Abstract
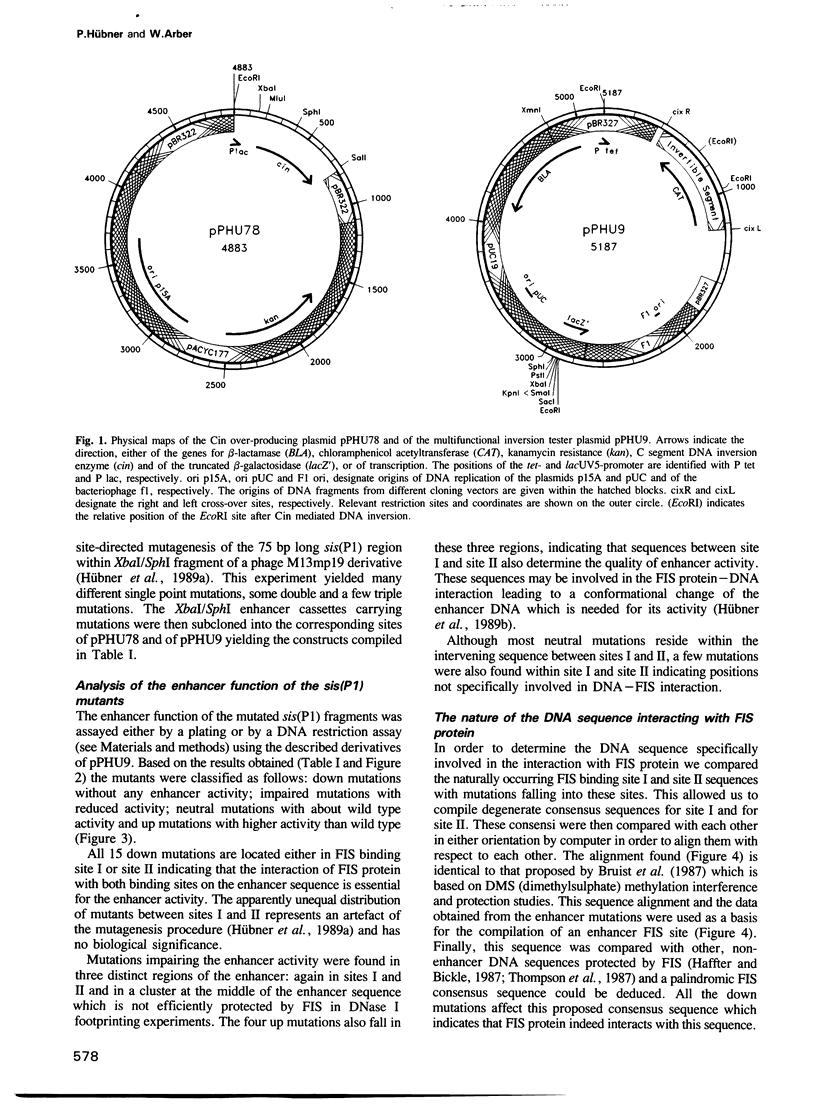
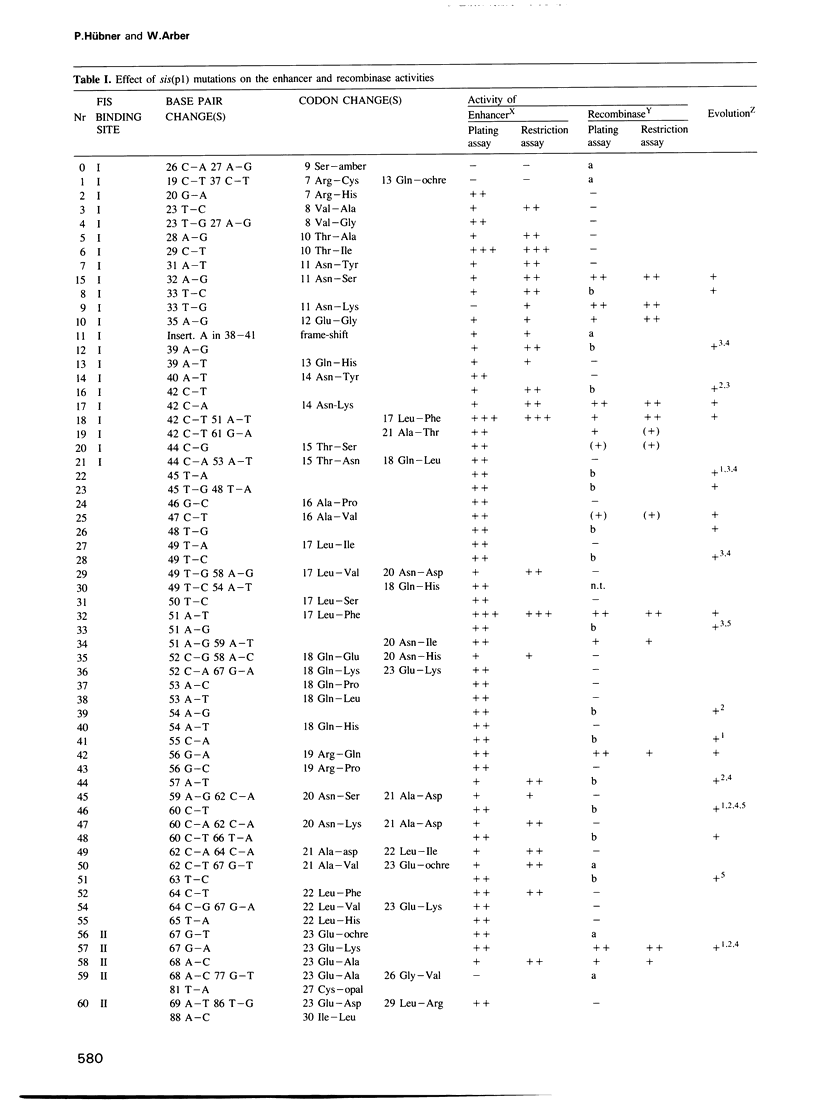
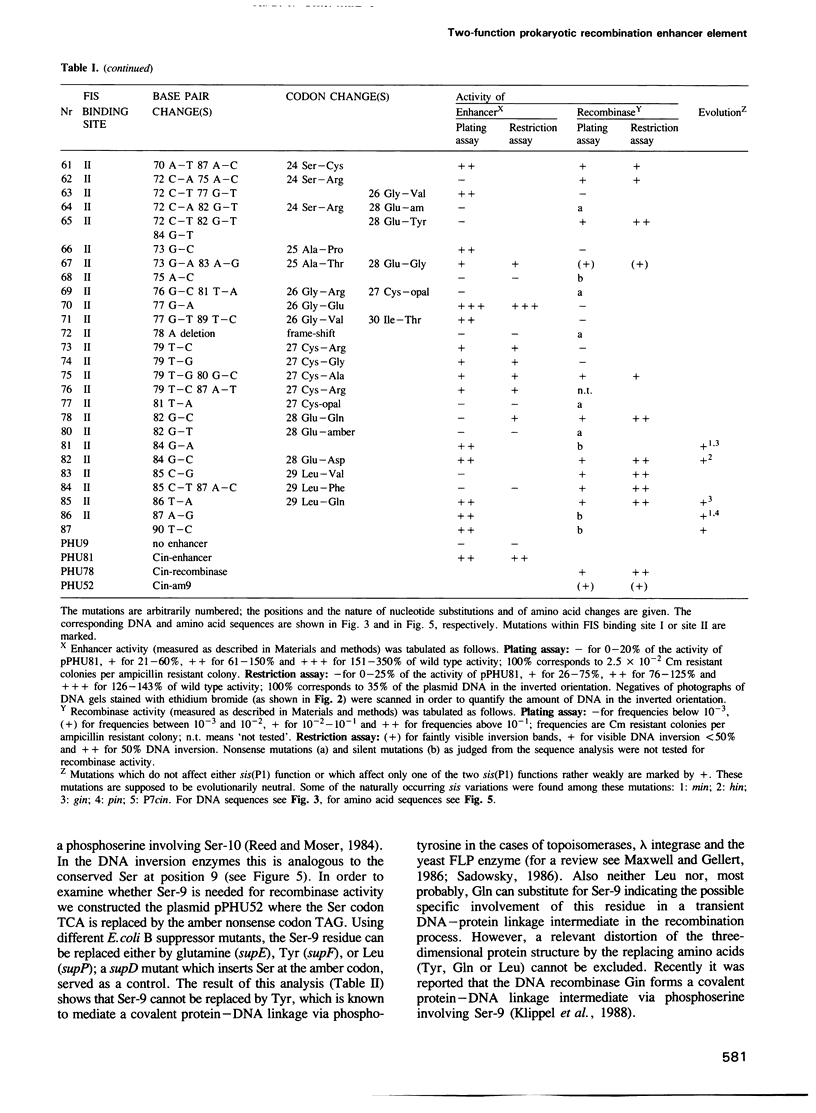
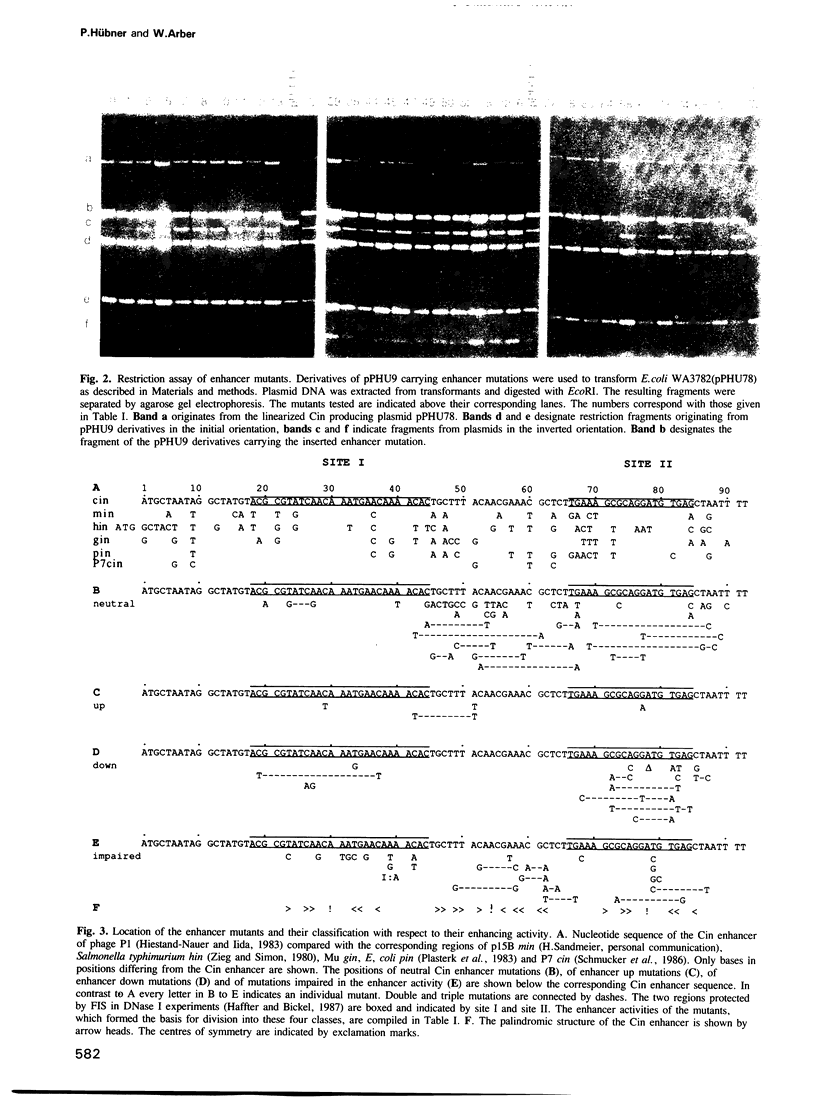
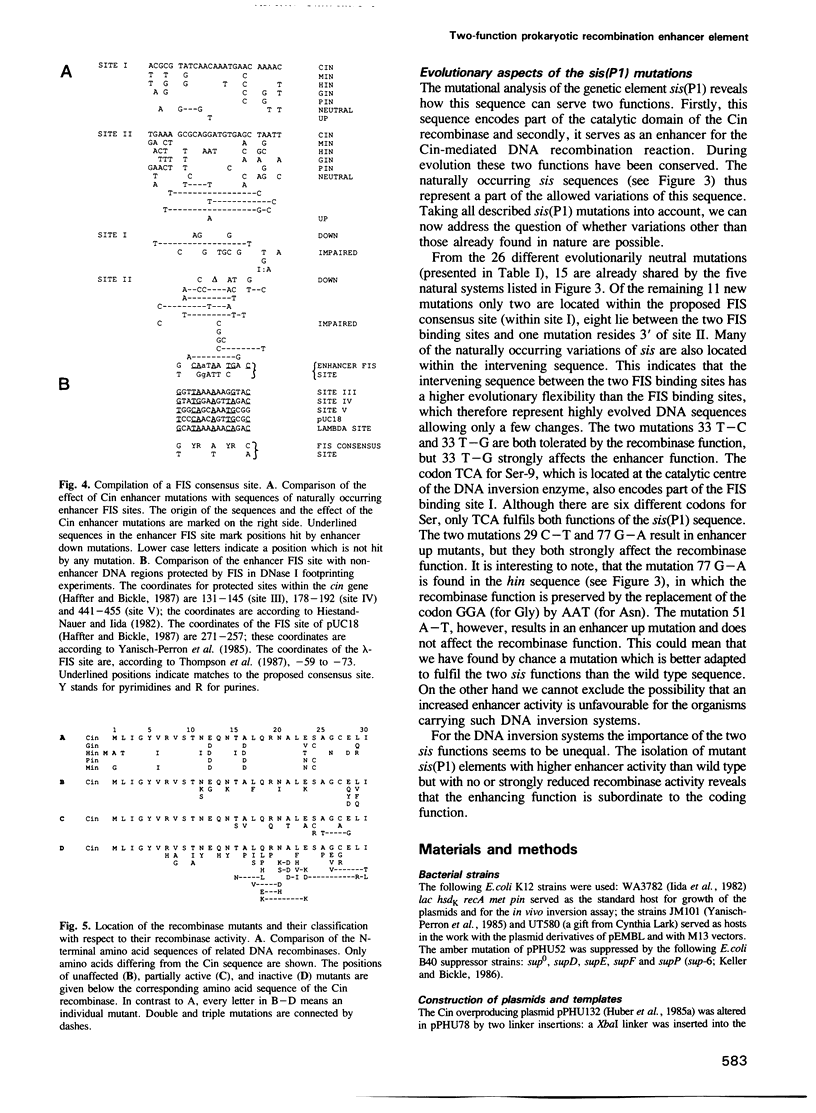
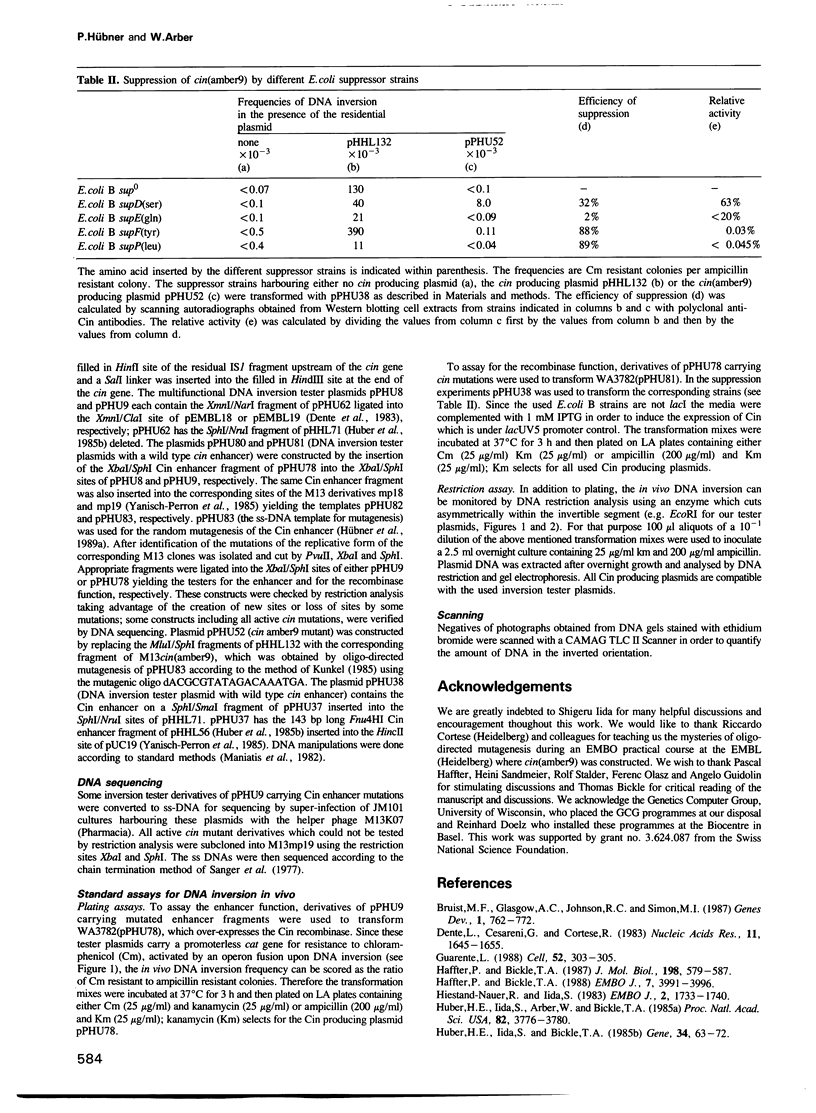
The site-specific DNA inversion system Cin encoded by the bacteriophage P1 consists of a recombinase, two inverted crossing-over sites and a recombinational enhancer. The latter approximately 75 bp long genetic element is bifunctional due to its location within the 5' part of the cin gene encoding the recombinase. In order to determine the essential nucleotides for each of its two biological functions we randomly mutated the recombinational enhancer sequence sis(P1) and analysed both functions of the mutants obtained. Three distinct regions of this sequence were found to be important for the enhancer activity. One of them occupies the middle third of the enhancer sequence and it can suffer a number of functionally neutral base substitutions, while others are detrimental. The other two regions occupy the two flanking thirds of the enhancer. They coincide with binding sites of the host-coded protein FIS (Factor for Inversion Stimulation) needed for efficient DNA inversion in vitro. These sequences appear to be highly evolved allowing only a few mutations without affecting either of the biological functions. Taking the effect of mutations within these FIS binding sites into account a consensus sequence for the interaction with FIS was compiled. This FIS consensus implies a palindromic structure for the recombinational enhancer. This is in line with the orientation independence of enhancer action with respect to the crossing-over sites.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bruist M. F., Glasgow A. C., Johnson R. C., Simon M. I. Fis binding to the recombinational enhancer of the Hin DNA inversion system. Genes Dev. 1987 Oct;1(8):762–772. doi: 10.1101/gad.1.8.762. [DOI] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. UASs and enhancers: common mechanism of transcriptional activation in yeast and mammals. Cell. 1988 Feb 12;52(3):303–305. doi: 10.1016/s0092-8674(88)80020-5. [DOI] [PubMed] [Google Scholar]
- Haffter P., Bickle T. A. Enhancer-independent mutants of the Cin recombinase have a relaxed topological specificity. EMBO J. 1988 Dec 1;7(12):3991–3996. doi: 10.1002/j.1460-2075.1988.tb03287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P., Bickle T. A. Purification and DNA-binding properties of FIS and Cin, two proteins required for the bacteriophage P1 site-specific recombination system, cin. J Mol Biol. 1987 Dec 20;198(4):579–587. doi: 10.1016/0022-2836(87)90201-4. [DOI] [PubMed] [Google Scholar]
- Hiestand-Nauer R., Iida S. Sequence of the site-specific recombinase gene cin and of its substrates serving in the inversion of the C segment of bacteriophage P1. EMBO J. 1983;2(10):1733–1740. doi: 10.1002/j.1460-2075.1983.tb01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H. E., Iida S., Arber W., Bickle T. A. Site-specific DNA inversion is enhanced by a DNA sequence element in cis. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3776–3780. doi: 10.1073/pnas.82.11.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H. E., Iida S., Bickle T. A. Expression of the bacteriophage P1 cin recombinase gene from its own and heterologous promoters. Gene. 1985;34(1):63–72. doi: 10.1016/0378-1119(85)90295-1. [DOI] [PubMed] [Google Scholar]
- Iida S. Bacteriophage P1 carries two related sets of genes determining its host range in the invertible C segment of its genome. Virology. 1984 Apr 30;134(2):421–434. doi: 10.1016/0042-6822(84)90309-x. [DOI] [PubMed] [Google Scholar]
- Iino T., Kutsukake K. Trans-acting genes of bacteriophages P1 and Mu mediate inversion of a specific DNA segment involved in flagellar phase variation of Salmonella. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):11–16. doi: 10.1101/sqb.1981.045.01.004. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Ball C. A., Pfeffer D., Simon M. I. Isolation of the gene encoding the Hin recombinational enhancer binding protein. Proc Natl Acad Sci U S A. 1988 May;85(10):3484–3488. doi: 10.1073/pnas.85.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Glasgow A. C., Simon M. I. Spatial relationship of the Fis binding sites for Hin recombinational enhancer activity. Nature. 1987 Oct 1;329(6138):462–465. doi: 10.1038/329462a0. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Simon M. I. Hin-mediated site-specific recombination requires two 26 bp recombination sites and a 60 bp recombinational enhancer. Cell. 1985 Jul;41(3):781–791. doi: 10.1016/s0092-8674(85)80059-3. [DOI] [PubMed] [Google Scholar]
- Kahmann R., Rudt F., Koch C., Mertens G. G inversion in bacteriophage Mu DNA is stimulated by a site within the invertase gene and a host factor. Cell. 1985 Jul;41(3):771–780. doi: 10.1016/s0092-8674(85)80058-1. [DOI] [PubMed] [Google Scholar]
- Kamp D., Chow L. T., Broker T. R., Kwoh D., Zipser D., Kahmann R. Site-specific recombination in phage mu. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1159–1167. doi: 10.1101/sqb.1979.043.01.131. [DOI] [PubMed] [Google Scholar]
- Kanaar R., van de Putte P., Cozzarelli N. R. Gin-mediated DNA inversion: product structure and the mechanism of strand exchange. Proc Natl Acad Sci U S A. 1988 Feb;85(3):752–756. doi: 10.1073/pnas.85.3.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller B., Bickle T. A. The nucleotide sequence of gene 21 of bacteriophage T4 coding for the prohead protease. Gene. 1986;49(2):245–251. doi: 10.1016/0378-1119(86)90285-4. [DOI] [PubMed] [Google Scholar]
- Klippel A., Mertens G., Patschinsky T., Kahmann R. The DNA invertase Gin of phage Mu: formation of a covalent complex with DNA via a phosphoserine at amino acid position 9. EMBO J. 1988 Apr;7(4):1229–1237. doi: 10.1002/j.1460-2075.1988.tb02935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C., Kahmann R. Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J Biol Chem. 1986 Nov 25;261(33):15673–15678. [PubMed] [Google Scholar]
- Koch C., Vandekerckhove J., Kahmann R. Escherichia coli host factor for site-specific DNA inversion: cloning and characterization of the fis gene. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4237–4241. doi: 10.1073/pnas.85.12.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell A., Gellert M. Mechanistic aspects of DNA topoisomerases. Adv Protein Chem. 1986;38:69–107. doi: 10.1016/s0065-3233(08)60526-4. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Brinkman A., van de Putte P. DNA inversions in the chromosome of Escherichia coli and in bacteriophage Mu: relationship to other site-specific recombination systems. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5355–5358. doi: 10.1073/pnas.80.17.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., Van de Putte P. Genetic switches by DNA inversions in prokaryotes. Biochim Biophys Acta. 1984 Jun 16;782(2):111–119. doi: 10.1016/0167-4781(84)90013-7. [DOI] [PubMed] [Google Scholar]
- Reed R. R., Moser C. D. Resolvase-mediated recombination intermediates contain a serine residue covalently linked to DNA. Cold Spring Harb Symp Quant Biol. 1984;49:245–249. doi: 10.1101/sqb.1984.049.01.028. [DOI] [PubMed] [Google Scholar]
- Sadowski P. Site-specific recombinases: changing partners and doing the twist. J Bacteriol. 1986 Feb;165(2):341–347. doi: 10.1128/jb.165.2.341-347.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker R., Ritthaler W., Stern B., Kamp D. DNA inversion in bacteriophage Mu: characterization of the inversion site. J Gen Virol. 1986 Jun;67(Pt 6):1123–1133. doi: 10.1099/0022-1317-67-6-1123. [DOI] [PubMed] [Google Scholar]
- Silverman M., Zieg J., Mandel G., Simon M. Analysis of the functional components of the phase variation system. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):17–26. doi: 10.1101/sqb.1981.045.01.005. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Moitoso de Vargas L., Koch C., Kahmann R., Landy A. Cellular factors couple recombination with growth phase: characterization of a new component in the lambda site-specific recombination pathway. Cell. 1987 Sep 11;50(6):901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zieg J., Simon M. Analysis of the nucleotide sequence of an invertible controlling element. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4196–4200. doi: 10.1073/pnas.77.7.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]




