Abstract
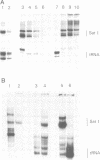
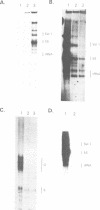
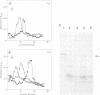
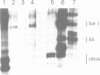
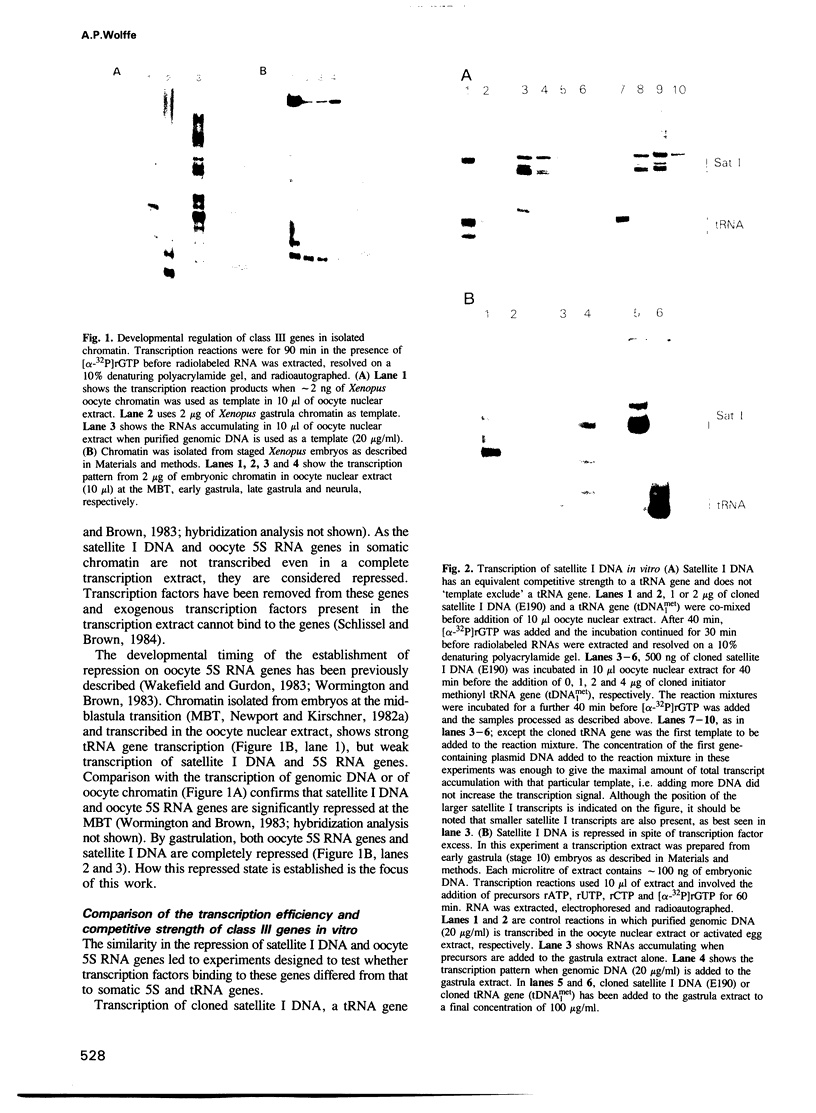
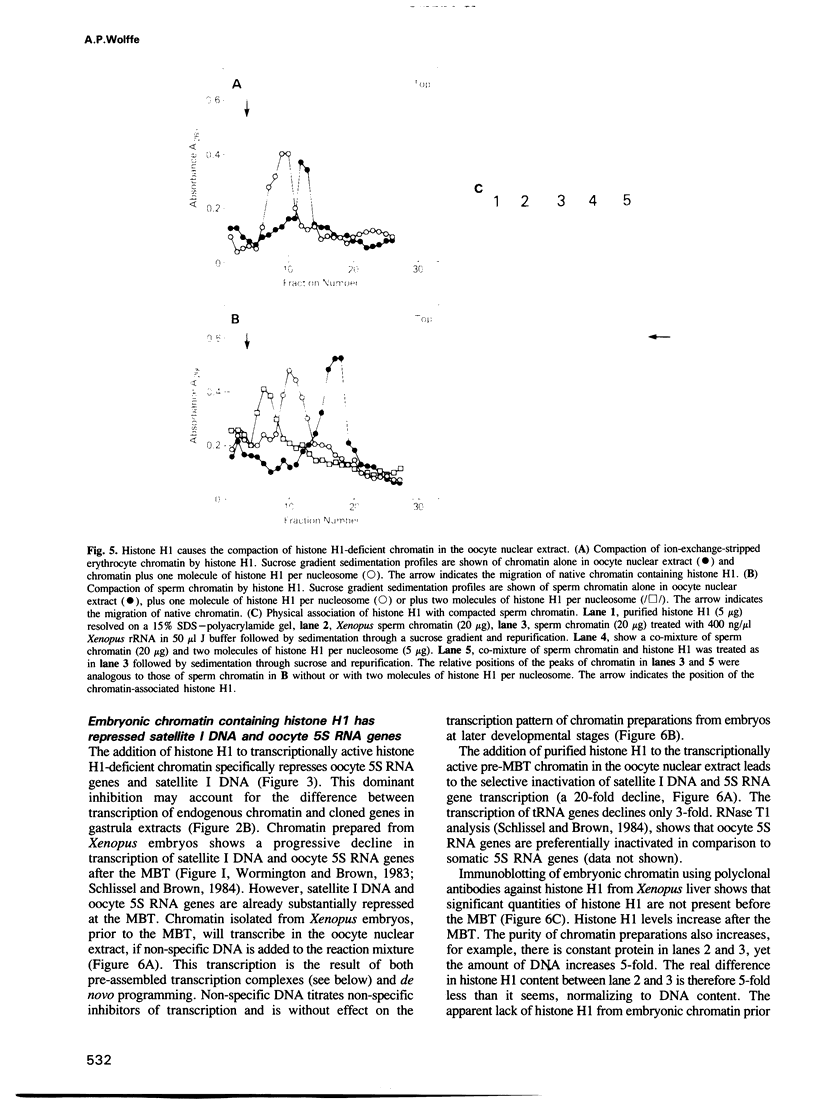
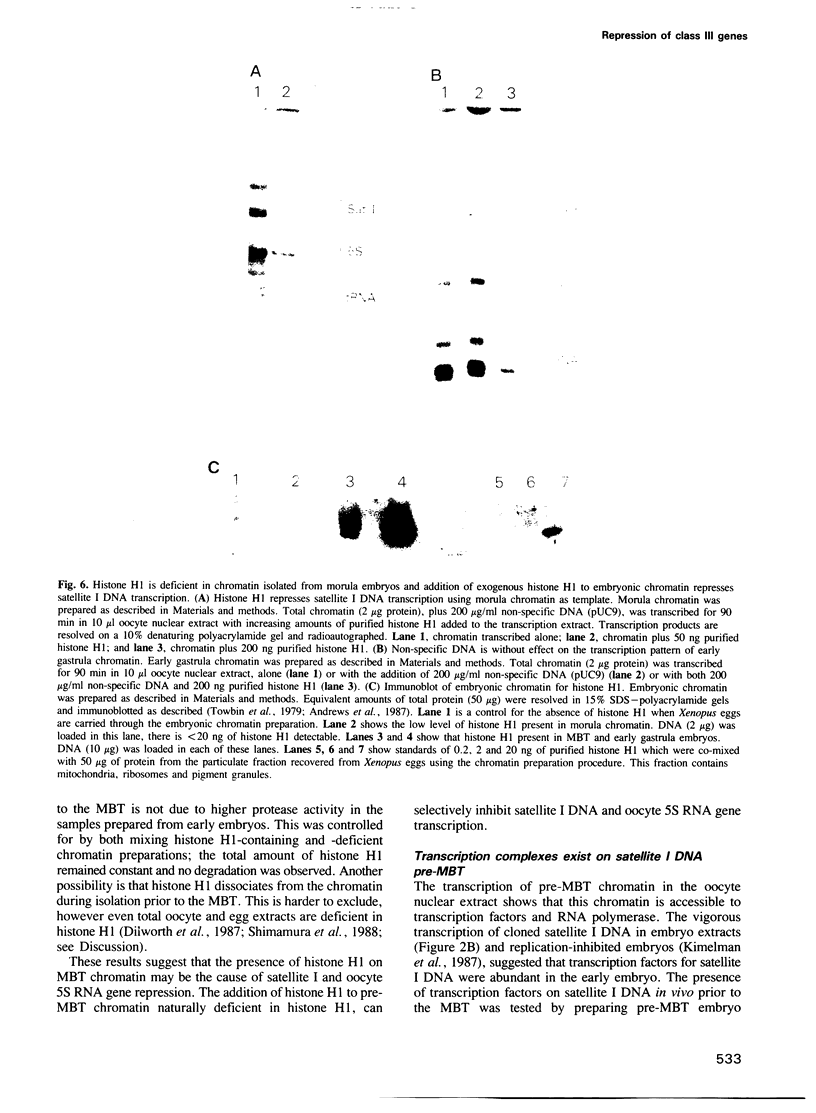
The genome of Xenopus laevis contains two large families of class III genes (oocyte 5S RNA and satellite I DNA) that are repressed in somatic cells. Both gene families are actively transcribed in a soluble extract of X.laevis oocyte nuclei, using chromatin deficient in histone H1 as a template. The addition of histone H1, to this transcriptionally active chromatin, results in the dominant and selective repression of oocyte 5S RNA genes and satellite I DNA. Somatic 5S RNA genes remain active following histone H1 addition. Changes in chromatin structure could have a dominant role in the regulation of class III gene expression during Xenopus embryogenesis.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackerman E. J. Molecular cloning and sequencing of OAX DNA: an abundant gene family transcribed and activated in Xenopus oocytes. EMBO J. 1983;2(8):1417–1422. doi: 10.1002/j.1460-2075.1983.tb01600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J., Cowling G. J., Harborne N., Cattini P., Craigie R., Gould H. Regulation of the higher-order structure of chromatin by histones H1 and H5. J Cell Biol. 1981 Aug;90(2):279–288. doi: 10.1083/jcb.90.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. The structure of histone H1 and its location in chromatin. Nature. 1980 Dec 25;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- Andrews D. L., Millstein L., Hamkalo B. A., Gottesfeld J. M. Competition between Xenopus satellite I sequences and Pol III genes for stable transcription complex formation. Nucleic Acids Res. 1984 Oct 25;12(20):7753–7769. doi: 10.1093/nar/12.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews M. T., Brown D. D. Transient activation of oocyte 5S RNA genes in Xenopus embryos by raising the level of the trans-acting factor TFIIIA. Cell. 1987 Nov 6;51(3):445–453. doi: 10.1016/0092-8674(87)90640-4. [DOI] [PubMed] [Google Scholar]
- Birkenmeier E. H., Brown D. D., Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978 Nov;15(3):1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- Blow J. J., Laskey R. A. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986 Nov 21;47(4):577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D. F., Wormington W. M., Brown D. D. Stable transcription complexes of Xenopus 5S RNA genes: a means to maintain the differentiated state. Cell. 1982 Feb;28(2):413–421. doi: 10.1016/0092-8674(82)90359-2. [DOI] [PubMed] [Google Scholar]
- Brown D. D. How a simple animal gene works. Harvey Lect. 1980;76:27–44. [PubMed] [Google Scholar]
- Brown D. D., Wensink P. C., Jordan E. Purification and some characteristics of 5S DNA from Xenopus laevis. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3175–3179. doi: 10.1073/pnas.68.12.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A., Kimura T., Gould H., Allan J. Perturbation of chromatin structure in the region of the adult beta-globin gene in chicken erythrocyte chromatin. J Mol Biol. 1987 Jan 5;193(1):57–70. doi: 10.1016/0022-2836(87)90626-7. [DOI] [PubMed] [Google Scholar]
- Ciliberto G., Castagnoli L., Cortese R. Transcription by RNA polymerase III. Curr Top Dev Biol. 1983;18:59–88. doi: 10.1016/s0070-2153(08)60579-7. [DOI] [PubMed] [Google Scholar]
- Clarkson S. G., Birnstiel M. L., Purdom I. F. Clustering of transfer RNA genes of Xenopus laevis. J Mol Biol. 1973 Sep 15;79(2):411–429. doi: 10.1016/0022-2836(73)90014-4. [DOI] [PubMed] [Google Scholar]
- Clarkson S. G., Kurer V., Smith H. O. Sequence organization of a cloned tDNA met fragment from Xenopus laevis. Cell. 1978 Jul;14(3):713–724. doi: 10.1016/0092-8674(78)90253-2. [DOI] [PubMed] [Google Scholar]
- Cozzarelli N. R., Gerrard S. P., Schlissel M., Brown D. D., Bogenhagen D. F. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983 Oct;34(3):829–835. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- Dilworth S. M., Black S. J., Laskey R. A. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 1987 Dec 24;51(6):1009–1018. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- Dumont J. N. Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J Morphol. 1972 Feb;136(2):153–179. doi: 10.1002/jmor.1051360203. [DOI] [PubMed] [Google Scholar]
- Fisher E. A., Felsenfeld G. Comparison of the folding of beta-globin and ovalbumin gene containing chromatin isolated from chicken oviduct and erythrocytes. Biochemistry. 1986 Dec 2;25(24):8010–8016. doi: 10.1021/bi00372a033. [DOI] [PubMed] [Google Scholar]
- Flynn J. M., Woodland H. R. The synthesis of histone H1 during early amphibian development. Dev Biol. 1980 Mar;75(1):222–230. doi: 10.1016/0012-1606(80)90157-8. [DOI] [PubMed] [Google Scholar]
- Forbes D. J., Kornberg T. B., Kirschner M. W. Small nuclear RNA transcription and ribonucleoprotein assembly in early Xenopus development. J Cell Biol. 1983 Jul;97(1):62–72. doi: 10.1083/jcb.97.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford P. J., Southern E. M. Different sequences for 5S RNA in kidney cells and ovaries of Xenopus laevis. Nat New Biol. 1973 Jan 3;241(105):7–12. doi: 10.1038/newbio241007a0. [DOI] [PubMed] [Google Scholar]
- Glikin G. C., Ruberti I., Worcel A. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell. 1984 May;37(1):33–41. doi: 10.1016/0092-8674(84)90298-8. [DOI] [PubMed] [Google Scholar]
- Gurdon J. B., Dingwall C., Laskey R. A., Korn L. J. Developmental inactivity of 5S RNA genes persists when chromosomes are cut between genes. Nature. 1982 Oct 14;299(5884):652–653. doi: 10.1038/299652a0. [DOI] [PubMed] [Google Scholar]
- Hannon R., Bateman E., Allan J., Harborne N., Gould H. Control of RNA polymerase binding to chromatin by variations in linker histone composition. J Mol Biol. 1984 Nov 25;180(1):131–149. doi: 10.1016/0022-2836(84)90434-0. [DOI] [PubMed] [Google Scholar]
- Hannon R., Richards E. G., Gould H. J. Facilitated diffusion of a DNA binding protein on chromatin. EMBO J. 1986 Dec 1;5(12):3313–3319. doi: 10.1002/j.1460-2075.1986.tb04645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland R. M., Laskey R. A. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell. 1980 Oct;21(3):761–771. doi: 10.1016/0092-8674(80)90439-0. [DOI] [PubMed] [Google Scholar]
- Hennen S. Influence of spermine and reduced temperature on the ability of transplanted nuclei to promote normal development in eggs of Rana pipiens. Proc Natl Acad Sci U S A. 1970 Jul;66(3):630–637. doi: 10.1073/pnas.66.3.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C., Tata J. R. Template-engaged and free RNA polymerases during Xenopus erythroid cell maturation. Dev Biol. 1978 Aug;65(2):496–507. doi: 10.1016/0012-1606(78)90044-1. [DOI] [PubMed] [Google Scholar]
- Jahn D., Wingender E., Seifart K. H. Transcription complexes for various class III genes differ in parameters of formation and stability towards salt. J Mol Biol. 1987 Jan 20;193(2):303–313. doi: 10.1016/0022-2836(87)90221-x. [DOI] [PubMed] [Google Scholar]
- Kimelman D., Kirschner M., Scherson T. The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell. 1987 Feb 13;48(3):399–407. doi: 10.1016/0092-8674(87)90191-7. [DOI] [PubMed] [Google Scholar]
- Labarca C., Paigen K. A simple, rapid, and sensitive DNA assay procedure. Anal Biochem. 1980 Mar 1;102(2):344–352. doi: 10.1016/0003-2697(80)90165-7. [DOI] [PubMed] [Google Scholar]
- Lam B. S., Carroll D. Tandemly repeated DNA sequences from Xenopus laevis. I. Studies on sequence organization and variation in satellite 1 DNA (741 base-pair repeat). J Mol Biol. 1983 Apr 25;165(4):567–585. doi: 10.1016/s0022-2836(83)80267-8. [DOI] [PubMed] [Google Scholar]
- Lassar A. B., Martin P. L., Roeder R. G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983 Nov 18;222(4625):740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- Lohka M. J., Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983 May 13;220(4598):719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- Losa R., Thoma F., Koller T. Involvement of the globular domain of histone H1 in the higher order structures of chromatin. J Mol Biol. 1984 Jun 5;175(4):529–551. doi: 10.1016/0022-2836(84)90183-9. [DOI] [PubMed] [Google Scholar]
- Marekov L. N., Beltchev B. Selective removal of histone H1 from chromatin at low salt concentration. Anal Biochem. 1981 Jul 15;115(1):93–96. doi: 10.1016/0003-2697(81)90529-7. [DOI] [PubMed] [Google Scholar]
- McConkey G. A., Bogenhagen D. F. TFIIIA binds with equal affinity to somatic and major oocyte 5S RNA genes. Genes Dev. 1988 Feb;2(2):205–214. doi: 10.1101/gad.2.2.205. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Nickol J. M., Felsenfeld G., Rau D. C. Histone hyperacetylation has little effect on the higher order folding of chromatin. Nucleic Acids Res. 1983 Jun 25;11(12):4065–4075. doi: 10.1093/nar/11.12.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millstein L., Eversole-Cire P., Blanco J., Gottesfeld J. M. Differential transcription of Xenopus oocyte and somatic-type 5 S genes in a Xenopus oocyte extract. J Biol Chem. 1987 Dec 15;262(35):17100–17110. [PubMed] [Google Scholar]
- Newport J., Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982 Oct;30(3):675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Newport J., Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982 Oct;30(3):687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Millstein L., Eversole-Cire P., Gottesfeld J. M., Varshavsky A. Transcriptionally inactive oocyte-type 5S RNA genes of Xenopus laevis are complexed with TFIIIA in vitro. Mol Cell Biol. 1987 Oct;7(10):3503–3510. doi: 10.1128/mcb.7.10.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R. C., Doering J. L., Brown D. D. Characterization of two xenopus somatic 5S DNAs and one minor oocyte-specific 5S DNA. Cell. 1980 May;20(1):131–141. doi: 10.1016/0092-8674(80)90241-x. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Searles M. A., Simpson R. T. Crystals of a nucleosome core particle containing defined sequence DNA. J Mol Biol. 1988 Jan 5;199(1):161–170. doi: 10.1016/0022-2836(88)90386-5. [DOI] [PubMed] [Google Scholar]
- Risley M. S., Eckhardt R. A., Mann M., Kasinsky H. E. Determinants of sperm nuclear shaping in the genus Xenopus. Chromosoma. 1982;84(4):557–569. doi: 10.1007/BF00292855. [DOI] [PubMed] [Google Scholar]
- Schlissel M. S., Brown D. D. The transcriptional regulation of Xenopus 5s RNA genes in chromatin: the roles of active stable transcription complexes and histone H1. Cell. 1984 Jul;37(3):903–913. doi: 10.1016/0092-8674(84)90425-2. [DOI] [PubMed] [Google Scholar]
- Segall J., Matsui T., Roeder R. G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980 Dec 25;255(24):11986–11991. [PubMed] [Google Scholar]
- Setzer D. R., Brown D. D. Formation and stability of the 5 S RNA transcription complex. J Biol Chem. 1985 Feb 25;260(4):2483–2492. [PubMed] [Google Scholar]
- Shastry B. S., Ng S. Y., Roeder R. G. Multiple factors involved in the transcription of class III genes in Xenopus laevis. J Biol Chem. 1982 Nov 10;257(21):12979–12986. [PubMed] [Google Scholar]
- Shimamura A., Tremethick D., Worcel A. Characterization of the repressed 5S DNA minichromosomes assembled in vitro with a high-speed supernatant of Xenopus laevis oocytes. Mol Cell Biol. 1988 Oct;8(10):4257–4269. doi: 10.1128/mcb.8.10.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R. H., Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979 Feb;6(2):689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Spadafora C., Oudet P., Chambon P. Rearrangement of chromatin structure induced by increasing ionic strength and temperature. Eur J Biochem. 1979 Oct;100(1):225–235. doi: 10.1111/j.1432-1033.1979.tb02053.x. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T. Influence of histone H1 on chromatin structure. Cell. 1977 Sep;12(1):101–107. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Ilyin Y. V., Georgiev G. P. Very long stretches of free DNA in chromatin. Nature. 1974 Aug 16;250(467):602–606. doi: 10.1038/250602a0. [DOI] [PubMed] [Google Scholar]
- Wakefield L., Ackerman E., Gurdon J. B. The activation of RNA synthesis by somatic nuclei injected into amphibian oocytes. Dev Biol. 1983 Feb;95(2):468–475. doi: 10.1016/0012-1606(83)90048-9. [DOI] [PubMed] [Google Scholar]
- Wakefield L., Gurdon J. B. Cytoplasmic regulation of 5S RNA genes in nuclear-transplant embryos. EMBO J. 1983;2(9):1613–1619. doi: 10.1002/j.1460-2075.1983.tb01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. A., Jared D. W., Dumont J. N., Sega M. W. Protein incorporation by isolated amphibian oocytes. 3. Optimum incubation conditions. J Exp Zool. 1973 Jun;184(3):321–333. doi: 10.1002/jez.1401840305. [DOI] [PubMed] [Google Scholar]
- Wegnez M., Monier R., Denis H. Sequence heterogeneity of 5 S RNA in Xenopus laevis. FEBS Lett. 1972 Sep 1;25(1):13–20. doi: 10.1016/0014-5793(72)80443-5. [DOI] [PubMed] [Google Scholar]
- Widmer R. M., Lezzi M., Koller T. Structural transition in inactive Balbiani ring chromatin of Chironomus during micrococcus nuclease digestion. EMBO J. 1987 Mar;6(3):743–748. doi: 10.1002/j.1460-2075.1987.tb04816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P., Brown D. D. DNA replication in vitro erases a Xenopus 5S RNA gene transcription complex. Cell. 1986 Oct 24;47(2):217–227. doi: 10.1016/0092-8674(86)90444-7. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Brown D. D. Developmental regulation of two 5S ribosomal RNA genes. Science. 1988 Sep 23;241(4873):1626–1632. doi: 10.1126/science.241.4873.1626. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Brown D. D. Differential 5S RNA gene expression in vitro. Cell. 1987 Dec 4;51(5):733–740. doi: 10.1016/0092-8674(87)90096-1. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P., Jordan E., Brown D. D. A bacteriophage RNA polymerase transcribes through a Xenopus 5S RNA gene transcription complex without disrupting it. Cell. 1986 Feb 14;44(3):381–389. doi: 10.1016/0092-8674(86)90459-9. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P. Transcription fraction TFIIIC can regulate differential Xenopus 5S RNA gene transcription in vitro. EMBO J. 1988 Apr;7(4):1071–1079. doi: 10.1002/j.1460-2075.1988.tb02915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormington W. M., Bogenhagen D. F., Jordan E., Brown D. D. A quantitative assay for Xenopus 5S RNA gene transcription in vitro. Cell. 1981 Jun;24(3):809–817. doi: 10.1016/0092-8674(81)90106-9. [DOI] [PubMed] [Google Scholar]
- Wormington W. M., Brown D. D. Onset of 5 S RNA gene regulation during Xenopus embryogenesis. Dev Biol. 1983 Sep;99(1):248–257. doi: 10.1016/0012-1606(83)90273-7. [DOI] [PubMed] [Google Scholar]
- van Dongen W. M., Moorman A. F., Destrée O. H. The accumulation of the maternal pool of histone H1A during oogenesis in Xenopus laevis. Cell Differ. 1983 May;12(5):257–264. doi: 10.1016/0045-6039(83)90021-0. [DOI] [PubMed] [Google Scholar]