi: 10.1016/0092-8674(85)90062-5. [DOI] [PubMed] [Google Scholar]
- Cathala G., Savouret J. F., Mendez B., West B. L., Karin M., Martial J. A., Baxter J. D. A method for isolation of intact, translationally active ribonucleic acid. DNA. 1983;2(4):329–335. doi: 10.1089/dna.1983.2.329. [DOI] [PubMed] [Google Scholar]
- Cole G. J., Loewy A., Cross N. V., Akeson R., Glaser L. Topographic localization of the heparin-binding domain of the neural cell adhesion molecule N-CAM. J Cell Biol. 1986 Nov;103(5):1739–1744. doi: 10.1083/jcb.103.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham B. A., Hemperly J. J., Murray B. A., Prediger E. A., Brackenbury R., Edelman G. M. Neural cell adhesion molecule: structure, immunoglobulin-like domains, cell surface modulation, and alternative RNA splicing. Science. 1987 May 15;236(4803):799–806. doi: 10.1126/science.3576199. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson G., Gower H. J., Barton C. H., Prentice H. M., Elsom V. L., Moore S. E., Cox R. D., Quinn C., Putt W., Walsh F. S. Human muscle neural cell adhesion molecule (N-CAM): identification of a muscle-specific sequence in the extracellular domain. Cell. 1987 Sep 25;50(7):1119–1130. doi: 10.1016/0092-8674(87)90178-4. [DOI] [PubMed] [Google Scholar]
- Edelman G. M. Cell adhesion molecules in the regulation of animal form and tissue pattern. Annu Rev Cell Biol. 1986;2:81–116. doi: 10.1146/annurev.cb.02.110186.000501. [DOI] [PubMed] [Google Scholar]
- Edelman G. M., Hoffman S., Chuong C. M., Thiery J. P., Brackenbury R., Gallin W. J., Grumet M., Greenberg M. E., Hemperly J. J., Cohen C. Structure and modulation of neural cell adhesion molecules in early and late embryogenesis. Cold Spring Harb Symp Quant Biol. 1983;48(Pt 2):515–526. doi: 10.1101/sqb.1983.048.01.056. [DOI] [PubMed] [Google Scholar]
- Edelman G. M. Morphoregulatory molecules. Biochemistry. 1988 May 17;27(10):3533–3543. doi: 10.1021/bi00410a001. [DOI] [PubMed] [Google Scholar]
- Gennarini G., Hirsch M. R., He H. T., Hirn M., Finne J., Goridis C. Differential expression of mouse neural cell-adhesion molecule (N-CAM) mRNA species during brain development and in neural cell lines. J Neurosci. 1986 Jul;6(7):1983–1990. doi: 10.1523/JNEUROSCI.06-07-01983.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goridis C., Deagostini-Bazin H., Hirn M., Hirsch M. R., Rougon G., Sadoul R., Langley O. K., Gombos G., Finne J. Neural surface antigens during nervous system development. Cold Spring Harb Symp Quant Biol. 1983;48(Pt 2):527–537. doi: 10.1101/sqb.1983.048.01.057. [DOI] [PubMed] [Google Scholar]
- Goridis C., Hirn M., Santoni M. J., Gennarini G., Deagostini-Bazin H., Jordan B. R., Kiefer M., Steinmetz M. Isolation of mouse N-CAM-related cDNA: detection and cloning using monoclonal antibodies. EMBO J. 1985 Mar;4(3):631–635. doi: 10.1002/j.1460-2075.1985.tb03676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. K., Rutishauser U. Visualization of neural cell adhesion molecule by electron microscopy. J Cell Biol. 1987 Jun;104(6):1579–1586. doi: 10.1083/jcb.104.6.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W. The RNA lariat: a new ring to the splicing of mRNA precursors. Cell. 1984 Dec;39(3 Pt 2):423–425. doi: 10.1016/0092-8674(84)90449-5. [DOI] [PubMed] [Google Scholar]
- Kornblihtt A. R., Umezawa K., Vibe-Pedersen K., Baralle F. E. Primary structure of human fibronectin: differential splicing may generate at least 10 polypeptides from a single gene. EMBO J. 1985 Jul;4(7):1755–1759. doi: 10.1002/j.1460-2075.1985.tb03847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S. E., Thompson J., Kirkness V., Dickson J. G., Walsh F. S. Skeletal muscle neural cell adhesion molecule (N-CAM): changes in protein and mRNA species during myogenesis of muscle cell lines. J Cell Biol. 1987 Sep;105(3):1377–1386. doi: 10.1083/jcb.105.3.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens G. C., Edelman G. M., Cunningham B. A. Organization of the neural cell adhesion molecule (N-CAM) gene: alternative exon usage as the basis for different membrane-associated domains. Proc Natl Acad Sci U S A. 1987 Jan;84(1):294–298. doi: 10.1073/pnas.84.1.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell L. M., Wallis S. C., Pease R. J., Edwards Y. H., Knott T. J., Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987 Sep 11;50(6):831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- Ruppert C., Goldowitz D., Wille W. Proto-oncogene c-myc is expressed in cerebellar neurons at different developmental stages. EMBO J. 1986 Aug;5(8):1897–1901. doi: 10.1002/j.1460-2075.1986.tb04442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser U., Acheson A., Hall A. K., Mann D. M., Sunshine J. The neural cell adhesion molecule (NCAM) as a regulator of cell-cell interactions. Science. 1988 Apr 1;240(4848):53–57. doi: 10.1126/science.3281256. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni M. J., Barthels D., Barbas J. A., Hirsch M. R., Steinmetz M., Goridis C., Wille W. Analysis of cDNA clones that code for the transmembrane forms of the mouse neural cell adhesion molecule (NCAM) and are generated by alternative RNA splicing. Nucleic Acids Res. 1987 Nov 11;15(21):8621–8641. doi: 10.1093/nar/15.21.8621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. M., Feagin J. E., Stuart K., Simpson L. Editing of kinetoplastid mitochondrial mRNAs by uridine addition and deletion generates conserved amino acid sequences and AUG initiation codons. Cell. 1988 May 6;53(3):401–411. doi: 10.1016/0092-8674(88)90160-2. [DOI] [PubMed] [Google Scholar]
- Small S. J., Shull G. E., Santoni M. J., Akeson R. Identification of a cDNA clone that contains the complete coding sequence for a 140-kD rat NCAM polypeptide. J Cell Biol. 1987 Nov;105(5):2335–2345. doi: 10.1083/jcb.105.5.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Frelinger A. L., 3rd, Rutishauser U. Topography of N-CAM structural and functional determinants. I. Classification of monoclonal antibody epitopes. J Cell Biol. 1986 Nov;103(5):1721–1727. doi: 10.1083/jcb.103.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbur W. J., Lipman D. J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci U S A. 1983 Feb;80(3):726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. K., Goridis C., Akeson R. Individual neural cell types express immunologically distinct N-CAM forms. J Cell Biol. 1985 Jul;101(1):36–42. doi: 10.1083/jcb.101.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]