Abstract
Dendritic cells (DCs) operate as the link between innate and adaptive immunity. Their expression of pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs) and C-type lectin receptors (CLRs), enables antigen recognition and mediates appropriate immune responses. Distinct subsets of human DCs have been identified; however their expression of PRRs is not fully clarified. Expressions of CLRs by DC subpopulations, in particular, remain elusive. This study aimed to identify and compare PRR expressions on human blood DC subsets, including CD1c+, CD141+ and CD16+ myeloid DCs and CD123+ plasmacytoid DCs, in order to understand their capacity to recognize different antigens as well as their responsiveness to PRR-directed targeting. Whole blood was obtained from 13 allergic and six non-allergic individuals. Mononuclear cells were purified and multi-colour flow cytometry was used to assess the expression of 10 CLRs and two TLRs on distinct DC subsets. PRR expression levels were shown to differ between DC subsets for each PRR assessed. Furthermore, principal component analysis and random forest test demonstrated that the PRR profiles were discriminative between DC subsets. Interestingly, CLEC9A was expressed at lower levels by CD141+ DCs from allergic compared with non-allergic donors. The subset-specific PRR expression profiles suggests individual responsiveness to PRR-targeting and supports functional specialization.
Keywords: C-type lectin receptors, human dendritic cell subsets, pattern recognition receptors, Toll-like receptors
Introduction
Dendritic cells (DCs) are professional antigen-presenting cells that induce immunity upon detection of pathogens, while maintaining tolerance in response to innocuous molecules.1 Recognition of microbes is enabled by the expression of pattern recognition receptors (PRRs) that bind conserved pathogenic structures associated with various microorganisms including bacteria, viruses, parasites and fungi.2,3 Subsequently, DCs become equipped to induce antigen-specific T-cell responses.
The DCs are equipped with an array of PRRs, including several Toll-like receptors (TLRs) and C-type lectin receptors (CLRs). Triggering of TLRs induces pro-inflammatory signalling pathways and DC maturation.2 Similarly, some CLRs are capable of inducing signalling cascades that induce DC activation (reviewed in ref. 4). In addition, CLRs can modulate signalling induced by TLRs. Furthermore, in contrast to TLRs, CLRs can mediate antigen internalization.5,6 Each PRR can recognize several different pathogens and also, several PRRs can be engaged by a specific pathogen. Hence, the overall PRR expression enables a tailored immune activation in response to specific pathogens.
The CLRs are attractive targets for antigen delivery and immune response modulation. In vivo targeting of CLRs has been shown to induce antigen-specific T-cell responses in mice.7–9 Furthermore, in vitro studies on human DCs have demonstrated that antigens targeted to, for example, DC immunoreceptor (DCIR), CD205/DEC205, CD209/dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin 1 (DC-SIGN), C-type lectin domain family 9A (CLEC9A) and CD301/ C-type lectin superfamily member 14 (CLECSF14) result in antigen presentation to T cells.10–15 Clinical trials targeting CD205/DEC205 and CD206/macrophage mannose receptor 1 (MR) in vivo are on-going to treat patients with cancer and HIV infection,16–19 but results from these studies have not yet been reported. Nevertheless, immunization of non-human primates with HIV antigen targeted to CD205/DEC205 induced T-cell immunity,20 whereas CD301/CLECSF14-targeted antigen has been shown to induce suppressive T-cell responses.12 Taken together, CLRs are promising targets for induction/boosting of inadequate responses as well as for inhibition of detrimental immune responses.
Four DC subsets have been identified in human blood including CD123+ plasmacytoid DCs (pDCs) as well as CD1c+, CD141+ and CD16+ myeloid DCs (mDCs),21 and data supporting functional specialization have emerged. For example, pDCs are important producers of interferon-α in response to viruses22 and have additionally been implicated in the induction of tolerance.23 CD1c+ mDCs are potent T-cell stimulators in mixed lymphocyte reactions,21 whereas CD141+ DCs are superior at cross-presenting viral antigens from necrotic cells24 and CD16+ DCs are thought to have a pro-inflammatory role.25 Although the extent to which blood DC subsets reflect tissue DCs is not fully clarified, blood DCs are considered suitable for studies of human DC biology.26 Levels of PRR mRNA in blood DC subsets have been investigated,25,27 whereas data on PRR protein levels is very scarce. As protein levels can be explained by mRNA levels to only about 50% in multicellular organisms,28 assessments of PRR protein expression by human DC subsets are warranted.
The involvement of specific DC subsets in allergic responses is not well understood. Reports have suggested that pDCs, CD1c+ DCs as well as CD141+ DCs are associated with allergic diseases.29–32 Several PRRs have recently been implicated in the process. For example, CD284/TLR4, CD209/DC-SIGN, CD206/MR and dendritic cell-associated C-type lectin 2 (Dectin-2) have been shown to interact with allergens such as house dust mite, cat, dog and peanut, and have additionally been implicated in allergen-specific T helper type 2 polarization.33–40 Delineating surface expression of allergen-interacting PRRs by distinct DC subsets can provide insight into their capacity to interact with allergens.
In this study, we used multi-colour flow cytometry to analyse the expression of 10 CLRs and two TLRs on distinct blood DC subsets from allergic and non-allergic donors. The PRR expressions were analysed with regards to allergic status as well as to study distinctions among specific DC subsets to understand their capacity to recognize antigens and their responsiveness to PRR-targeting for modulation of immunological responses.
Materials and methods
Subjects and staining procedures
Nineteen donors participated in the study, of whom six were non-allergic and 13 were diagnosed with allergic rhinitis. Allergic donors tested positive for one or several respiratory allergens in skin prick tests (Table 1) and had a history of allergic rhinitis. Samples were taken outside the pollen season and all donors experienced no or low levels of symptoms (nasal congestion and runny nose symptoms scored on a 0–9 scale, see Table 1). The study was approved by the regional ethics committee and informed consent was obtained.
Table 1.
Donor characteristics
| Donor | Status | Sensitizations (skin prick test) | Symptom score1 (0–9) | Sex | Age |
|---|---|---|---|---|---|
| Don 1 | Allergic | Birch, Timothy grass | 1 | F | 38 |
| Don 2 | Allergic | Birch, Timothy grass, Cat | 0 | M | 30 |
| Don 3 | Allergic | Birch, Timothy grass | 0 | M | 30 |
| Don 4 | Allergic | Birch, Timothy grass, House dust mite, Cat, Dog | 0 | F | 36 |
| Don 5 | Allergic | House dust mite, Mould (Aspergillus), Cat, Dog | 4 | M | 23 |
| Don 6 | Allergic | Birch, Mugwort, House dust mite, Moulds (Cladosporium,Alternaria) | 1 | F | 41 |
| Don 7 | Allergic | Birch, Timothy grass, Mugwort | 2 | M | 27 |
| Don 8 | Allergic | Birch, Timothy grass, Cat, Dog, Horse | 0 | F | 24 |
| Don 9 | Allergic | Birch, Timothy grass, Mugwort, Dog, Cat | 0 | M | 25 |
| Don 10 | Allergic | Birch, Timothy grass, Mugwort, Dog | 0 | F | 30 |
| Don 11 | Allergic | Birch, Timothy grass | 0 | M | 23 |
| Don 12 | Allergic | House dust mite, Birch | 2 | F | 24 |
| Don 13 | Allergic | Birch, Timothy grass, House dust mite, Cat, Dog | 0 | F | 24 |
| Don 14 | Non-allergic | – | 0 | M | 30 |
| Don 15 | Non-allergic | – | 0 | F | 42 |
| Don 16 | Non-allergic | – | 0 | F | 41 |
| Don 17 | Non-allergic | – | 0 | M | 39 |
| Don 18 | Non-allergic | – | 0 | F | 51 |
| Don 19 | Non-allergic | – | 0 | M | 35 |
Nasal congestion and runny nose symptoms scored on a 0–9 scale.
Sample preparation and staining procedures
Peripheral blood mononuclear cells were isolated from whole blood by Lymphoprep (Medinor, Lidingö, Sweden) density gradient centrifugation and washed twice before antibody staining. Selection of which PRRs to include in the analysis was based on DC subset-specific gene expression profiles,27 availability of commercial PRR-specific antibodies and on PRR involvement in allergic responses33–40 (PRR antibodies are listed in Table 2). For the most part, antibodies against PRRs were conjugated with FITC, Alexa Fluor 488 or phycoerythrin (PE). Unconjugated antibodies against PRRs were detected using either rabbit anti-mouse PE (Dako Cytomation, Glostrup, Denmark) or goat anti-rabbit FITC (BD Biosciences, San Jose, CA). To identify DC subsets, samples were additionally stained with an antibody cocktail including CD1c-PE, CD141-biotin (Miltenyi Biotec, Bergisch Gladbach, Germany), HLA-DR-Peridinin chlorophyll protein (PerCp)-Cy5.5 (Biolegend, San Diego, CA), CD3-allophycocyanin (APC), CD14-APC and CD19-APC (Invitrogen, Carlsbad, CA) or a cocktail including CD123-biotin, CD16-PE-Cy7 (BD Biosciences), HLA-DR-PerCp-Cy5.5, CD3-PE (BD Biosciences), CD14-PE and CD19-PE (Dako). The PRRs targeted with PE-conjugated antibodies were instead stained with antibody cocktails including HLA-DR-PerCp-Cy5.5 and either CD123-biotin, CD16-PE-Cy7, CD3-FITC (BD Biosciences), CD14-FITC (Invitrogen) and CD19-FITC (Dako) or CD1c-FITC, CD141-biotin, CD3-APC, CD14-APC and CD19-APC. Biotin-labelling was subsequently detected with streptavidin-APC-Cy7 (BD Biosciences). Guidelines for accurately setting the boundary gates for positivity in multi-colour flow cytometry samples were followed.41 Importantly, for every donor and each PRR staining, fluoroscence minus one (FMO) controls were applied by staining samples identically except for the PRR antibodies which were excluded. FMO stainings are recommended as controls in multi-colour flow cytometry instead of isotype staining because the remaining fluorochromes generally give rise to higher background signal than unspecific binding and because perfect matching of the isotype antibody is virtually impossible.42 Additionally, unspecific binding was minimized by blocking all samples with mouse IgG antibodies, either before adding any antibodies, or after staining with secondary antibodies in cases where indirect stainings were performed. Washing was performed between each staining step and cells were subsequently resuspended carefully to ensure accurate antibody dilutions and either the same lot of antibodies was used for all stainings or new lots were compared to confirm identical performance. Samples were run on FACS Canto (BD Bioscience) and the flow cytometer performance was checked daily using Cytometer Setting and Tracking beads (BD Bioscience). Equal performance from time to time was ensured by using the Application Setting feature of the instrument.
Table 2.
Antibodies used to assess pattern recognition receptor expressions
| Antibody | Company |
|---|---|
| CD282/TLR2-PE | Biolegend, San Diego, CA |
| TLR4-PE | Biolegend |
| CLECSF14-Alexa 488 | Dendritics, Lyon, France |
| CD206-PE | Beckman Coulter, Brea, CA |
| CD207-PE | Beckman Coulter |
| CD205-FITC | eBioscience, San Diego, CA |
| Dectin-1-FITC | AbD Serotec, Kidlington, UK |
| CLECSF6 | RnD Systems, Minneapolis, MN |
| Dectin-2 | RnD Systems |
| MRC2 | Abcam, Cambridge, UK |
| CLEC9A | RnD Systems |
| CD209 | Abcam |
Analyses
Samples were analysed using FCS express (De Novo Software, Los Angeles, CA) and DC subsets were gated as outlined in Fig. 1. For each donor, the signals from PRR staining and FMO controls were analysed for the specific DC subsets (representative analyses shown in the Supporting information, Fig. S1). Boundary for positivity was based on FMO control samples (stained identically but without the respective PRR antibody), and net percentage of positive cells was calculated. Based on the results, PRR profiles were outlined for each DC subset. To investigate whether or not the overall PRR profiles were distinct in allergic and non-allergic subjects, the two groups were compared using a Support Vector Machine (SVM)43 and the leave-one-out cross-validation procedure.44 Briefly, the leave-one-out method uses all but one donor profiles to train the SVM and then tests the SVM using the left out profile. A decision value is assigned by the SVM and the procedure is repeated until each profile has been used as the test profile once. The decision values are used to create a Receiver operating characteristic curve, a graphical plot where the area under the curve (AUC) corresponds to the ability of the SVM to discriminate between the allergic and non-allergic donors, and AUC = 1 reflects perfect separation. In addition, individual comparisons of non-allergic versus allergic subjects were performed for each PRR and each DC subset using Mann–Whitney test in graphpad prism software (GraphPad Software, La Jolla, CA). In these comparisons, six non-allergic donors were used in all cases and 13 allergic donors for all PRRs except CD280/MRC2 (n = 12), CLEC9A, CD282/TLR2, CD284/TLR4 (n = 10) and Dectin-2 (n = 9). In cases where allergic and non-allergic donors were shown to express PRRs similarly (i.e. all PRRs except CLEC9A), further analyses were performed with the entire set of donors as one group, whereas only non-allergic donors were used otherwise. Next, differences in PRR expression between individual DC subsets were assessed in graphpad using analysis of variance followed by Bonferroni correction for multiple testing. Furthermore, whether or not the PRR profiles were distinctive for specific DC subsets and able to discriminate between the subsets was assessed using principal component analysis (PCA). Briefly, PCA transforms a large set of parameters (expressions of 12 PRRs in this case) into three summary variables (main components), which are illustrated as three axes (described in more detail in ref. 45). PCA is commonly used to analyse multi-dimensional data (reviewed in ref. 46) because it creates a three-dimensional image that can instantly be interpreted in terms of inter-relationships among samples. In addition, a random forest algorithm47 was used as an additional method to test whether DC subsets display distinct PRR profiles. The random forest algorithm predicts the subset origin of a PRR profile based on the remaining profiles. This is repeated once for each profile and results are presented in a table where perfect prediction would give the total number of donors (n = 19) along the diagonal.
Figure 1.
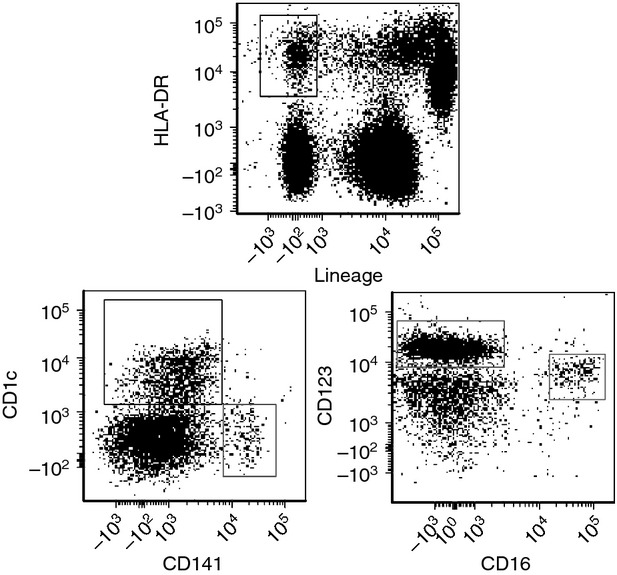
Gating strategy for identification of distinct dendritic cell (DC) subsets in human blood. Mononuclear cells were gated for viable cells in forward and side scatter, and DC subsets were thereafter identified within Lin-HLA-DR+ gated cells (upper dotplot) based on their unique expression of CD123, CD16, CD1c or CD141, respectively (lower dot plots).
Results
Allergic and non-allergic subjects exhibit non-discriminative PRR profiles
To analyse whether or not blood DC subsets (identified as shown in Fig. 1) from allergic and non-allergic donors display different PRR profiles, the SVM algorithm and the leave-one-out cross-validation procedure43,44 were applied for each DC subset. A value of AUC = 1 corresponds to perfect discrimination between the two groups, but as shown in Fig. 2, none of the DC subsets obtained AUC values close to one. Hence, the overall PRR expression profiles of DC subsets were similar for allergic and non-allergic individuals.
Figure 2.
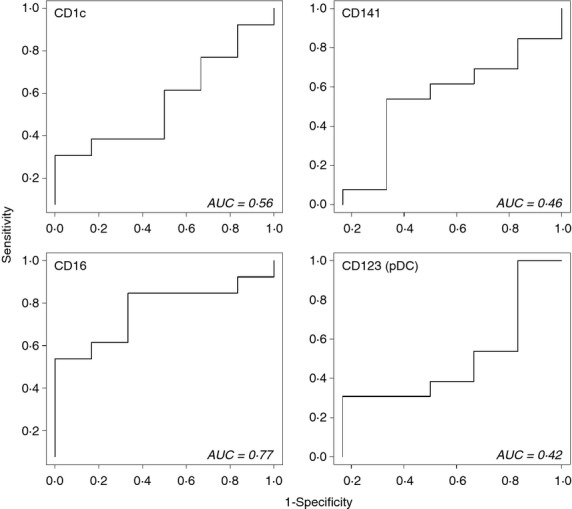
Pattern recognition receptor (PRR) expression profiles of dendritic cell (DC) subsets do not differ between allergic individuals and non-allergic controls. Expression of 12 PRRs on distinct DC subsets were analysed and compared between allergic (n = 13) and non-allergic (n = 6) individuals using a support vector machine (SVM) algorithm and leave-one-out cross-validation procedure. The decision values obtained from the comparisons are illustrated in receiver operating characteristic curves with the area under the curve (AUC) value corresponding to the ability of the SVM to discriminate between the allergic and non-allergic donors. An AUC value of one corresponds to perfect separation between groups, but values close to one were not obtained for any of the DC subsets and allergic and non-allergic individuals therefore express similar PRR profiles.
CLEC9A is expressed at lower levels on CD141+ DCs from allergic subjects
In addition to comparing overall PRR profiles of allergic and non-allergic subjects, expression of individual PRRs by distinct DC subsets was analysed and compared between the two groups using the Mann–Whitney U-test (Fig 3, see Supporting information, Fig. S2). Interestingly, the frequency of CD141+ DCs showing positivity for CLEC9A was lower on cells from allergic participants compared with non-allergic controls and the difference was shown to be statistically significant (Fig. 3). For all other PRRs, expressions on DC subsets were similar between allergic and non-allergic subjects. For PRRs where no differences were detected between allergic and non-allergic donors, the two groups were merged for further analysis and when CLEC9A data were included, only healthy donors were used.
Figure 3.
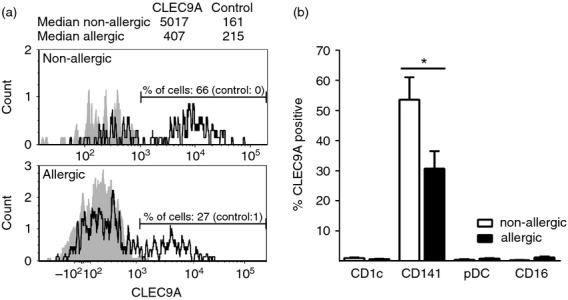
Frequency of CD141+ dendritic cells (DCs) expressing CLEC9A differed in allergic donors compared with non-allergic donors.(a) CLEC9A stainings (black line) as compared with fluorescence minus one (FMO) controls (grey areas) for CD141+ DCs from a representative non-allergic and allergic individual, respectively. The % of CLEC9A-positive cells (boundary for positivity set based on FMO controls) and median intensities of all CD141+ DCs are presented. (b) Average CLEC9A expression on distinct DC subsets calculated based on net positive cells (CLEC9A stainings minus FMO controls) of all donors (ten allergic and six non-allergic). Statistical analysis performed using Mann–Whitney U-test.* = P < 0·05.
Expressions of PRRs differ between DC subsets
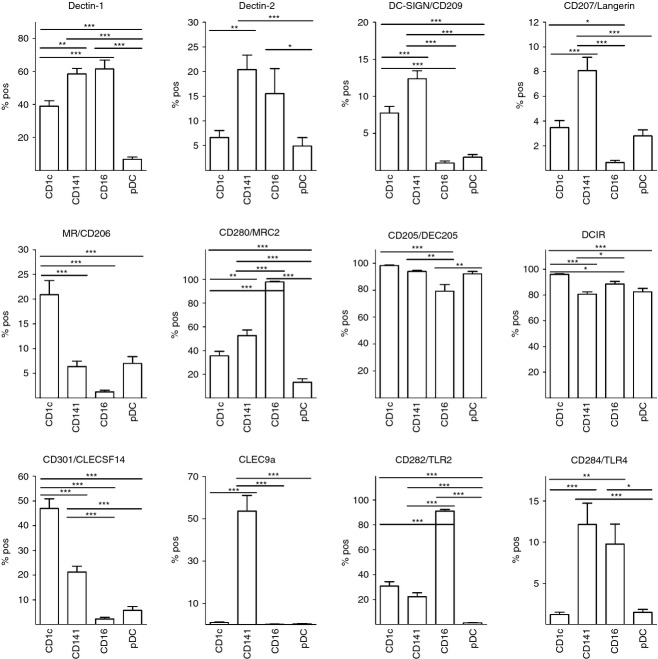
The present study demonstrates the expression of each of the CLRs and TLRs investigated by one or more DC subsets (Fig. S1, Figs 4 and 5) and in several cases, expression was shown for the first time. For example, we demonstrate for the first time that CD301/CLECSF14 is expressed preferentially by CD1c+ DCs and by a smaller proportion of CD141+ DCs. Also, the entire population of CD16+ DCs were positive for CD280/MRC2, whereas half of the CD141+ DCs and almost half of CD1c+ DCs showed CD280/MRC2 expression. Furthermore, Dectin-2 was shown to be expressed by a fifth of the CD141+ DCs and by smaller proportions of CD16+ and CD1c+ DCs. Importantly, statistical analysis demonstrated that one or more DC subsets expressed a significantly different frequency of positive cells compared with other subsets, for each of the PRRs investigated (Fig. 4). CD205/DEC205 and DCIR were expressed on a majority of each of the DC subpopulations, but statistically significant differences were still detected between subsets. An overall low percentage of DC subsets expressed CD209/DC-SIGN, CD207/Langerin and CD284/TLR4, with CD141+ DCs showing the greatest positive proportion with a maximum of 15%. Although transcriptional profiles of CD1c+ and CD141+ DCs showed extensive similarities in a previous study,27 the PRR protein profiles presented in this study demonstrate that these subsets express specific PRRs to different extents and in many cases, statistical significance was shown (Fig. 4). For example, in line with previously published data, blood DC expression of CLEC9A was shown to be restricted to the CD141+ subpopulation.24 In addition, CD1c+ showed higher frequency of cells positive for CD206/MR, DCIR and CD301/CLECSF14, compared with CD141+ DCs, whereas higher percentage of the CD141+ DC subpopulation was positive for Dectin-1, Dectin-2, CD209/DC-SIGN, CD207/Langerin, CD280/MRC2, CD284/TLR4 and CLEC9A. As evident from the expression profiles for each DC subset (Fig. 5), an average of at least 20% of the CD141+ DCs showed positivity for eight out of twelve PRRs investigated, whereas the corresponding figures for CD1c+ DCs, CD16+ DCs and pDCs were seven, five and two out of twelve PRRs, respectively.
Figure 4.
Dendritic cell (DC) subsets express pattern recognition receptors (PRRs) differently. Expression of 12 different PRRs on distinct DC subsets was analysed and % positive cells was compared between DC subsets using analysis of variance followed by Bonferonni correction for multiple testing. n = 19 for all PRRs except CD280/MRC2: n = 18; CD282/TLR2 and CD284/TLR4: n = 16; Dectin-2: n = 15; CLEC9A: n = 6 (healthy donors only). *P < 0·05; **P < 0·01; ***P < 0·005.
Figure 5.
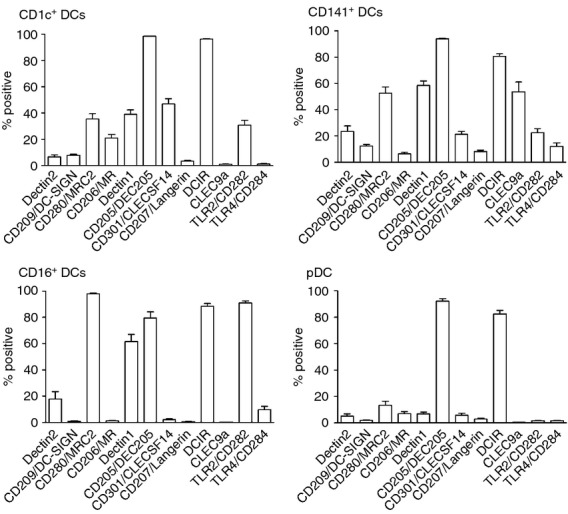
Pattern recognition receptor (PRR) expression profiles of distinct dendritic cell (DC) subsets. Expressions of 12 PRRs on distinct DC subsets (defined by their expression of CD1c, CD141, CD16 and CD123, respectively) were analysed and % positive cells is illustrated in separate graphs for each DC subset. n = 19 for all PRRs except CD280/MRC2: n = 18; CD282/TLR2 and CD284/TLR4: n = 16; Dectin-2: n = 15, CLEC9A: n = 6 (healthy donors only).
Distinct DC subsets display discriminative PRR profiles
The significantly different PRR expressions among distinct DC subsets (Fig. 4) and the different shapes of the PRR profiles (Fig. 5) led to the hypothesis that DC subsets exhibit discriminative PRR profiles. To test this statistically, PCA was performed. Indeed, the distinct DC subsets were completely separated in the PCA plot created (Fig. 6). As an additional statistical analysis, a random forest test47 was applied. Similarly, the random forest classifier was able to predict the subset origin of all but one of the profiles created (see Supporting information, Fig. S3). The misclassified profile was a CD141+ DC profile of one donor, which was classified as a CD1c+ DC. Nevertheless, CD141+ and CD1c+ DCs were completely separated in the PCA plot and were correctly identified by the random forest classifier in 18 out of 19 cases, and the subsets can therefore be considered to express PRRs differently.
Figure 6.
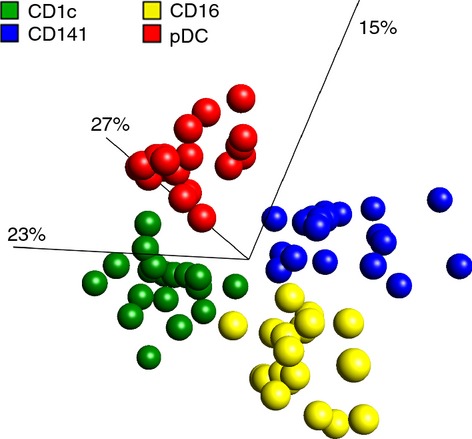
Dendritic cell (DC) subsets exhibit distinctive pattern recognition receptor (PRR) expression profiles. Principal component analysis (PCA) of PRR expression profiles from distinct DC subsets demonstrated clustering according to the DC subset from which the PRR profile was obtained. PCA condensed each DC subset donor profile, made from 12 PRR expressions, into three main components (which correspond to the axes). In this way, each dot corresponds to a distinct DC subset profile of a specific donor and in total 19 donors were used. Numbers in brackets correspond to percentage of total variation contained within each component.
Discussion
This study is the most thorough investigation undertaken so far to assess and compare PRR protein expressions by distinct human blood DC subsets. It originally demonstrates the protein expressions of several CLRs by distinct DC subsets, including Dectin-2, CD301/CLECSF14 and CD280/MRC2. Furthermore, it shows that PRR expression by DC subsets is highly selective and that CLEC9A expression by CD141+ DCs discriminates between allergic and non-allergic individuals. The results support differential functionality of human DC subsets and are relevant in the context of therapeutic PRR-targeting.
The present study demonstrates differential PRR surface expressions by the individual DC subsets. Although previous reports have investigated and compared PRR mRNA expression by DC subsets,25,27 the present study extends current knowledge by demonstrating novel PRR expressions, presenting PRR expressions at a protein level, as well as by comparing DC subsets statistically with regards to PRR expressions at a protein level. Previous data on CD284/TLR4 expression by CD141+ DCs is contradictory as a lack, as well as the presence, of CD284/TLR4 mRNA is suggested.24,25 In this study, CD284/TLR4 surface expression was detected on a proportion of CD141+ DCs. In agreement, pulmonary CD141+ DCs have indeed been shown to express CD284/TLR4 protein and respond to CD284/TLR4 stimulation.48 MacDonald et al.21 investigated protein expression of a limited set of PRRs on individual DC subsets and similarly to their data, CD205/DEC205 was detected on all blood DC subsets. By contrast, in the present study minor proportions of DC subsets were shown to be positive for CD206/MR, CD209/DC-SIGN and CD207/Langerin. The current study examined the proportions of positive cells, allowing minor expressions to become discernible, instead of presenting results for the entire population,21 which can explain the discrepancies. In addition to presenting PRR protein expressions by DC subsets, the presented data provide statistical evidence that PRR expression profiles are subset-specific and the differential PRR protein expressions support functional specialization among DC subsets.
All DC subsets were shown to express an array of different PRRs. A pathogen generally interacts with several different PRRs (reviewed in refs 2,3), suggesting that a specific cell responds to a pathogen via several PRRs and that the ultimate outcome will be balanced by the overall PRR profile. Moreover, all PRRs except CLEC9A were detected on more than one DC subset, indicating that a mixture of DC subsets responds to a specific PRR ligand. Furthermore, the PRR profiles indicate that response to, for example, bacterial, viral or fungal pathogens is not DC subset specific. For example, all DC subsets were shown to express PRRs recognizing fungal antigens, such as CD282/TLR2 and/or Dectin-1,3,49 although the Dectin-1-positive portion of pDCs was low. Also, interactions with viral and bacterial ligands can be expected by all mDC subsets as each subset expressed to some extent one or more receptors of CD209/DC-SIGN, CD206/MR, CD207/Langerin, CD282/TLR2 and CD284/TLR4.3,49 Although the ability to bind fungal, viral and bacterial pathogens appears to apply to several DC subsets, PRRs have their specificity and bind specific species.3,49 As PRR expression profiles were shown to differ between the distinct mDC subsets, their relative responsiveness to specific pathogen species may differ. Furthermore, the proportion of cells expressing a specific PRR profile is subset-specific, which is evident from the PCA as well as the random forest test, suggesting that a particular composition of DC subsets is enabled to respond to a specific pathogen. Considering that DC subsets have different functionalities,21–25 the unique profiles of PRR expression levels may have evolved to fine-tune immune responses and enable eradication of pathogens. In summary, the results of the present study suggest that the overall immune response is balanced both on a specific cell level, based on the PRR profile of individual cells, as well as on a DC subset level, depending on the composition of DC subsets expressing the PRRs interacting with a specific antigen.
The CLRs are attractive targets for antigen delivery in order to modulate immune responses, and in vivo targeting of CLRs in humans is currently being investigated.16 Two strategies currently under investigation are antigen delivery via CD205/DEC205 for induction of anti-HIV responses and antigen targeting to CD206/MR for treatment of cancer.16–19 Based on the PRR profiles demonstrated in this study, in vivo targeting of CD205/DEC205 in humans can be expected to affect all four DC subsets. In vitro, antigen delivered to CD205/DEC205 on monocyte-derived DCs, a model for mDCs,50 has been shown to mediate MHC class II presentation,10 whereas it has been shown to inhibit TLR-induced interferon-α production with concurrent T-cell activation by human pDCs.51 However, which subsets of T cells were induced was not presented in these studies and it is therefore unclear if CD205/DEC205-targeting on DCs induces suppressive or stimulatory T-cell responses. In contrast to CD205/DEC205, the PRR profiles indicate that CD206/MR-directed delivery in vivo would preferentially target CD1c+ DCs, and if these behave in line with monocyte-derived DCs,52 an immunosuppressive phenotype can be expected upon CD206/MR cross-linking. Hence, using CD206/MR for antigen delivery may benefit from additional stimulation to provoke immune activation. Targeting of CLR to inhibit detrimental immune responses has, to our knowledge not yet been assessed in vivo in humans. However, targeting CD301/CLECSF14 in non-human primates was recently shown to induce antigen-specific CD4+ T cells with a suppressive phenotype12 and according to the PRR profiles shown in this study, CD301/CLECSF14-directed delivery in humans can be expected to preferentially target CD1c+ DCs. Alternatively, pDC targeting may be the most effective strategy to inhibit detrimental immune responses as reports indicate an intrinsic capacity of this subset to induce regulatory T cells53 and because pDCs are implicated in tolerance induction.23 In this study, pDCs were shown to primarily express DCIR and CD205/DEC205, but none of these were exclusively expressed by pDCs and further studies are required to characterize the overall outcome upon targeting of these receptors. Taken together, CLR-directed antigen delivery is a promising approach to induce as well as to inhibit immune responses and the PRR profiles demonstrated in this study give clues to which human DC subsets are affected depending on the specific PRR that is targeted.
Expressions of PRRs by distinct DC subsets can give insights into their ability to interact with allergens as several CLRs have been implicated in this process.33,37,38,54 The significantly larger proportion of CD1c+ DCs shown to express CD206/MR indicates that, in comparison to other DC subsets, CD1c+ DCs are more prone to CD206/MR-mediated interaction with house dust mite, cat, dog, cockroach and peanut allergens.37 In contrast, CD141+ DCs and CD16+ DCs showed the largest proportions of cells positive for Dectin-2 and CD284/TLR4, suggesting a pronounced capability to interact with house dust mite via these receptors.33,38 Moreover, small proportions of CD141+ and CD1c+ DCs expressed CD209/DC-SIGN, indicating an ability to interact with peanut allergen.54 Based on the PRR expressions presented in the current study, it would be interesting to perform functional studies to understand the roles of the specific PRRs on DC subsets in allergic responses. For example, the influence of the receptors on allergen internalization and allergen-specific T-cell activation by the specific DC subset could be assessed. Regarding overall PRR expression on DC subsets from allergic and non-allergic donors, no overall differences were found, as evident from the SVM assessment (Fig. 2). However, CLEC9A showed reduced expression on CD141+ DCs from allergic donors compared with non-allergic donors. Hence, CD141+ DCs from allergic and non-allergic donors can be expected to interact with CLEC9A ligands to different extents and allergic donors may show a diminished response to CLEC9A targeted antigen delivery.9 Altogether, distinct DC subsets display allergy-associated PRRs to different extents, suggesting that they have different capacities to interact with allergens.
In conclusion, expression of twelve different PRRs, including ten CLRs and two TLRs, was demonstrated on distinct human blood DC subsets. For each PRR, statistically significant differences were demonstrated between DC subsets when comparing the proportion of positive cells. Additionally, selective expression of CLEC9A by CD141+ DCs was confirmed. Furthermore, CLEC9A showed lower expression on cells from allergic donors compared with non-allergic donors. Importantly, each DC subset was shown to exhibit a distinctive PRR expression profile, suggesting that a fixed composition of DC subsets is responding to a specific antigen. Altogether, the presented results suggest subset-specific responsiveness to pathogens, supporting functional specialization, as well as subset-specific responsiveness to PRR-directed targeting for treatment of immunological disorders.
Acknowledgments
The study was supported by grants from Signhild Engkvists foundation, Faculty of Engineering, Lund University (LTH) and Swedish Research Council.
Glossary
- APC
allophycocyanin
- AUC
area under the curve
- CLEC
C-type lectin domain family
- CLECSF
C-type lectin superfamily member
- CLR
C-type lectin receptors
- DC
dendritic cell
- DCIR
dendritic cell immunoreceptor
- DC-SIGN
dendritic cell-specific ICAM-3-grabbing non-integrin 1
- Dectin
dendritic cell-associated C-type lectin
- FMO
fluorescence minus one
- mDC
myeloid dendritic cell
- MR
macrophage mannose receptor 1
- PCA
principal Component Analysis
- pDC
plasmacytoid dendritic cell
- PE
phycoerythrin
- PerCP
peridinin chlorophyll protein
- PRR
pattern recognition receptors
- SVM
support vector machine
- TLR
Toll-like receptor
Disclosures
The authors declare no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Pattern recognition receptor expression by dendritic cell subsets.
Dendritic cell subsets from allergic and nonallergic donors express 11 pattern recognition receptors similarly.
Dendritic cell subsets exhibit distinctive pattern recognition receptor expression profiles.
References
- 1.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–50. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Osorio F, Reis e Sousa C. Myeloid C-type lectin receptors in pathogen recognition and host defense. Immunity. 2011;34:651–64. doi: 10.1016/j.immuni.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 4.Geijtenbeek TB, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol. 2009;9:465–79. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engering AJ, Cella M, Fluitsma DM, Hoefsmit EC, Lanzavecchia A, Pieters J. Mannose receptor mediated antigen uptake and presentation in human dendritic cells. Adv Exp Med Biol. 1997;417:183–7. doi: 10.1007/978-1-4757-9966-8_31. [DOI] [PubMed] [Google Scholar]
- 6.Mahnke K, Guo M, Lee S, Sepulveda H, Swain SL, Nussenzweig M, Steinman RM. The dendritic cell receptor for endocytosis, DEC-205, can recycle and enhance antigen presentation via major histocompatibility complex class II-positive lysosomal compartments. J Cell Biol. 2000;151:673–84. doi: 10.1083/jcb.151.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonifaz LC, Bonnyay DP, Charalambous A, et al. In vivo targeting of antigens to maturing dendritic cells via the DEC-205 receptor improves T cell vaccination. J Exp Med. 2004;199:815–24. doi: 10.1084/jem.20032220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He LZ, Crocker A, Lee J, et al. Antigenic targeting of the human mannose receptor induces tumor immunity. J Immunol. 2007;178:6259–67. doi: 10.4049/jimmunol.178.10.6259. [DOI] [PubMed] [Google Scholar]
- 9.Joffre OP, Sancho D, Zelenay S, Keller AM, Reis e Sousa C. Efficient and versatile manipulation of the peripheral CD4+ T-cell compartment by antigen targeting to DNGR-1/CLEC9A. Eur J Immunol. 2010;40:1255–65. doi: 10.1002/eji.201040419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birkholz K, Schwenkert M, Kellner C, et al. Targeting of DEC-205 on human dendritic cells results in efficient MHC class II-restricted antigen presentation. Blood. 2010;116:2277–85. doi: 10.1182/blood-2010-02-268425. [DOI] [PubMed] [Google Scholar]
- 11.Klechevsky E, Flamar AL, Cao Y, et al. Cross-priming CD8+ T cells by targeting antigens to human dendritic cells through DCIR. Blood. 2010;116:1685–97. doi: 10.1182/blood-2010-01-264960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li D, Romain G, Flamar AL, et al. Targeting self-and foreign antigens to dendritic cells via DC-ASGPR generates IL-10-producing suppressive CD4+ T cells. J Exp Med. 2012;209:109–21. doi: 10.1084/jem.20110399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer-Wentrup F, Benitez-Ribas D, Tacken PJ, Punt CJ, Figdor CG, de Vries IJ, Adema GJ. Targeting DCIR on human plasmacytoid dendritic cells results in antigen presentation and inhibits IFN-α production. Blood. 2008;111:4245–53. doi: 10.1182/blood-2007-03-081398. [DOI] [PubMed] [Google Scholar]
- 14.Schreibelt G, Klinkenberg LJ, Cruz LJ, et al. The C-type lectin receptor CLEC9A mediates antigen uptake and (cross-)presentation by human blood BDCA3+ myeloid dendritic cells. Blood. 2012;119:2284–92. doi: 10.1182/blood-2011-08-373944. [DOI] [PubMed] [Google Scholar]
- 15.Tacken PJ, de Vries IJ, Gijzen K, et al. Effective induction of naive and recall T-cell responses by targeting antigen to human dendritic cells via a humanized anti-CD209/DC-SIGN antibody. Blood. 2005;106:1278–85. doi: 10.1182/blood-2005-01-0318. [DOI] [PubMed] [Google Scholar]
- 16.Tacken PJ, Figdor CG. Targeted antigen delivery and activation of dendritic cells in vivo: steps towards cost effective vaccines. Semin Immunol. 2011;23:12–20. doi: 10.1016/j.smim.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 17. ClinicalTrials.gov_Identifier: NCT01127464. DCVax Plus Poly ICLC in Healthy Volunteers.
- 18. ClinicalTrials.gov_IdentifierNCT01522820. Vaccine Therapy With or Without Sirolimus in Treating Patients With NY-ESO-1 Expressing Solid Tumors.
- 19. ClinicalTrials.gov_Identifier: NCT00648102. Phase I Study of CDX-1307, hCG-B Vaccine, for Patients With Incurable, Locally Advanced or Metastatic Breast, Colorectal, Pancreatic, Bladder or Ovarian Cancer (CDX1307-02)
- 20.Flynn BJ, Kastenmuller K, Wille-Reece U, et al. Immunization with HIV Gag targeted to dendritic cells followed by recombinant New York vaccinia virus induces robust T-cell immunity in nonhuman primates. Proc Natl Acad Sci USA. 2011;108:7131–6. doi: 10.1073/pnas.1103869108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacDonald KP, Munster DJ, Clark GJ, Dzionek A, Schmitz J, Hart DN. Characterization of human blood dendritic cell subsets. Blood. 2002;100:4512–20. doi: 10.1182/blood-2001-11-0097. [DOI] [PubMed] [Google Scholar]
- 22.Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, Ho S, Antonenko S, Liu YJ. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–7. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 23.Hadeiba H, Lahl K, Edalati A, et al. Plasmacytoid dendritic cells transport peripheral antigens to the thymus to promote central tolerance. Immunity. 2012;36:438–50. doi: 10.1016/j.immuni.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jongbloed SL, Kassianos AJ, McDonald KJ, et al. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J Exp Med. 2010;207:1247–60. doi: 10.1084/jem.20092140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piccioli D, Tavarini S, Borgogni E, et al. Functional specialization of human circulating CD16 and CD1c myeloid dendritic-cell subsets. Blood. 2007;109:5371–9. doi: 10.1182/blood-2006-08-038422. [DOI] [PubMed] [Google Scholar]
- 26.Mittag D, Proietto AI, Loudovaris T, Mannering SI, Vremec D, Shortman K, Wu L, Harrison LC. Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J Immunol. 2011;186:6207–17. doi: 10.4049/jimmunol.1002632. [DOI] [PubMed] [Google Scholar]
- 27.Lindstedt M, Lundberg K, Borrebaeck CA. Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J Immunol. 2005;175:4839–46. doi: 10.4049/jimmunol.175.8.4839. [DOI] [PubMed] [Google Scholar]
- 28.de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol BioSyst. 2009;5:1512–26. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jahnsen FL, Lund-Johansen F, Dunne JF, Farkas L, Haye R, Brandtzaeg P. Experimentally induced recruitment of plasmacytoid (CD123high) dendritic cells in human nasal allergy. J Immunol. 2000;165:4062–8. doi: 10.4049/jimmunol.165.7.4062. [DOI] [PubMed] [Google Scholar]
- 30.Jahnsen FL, Moloney ED, Hogan T, Upham JW, Burke CM, Holt PG. Rapid dendritic cell recruitment to the bronchial mucosa of patients with atopic asthma in response to local allergen challenge. Thorax. 2001;56:823–6. doi: 10.1136/thorax.56.11.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kayserova J, Zentsova-Jaresova I, Budinsky V, et al. Selective increase in blood dendritic cell antigen-3-positive dendritic cells in bronchoalveolar lavage fluid in allergic patients. Scand J Immunol. 2012;75:305–13. doi: 10.1111/j.1365-3083.2011.02649.x. [DOI] [PubMed] [Google Scholar]
- 32.Lundberg K, Greiff L, Borrebaeck CA, Lindstedt M. FcεRI levels and frequencies of peripheral blood dendritic cell populations in allergic rhinitis. Hum Immunol. 2010;71:931–3. doi: 10.1016/j.humimm.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Barrett NA, Maekawa A, Rahman OM, Austen KF, Kanaoka Y. Dectin-2 recognition of house dust mite triggers cysteinyl leukotriene generation by dendritic cells. J Immunol. 2009;182:1119–28. doi: 10.4049/jimmunol.182.2.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deslee G, Charbonnier AS, Hammad H, et al. Involvement of the mannose receptor in the uptake of Der p 1, a major mite allergen, by human dendritic cells. J Allergy Clin Immunol. 2002;110:763–70. doi: 10.1067/mai.2002.129121. [DOI] [PubMed] [Google Scholar]
- 35.Hsu SC, Chen CH, Tsai SH, et al. Functional interaction of common allergens and a C-type lectin receptor, dendritic cell-specific ICAM3-grabbing non-integrin (DC-SIGN), on human dendritic cells. J Biol Chem. 2010;285:7903–10. doi: 10.1074/jbc.M109.058370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang HJ, Lin YL, Liu CF, Kao HF, Wang JY. Mite allergen decreases DC-SIGN expression and modulates human dendritic cell differentiation and function in allergic asthma. Mucosal Immunol. 2011;4:519–27. doi: 10.1038/mi.2011.17. [DOI] [PubMed] [Google Scholar]
- 37.Royer PJ, Emara M, Yang C, et al. The mannose receptor mediates the uptake of diverse native allergens by dendritic cells and determines allergen-induced T cell polarization through modulation of IDO activity. J Immunol. 2010;185:1522–31. doi: 10.4049/jimmunol.1000774. [DOI] [PubMed] [Google Scholar]
- 38.Hammad H, Chieppa M, Perros F, Willart MA, Germain RN, Lambrecht BN. House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nat Med. 2009;15:410–6. doi: 10.1038/nm.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett NA, Rahman OM, Fernandez JM, et al. Dectin-2 mediates Th2 immunity through the generation of cysteinyl leukotrienes. J Exp Med. 2011;208:593–604. doi: 10.1084/jem.20100793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li DQ, Zhang L, Pflugfelder SC, et al. Short ragweed pollen triggers allergic inflammation through Toll-like receptor 4-dependent thymic stromal lymphopoietin/OX40 ligand/OX40 signaling pathways. J Allergy Clin Immunol. 2011;128:1318–25. doi: 10.1016/j.jaci.2011.06.041. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herzenberg LA, Tung J, Moore WA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol. 2006;7:681–5. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- 42.Maecker HT, Trotter J. Flow cytometry controls, instrument setup, and the determination of positivity. Cytometry A. 2006;69:1037–42. doi: 10.1002/cyto.a.20333. [DOI] [PubMed] [Google Scholar]
- 43.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–97. [Google Scholar]
- 44.Arlot S, Celisse A. A survey of cross-validation procedures for model selection. Stat Surv. 2010;4:40–79. [Google Scholar]
- 45.Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA. 2000;97:10101–6. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Genser B, Cooper PJ, Yazdanbakhsh M, Barreto ML, Rodrigues LC. A guide to modern statistical analysis of immunological data. BMC Immunol. 2007;8:27. doi: 10.1186/1471-2172-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 48.Demedts IK, Bracke KR, Maes T, Joos GF, Brusselle GG. Different roles for human lung dendritic cell subsets in pulmonary immune defense mechanisms. Am J Respir Cell Mol Biol. 2006;35:387–93. doi: 10.1165/rcmb.2005-0382OC. [DOI] [PubMed] [Google Scholar]
- 49.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 50.Nozohoor S, Sjogren J, Ivert T, Hoglund P, Nilsson J. Validation of a modified EuroSCORE risk stratification model for cardiac surgery: the Swedish experience. Eur J Cardiothorac Surg. 2011;40:185–91. doi: 10.1016/j.ejcts.2010.10.040. [DOI] [PubMed] [Google Scholar]
- 51.Tel J, Benitez-Ribas D, Hoosemans S, et al. DEC-205 mediates antigen uptake and presentation by both resting and activated human plasmacytoid dendritic cells. Eur J Immunol. 2011;41:1014–23. doi: 10.1002/eji.201040790. [DOI] [PubMed] [Google Scholar]
- 52.Chieppa M, Bianchi G, Doni A, et al. Cross-linking of the mannose receptor on monocyte-derived dendritic cells activates an anti-inflammatory immunosuppressive program. J Immunol. 2003;171:4552–60. doi: 10.4049/jimmunol.171.9.4552. [DOI] [PubMed] [Google Scholar]
- 53.Moseman EA, Liang X, Dawson AJ, et al. Human plasmacytoid dendritic cells activated by CpG oligodeoxynucleotides induce the generation of CD4+ CD25+ regulatory T cells. J Immunol. 2004;173:4433–42. doi: 10.4049/jimmunol.173.7.4433. [DOI] [PubMed] [Google Scholar]
- 54.Shreffler WG, Castro RR, Kucuk ZY, et al. The major glycoprotein allergen from Arachis hypogaeaAra h 1, is a ligand of dendritic cell-specific ICAM-grabbing nonintegrin and acts as a Th2 adjuvant in vitro. J Immunol. 2006;177:3677–85. doi: 10.4049/jimmunol.177.6.3677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pattern recognition receptor expression by dendritic cell subsets.
Dendritic cell subsets from allergic and nonallergic donors express 11 pattern recognition receptors similarly.
Dendritic cell subsets exhibit distinctive pattern recognition receptor expression profiles.