Abstract
CD169-positive macrophages in the marginal zone of the spleen and subcapsular sinus of lymph nodes play an important role as gatekeepers, strategically located to capture pathogens. Here we identified a population of CD169-positive macrophages in the colon and investigated which factors influenced their development. Murine colonic CD115+ F4/80lo CD11clo macrophages expressing CD169 were present in the lamina propria, mainly surrounding the crypts. In spite of the high levels of bacterial flora in the colon and the importance of Toll-like receptor signalling in mucosal homeostasis, the presence of CD169+ macrophages was not affected in mice that were deficient in MyD88-mediated Toll-like receptor signalling and in mice in which the bacterial flora was eradicated. Whereas the development of splenic CD169+ macrophages was dependent on lymphotoxin α, colonic CD169+ macrophages were present in normal numbers in lymphotoxin α-deficient mice. In contrast, reduced numbers of CD169+ macrophages were found in the colon of mice deficient in vitamin A, whereas CD169+ macrophages in the spleen were unaffected. In conclusion, we identified a new macrophage subset in the lamina propria of the colon characterized by the expression of CD169. Its differentiation, unlike CD169+ macrophages in lymphoid organs, is independent of lymphotoxin α signalling, but requires vitamin A.
Keywords: development, intestine, Siglec-1, spleen, vitamin A
Introduction
CD169 (also known as sialoadhesin, Siglec-1) is a cell surface sialic-acid-binding receptor that belongs to the family of sialic-acid-binding immunoglobulin-like lectins (Siglecs). CD169 is an immunoglobulin superfamily member with 17 immunoglobulin-like domains that preferentially binds α2-3 and α2-6-linked sialic acid moieties on other cells or microbial surfaces. Monovalent ligand binding by CD169 is of very low affinity. However, multivalent binding of ligands is thought to create a high enough avidity to induce a biological response.1,2
CD169 is expressed by specific macrophage subsets in secondary lymphoid organs, and also bone marrow, liver and lung were shown to contain CD169+ macrophages.3–5 In lymph nodes both macrophages lining the subcapsular sinus and the medulla highly express CD169, while in the murine spleen CD169 is highly expressed by marginal metallophilic macrophages that line the white pulp in the marginal zone. In both lymphoid organs, the CD169-expressing macrophages are located at the entry site of lymph or blood and close to T-and B-lymphocyte areas. Therefore, they are ideally located to affect innate and adaptive immune responses upon an encounter with pathogens brought in via the lymph or blood.6–8 Indeed, lymph node CD169+ macrophages were shown to be involved in the uptake and presentation of antigens to B cells,9–12 innate lymphoid cells, and natural killer T and natural killer cells.13–16 Splenic CD169+ macrophages facilitated the generation of CD8+ cytotoxic T-cell responses.17 Recent studies have indicated that CD169+ macrophages show permissiveness for local viral replication, which ensures the presence of enough antigen to activate an effective adaptive immune response,18 but prevent viral and bacterial spread into the central nervous system and the systemic circulation via the activation of innate immune responses.19,20 In contrast, splenic CD169+ macrophages are scavengers of dead cell debris and are implicated in the induction of tolerance against these cellular remnants.21,22 Apparently, CD169+ macrophages exhibit diverse functions in both the innate and adaptive immune systems and as gatekeepers can prevent further infection by pathogens.
The colon, which is a potential entry site for microbial or food-derived antigens, contains multiple subsets of macrophages. Under steady-state conditions these macrophages are the most abundant type of mononuclear phagocyte present in the lamina propria. Although they are highly phagocytic and have high bactericidal activity, colonic macrophages hardly produce any pro-inflammatory cytokines23 and have poor T-cell priming capacity.24,25 Instead, colonic macrophages produce large amounts of interleukin-10, which makes them important for maintenance of immune tolerance, because interleukin-10 is necessary for the induction, local expansion and maintenance of FoxP3 expression in regulatory T cells.24–27 So far, two different colonic macrophage subsets have been identified, which are characterized by the phenotypes F4/80+ CD11c+ CD11b+ and F4/80+ CD11c− CD11b+. The CD11c+ macrophage subset is localized in close proximity to the epithelial layer, whereas the CD11c− macrophage subset is scattered throughout the lamina propria.25
Because of the typical gatekeeper function of CD169+ macrophages in lymph nodes and spleen, we investigated whether a similar type of CD169+ macrophage could be detected in the colon in which continuous interaction with the intestinal microflora takes place. CD169+ macrophages were detected in the lamina propria of the colon, mainly surrounding the epithelial crypts. Whereas surface expression of various markers was similar on colonic CD169+ macrophages and splenic CD169+ macrophages, they differed substantially in their differentiation requirements. In contrast to splenic CD169+ macrophages, colonic CD169+ macrophages depended on vitamin A for their proper development, but did not require lymphotoxin-α (LTα) signalling. These data show that the colonic lamina propria contains a distinct population of CD169+ macrophages, which are unique in their requirements for development.
Materials and methods
Mice
C57BL/6, myeloid differentiation antigen 88 (MyD88) knockout (ko) and lymphotoxin-α knockout (LTαko) mice were bred and maintained at the animal facility of the VU University Medical Centre in Amsterdam. Both male and female mice between 8 and 12 weeks old were used in the experiments described. All experiments were approved by the animal experimentation ethics committee of the VU University Medical Centre and according to local and governmental regulations.
To eradicate the intestinal microflora of mice, C57BL/6 mice were treated for the indicated time periods with an antibiotics cocktail containing streptomycin (5 g/l), colistin (1 g/l), ampicillin (1 g/l) and sucrose (2·5%) (all ordered from Sigma-Aldrich, Zwijndrecht, the Netherlands) in their drinking water.28 Faeces were collected during the experiment to assess the effectiveness of the treatment by determining the amount of 16S bacterial DNA.
Pregnant C57BL/6 females (Charles River Laboratories, Maastricht, the Netherlands) were fed with vitamin A-deficient (0 IU vitamin A/kg) or control (4000 IU vitamin A/kg) AIN-93M diet produced with vitamin-free casein (MP Biomedicals, Solon, OH or Research Diets Services, Wijk bij Duurstede, the Netherlands) from day 7·5 to 10·5 of pregnancy. Pups were weaned at week 5 and kept on their specific diet until they were killed at week 9–14. Animals were kept under specific pathogen-free conditions.
To induce an acute form of colitis, mice were supplied with 2% dextran sulphate sodium in drinking water ad libitum, which was changed on a daily basis for 7 days.
Antibodies and flow cytometric analysis
The following antibodies were used for flow cytometric analysis: anti-CD169-Alexa Fluor® 488 (clone Ser4, 1 : 800), anti-CD11b-phycoerythrin (PE)-Cy7 (clone M1/70, 0·4 μg/ml; eBioscience, Vienna, Austria), anti-F4/80-PE (clone BM8, 4 μg/ml), anti-CD11c-PE or-allophycocyanin (clone N418, 1 μg/ml; eBioscience), anti-MHC II(I-A/I-E)-Alexa Fluor® 647 (clone M5/114,1 : 200), CD64-PE (clone X54-5/7.1, 1 μg/ml; Biolegend, Fell, Germany), CD135-PE (clone A2F10, 2 μg/ml; eBioscience), CD103-PE (clone M290, 1 μg/ml; BD Biosciences), intercellular adhesion molecule (ICAM)-PE (clone YN1/1.7.4, 1 μg/ml; eBioscience), vascular cell adhesion molecule (VCAM)-PE (clone 429, 1 μg/ml; eBioscience), SYTOX® Blue nucleic acid stain and Fixable Near-Infrared Live/Dead stain (Invitrogen Life Technologies, Breda, the Netherlands). Cells were incubated with 2.4G2 supernatant to block aspecific binding. For flow cytometric analysis a CyAn ADP flow cytometer (Beckmann Coulter, Woerden, the Netherlands) was used. Flow cytometry data were analysed using flowjo 9.2 Software (Treestar, Ashland, OR).
Preparation of splenic cell suspensions for flow cytometry
Single cell suspensions of spleen were obtained by Liberase TL-mediated enzymatic digestion for 30 min at 37° with constant stirring (1 Wünsch unit/ml; Roche Diagnostics GmbH, Mannheim, Germany) after which erythrocytes were lysed using a buffer containing 0·15 m NH4Cl, 10 mm KHCO3 and 0·1 mm EDTA in PBS.
Preparation of colonic cell suspensions for flow cytometry
Colons were dissected, opened longitudinally, washed in PBS and cut into pieces of ˜ 2 mm. Faecal contents were removed by washing in Hanks’ balanced salt solution (HBSS) buffer (without Ca2+ and Mg2+) containing 15 mm HEPES and 250 μg/ml gentamicin. Mucus and epithelial cells were removed from the tissue by incubating the tissue pieces in HBSS buffer (without Ca2+ and Mg2+) containing 5 mm EDTA, 1 μm dithiothreitol, 14 mm 2-mercaptoethanol, 10% fetal calf serum, 15 mm HEPES, 0·5% penicillin-streptomycin, twice, for 15 min each time at 37° under constant stirring. Subsequently, the colonic segments were cut into smaller pieces and digested in HBSS buffer containing 15 mm HEPES, 10% heat-inactivated fetal calf serum, 200 μg/ml DNAse (Roche Diagnostics GmbH) and 150 μg/ml liberase™ (Roche Diagnostics GmbH) for 15 min at 37° under constant stirring. Cells were washed twice in HBSS buffer containing calcium and magnesium, 15 mm HEPES and 250 μg/ml gentamicin and filtered through a 70-μm cell strainer. CD45+ cells were purified from the cell suspensions by immunomagnetic cell separation using anti-CD45-biotin (clone 30-F11; eBioscience) and the EasySep mouse biotin positive selection kit (StemCell Technologies, Grenoble, France).
Immunofluorescence microscopy
Colonic and splenic tissues were snap-frozen in tissue-TEK on dry ice. Sections were cut (colon: 8 μm, spleen: 5 μm), fixed for 10 min in acetone, and stained with the following antibodies as indicated: anti-CD169-Alexa Fluor® 488 (clone Ser4, 1 : 800), anti-CD115-biotin (clone AFS98, 5 μg/ml; eBioscience), anti-F4/80 (clone BM8, 4 μg/ml), goat anti-rat IgG (H+L)-Alexa Fluor® 647 (5 μg/ml; Molecular Probes, Invitrogen Life Technologies), anti-CD11b-PE (clone M1/70, 0·4 μg/ml; eBioscience), anti-CD11c-PE (clone N418, 1 μg/ml; eBioscience), anti-CD64-PE (clone X54-5/7.1, 1 μg/ml; Biolegend), MadCAM-Alexa Fluor® 555 (clone MECA367, 1 : 200), anti-αSMA-(clone 1A4, 1 : 500; Sigma-Aldrich). Staining with isotype antibodies or secondary antibodies only was used for negative controls. Staining was analysed using a DM6000 Leica immunofluorescence microscope (Leica microsystems, Wetzlar, Germany).
RNA isolation and real-time PCR
Colon and spleen tissues were homogenized in TRIzol (Invitrogen Life Technologies), and phenol/chloroform extraction was performed using Phase Lock Heavy Gel tubes (Eppendorf, Nijmegen, the Netherlands), followed by consecutive isopropanol and ethanol precipitation. The concentration of isolated RNA, dissolved in RNAse free water, was measured using a Nanodrop Spectrophotometer (Nanodrop Technologies, Wilmington, DE). An RNA input of 1 μg for colons and 5 μg for spleens was used for cDNA synthesis using a RevertAid™ First-strand cDNA synthesis kit (Fermentas Life Sciences, Burlington, ON, Canada), according to the manufacturer's protocol. The resulting cDNA was used for analysis by RT-PCR.
Expression levels of mRNA were determined using an ABI Prism 7900HT Sequence Detection System (PE Applied Biosystems, Foster City, CA). Reactions were performed in a total reaction volume of 10 μl, containing cDNA, 300 nm forward primer, 300 nm reverse primer and SYBR Green PCR Mastermix (PE Applied Biosystems). Primers used for analysis of mRNA expression were designed using OligoExplorer1.2 software (http://www.GeneLink.com) and synthesized by Invitrogen Life Technologies. Primer sequences: CD169-forward: 5′-CCAGGCTGGATTTGGATACCT-3′, CD169-reverse: 5′-ACGTGGCACAAGA GGATGC-3′, hypoxanthine–guanine phosphoribosyltransferase (HPRT)-forward: 5′-CCTAAGATGAGCGCAAGTTGAA-3′, HPRT-reverse: 5′-CCAC AGGACTAGAACACCTGCTAA-3′. A standard curve was generated using pooled lymph node tissue to correct for primer efficiency. Quantities of mRNA were normalized to housekeeping gene HPRT.
Statistical evaluation
Values are expressed as the mean ± SEM. An analysis of variance was used to test for statistical significant differences between three or more groups. A Student's t-test was performed when comparing two samples. A P-value of < 0·05 was considered statistically significant. *P < 0·05, **P < 0·01, ***P < 0·001.
Results
CD169+ macrophages in the colon
Recent literature demonstrates the existence of multiple subtypes of macrophages in the colon,25 which serve important functions in the maintenance of local immune homeostasis in the presence of microorganisms and food components in the intestinal lumen. In lymphoid organs CD169+ macrophages are present as gatekeepers at the sites where antigens enter.6–8 The CD169 molecule is thought to play a role in pathogen recognition and possibly supports the interaction of the macrophages with other cells from the immune system.2 To investigate whether in the colon CD169-expressing macrophages are present that may perform similar functions in the intestines, colon sections were stained for CD169 and analysed by immunofluorescence microscopy.
CD169+ cells were detected in the colonic lamina propria, mainly surrounding the intestinal epithelial crypts, but they were absent from local lymphoid structures (Fig. 1a,b). Few CD169+ macrophages were present in the colonic muscularis mucosae, submucosa and muscularis externa, which layers were visualized by staining for α-smooth muscle actin, compared with the lamina propria (Fig. 1c). During colonic inflammation caused by the addition of dextran sulphate sodium to the drinking water for 7 days, more CD169+ cells were observed (Fig. 1d).
Figure 1.
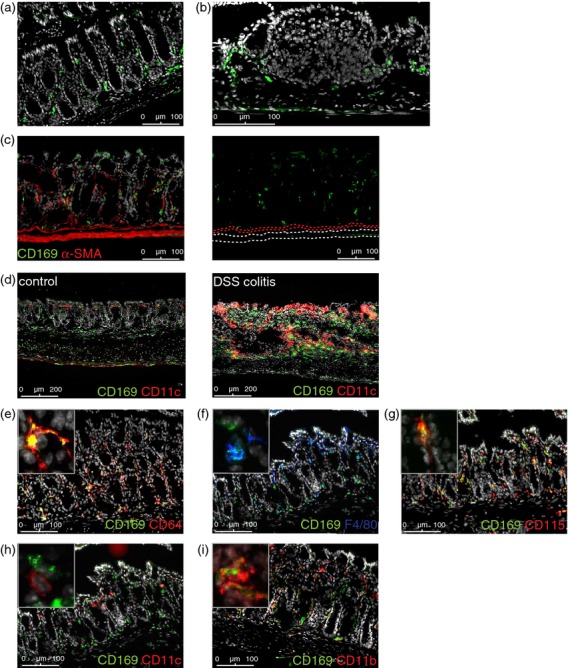
CD169+ macrophages in the colon. (a–c) Representative immunofluorescence stainings of C57BL/6 wild-type (Wt) colon sections, stained for CD169, nuclei (Dapi) (a–c) and α-smooth muscle actin (c). (d) Representative immunofluorescence stainings of colon sections from mice with acute dextran sulphate sodium-induced colitis and healthy controls (n = 2), stained for CD169, CD11c and nuclei (Dapi). (e–i) Immunofluorescent stainings of C57BL/6 Wt colon sections, stained for CD169, nuclei (Dapi), and CD64, F4/80, CD115, CD11c, or CD11b as indicated. The panel in the upper left corner shows a 5× magnification of a CD169+ cell.
The CD169+ cells had an elongated appearance and showed dendritic extensions, matching a macrophage or dendritic cell (DC)-like phenotype. Co-staining the sections showed co-localization of CD169 with macrophage markers CD64, CD115 (macrophage colony-stimulating factor receptor) and to a lesser extent F4/80 (Fig. 1e–g), but not with the DC marker CD11c (Fig. 1h), indicating that the colon contains CD169+ macrophages, which in addition express CD11b (Fig. 1i).
The colonic CD169+ macrophages were further analysed by flow cytometry and compared with splenic CD169+ macrophages to determine similarities between the two CD169+ macrophage populations. Colonic CD169+ macrophages were gated as live CD45+ CD11b+ CD169+ cells (Fig. 2a), whereas splenic CD169+ macrophages were gated as live autofluorescent CD169+ cells (Fig. 2b). Confirming the immunofluorescence analysis, also flow cytometric analysis showed that the colonic CD169+ cells highly expressed macrophage marker CD64 and low levels of F4/80 (Fig. 2c). In addition, no expression of DC markers CD103, CD135 and low expression of CD11c were detected on CD169+ cells (Fig. 2c). These data further supported our conclusion that the CD169+ cells were macrophages. In addition, the cells expressed high levels of MHC class II and ICAM-1, but no VCAM-1.
Figure 2.
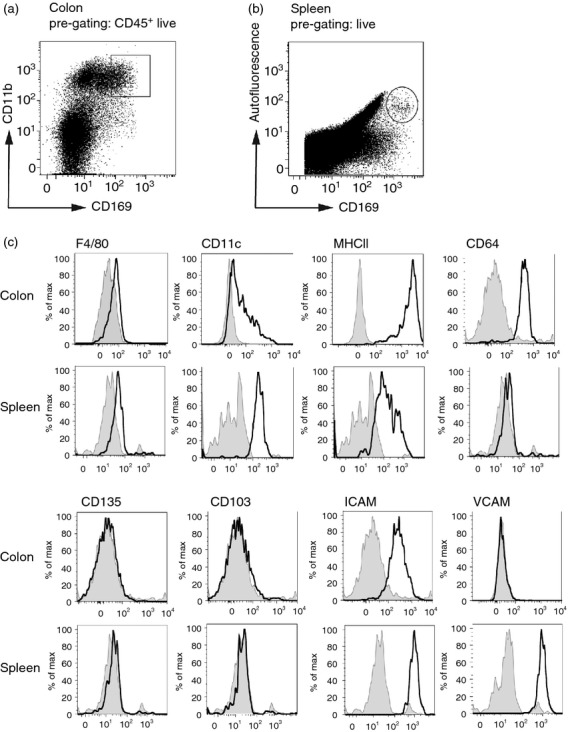
Flow cytometric analysis of CD169+ macrophages. (a) Flow cytometric gating strategy of colonic live CD169+ macrophages from CD45+ single cell colon suspensions. (b) Flow cytometric gating strategy of splenic live, autofluorescent CD169+ macrophages from single cell spleen suspensions. (c) Colonic and splenic CD169+ macrophages from single cell suspensions of C57BL/6 wild-type (Wt) colons and spleens were analysed for the expression of the different indicated markers by flow cytometry. Isotype controls are depicted in grey.
Comparison of the colonic CD169+ macrophages to splenic CD169+ macrophages showed many similarities. Both colonic and splenic cells expressed low levels of F4/80 and CD11c, but not the DC markers CD103 and CD135. In addition, both cell types expressed MHC class II and ICAM-1 (Fig. 2c). CD64 was recently characterized as a marker to discriminate macrophages from DCs in the intestine.29 Indeed, we observed CD64 expression on CD169+ macrophages in the colon, but splenic CD169+ macrophages apparently do not express this molecule (Fig. 2c). VCAM-1 was only expressed by splenic CD169+ macrophages, not by colonic CD169+ macrophages.
Lymphotoxin-α is dispensable for colonic CD169+ macrophage development
Since surface marker expression of colonic CD169+ macrophages and splenic CD169+ macrophages was comparable, we wondered whether their differentiation requirements also show similarities. Presence and function of CD169+ macrophages in the spleen has been shown to depend on lymphotoxin β receptor (LTβR) signalling, induced by B-cell surface-expressed LTα1β2.30–32 To investigate whether colonic CD169+ macrophages depend on LTα1β2 in a similar way, CD169 mRNA expression levels were measured in the colons of wild-type (Wt) and LTα-deficient (i.e. LTαko) mice. Whereas CD169 mRNA levels were significantly decreased in spleens of LTαko mice, they were unchanged in LTαko colons (Fig. 3a). In addition, flow cytometric analysis of CD169+ macrophages from spleen and colon of Wt and LTαko mice indicated a significant decrease of these cells in spleens of LTαko mice compared with Wt mice, whereas their numbers remained unchanged in the colons (Fig. 3b). Also, immunofluorescent imaging showed no differences in the amount or localization of CD169+ macrophages in Wt and LTαko colons, whereas CD169+ macrophages in the splenic marginal zone were absent in LTαko mice, compared with Wt (Fig. 3c). In conclusion, these data show that in contrast to splenic CD169+ macrophages, which are known to depend on LTα-induced signalling for their presence, LTα signalling is dispensable for the presence of colonic CD169+ macrophages.
Figure 3.
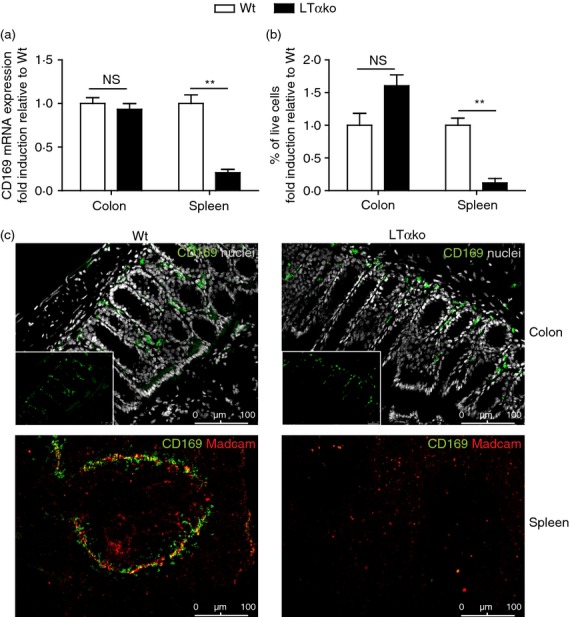
Lymphotoxin α (LTα) is dispensable for colonic CD169+ macrophage development. (a) CD169 mRNA expression was determined in total colon and spleen homogenates of C57BL/6 wild-type (Wt) and LTα knockout (ko) mice by RT-PCR (colon n = 3, spleen n = 5 Wt, n = 3 LTαko). Expression levels were calculated as relative amounts normalized to housekeeping gene HPRT and shown here as fold induction compared with the Wt expression level in that organ. (b) Flow cytometric analysis of the relative amount of CD169+ colonic macrophages and splenic CD169+ macrophages in single cell suspensions of colon and spleen from Wt and LTαko mice (n = 3). (c) Immunofluorescence stainings of Wt and LTαko colon and spleen sections, stained for CD169 (green) and nuclei (grey, colon) or CD169 (green) and Madcam+ marginal sinus-lining cells (red, spleen). Small inserts in colon pictures show CD169 staining only.
Colonic CD169+ macrophage differentiation is independent of intestinal microflora
Characteristic of the colonic microenvironment is the high load of bacteria that is present nearby in the intestinal lumen. This microflora is essential for shaping and a proper functioning of the intestinal immune system and influences the presence of colonic macrophages.33,34 One way in which bacterial products are recognized by host cells is through Toll-like receptors, which signal via adapter protein MyD88.35 To investigate whether the intestinal microflora affects the presence of CD169+ macrophages locally in the colon through Toll-like receptor signalling, CD169 mRNA expression levels were determined in colons and spleens of Wt and MyD88ko mice. Neither colons nor spleens of Wt and MyD88ko mice showed any differences in CD169 mRNA expression (Fig. 4a). In addition, flow cytometry and immunofluorescence imaging showed no difference in the number, presence and localization of CD169+ macrophages between Wt and MyD88ko mice in both organs (Fig. 4b,c).
Figure 4.
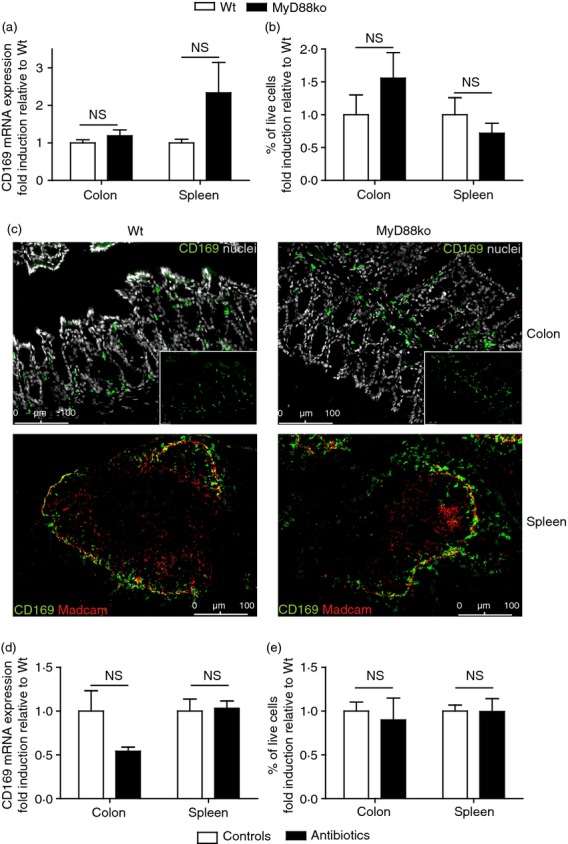
Presence of colonic CD169+ macrophages is independent of intestinal microflora. (a) CD169 mRNA expression was determined in total colon and spleen homogenates of C57BL/6 wild-type (Wt) and MyD88 knockout (ko) mice by RT-PCR (colon n = 4, spleen n = 5 Wt n = 3 MyD88ko). Expression levels were calculated as relative amounts compared with housekeeping gene HPRT and are shown here as fold induction compared with the Wt expression level. (b) Flow cytometric analysis of the amount of CD169+ colonic macrophages and splenic CD169+ macrophages in single cell suspensions of colon and spleen from Wt and MyD88ko mice (colon n = 3, spleen n = 8 Wt, n = 5 MyD88ko). (c) Immunofluorescence stainings of Wt and MyD88ko colon and spleen sections, stained for CD169 (green) and nuclei (grey, colon) or CD169 (green) and Madcam+ marginal sinus-lining cells (red, spleen). Small inserts in colon pictures show CD169 staining only. (d) CD169 mRNA expression was determined in total colon and spleen homogenates of C57BL/6 control and antibiotics-treated mice by RT-PCR (colon n = 5, spleen n = 4 Wt n = 3 antibiotics-treated mice). Expression levels were calculated as relative amounts compared with housekeeping gene HPRT and shown here as fold induction compared with the Wt expression level. (e) Flow cytometric analysis of the amount of CD169+ colonic macrophages and splenic CD169+ macrophages in single cell suspensions of colon and spleen from control and antibiotics-treated mice (colon n = 3, spleen n = 5).
To investigate whether other microflora-derived signals affect the presence of CD169+ macrophages in colon, mice were administered antibiotics in their drinking water for 33 days, after which the presence of CD169+ macrophages was determined in colons and spleens and compared with tissues from control animals. CD169 mRNA expression was similar in both colons and spleens of control and antibiotics-treated animals (Fig. 4d), as were the numbers of CD169+ macrophages measured in both organs by flow cytometry (Fig. 4e).
In conclusion, the presence of CD169+ macrophages in colon and spleen appeared to be independent of the intestinal microflora and MyD88-dependent signalling.
Colonic CD169+ macrophages depend on vitamin A
Vitamin A is essential for maintenance of immune homeostasis in the intestine. It is converted into its active metabolite retinoic acid by retinaldehyde dehydrogenase expressed by the intestinal epithelium and by immune cells like DCs and macrophages.36–39 To investigate whether vitamin A affects the presence of CD169+ macrophages in the colon, vitamin A-deficient mice were generated. CD169 mRNA expression was significantly decreased in the colons of vitamin A-deficient mice, compared with those of vitamin A control mice, whereas no difference was observed between spleens of both types of mice (Fig. 5a). In addition, flow cytometric and immunofluorescence staining showed a marked decrease in number of CD169+ macrophages in the colon of vitamin A-deficient mice, whereas no difference was observed in the spleen (Fig. 5b,c). These data show that, in contrast to splenic CD169+ macrophages, vitamin A is essential for the proper development and differentiation of colonic CD169+ macrophages.
Figure 5.
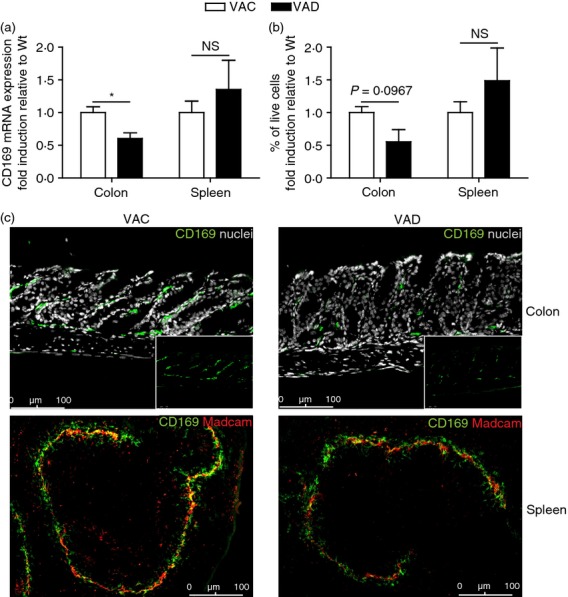
Colonic CD169+ macrophages depend on vitamin A. (a) CD169 mRNA expression was determined in total colon and spleen homogenates of C57BL/6 vitamin A control (VAC) and vitamin A deficient (VAD) mice by RT-PCR (colon n = 5, spleen n = 3). Expression levels were calculated as relative amounts compared with housekeeping gene HPRT and shown here as fold induction compared with the Wt expression level. (b) Flow cytometric analysis of the amount of CD169+ colonic macrophages and splenic CD169+ macrophages in single cell suspensions of colon and spleen from VAC and VAD mice (colon n = 3, spleen n = 4–5). (c) Immunofluorescence stainings of VAC and VAD colon and spleen sections, stained for CD169 (green) and nuclei (grey, colon) or CD169 (green) and Madcam+ marginal sinus-lining cells (red, spleen). Small inserts in colon pictures show CD169 staining only.
Discussion
Our studies demonstrate the presence of a previously unrecognized CD169+ macrophage subset present in the colonic lamina propria. This macrophage subset is, in contrast to splenic CD169+ macrophages, not dependent on LTα signalling for its development. Also microflora-induced signalling did not affect the presence of the CD169+ macrophage subset in the colon. In contrast, vitamin A deficiency severely reduced levels of colonic CD169+ macrophages while splenic CD169+ macrophages remained unaffected. These data show striking local differences in the requirements for differentiation of colonic and splenic CD169+ macrophages.
Although the presence of CD169+ macrophages in the outer muscle layer of the intestines has been reported,40 the presence in the colonic lamina propria is remarkable. Evidence of a human counterpart of the colonic lamina propria CD169+ macrophages that we have described here, was previously documented.41
Because the CD169+ macrophages show low expression of both F4/80 and CD11c, they do not fit into the colonic macrophage classification introduced by Rivollier et al.,25 which describes two different tolerogenic macrophage subsets that are either F4/80+ CD11c− or F4/80+ CD11c+. Therefore the discovery of CD169+ macrophages in the colonic lamina propria might add a new macrophage subset to this classification. Whether this subset, like the other two colonic macrophage subsets in the lamina propria, serves a tolerogenic function, or whether it, like the splenic and lymph node CD169+ macrophages, is involved in the generation of innate and adaptive immune responses remains to be shown. Interestingly, CD169+ macrophages in the central nervous system were shown to directly bind to sialic acids on regulatory T cells, thereby suppressing their expansion during experimental autoimmune encephalomyelitis.42 This raises the possibility that colonic CD169+ macrophages may exert similar pro-inflammatory functions in the intestine, but further studies will be necessary to elucidate their specific role in mucosal immunity.
Comparison of the colonic CD169+ macrophages with splenic CD169+ macrophages revealed highly similar surface marker expression. However, both cell types contrasted in their differentiation requirements. Whereas it was previously shown that splenic CD169+ macrophages depend on LTβR-signalling, induced by LTα1β2 expressed on B cells,30–32 this signalling pathway induced by LTα appeared to be dispensable for CD169+ macrophages in the colonic lamina propria. Although LTβR-induced signalling in the colon does serve important functions in the organization of lymphoid tissue and in the induction of IgA production,43–45 the absence of B cells in close proximity to the CD169+ macrophages in the colonic lamina propria suggests that B-cell-derived LTα1β2–LTβR signalling is not essential for colonic CD169 expression. In addition, our data indicate that LTα1β2 provided by other cell types such as innate lymphoid cells is not driving CD169 macrophage differentiation. However, other members of the tumour necrosis factor superfamily, such as LIGHT, which also binds to the LTβR, could possibly be involved in the generation of CD169+ macrophages.45,46
Instead of being LT-dependent, the presence of CD169+ macrophages in the colon was shown to be vitamin A-dependent. Vitamin A is converted into its active metabolite retinoic acid by intestinal epithelium-expressed and DC-expressed retinaldehyde dehydrogenase enzymes. Retinoic acid serves very important roles in the maintenance of intestinal immune homeostasis in the intestine, such as the generation of regulatory T cells47–52 and the induction of IgA class switching.53 Recently, retinoic acid was shown to be important for CXCL13 expression during lymph node development.54 Interestingly, CD169+ macrophages are absent in the spleens of mice that are deficient for the chemokine CXCL13.32 In the spleen, the expression of this chemokine is normally induced in follicular stromal cells by LTα1β2 signalling.55 The possibility that in the colonic lamina propria, retinoic acid instead of LTα1β2 is responsible for the induction of CXCL13 expression and possibly the presence of CD169+ macrophages could be excluded, because no difference in CXCL13 mRNA expression levels between control and vitamin A-deficient colons could be observed (data not shown).
In conclusion, we identified a vitamin A-dependent CD169+ macrophage subset in the lamina propria of the colon. Future research will be aimed at unravelling the function of this particular macrophage subset in innate and adaptive immune responses of the mucosal immune system.
Acknowledgments
This work was supported by grants from the Dutch Scientific Research program (NWO grants 854.10.005 to REM and 836.08.003 to JMMdH). We thank Erwin van Gelderop, Carla Prins and Rika van der Laan from VU University, Amsterdam, the Netherlands for their care of the animals used in this study.
Glossary
- DC
dendritic cell
- HBSS
Hanks’ balanced salt solution
- HPRT
hypoxanthine–guanine phosphoribosyltransferase
- ICAM
intercellular adhesion molecule
- ko
knockout
- LTα
lymphotoxin α
- LTβR
lymphotoxin-β receptor
- MyD88
myeloid differentiation antigen 88
- PE
phycoerythrin
- VCAM
vascular cell adhesion molecule
- Wt
wild-type
Author contributions
IHH designed and performed experiments, analysed data and wrote the manuscript; MRB designed and performed experiments and analysed data; HV was consulted for and contributed to experimental design of flow cytometric analysis of spleen CD169+ macrophages; KV performed RT-PCR experiments and supported animal experiments; EB performed immunofluorescence experiments and supported animal experiments; BO performed dextran sulphate sodium colitis experiments; REM was consulted for vitamin A-related experiments, contributed to experimental design of vitamin A deficiency mouse model and provided LTαko mice; GK was a principal investigator and was consulted for conceptual support; JMMdH was a principal investigator, designed the research and contributed to finalizing the manuscript.
Disclosures
The authors declare that they have no conflicts of interest.
References
- 1.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–66. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 2.Klaas M, Crocker PR. Sialoadhesin in recognition of self and non-self. Semin Immunopathol. 2012;34:353–64. doi: 10.1007/s00281-012-0310-3. [DOI] [PubMed] [Google Scholar]
- 3.Chow A, Lucas D, Hidalgo A, et al. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J Exp Med. 2011;208:261–71. doi: 10.1084/jem.20101688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crocker PR, Gordon S. Properties and distribution of a lectin-like hemagglutinin differentially expressed by murine stromal tissue macrophages. J Exp Med. 1986;164:1862–75. doi: 10.1084/jem.164.6.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ducreux J, Crocker PR, Vanbever R. Analysis of sialoadhesin expression on mouse alveolar macrophages. Immunol Lett. 2009;124:77–80. doi: 10.1016/j.imlet.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 6.den Haan JM, Kraal G. Innate immune functions of macrophage subpopulations in the spleen. J Innate Immun. 2012;4:437–45. doi: 10.1159/000335216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray EE, Cyster JG. Lymph node macrophages. J Innate Immun. 2012;4:424–36. doi: 10.1159/000337007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez-Pomares L, Gordon S. CD169+ macrophages at the crossroads of antigen presentation. Trends Immunol. 2012;33:66–70. doi: 10.1016/j.it.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Carrasco YR, Batista FD. B cells acquire particulate antigen in a macrophage-rich area at the boundary between the follicle and the subcapsular sinus of the lymph node. Immunity. 2007;27:160–71. doi: 10.1016/j.immuni.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Junt T, Moseman EA, Iannacone M, et al. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature. 2007;450:110–4. doi: 10.1038/nature06287. [DOI] [PubMed] [Google Scholar]
- 11.Phan TG, Grigorova I, Okada T, Cyster JG. Subcapsular encounter and complement-dependent transport of immune complexes by lymph node B cells. Nat Immunol. 2007;8:992–1000. doi: 10.1038/ni1494. [DOI] [PubMed] [Google Scholar]
- 12.Phan TG, Green JA, Gray EE, Xu Y, Cyster JG. Immune complex relay by subcapsular sinus macrophages and noncognate B cells drives antibody affinity maturation. Nat Immunol. 2009;10:786–93. doi: 10.1038/ni.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barral P, Polzella P, Bruckbauer A, et al. CD169+ macrophages present lipid antigens to mediate early activation of iNKT cells in lymph nodes. Nat Immunol. 2010;11:303–12. doi: 10.1038/ni.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coombes JL, Han SJ, van Rooijen N, Raulet DH, Robey EA. Infection-induced regulation of natural killer cells by macrophages and collagen at the lymph node subcapsular sinus. Cell Rep. 2012;2:124–35. doi: 10.1016/j.celrep.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia Z, Lemaitre F, van Rooijen N, et al. Subcapsular sinus macrophages promote NK cell accumulation and activation in response to lymph-borne viral particles. Blood. 2012;120:4744–50. doi: 10.1182/blood-2012-02-408179. [DOI] [PubMed] [Google Scholar]
- 16.Pak-Wittel MA, Yang L, Sojka DK, Rivenbark JG, Yokoyama WM. Interferon-γ mediates chemokine-dependent recruitment of natural killer cells during viral infection. Proc Natl Acad Sci USA. 2013;110:E50–9. doi: 10.1073/pnas.1220456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Backer R, Schwandt T, Greuter M, et al. Effective collaboration between marginal metallophilic macrophages and CD8+ dendritic cells in the generation of cytotoxic T cells. Proc Natl Acad Sci USA. 2010;107:216–21. doi: 10.1073/pnas.0909541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honke N, Shaabani N, Cadeddu G, et al. Enforced viral replication activates adaptive immunity and is essential for the control of a cytopathic virus. Nat Immunol. 2012;13:51–7. doi: 10.1038/ni.2169. [DOI] [PubMed] [Google Scholar]
- 19.Iannacone M, Moseman EA, Tonti E, et al. Subcapsular sinus macrophages prevent CNS invasion on peripheral infection with a neurotropic virus. Nature. 2010;465:1079–83. doi: 10.1038/nature09118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kastenmuller W, Torabi-Parizi P, Subramanian N, Lammermann T, Germain RN. A spatially-organized multicellular innate immune response in lymph nodes limits systemic pathogen spread. Cell. 2012;150:1235–48. doi: 10.1016/j.cell.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGaha TL, Chen Y, Ravishankar B, van RN, Karlsson MC. Marginal zone macrophages suppress innate and adaptive immunity to apoptotic cells in the spleen. Blood. 2011;117:5403–12. doi: 10.1182/blood-2010-11-320028. [DOI] [PubMed] [Google Scholar]
- 22.Miyake Y, Asano K, Kaise H, Uemura M, Nakayama M, Tanaka M. Critical role of macrophages in the marginal zone in the suppression of immune responses to apoptotic cell-associated antigens. J Clin Invest. 2007;117:2268–78. doi: 10.1172/JCI31990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smythies LE, Sellers M, Clements RH, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005;115:66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol. 2007;8:1086–94. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 25.Rivollier A, He J, Kole A, Valatas V, Kelsall BL. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J Exp Med. 2012;209:139–55. doi: 10.1084/jem.20101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hadis U, Wahl B, Schulz O, et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 2011;34:237–46. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Murai M, Turovskaya O, Kim G, et al. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat Immunol. 2009;10:1178–84. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sawa S, Lochner M, Satoh-Takayama N, et al. RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol. 2011;12:320–6. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 29.Tamoutounour S, Henri S, Lelouard H, et al. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur J Immunol. 2012;42:3150–66. doi: 10.1002/eji.201242847. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto M, Mariathasan S, Nahm MH, Baranyay F, Peschon JJ, Chaplin DD. Role of lymphotoxin and the type I TNF receptor in the formation of germinal centers. Science. 1996;271:1289–91. doi: 10.1126/science.271.5253.1289. [DOI] [PubMed] [Google Scholar]
- 31.Moseman EA, Iannacone M, Bosurgi L, et al. B cell maintenance of subcapsular sinus macrophages protects against a fatal viral infection independent of adaptive immunity. Immunity. 2012;36:415–26. doi: 10.1016/j.immuni.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu P, Wang Y, Chin RK, et al. B cells control the migration of a subset of dendritic cells into B cell follicles via CXC chemokine ligand 13 in a lymphotoxin-dependent fashion. J Immunol. 2002;168:5117–23. doi: 10.4049/jimmunol.168.10.5117. [DOI] [PubMed] [Google Scholar]
- 33.Diehl GE, Longman RS, Zhang JX, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX3CR1hi cells. Nature. 2013;494:116–20. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–69. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 35.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 36.Bhat PV. Retinal dehydrogenase gene expression in stomach and small intestine of rats during postnatal development and in vitamin A deficiency. FEBS Lett. 1998;426:260–2. doi: 10.1016/s0014-5793(98)00355-x. [DOI] [PubMed] [Google Scholar]
- 37.Frota-Ruchon A, Marcinkiewicz M, Bhat PV. Localization of retinal dehydrogenase type 1 in the stomach and intestine. Cell Tissue Res. 2000;302:397–400. doi: 10.1007/s004410000281. [DOI] [PubMed] [Google Scholar]
- 38.Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–38. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Thomas S, Prabhu R, Balasubramanian KA. Retinoid metabolism in the rat small intestine. Br J Nutr. 2005;93:59–63. doi: 10.1079/bjn20041306. [DOI] [PubMed] [Google Scholar]
- 40.Mikkelsen HB, Larsen JO, Froh P, Nguyen TH. Quantitative assessment of macrophages in the muscularis externa of mouse intestines. Anat Rec (Hoboken) 2011;294:1557–65. doi: 10.1002/ar.21444. [DOI] [PubMed] [Google Scholar]
- 41.Hartnell A, Steel J, Turley H, Jones M, Jackson DG, Crocker PR. Characterization of human sialoadhesin, a sialic acid binding receptor expressed by resident and inflammatory macrophage populations. Blood. 2001;97:288–96. doi: 10.1182/blood.v97.1.288. [DOI] [PubMed] [Google Scholar]
- 42.Wu C, Rauch U, Korpos E, et al. Sialoadhesin-positive macrophages bind regulatory T cells, negatively controlling their expansion and autoimmune disease progression. J Immunol. 2009;182:6508–16. doi: 10.4049/jimmunol.0804247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Futterer A, Mink K, Luz A, Kosco-Vilbois MH, Pfeffer K. The lymphotoxin β receptor controls organogenesis and affinity maturation in peripheral lymphoid tissues. Immunity. 1998;9:59–70. doi: 10.1016/s1074-7613(00)80588-9. [DOI] [PubMed] [Google Scholar]
- 44.Kang HS, Chin RK, Wang Y, et al. Signaling via LTβR on the lamina propria stromal cells of the gut is required for IgA production. Nat Immunol. 2002;3:576–82. doi: 10.1038/ni795. [DOI] [PubMed] [Google Scholar]
- 45.Steinberg MW, Shui JW, Ware CF, Kronenberg M. Regulating the mucosal immune system: the contrasting roles of LIGHT, HVEM, and their various partners. Semin Immunopathol. 2009;31:207–21. doi: 10.1007/s00281-009-0157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ware CF. Network communications: lymphotoxins, LIGHT, and TNF. Annu Rev Immunol. 2005;23:787–819. doi: 10.1146/annurev.immunol.23.021704.115719. [DOI] [PubMed] [Google Scholar]
- 47.Benson MJ, Pino-Lagos K, Rosemblatt M, Noelle RJ. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J Exp Med. 2007;204:1765–74. doi: 10.1084/jem.20070719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coombes JL, Siddiqui KR, Rancibia-Carcamo CV, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–64. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elias KM, Laurence A, Davidson TS, et al. Retinoic acid inhibits Th17 polarization and enhances FoxP3 expression through a Stat-3/Stat-5 independent signaling pathway. Blood. 2008;111:1013–20. doi: 10.1182/blood-2007-06-096438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hill JA, Hall JA, Sun CM, et al. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi cells. Immunity. 2008;29:758–70. doi: 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang SG, Lim HW, Andrisani OM, Broxmeyer HE, Kim CH. Vitamin A metabolites induce gut-homing FoxP3+ regulatory T cells. J Immunol. 2007;179:3724–33. doi: 10.4049/jimmunol.179.6.3724. [DOI] [PubMed] [Google Scholar]
- 52.Mucida D, Park Y, Kim G, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–60. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 53.Tokuyama H, Tokuyama Y. Retinoic acid induces the expression of germ-line C α transcript mainly by a TGF-β-independent mechanism. Cell Immunol. 1997;176:14–21. doi: 10.1006/cimm.1996.1069. [DOI] [PubMed] [Google Scholar]
- 54.van de Pavert SA, Olivier BJ, Goverse G, et al. Chemokine CXCL13 is essential for lymph node initiation and is induced by retinoic acid and neuronal stimulation. Nat Immunol. 2009;10:1193–9. doi: 10.1038/ni.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ngo VN, Korner H, Gunn MD, et al. Lymphotoxin α/β and tumor necrosis factor are required for stromal cell expression of homing chemokines in B and T cell areas of the spleen. J Exp Med. 1999;189:403–12. doi: 10.1084/jem.189.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]


