Summary
When Escherichia coli grows in the presence of DNA-damaging agents such as methyl methanesulfonate (MMS), absence of the full-length form of Translation Initiation Factor 2 (IF2-1) or deficiency in helicase activity of replication restart protein PriA leads to a considerable loss of viability. MMS sensitivity of these mutants was contingent on the stringent response alarmone (p)ppGpp being at low levels. While zero levels (ppGpp0) greatly aggravated sensitivity, high levels promoted resistance. Moreover, M+ mutations, which suppress amino acid auxotrophy of ppGpp0 strains and which have been found to map to RNA polymerase subunits, largely restored resistance to IF2-1- and PriA helicase-deficient mutants. The truncated forms IF2-2/3 played a key part in inducing especially severe negative effects in ppGpp0 cells when restart function priB was knocked out, causing loss of viability and severe cell filamentation, indicative of SOS induction. Even a strain with the wild-type infB allele exhibited significant filamentation and MMS sensitivity in this background whereas mutations that prevent expression of IF2-2/3 essentially eliminated filamentation and largely restored MMS resistance. The results suggest different influences of IF2-1 and IF2-2/3 on the replication restart system depending on (p)ppGpp levels, each having the capacity to maximize survival under differing growth conditions.
Keywords: IF2, (p)ppGpp, replication restart, replication-transcription conflicts, phage Mu transposition, PriA helicase
Introduction
Successful chromosomal replication during cell growth requires mechanisms not only for initiating DNA replication coordinated with growth rate but also for ensuring effective remedies for accidents of replication forks. Regulation of the latter is most likely as important as the former since the types of impediments encountered by replication forks will vary under diverse conditions, whether the cells are in exponential growth phase or stationary phase, in the presence of DNA-damaging agents, or under a stress condition such as nutrient deprivation. Moreover, the optimal mechanisms for coping with accidents under each condition and the risks associated with each are likely to differ.
In Escherichia coli various recombination, repair, and replication restart functions effectively collaborate to cope with conflicts arising with DNA replication, eventually repairing the template and restarting replication (Atkinson & McGlynn, 2009, Heller & Marians, 2006b, Michel et al., 2004, Mirkin & Mirkin, 2007). Conflicts of DNA replication with transcription are especially problematic in exponentially growing cells as stalled transcription complexes accumulate at DNA lesions, but the stringent response induced by nutrient stress can help to diminish such conflicts (Mahdi et al., 2012, McGlynn & Lloyd, 2000, Tehranchi et al., 2010, Trautinger et al., 2005). A key event in the stringent response is the elevation of the signaling effector nucleotides pppGpp and ppGpp, the latter being the predominant and more stable form derived from the former. (See (Potrykus & Cashel, 2008) for a review; for simplicity, the alarmones will be predominantly referred to as ppGpp although the collective term (p)ppGpp may be applicable in some cases. Generally, ppGpp has been found to be more potent than pppGpp in growth inhibition, and it is more potent in processes such as inhibition of transcription of rRNA in the presence of stringent response regulator protein DksA (Mechold et al., 2013).) When ppGpp is absent, the ability of cells to overcome DNA damage can be compromised: for example, UV sensitivity of a mutant lacking the RuvABC Holliday junction resolvase activity is aggravated while its survival is greatly enhanced at elevated ppGpp levels (McGlynn & Lloyd, 2000). At 0 ppGpp levels, multiple RNA polymerase (RNAP) molecules can become stalled at DNA lesions and accumulate as transcription arrays, which impede replication forks, and DksA, RNAP elongation factors GreA/B, elevated ppGpp, and certain RNAP mutations can diminish the accumulation of RNAP arrays to reduce transcription-DNA replication conflicts (Tehranchi et al., 2010, Trautinger et al., 2005).
Recent evidence has indicated an important influence of translation initiation factor 2 (IF2) and its various molecular weight forms on replication restart and the cell’s ability to grow in the presence of DNA-damaging agents such as methyl methanesulfonate (MMS) (Madison et al., 2012). The infB gene encodes the full-length form of 97.3 kDa (IF2-1) and two truncated forms of 79.7 kDa (IF2-2) and 78.8 kDa (IF2-3), the latter two isoforms arising from two internal, in-frame initiation codons (Laursen et al., 2002, Nyengaard et al., 1991). The infB(del1) allele, which results in the loss of IF2-1, dramatically reduces the cell’s viability on MMS plates even though the infB(del1) allele does not affect cell viability if cells are first exposed to MMS and then allowed to recover and grow in its absence. In contrast, infB(del2/3), which prevents expression of the two truncated forms IF2-2 and IF2-3 (IF2-2/3), supports high viability when grown on MMS plates. (Although del2/3 is not a deletion mutation, consisting of mutations that inactivate the two internal start codons to prevent expression of IF2-2/3, it is so named to simplify nomenclature as a contrast to del1, which deletes the N-terminal coding region for IF2-1.) The restart allele priA300, which introduces the K230R amino acid replacement to restart protein PriA to inactivate its helicase activity (Zavitz & Marians, 1992), elicits the same characteristic for MMS sensitivity as the infB(del1) allele, and these two alleles are epistatic with each other for this phenotype (Madison et al., 2012). That is, IF2-1 and PriA helicase appear to function in a common pathway that allows growth with high viability in the presence of DNA-damaging agents.
Genetic analysis has indicated the presence of two major restart pathways. This has initially been indicated by the finding that the knockout of restart function priB or priC alone has only a small effect whereas the knockout of both results in severe negative effects including low viability, severe sensitivity to DNA-damaging agents, and constant SOS induction, a phenotype that is even more severe than that of the priA or dnaT knockout (McCool et al., 2004, Sandler, 2000, Sandler et al., 1999). PriA binds to D-loops and forked DNA structures at which DNA synthesis can be initiated (Jones & Nakai, 1999, McGlynn et al., 1997). PriB promotes formation of a complex of DNA-bound PriA with DnaT (Liu et al., 1996), a process that leads to loading of the major helicase DnaB onto the lagging strand template of the fork (Heller & Marians, 2005a, Jones & Nakai, 1999). PriC, on the other hand, can load DnaB onto DNA in vitro from the DnaB-DnaC complex without PriA, PriB, and DnaT (Heller & Marians, 2005a), with Rep or PriA helicase apparently playing an essential role for PriC-dependent pathways (Sandler, 2000, Sandler et al., 2001). Thus, biochemical analysis indicates two major systems for loading DnaB: one dependent on PriA/PriB/DnaT (the PriA-PriB pathway) and the other on PriC (PriA-PriC and Rep-PriC pathways, hereafter called the PriC system).
Deficiencies in the restart systems can lead to consequences not only from the inability to recover from replication fork accidents but also from the cellular response to these accidents (Sassanfar & Roberts, 1990). The latter includes the expression of the SOS gene sulA, which encodes a cell division inhibitor causing cell filamentation and loss of viability (Gottesman et al., 1981). Knockout of either priA or dnaT function results in a very severe negative phenotype (Lee & Kornberg, 1991, McCool et al., 2004, Nurse et al., 1991). The combination of the priB knockout and the priA300 allele, which inactivates helicase activity but not PriA’s function of loading DnaB onto DNA (Zavitz & Marians, 1992), elicits a severe negative phenotype comparable to that of the priA knockout whereas either allele alone has a much smaller effect (Sandler et al., 2001). Thus, PriA helicase appears to play a critical role in the PriC system even though either Rep or PriA helicases can support this system in vitro (Heller & Marians, 2005b). PriB generally has a considerable impact under genetic backgrounds and conditions where stalling of replication forks would be frequent, with PriA helicase playing a significant role as well in some cases (Flores et al., 2002, Grompone et al., 2003, Madison et al., 2012, Mahdi et al., 2012).
The epistatic relationship of infB(del1) and priA300 mutations (Madison et al., 2012) raises the question of how IF2 function might influence or interact with restart functions. Bacteriophage Mu replication by transposition (see ref. (Chaconas & Harshey, 2002) for a review), which relies on the cellular restart systems (Jones & Nakai, 1997), provides one possible clue to a function for PriA helicase related to IF2 function. In the in vitro reaction, IF2 plays an important role in the transition from strand exchange that integrate Mu DNA into target DNA and the assembly of the replisome on Mu DNA. In this process, proteins bound to the Mu ends (strand transfer complex) are progressively remodeled to a replisome, and IF2 not only plays a critical role for removing protein impediments that prevent binding of replication proteins to Mu DNA but also prevents replisome assembly until it is removed from DNA by PriA helicase action (North et al., 2007, North & Nakai, 2005).
In this paper we examine the varying effects of truncated and full-length IF2 isoforms, depending on levels of ppGpp. E. coli, other members of the Enterobacteriaceae family, and Bacillus subtilis have been characterized to express more than one molecular weight isoform of IF2 (Hubert et al., 1992, Laursen et al., 2002, Nyengaard et al., 1991). The function of having different isoforms has been largely a mystery. No clear differences in translation initiation activity are evident, for even a 65-kDa C-terminal fragment present in both IF2-1 and IF2-2/3 has been found to have all translation initiation activities at a specific activity essentially equivalent to that of IF2-1 (Cenatiempo et al., 1987). However, expressing IF2-1 or IF2-2/3 without the other from single-copy alleles can cause cold sensitivity at temperatures below 42°C; for example, the doubling time is increased as much as two fold over that of the wild-type strain at 37°C in LB medium (Sacerdot et al., 1992). Such single-copy infB(del1) and infB(del2/3) alleles are known to maintain the respective isoforms at 60–70% the level of total IF2 in wild-type cells (Sacerdot et al., 1992), in which intracellular IF2 concentrations can be reduced to 50% with little effect on growth rate (Cole et al., 1987). Here, we demonstrate that IF2-1 and IF2-2/3 have differing effects on cell survival, depending on ppGpp levels, by affecting function(s) closely related to replication restart. We demonstrate how each isoform can have differing impacts, whether positive or negative, depending upon the ppGpp levels and the status of restart functions.
Results
Use of relA and spoT alleles that progressively alter cellular levels of ppGpp
To examine the sensitivity of various mutants to DNA-damaging agents at ppGpp levels from 0 to high, we made use of various relA and spoT mutants that have different basal levels of (p)ppGpp in exponential growth phase (Sarubbi et al., 1988). The RelA protein synthesizes most of the pppGpp, and the second protein SpoT has a lower level of synthetic activity and also has the hydrolase activity that breaks down the nucleotides. The del(relA) del(spoT), del(relA) SpoT+, RelA+ SpoT+, del(relA) spoT202, and del(relA) spoT203 have 0 to very high basal levels of ppGpp in ascending order. To simplify nomenclature, we will be referring to these genetic backgrounds as corresponding to 0 (or ppGpp0), low (lo; 1X level of ppGpp), wild type (wt; 1–2X levels), high (hi; 4.5–6X), and very high (hi*; 6–8X) ppGpp, respectively (Sarubbi et al., 1988, Slominska et al., 1999). The del(relA) del(spoT) strain (ppGpp0) lacks all capacity to synthesize ppGpp (Xiao et al., 1991) while the RelA+ SpoT+ strain (wt) in exponential phase and the del(relA) single mutant (low) have relatively low levels of ppGpp. Although the methods for quantifying basal ppGpp levels in these strains have yielded varying absolute values in different laboratories (Sarubbi et al., 1988, Slominska et al., 1999), what has been clear is the relative ppGpp levels in these strains. Sarubbi et al. (1988) have demonstrated that the relative ppGpp levels present in the lo to hi* strains are clearly indicated by their growth rates (also reflected by colony size on LB plates), which we verified after strain construction (see Experimental Procedures; for example, see Figure S1A). For our analysis, the relative change in phenotype as ppGpp levels change from 0 to higher levels in a given genetic background is what is relevant.
Enhanced sensitivity of ppGpp0 strains for growth in the presence of DNA-damaging agents, suppressed by mutations that suppress amino acid auxotrophy of ppGpp0 mutants
We first asked whether 0 or low levels of ppGpp could dramatically affect growth of infB(wt) priA(wt) cells in the presence of 6 mM MMS or 10 μM nitrofurazone (NFZ), comparing this with the effect of UV irradiation. MMS methylates nitrogen atoms in purine residues, and the major error-free mode of repair is through base excision repair or direct reversal of alkylated bases (Sikora et al., 2010), in contrast to the pyrimidine dimers formed by UV irradiation and repaired by nucleotide excision repair (NER). NFZ causes lesions by introducing N2-deoxyguanosine adducts, which are predominantly repaired by NER (Ona et al., 2009). All three agents create DNA lesions capable of stalling both DNA replication and transcription.
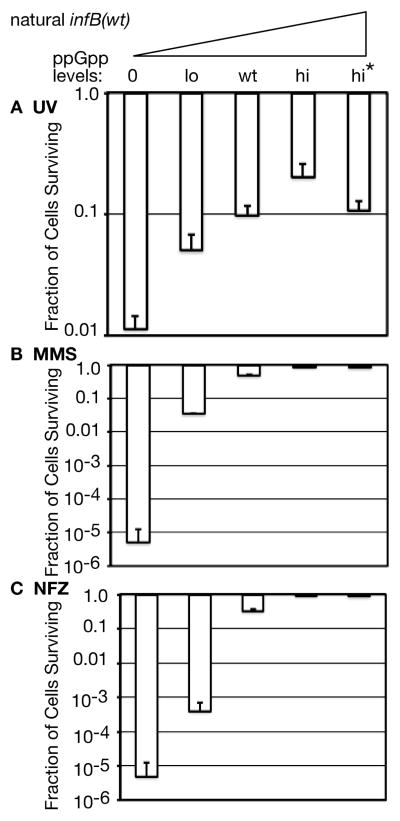
Cells progressively became more resistant to UV light as ppGpp levels were elevated from 0 to high and very high (Figure 1A). The enhanced UV sensitivity at 0 ppGpp levels and increased resistance at elevated ppGpp levels has been previously observed (McGlynn & Lloyd, 2000). Changes in UV sensitivity at varying ppGpp levels were nevertheless modest; the ppGpp0 strain had a 10-fold lower viability at 68 J/m2 irradiation than the corresponding strain with wt ppGpp levels. However, there can be a sharp contrast between UV survival, in which cells are allowed to recover and grow after irradiation, and viability on MMS plates in which cells must grow in the presence of a DNA-damaging agent. Such is the case with both infB(del1) and priA300 mutants, which exhibit little UV sensitivity over wild type but have very low viability on MMS plates (Madison et al., 2012). And indeed, viability of the ppGpp0 strain (wild type for infB and restart functions) was reduced 5 orders of magnitude on MMS plates while viability at wt ppGpp levels approached 100% (Figure 1B). Viability of these strains on plates containing NFZ was similar (Figure 1C), with the strain that has lo ppGpp levels exhibiting greater sensitivity to NFZ than to MMS. These results indicate that the absence of ppGpp can severely affect the ability of the cells to grow in the presence of DNA-damaging agents. It has been demonstrated that ppGpp effectively reduces the accumulation of stalled RNAP arrays that can cause conflicts with DNA replication (Trautinger et al., 2005). Thus there is a prominent possibility that such arrays accumulating at 0 to lo ppGpp levels cause the high sensitivity to MMS and NFZ.
Figure 1. Impact of ppGpp on survival after UV irradiation and growth in the presence of DNA-damaging agents.
A. UV sensitivity. Irradiation was at 254 nm at 68 J/m2. The surviving cells were scored on LB plates.
B. MMS sensitivity.
C. NFZ sensitivity.
Plating was on LB plates with or without 6 mM MMS or 10 μM NFZ. The genotype associated with each ppGpp level is described in the text. Strains used in this analysis are, in order of ascending ppGpp levels: GTN704, GTN701, GTN932, GTN722, GTN723. As indicated under Experimental Procedures, all results are reported as the mean of 3 experiments ± SD.
Enhanced sensitivity of ppGpp0 strains for growth in the presence of DNA-damaging agents is largely attributed to expression of the truncated forms IF2-2/3
As the infB gene strongly affects the cell’s ability to grow in the presence of MMS, we examined the effect of infB alleles that knock out the expression of the full-length form IF2-1 (infB(del1)) or the truncated forms IF2-2/3 (infB(del2/3)) at varying ppGpp levels. For this purpose we used infB mutants in which the natural infB has been knocked out (del(infB)1::tet) and a single-copy infB allele has been inserted into a second site as part of the nusA infB operon (flgJ::<nusA infB(wt, del1, or del2/3)>) (Madison et al., 2012). (We will refer to this second site allele as “<infB(wt, del1, or del2/3)>”, and this will also indicate that the natural wild-type infB allele has been deleted.) The <infB(del1)> and <infB(del2/3)> cells still exhibited the slowing of growth rates in the hi versus the wt ppGpp background (Figure S1C-E).
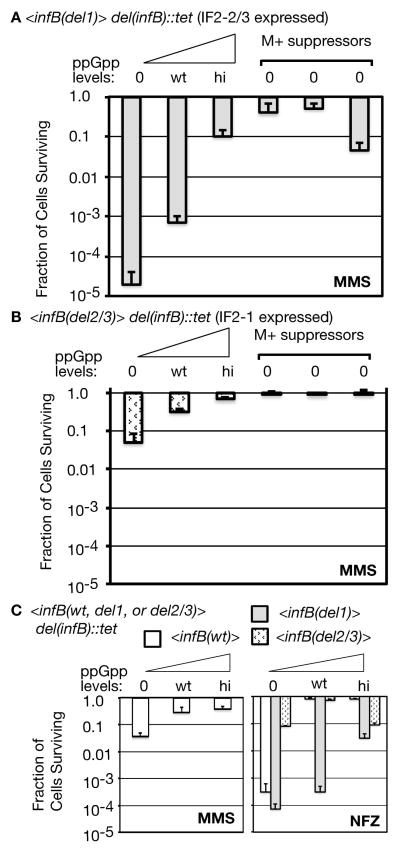
We found that viability of the ppGpp0 <infB(del1)> strain on MMS plates (Figure 2A) generally matched that of the ppGpp0 strain with the natural infB(wt) (Figure 1B) whereas the ppGpp0<infB(del2/3)> strain had nearly 4 orders of magnitude higher viability. At wt ppGpp levels the <infB(del1)> strain still exhibited significant sensitivity whereas the <infB(del2/3)> strain was largely resistant as previously determined (Madison et al., 2012). That is, the expression of IF2-2/3 without IF2-1 causes significant MMS sensitivity at lo and wt ppGpp levels while expression of IF2-1 without IF2-2/3 largely restores resistance. The <infB(del1)> strain also gained considerable resistance to MMS at hi ppGpp levels. We also isolated three independent M+ suppressor mutants of the ppGpp0 <infB(del1)> strain and examined their MMS sensitivity. These are suppressors of the amino acid auxotrophy of double relA spoT knockout (Xiao et al., 1991), mutations that so far have been found to map exclusively to RNAP subunit genes rpoB, rpoC, and rpoD (Cashel et al., 1996, Murphy & Cashel, 2003). The M+ suppressor mutations increased the viability of the ppGpp0 <infB(del1)> strain on MMS plates by 4 to 5 orders of magnitude, viability that was equivalent to or greater than that of the <infB(del1)> strain with hi ppGpp (Figure 2A). While the ppGpp0 <infB(del2/3)> strain, which cannot express IF2-2/3, has a low level of sensitivity with 10% of the cells viable on MMS plates, either hi ppGpp or the M+ suppressors could restore essentially 100% viability in MMS (Figure 2B). As some proteolytic degradation of IF2-1 could potentially yield fragments with activity analogous to IF2-2/3, we of course cannot rule out the possibility that the low level of MMS sensitivity of the ppGpp0 <infB(del2/3)> strain could be attributed to these fragments. What is clear is that blocking IF2-2/3 expression largely abrogates the extreme sensitivity of the ppGpp0 cell for growth on MMS plates.
Figure 2. Sensitivity of cells to MMS and NFZ as influenced by IF2 forms and ppGpp levels and suppressed by M+ suppressors.
A. MMS sensitivity of the <infB(del1)> strain.
B. MMS sensitivity of the <infB(del2/3)> strain.
C. MMS sensitivity of the <infB(wt)> strain and comparison of NFZ sensitivity of <infB> mutants.
The specific strains, listed in ascending ppGpp for each group, are <infB(del1)>: GTN1701, GTN1114, GTN1704; <infB(del2/3)>: GTN1702, GTN1115, GTN1705; <infB(wt)>: GTN1700, GTN1050, GTN1703. Three M+ suppressors of GTN1701 and GTN1702 were independently isolated for the analysis shown in panel A and B, respectively.
The high MMS and NFZ sensitivity of the ppGpp0 strain with the natural infB(wt) gene (Figure 1B and C) suggests that it is the expression of IF2-2/3, not the absence of IF2-1, that causes sensitivity. However, results with the <infB(wt)> strain (i.e., the strain with the wild-type nusA infB operon inserted at flgJ and the natural infB gene deleted) did not necessarily confirm this. The <infB(wt)> strain does not always behave exactly like the wild-type strain with the natural infB gene as we have found previously (Madison et al., 2012). In the ppGpp0 background, the <infB(wt)> strain had viability on MMS plates similar to that of the <infB(del2/3)> strain (Figure 2C left panel, cf. with panel B), not the extreme sensitivity displayed by the ppGpp0 strain with the natural infB (Figure 1B) or with <infB(del1)> (Figure 2A). On the other hand, high sensitivity of the ppGpp0 <infB(wt)> strain to NFZ is indeed similar to that of the <infB(del1)> strain (Figure 2C, right panel) and also similar to that of the strain with the natural infB(wt) (Figure 1C). Thus, it is not clear whether IF2-2/3 has the dominant effect in being toxic and promoting high sensitivity to MMS and NFZ in the ppGpp0 background or if IF2-1 has the dominant effect to promote resistance. The balance of full-length (IF2-1) and truncated (IF2-2/3) forms synthesized from the infB transcript, for example, may differ depending on whether it is transcribed from <infB(wt)> or the natural infB gene, and the balance from <infB(wt)> may be optimal for conferring relatively high resistance to MMS but not to NFZ. What is clear, however, is that the presence of IF2-2/3 is necessary to exhibit the very high sensitivity to MMS and NFZ in the ppGpp0 background.
The negative effects of expressing IF2-2/3 can be exacerbated by deficiency in restart function priB
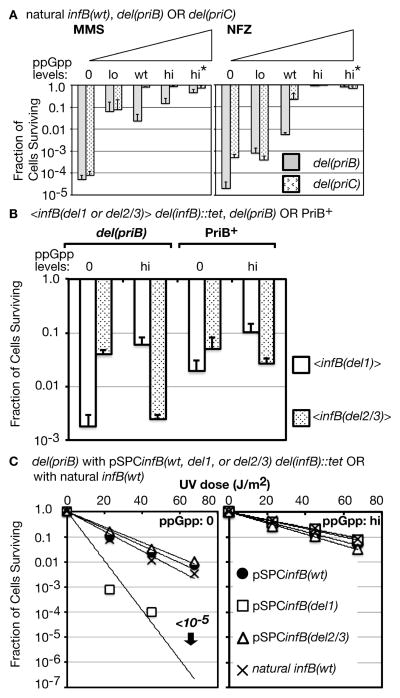
The absence of PriB is known to cause significant loss of viability under growth conditions and genetic backgrounds that promote replication arrest at high frequency (Flores et al., 2002, Grompone et al., 2003, Madison et al., 2012, Mahdi et al., 2012). Moreover, infB(del1) is synergistic with the priB knockout in increasing UV sensitivity (Madison et al., 2012). We therefore examined how the del(priB)302 allele affects sensitivity to growth on MMS and NFZ plates at varying ppGpp levels. At 0 and lo ppGpp levels, the del(priB) allele did not significantly increase sensitivity of cells to MMS or NFZ (Figure 3A; gray bars, cf. with Figure 1B and C). It is convenient to compare these results with MMS and NFZ sensitivity of del(priC) strains (Figure 3A, stippled bars), which generally yielded results that were quite similar to those of PriB+ PriC+ cells. At wt ppGpp levels, del(priB) cells exhibited significantly higher sensitivity than del(priC) and PriB+ PriC+ cells, but higher ppGpp levels conferred resistance. These results are consistent with those of Mahdi et al. (2012), who have reported that the priB knockout in the wt ppGpp background elicits moderate sensitivity for growth on media containing mitomycin C. Moreover, they demonstrated that mutations in RNAP subunits, mutations (rpo*) that are a subclass of M+ mutations (McGlynn & Lloyd, 2000), could suppress the DNA-damage sensitivity of del(priB) cells, consistent with our observation that elevated ppGpp could suppress MMS sensitivity caused by del(priB). Both ppGpp and rpo* mutations are known to diminish backed-up arrays of transcription complexes stalled at DNA lesions (Trautinger et al., 2005).
Figure 3. Very high sensitivity to DNA-damaging agents in the presence of truncated IF2 isoforms and the absence of both ppGpp and PriB.
A. MMS and NFZ sensitivity of del(priB) and del(priC) mutants at various ppGpp levels. The specific strains, listed in ascending ppGpp order, are del(priB): GTN703, GTN700, GTN394, GTN724, GTN726; del(priC): GTN1896, GTN1829, GTN1821, GTN1765, GTN1872.
B. Comparison of UV sensitivity of <infB> mutants in the del(priB) and PriB+ backgrounds. Irradiation with UV was at 68 J/m2. The specific strains (each pair having 0 and hi ppGpp, respectively) are <infB(wt)> del(priB): GTN1498, GTN1502; <infB(wt)>: GTN1700, GTN1703; <infB(del1)> del(priB): GTN1537, GTN1503; <infB(del1)>: GTN1701, GTN1704; <infB(del2/3)> del(priB): GTN1499, GTN1505; <infB(del2/3)>: GTN1702, GTN1705.
C. Comparison of UV sensitivity of del(priB) strains bearing the indicated plasmids. The specific strains (each pair having 0 and hi ppGpp, respectively) are pSPCinfB(wt): GTN1480, GTN1466; pSPCinfB(del1): GTN1610, GTN1541; pSPCinfB(del2/3): GTN1539, GTN1543; natural infB(wt): GTN1486, GTN1487.
As we shall discuss below, the del(priB) allele in combination with infB(del1) can have a significantly negative effect on the cell. However, the very low viability of infB(del1) cells on MMS and NFZ plates was not dramatically changed by the del(priB) mutation (Figure S2A). The presence of IF2-1 (<infB(del2/3)>) instead of IF2-2/3 (<infB(del1)>) still increased viability by several orders of magnitude in the del(priB) background. So did hi levels of ppGpp. These results suggest that the presence of IF2-1 and the absence of IF2-2/3 greatly alleviate transcription-DNA replication conflicts at 0 to wt ppGpp levels.
The combined effect of del(priB) and infB(del1) is more apparent when examining survival to UV irradiation. In the wt ppGpp background, del(priB) by itself has little effect on UV sensitivity (Sandler et al., 1999) but has a mildly synergistic effect with infB(del1) to increase UV sensitivity (Madison et al., 2012). However, we found that this synergistic effect is dependent on the ppGpp levels. In the ppGpp0 background the del(priB) allele mildly enhanced UV sensitivity of the <infB(del1)> mutant (Figure 3B; cf. white bars at 0 levels of ppGpp in the del(priB) and PriB+ backgrounds) but not of the <infB(del2/3)> mutant (cf. stippled bars at 0 levels of ppGpp). In the hi ppGpp background, the del(priB) had no effect on the UV sensitivity of <infB(del1)> cells (cf. white bars at hi ppGpp levels). UV sensitivity of the <infB(del2/3)> mutant instead was increased by the del(priB) allele in this background (cf. stippled bars at hi ppGpp).
A question arises as to whether IF2-2/3 or IF2-1 expressed from the <infB(del1)> or <infB(del2/3)> allele, respectively, is limiting in the ppGpp0 background or hi ppGpp background, causing differing recovery from DNA-damaging agents during growth. Thus, we wished to examine the effect of the infB allele in a different context, especially harbored on a plasmid vector where expression of the protein is likely to be higher. A much greater effect of expressing IF2-2/3 alone was observed if the infB(del1) allele (as part of the nusA infB operon) was harbored as the sole allele on a pSC101-based vector (see Experimental Procedures), which is a plasmid of relatively low copy number (Stewart et al., 1999). In the del(priB) ppGpp0 background, strains bearing pSPCinfB(wt) and pSPCinfB(del2/3) were relatively resistant to UV light (68 J/m2), indistinguishable from the level of sensitivity of the isogenic strain with the natural infB(wt) on the chromosome; in contrast, the pSPCinfB(del1)-bearing strain was more sensitive to UV irradiation by greater than 3 orders of magnitude (Figure 3C, left panel). In contrast, all three plasmid-bearing del(priB) strains were relatively UV resistant at hi ppGpp, indistinguishable from the isogenic strain with the natural infB(wt) allele (Figure 3C, right panel). That is, IF2-1 expressed from the plasmid vector did not increase UV sensitivity in the hi ppGpp background whereas IF2-2/3 did in ppGpp0. These results indicate that IF2-2/3 expressed from the pSPCinfB(del1) plasmid has an inhibitory effect on recovery of the del(priB) ppGpp0 strain from UV irradiation, an effect that is reversed by the presence of high ppGpp levels.
The detrimental effect of IF2-2/3 expressed from pSPCinfB(del1) in the del(priB) strain was also apparent from its very low viability even in the absence of DNA damaging agents (5 × 106 cells per ml at OD600 of 0.4 compared with 1–2 × 108 cells per ml for the corresponding pSPCinfB(wt) and pSPCinfB(del2/3) strains). The detrimental effects of the multicopy infB(del1) however could be greatly alleviated by the presence of the single-copy <infBdel2/3> allele (Figure S2B, left panel), indicating that the presence of IF2-1 counteracts the effects of elevated IF2-2/3. It is notable that the presence of the multicopy infB(del1) caused a small increase in UV sensitivity to the <infB(del2/3)> ppGpp0 strain (Figure S2B, left panel, cf. pSPCinfB(del1) with the empty vector) but promoted higher resistance in the <infB(del2/3)>, hi ppGpp strain (Figure S2B, right panel). That is, while IF2-2/3 may have a detrimental effect at 0 ppGpp, it can contribute to increased cell survival after UV irradiation when ppGpp is at hi levels. At hi ppGpp, either the obstacles resulting from DNA damage are changed or alleviated, or IF2-2/3 itself may be changed in function or activity so that it can aid in cell recovery.
Cellular filamentation induced by the presence of IF2-2/3 and the absence of PriB
Disruption of DNA replication and the resulting SOS response can induce SulA expression, which halts cell division and causes cellular filamentation (Huisman et al., 1984). Restart mutants such as the priA knockout exhibit severe SOS induction even without DNA damage (Nurse et al., 1991). We have previously reported that the <infB(del1)> del(priB) combination leads to a low but significant level of sporadic SOS induction, with 0.13% of the cell population producing filaments of greater than 30 nm in length (Madison et al., 2012). We examined whether levels of ppGpp have a significant influence on the level of cell filamentation without DNA damage.
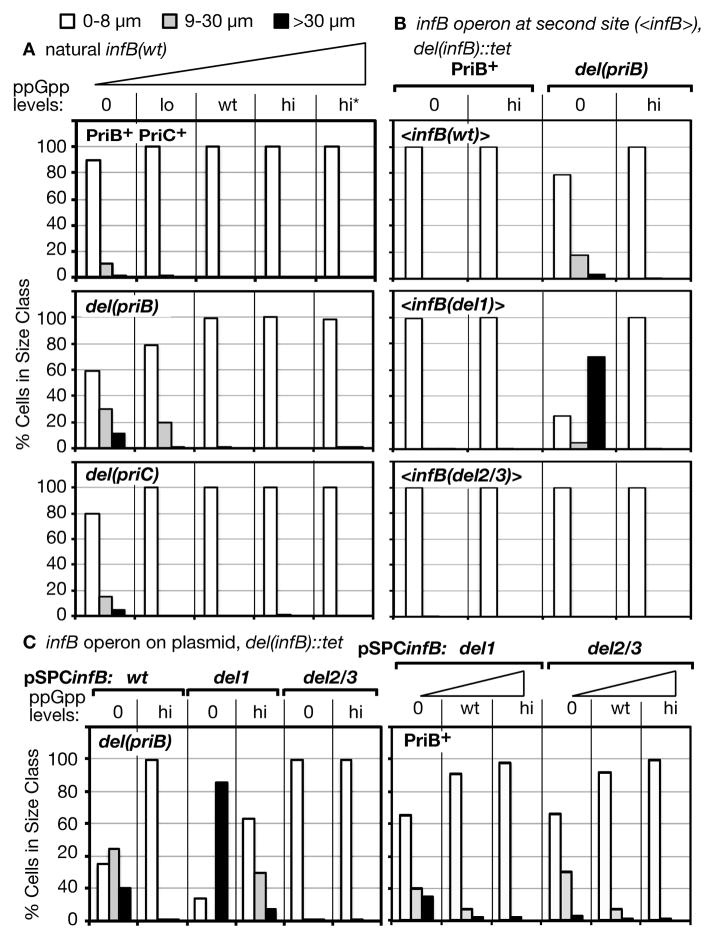
Exponentially growing cells with the natural infB(wt) allele in the PriB+ PriC+, del(priB), and del(priC) backgrounds displayed essentially no filamentation when levels of ppGpp were at wt or high levels (Figure 4A, 100% of cells ≤ 8 μm). In the absence of ppGpp, however, some filamentation could be detected in all three backgrounds, the greatest in the del(priB) cells and only a marginal amount in the PriB+ PriC+ cells. The tendency for ppGpp0 cells to form filaments has been previously reported (Xiao et al., 1991). In the case of the del(priB) cells, a lower but significant level of filamentation was also detected at lo ppGpp levels.
Figure 4. High cellular filamentation in the presence of truncated IF2 and in the absence of ppGpp.
Analysis of each strain grown in LB was conducted by examining 10,000 cells, which were scored and classified into one of three indicated size ranges by brightfield microscopy.
A. Size classes of PriB+ PriC+, del(priB), and del(priC) mutants at varying ppGpp levels. Strains, listed in order of ascending ppGpp levels for each group, are PriB+ PriC+: GTN704, GTN701, GTN932, GTN722, GTN723; del(priB): GTN703, GTN700, GTN394, GTN724, GTN726; del(priC): GTN1896, GTN1829, GTN1821, GTN1765, GTN1872.
B. Filamentation of <infB> mutants in the PriB+ and del(priB) backgrounds at 0 and high ppGpp. The strains from left to right for each group are <infB(wt)>: GTN1700, GTN1703, GTN1498, GTN1502; <infB(del1)>: GTN1701, GTN1704, GTN1537, GTN1503; <infB(del2/3)>: GTN1702, GTN1705, GTN1499, GTN1505.
C. Filamentation resulting from IF2-2/3 expressed from a plasmid vector in a del(priB) strain. Strains from left to right are GTN1480, GTN1466, GTN1610, GTN1541, GTN1539, GTN1543, GTN1698, GTN1943, GTN1679, GTN1675, GTN1957, GTN1681.
To determine whether the filamentation in the del(priB) ppGpp0 background could be further aggravated by knocking out IF2-1 and expressing only IF2-2/3 (infB(del1)), we compared the filamentation of <infB(wt)>, <infB(del1)>, and <infB(del2/3)> strains in the del(priB) ppGpp0 background (Figure 4B). Expression of IF2-2/3 only (<infB(del1)>) had an especially severe effect, causing extensive filamentation where approximately 70% of the cells were greater than 30 μm in length. The <infB(wt)> strain exhibited a much lower level of filamentation while the <infB(del2/3)> strain, which expresses no IF2-2/3, exhibited essentially no filamentation, indicating that the expression of IF2-2/3 is causing the filamentation. Moreover, the extensive filamentation of the <infB(del1)> strain was not observed with the PriB+ background (Figure 4B, PriB+). In the absence of PriB, the cell largely depends on the PriC system to maintain full viability without general SOS induction. The extensive filamentation exhibited by the <infB(del1)> del(priB) ppGpp0 strain suggests that the absence of ppGpp and the presence of only truncated IF2 forms can lead to either increased incidence of replication fork accidents or the inability to resolve them efficiently, leading to SOS induction. The results are also consistent with the previous conclusion that the presence of IF2-2/3 causes the enhanced sensitivity to DNA-damaging agents at 0 ppGpp levels. When its expression is blocked by the infB(del2/3) allele, no filamentation is observed. In this regard, it is worth noting that the strains with the second site <infB(wt)> allele exhibit much less filamentation than the corresponding strains with the natural infB(wt) allele (cf. Figure 4B, top panel, with 4A, top two panels). This is consistent with our finding that the <infB(wt)> ppGpp0 strain is more resistant to MMS than the analogous strain with the natural infB(wt). The possibility is that <infB(wt)> has a different balance of IF2-1 and IF2-2/3, favoring the former such that filamentation is not so severe.
When IF2-2/3 was expressed from a multi-copy plasmid (pSPCinfB(del1)) in the del(priB) ppGpp0 strain, filamentation was even more severe, with almost 90% the cells forming filaments of greater than 30 μm in length (Figure 4C, left panel, 0 ppGpp, del1). Clearly, the effect of high IF2-2/3 levels was most pronounced for the del(priB) ppGpp0 strain as the PriB+ version of this strain exhibited considerably lower levels of filamentation (Figure 4C, right panel, del1 at 0 ppGpp). Consistent with the conclusion that IF2-2/3 is causing the filamentation in the del(priB) strain, the analogous pSPCinfB(wt)-bearing strain, which expresses both IF2-1 and IF2-2/3, also exhibited extensive filamentation but at a lower level than the pSPCinfB(del1) strain while the analogous pSPCinfB(del2/3) strain exhibited essentially no filamentation (Figure 4C, left panel). In the two strains expressing IF2-2/3, hi ppGpp levels greatly suppressed filamentation although this was not sufficient to suppress severe filamentation of the pSPCinfB(del1) strain completely. Interestingly, although pSPCinfB(del1) caused less filamentation in the PriB+ ppGpp0 background, pSPCinfB(del2/3) that expresses IF2-1 caused significant filamentation as well (Figure 4C, right panel), in contrast to what was seen in the del(priB) ppGpp0 background. As we shall see, this effect was even more pronounced in the priA300 ppGpp0 background.
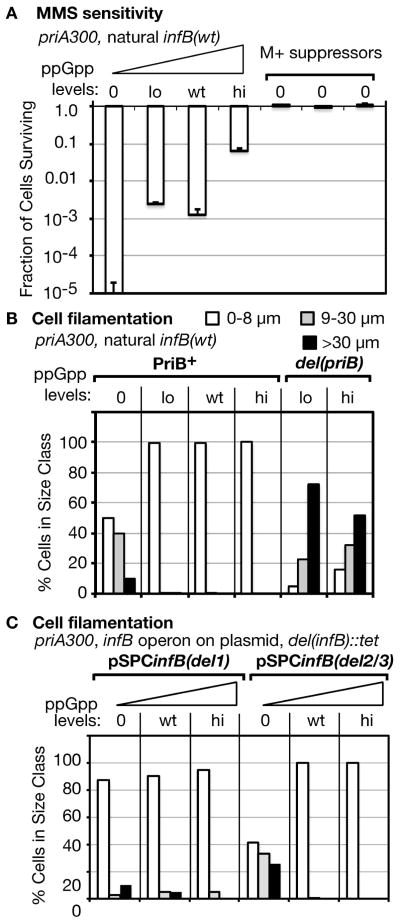
The effect of varying ppGpp levels and M+ suppressor mutations on the priA300 mutant is analogous to their effect on the infB(del1) mutant
As the priA300 allele is epistatic with infB(del1) in eliciting MMS sensitivity at wt ppGpp levels, the question arises whether ppGpp levels and M+ suppressor mutations affect the priA300 mutant in the same way they affect the infB(del1) mutant. Indeed, the loss of viability of the priA300 mutant on MMS plates could be aggravated by the ppGpp0 background while it could be largely suppressed by hi ppGpp (Figure 5A; 0 to hi). Moreover, when three M+ suppressor mutants of the priA300 ppGpp0 strain were independently isolated, all three were found to be essentially 100% resistant to MMS (Figure 5A; M+). The varying ppGpp levels affected the priA300 mutant’s sensitivity to NFZ in essentially an identical fashion (Figure S3). However, hi ppGpp had only a small effect in alleviating both MMS and NFZ resistance if the priA300 strain also had a del(priB) mutation (Figure S3, priB−).
Figure 5. Effect of ppGpp on the priA300 mutant.
A. Effect of ppGpp and M+ suppressor mutations on viability on MMS and NFZ plates. The strains are from left to right: GTN1867, GTN1713, GTN381, GTN1717. Three M+ suppressor mutants were independently isolated from GTN1867.
B. Effect of ppGpp levels on cellular filamentation of the priA300 mutant. Cells were analyzed as described in the legend to Figure 4. The strains are from left to right: GTN1867, GTN1713, GTN381, GTN1717, GTN1789, GTN1811.
C. Effect of IF2-1 and IF2-2/3 expressed from a plasmid vector on the cellular filamentation of the priA300 mutant. The strains are from left to right: GTN1860, GTN1858, GTN1856, GTN1800, GTN1757, GTN1759.
While the <infB(del1)> mutant exhibited severe filamentation in the ppGpp0 background only in combination with the del(priB) allele, the priA300 ppGpp0 mutant exhibited fairly severe filamentation without an accompanying mutant infB allele; however, even lo ppGpp levels completely eliminated any filamentation (Figure 5B, PriB+). In contrast, the priA300 del(priB) mutant exhibited severe filamentation even with the hi ppGpp background (Figure 5B, del(priB)), consistent with the high MMS and NFZ sensitivity of these cells (Figure S3) and with previous findings that cells lacking PriB are highly dependent on PriA helicase for normal viability (Sandler et al., 2001).
The results so far indicate that ppGpp0 cells are dependent on the presence of PriA helicase to avoid severe filamentation, with IF2-2/3 aggravating filamentation when PriB is absent. Could it be that when ppGpp is absent, IF2-2/3 preferentially supports pathway(s) that are not dependent on PriA helicase and tends to inhibit pathways that engage PriA helicase? Since PriA helicase plays a significant part in maintaining cell viability and preventing constant SOS induction with the PriC system (Sandler et al., 2001), this could be why the absence of PriB in infB(del1) ppGpp0 cells promotes a high level of filamentation. Additionally, since PriA helicase is needed for efficient growth on MMS and NFZ plates at 0 to wt ppGpp levels, the pathways favored by IF2-2/3 may not be robust enough to cope with frequent DNA damage needed to grow on these plates. If this is indeed case, IF2-2/3 provided from a plasmid vector should not have a detrimental effect in the priA300 mutant provided that it expresses PriB.
The pSPCinfB(del1) plasmid supplying the sole multicopy infB allele greatly alleviated filamentation of the priA300 ppGpp0 mutant (Figure 5C, pSPCinfB(del1), 0 ppGpp) instead of aggravating filamentation as it did in the del(priB) mutant. In contrast, pSPCinfB(del2/3) caused severe cell filamentation of this mutant, filamentation that was prevented when ppGpp was raised to wt levels (Figure 5C). Both the pSPCinfB(del1)- and pSPCinfB(del2/3)-bearing strains with the priA300 background had very low viability on MMS plates, the former (GTN1860) having survival of 0.16 ± 0.04% and the latter (GTN1800) 0.003 ± 0.003%. That is, the strain bearing pSPCinfB(del2/3) and expressing IF2-1 is even more sensitive to MMS than the one expressing IF2-2/3. The results indicate that under different circumstances, both IF2-1 and IF2-2/3 when expressed from a multicopy allele can have deleterious effects in ppGpp0 cells when one is expressed without the other—IF2-1 when PriA helicase is inactive (and PriB is present) and IF2-2/3 when PriB is absent. They suggest that these proteins inhibit recovery from replication fork accidents in distinct ways, inhibition that is overcome by action of PriA helicase or the presence of ppGpp.
Impact of IF2 forms and ppGpp levels on Mu development
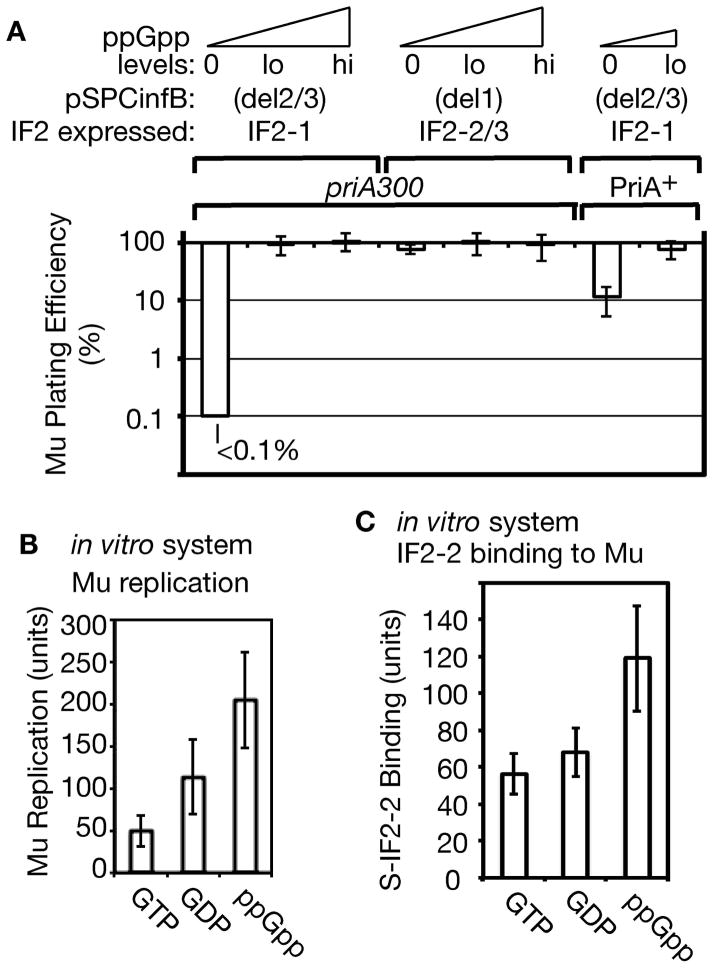
As bacteriophage Mu replication by transposition requires the host replication restart system, we asked whether it is differentially influenced by the various IF2 forms and is activated by elevated ppGpp levels. IF2-1 expressed from pSPCinfB(del2/3) in a priA300 ppGpp0 cell not only caused extensive filamentation (Figure 5C) but also diminished Mu plating efficiency to less than 0.1% whereas IF2-2/3 expressed from pSPCinfB(del1) had no effect on Mu plating efficiency in this background (Figure 6A, cf. pSPCinfB(del2/3) versus pSPCinfB(del1) for priA300 ppGpp0; it should be noted that the pSPCinfB(del2/3) priA300 ppGpp0 strain had a reasonable cell viability in the absence of DNA-damaging agents, with a viable count of 1 X 108 cells/ml in LB at OD600 of 0.4 and within the range of 1 to 2 X 108 cells/ml for most strains used in this work). In the PriA+ background, Mu plating efficiency on the pSPCinfB(del2/3) ppGpp0 strain was largely restored to 10%—to 100% with the lo ppGpp background (Figure 6A, PriA+, 0 and lo ppGpp levels). IF2-1 also caused no decrease in Mu plating efficiency on the priA300 strain that had lo levels of ppGpp (Figure 6A, IF2-1, priA300, and lo ppGpp).
Figure 6. Reduced Mu replication in the presence of truncated IF2 at 0 or low levels of ppGpp.
A. Phage Mucts62 plating efficiency on priA300 strains expressing IF2-1 or IF2-2/3 from plasmid vectors pSPCinfB(del2/3 or del1). Strains (from left to right): GTN1800, GTN1757, GTN1759, GTN1860, GTN1858, GTN1856, GTN1675, GTN1957.
B. Levels of Mu replication in vitro with IF2-2 and 1 mM GTP, GDP, and ppGpp. The amount of replication obtained without the added 1 mM guanine nucleotides was set to 100 units (approximately 25% of total Mu DNA replicated).
C. Effect of GTP, GDP, and ppGpp on the binding of S-IF2-2 to Mu DNA in vitro. The level of binding when no nucleotides were present is set to 100 units.
IF2-2/3 had negative effects on Mu replication in the ppGpp0 del(priB) background. The pSPCinfB(del1) strain with this background indeed plates Mu at very low efficiency, but it has growth properties that include very low viability as stated earlier such that the experiments were marred by its inability to form an even lawn of cells. We therefore compared Mu plating efficiency in single-copy <infB(wt)>, <infB(del1)>, and <infB(del2/3)> strains that are PriB+ or del(priB) at 0 or high ppGpp levels (Figure S4). The differences were not so much in the number of plaques produced but in the sizes of plaques. The size of plaques was most dramatically reduced with indicator strains that are ppGpp0 del(priB) <infB(wt or del1)—i.e., strains expressing IF2-2/3; there was barely any effect of del(priB) on the <infB(del2/3)> mutant, which does not express IF2-2/3 (Figure S4; cf. row 1 with 3). The hi ppGpp background generally improved the apparent Mu plating efficiency, and the del(priB) mutation had a smaller effect in decreasing plaque size in this background (Figure S4; cf. row 2 with 4). These results are consistent with an activating effect of ppGpp and an inhibitory effect of IF2-2/3 at 0 ppGpp levels when Mu development proceeds in the absence of PriB.
Effect of ppGpp on the in vitro Mu replication system
As we will describe more fully in the Discussion, ppGpp binds to a number of proteins to exert various effects. This includes IF2, whose G domain binds not only GTP but also ppGpp, the latter inhibiting it as a translation initiation factor (Milon et al., 2006). Thus, the question arises as to what effect the presence of ppGpp has on the reconstituted Mu replication system where IF2 plays an essential role. This system is reconstituted using IF2-2 by a PriA-PriC pathway (North et al., 2007), the PriC system where PriA helicase plays an essential role, and the effect of 1 mM GTP, GDP, or ppGpp was examined. The standard Mu replication system includes 0.1 mM GTP, which was present in all reactions. Thus, the guanine nucleotides were first mixed together with IF2-2 on ice (pre-mix) before addition of the strand transfer complex and other components of the replication system. The level of Mu cointegrates (the Mu transposition and replication product) produced in this reaction was highest with ppGpp and the lowest with GTP (Figure 6B).
Although the level of Mu replication was only about 4-fold higher with ppGpp than with GTP, this was most significant for the fact that ppGpp activates rather than inhibits the Mu replication reaction. To determine whether IF2 function in Mu replication is indeed not inhibited by the presence of ppGpp, we examined binding of S-tagged IF2-2 (S-IF2-2) to Mu DNA in the presence of the three guanine nucleotides at 1 mM. Following the disassembly of the Mu transpososome from the forked Mu ends created by Mu strand exchange, a new nucleoprotein assembly is formed, allowing the binding of IF2-2 (North et al., 2007). The presence of ppGpp allowed maximal binding of S-IF2-2 to Mu DNA at a level that was approximately 2 fold higher than the amount bound in the presence of GTP or GDP (Figure 6C). Once again, what was most relevant here was that ppGpp increased rather than decreased S-IF2-2 binding to Mu DNA. These results raise the possibility that IF2 and ppGpp may have a positive modulatory effect on the replication restart system, but more importantly, they indicate that the assembly of restart proteins under the influence of IF2 is not necessarily inhibited by ppGpp, which does inhibit its capacity to form translation initiation complexes (Milon et al., 2006).
Discussion
The functions of PriA helicase and IF2 isoforms relative to the effect of varying ppGpp levels and the M+ suppressor mutations
Both the infB(del1) mutation, which knocks out expression of IF2-1, and the priA300 mutation, which inactivates PriA helicase, cause sensitivity to DNA-damaging agents such as MMS in a specific way. While the cells readily recover from exposure to MMS if allowed to recover and grow in its absence, the mutants are not able to maintain high viability if they must grow in its presence (Madison et al., 2012). The results of the present work indicate that MMS sensitivity resulting from either mutation can be suppressed by elevated ppGpp levels. Alternately, M+ mutations, which suppress amino acid auxotrophy of the ppGpp0 mutant and which so far have all been found to map in genes encoding subunits of RNAP (Cashel et al., 1996, Murphy & Cashel, 2003), effectively suppress DNA damage sensitivity of both the priA300 and infB(del1) mutant with the ppGpp0 background. And even though the ppGpp0 mutant generally exhibited very high sensitivity to growth in the presence of DNA-damaging agents, the expression of only IF2-1 (and the absence of IF2-2/3) largely suppressed sensitivity.
In the absence of a DNA-damaging agent, expression of IF2-2/3 without IF2-1 in the del(priB) ppGpp0 background elicits a phenotype that would be consistent with the processing of the stalled fork being obstructed such that it cannot efficiently proceed to restart, causing accumulation of single-stranded DNA and SOS induction. IF2-1 does not elicit the same phenotype in this background; in the absence of IF2-2/3, the cells exhibit essentially no filamentation. Expression of IF2-2/3 from the multicopy pSPCinfB(del1) allele instead of the single-copy <infB(del1)> allele resulted in even more severe filamentation and low viability of del(priB) ppGpp0 cells, suggesting an inhibitory effect of IF2-2/3 rather than simply the absence of an activity only present in IF2-1. The presence of ppGpp, especially at high levels, prevents the detrimental effects of expressing only IF2-2/3 in the del(priB) cell, suggesting that it prevents the obstruction of replication from occurring at high frequency or endows IF2-2/3 with the capacity to better cope with accidents of replication forks.
Possible role of IF2 forms in promoting optimal recovery from DNA damage during growth depending on physiological condition
The different effects of IF2-1 and IF2-2/3 indicate distinct influences, dependent on status of restart functions and ppGpp levels, in how obstructions to DNA replication are created or how replication forks recover after arrest during cell growth. The effects we have observed would be compatible with a model in which IF2-2/3 in the ppGpp0 cell has a reduced capacity to promote recovery from replication fork accidents by PriA helicase-dependent mechanisms, even being inhibitory for PriC-dependent mechanisms that require the helicase. Elevated ppGpp from the stringent response alleviates this problem, either by making the repair, recombination, or restart system more active or by lessening the obstacles that the replication forks would encounter. Although ppGpp (and pppGpp) are bound by a number of proteins to exert various effects (see (Kanjee et al., 2012) for a review), the suppression of DNA-damage sensitivity of both infB(del1) and priA300 mutants by M+ mutations focus attention on two major suspects: IF2 and RNAP.
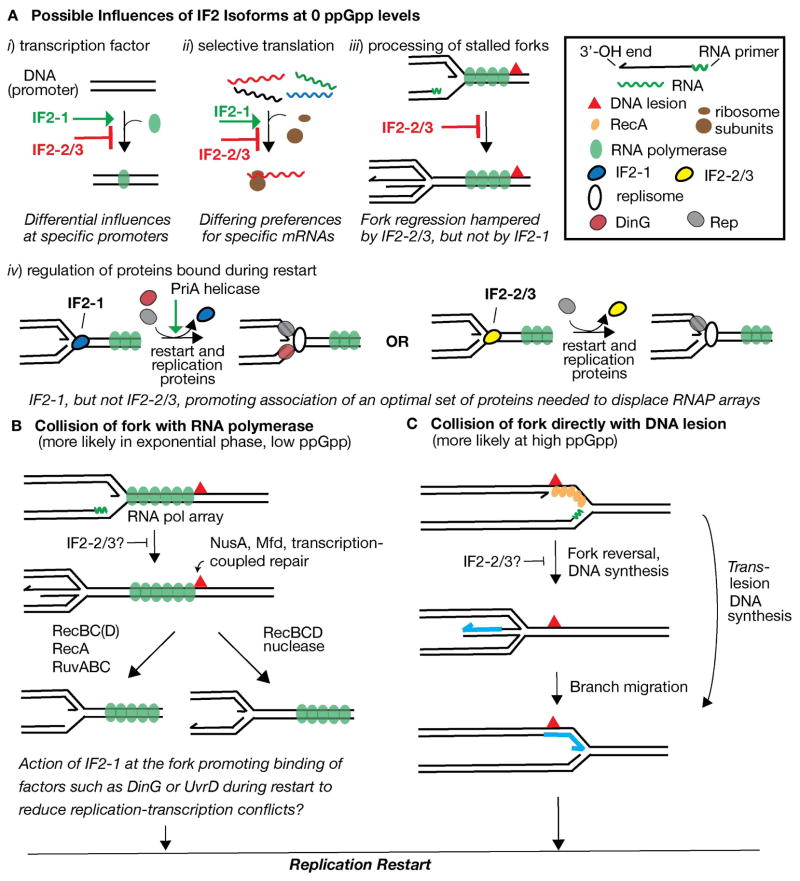
There are conceivably four general ways that IF2 forms can exert differing effects at 0 to low ppGpp levels to affect sensitivity to DNA-damaging agents. The two forms could affect gene expression differently either by acting as transcription factors (Figure 7A, i) or by influencing which mRNA transcripts are translated preferentially (ii), modulated by ppGpp levels. The expression of proteins promoted by IF2-1, but not by IF2-2/3, may enhance recovery of cells from the effects of MMS and NFZ during rapid growth. Alternatively, IF2-2/3 may be a more potent transcriptional activator at certain operons than IF2-1 (opposite of the regulation shown in Figure 7A, i), intensifying DNA-replication transcription conflicts. While it has been reported that IF2 can increase transcription of rRNA in vitro (Travers et al., 1980), it is not known if the various IF2 forms can have differential effects in promoting this process. Nor is it known if the IF2 forms can differentially influence which mRNA transcripts are chosen for translation to alter gene expression.
Figure 7. Model for the influence of ppGpp and IF2 forms in recovery from DNA damage.
Depiction of transcription-DNA replication conflicts and recovery of replication forks is based on models by Trautinger et al. (2005) and De Septenville et al. (2012).
A. Possible activities of IF2 and potential differences between IF2-1 and IF2-2/3 accounting for their differential effects. Activities are hypothesized for 0 levels ppGpp, where the function and activity of IF2 isoforms are potentially modulated by ppGpp. i) IF2 as transcription factor. IF2-1 and IF2-2/3 may have differential activity as transcription factors at various promoters. IF2-1 activates transcription at a promoter whereas IF2-2/3 may not have this activity and may even inhibit this process. Alternatively, the roles of IF2-1 and IF2-2/3 may be reversed. ii) IF2-1 and IF2-2/3 having differing preferences for mRNA transcripts, preferences that may be changed by ppGpp. iii) IF2-2/3 as inhibitor of fork regression. iv) After fork regression and DNA processing, IF2-1, but not IF2-2/3, promotes loading of accessory proteins (DinG as shown, UvrD, or other proteins) with the replisome during restart, enabling efficient replication through transcription obstacles. PriA helicase activity is hypothesized to have an activating effect on this process.
B. Events during DNA replication-transcription conflicts, most frequent during exponential growth and low ppGpp levels. NusA, Mfd: proteins involved in transcription-coupled repair (Cohen et al., 2010, Selby & Sancar, 1993).
C. Events after direct collision of fork with a DNA lesion, more frequent during nutrient stress and high ppGpp levels. See text for further details as well as alternative mechanisms.
The role of IF2 in promoting bacteriophage Mu replication by restart proteins also suggests possible activities of IF2 at DNA forks that result from replication arrest, acting on proteins bound to the fork and even binding there itself. Its activity as a molecular chaperone (Caldas et al., 2000) can potentially remodel protein assemblies at the fork, influencing what protein functions are active there. For example, if IF2-2/3, but not IF2-1, inhibited fork regression after replication arrest at transcription complexes (Figure 7A, iii), it may greatly diminish the cell’s capacity to cope with transcription-replication conflicts. Experiments examining the collisions of replication forks head on with transcription of an inverted rRNA operon have suggested such conflicts generally lead to fork regression, DNA processing that requires the RecBC functions and that creates a fork, and replication restart (De Septenville et al., 2012). (See Figure 7B. In this model, transcription is shown to be proceeding in the same direction as DNA replication. Various mechanisms for overcoming transcription-replication conflicts are reviewed in (Merrikh et al., 2012).) These results have suggested that fork regression and subsequent DNA processing are essential steps toward restarting replication forks better able to cope with a high level of conflict with transcription complexes (De Septenville et al., 2012). The disadvantage of having only IF2-2/3 available may be that it binds to the fork or does not enable proteins such as the replisome components to disassemble efficiently, thus interfering with fork regression and the subsequent assembly of a more resilient replisome. IF2-1 on the other hand may not bind at such an arrested fork or may bind but is readily removed; alternatively it may enable other proteins to be efficiently removed from the fork such that fork regression proceeds unimpeded.
A closely related model is one in which IF2-1 plays a more direct role in the assembly and activation of accessory proteins that render the replisome more resilient to transcription complexes (Figure 7A, iv). The accessory replicative helicases Rep, DinG, and UvrD enable the replisome to overcome transcriptional obstacles (Baharoglu et al., 2010, Boubakri et al., 2010, Guy et al., 2009). De Septenville et al. (2012) have proposed that a replisome with not only Rep but also a second accessory helicase (DinG or UvrD) is assembled during restart after replication arrest at transcription complexes, fork regression, and DNA processing, allowing the reassembled replisome to better displace formidable transcription obstacles. IF2-1 may play a role as molecular chaperone in promoting association of the second accessory helicase or other proteins that may render the replisome more resilient. PriA helicase may activate this process in conjunction with IF2-1; if it displaces IF2 bound to the fork as proposed for Mu replication, IF2-1 disassembly may be coupled to the binding of the accessory proteins. IF2-2/3 may not be able to promote association of the additional accessory proteins, thus leading to additional and frequent replication arrest. Similarly, in the absence of PriA helicase activity, the removal of IF2-1 from the fork and assembly of resilient replisomes may be inefficient.
Because of IF2’s role as translation factor and its potential influence on gene expression, the loss of IF2-1 or IF2-2/3 can have pleiotropic effects that lead to DNA-damage sensitivity or resistance, respectively, at 0 or low ppGpp levels. Thus, a combination of effects including those depicted in Figure 7A may be applicable. Nevertheless, from the characteristics of the infB(del1) and infB(del2/3) mutants, we favor a model in which full-length isoform allows efficient recovery of replication forks stalling at transcription arrays whereas the truncated isoforms are better suited to promoting recovery of forks colliding directly with DNA lesions (Figure 7B versus C, respectively). As pointed out by Trautinger et al. (2005), a significant difference between conditions of low and high ppGpp levels is that the replication fork is more likely to collide with a stalled transcription array at 0 or low levels and directly with the DNA lesion at high levels. Fork regression that follows collision of the fork with transcription complexes would disassemble the replisome and re-establish the fork away from the point of conflict (De Septenville et al., 2012) (Figure 7B), and the full reassembly of a replisome, which may be more resilient to conflict with transcription, would be required. Thus, if IF2-2/3 were unable to efficiently allow the assembly of such a resilient replisome, 0 and low ppGpp levels would lead to frequent replication arrest, a condition known to cause strong dependence on PriB (i.e., the PriA-PriB pathway) for cell viability (Flores et al., 2002, Grompone et al., 2003, Mahdi et al., 2012).
In contrast, when a replication fork collides directly with the DNA lesion (Figure 7C), the capacity to overcome a transcriptional obstacle is not needed, and replisome disassembly and fork regression do not necessarily occur. For example, the E. coli translesion DNA polymerases can exchange places with DNA polymerase III in the replisome without collapsing the fork, a switch that can be regulated by RecA, to enable DNA synthesis through the lesion (Figure 7C) (Indiani et al., 2009, Indiani et al., 2013). Even if translesion DNA synthesis is not catalyzed by the replisome, the DNA polymerase does not necessarily disassemble from the fork (see (Yeeles et al., 2013) for a review). When DNA polymerase III holoenzyme encounters a lesion on the leading strand template, the replisome can remain strongly associated with the fork, and leading strand synthesis can be reinitiated with the laying down of a primer downstream (Yeeles & Marians, 2011). And if DnaB helicase is disassembled, a PriC-dependent mechanism can reload it for reinitiation in this fashion (Heller & Marians, 2006a). That is, under conditions where forks are likely to collide directly with the DNA lesion rather than a transcription array, the need to reassemble a replisome may be relatively infrequent. The requirement for the PriA-PriB pathway, hypothesized to be the most robust one essential for replication restart at high frequency (Flores et al., 2002, Grompone et al., 2003), would then be reduced.
Differences between IF2-1 and IF2-2/3 in their ability to cope with frequent DNA damage during growth at various ppGpp levels may reflect different strategies to enhance cell survival whether the cell is in the growth or maintenance modes—what has been described as the directing of metabolic and energy resources to either reproduction or the preservation of the soma (Nystrom, 2004). IF2-2/3 may be especially important under conditions of stress such as when ppGpp levels are high. IF2-1 levels are known to be proportional to growth rate (Howe & Hershey, 1983), and while IF2-2/3 levels are generally equivalent under these conditions (Sacerdot et al., 1992), the ratio of IF2-2/3 to IF2-1 rises in response to cold shock (Giuliodori et al., 2004). Under nutrient limitation or cold shock, growth rate and DNA replication would be at relatively low levels, and replication forks may also stall at a high frequency even without DNA-damaging agents due to obstacles such as the dearth of nucleotides that halt DNA synthesis. At high ppGpp levels new rounds of replication at oriC and chromosomal segregation are prevented (Ferullo & Lovett, 2008, Levine et al., 1995, Levine et al., 1991, Schreiber et al., 1995, Zyskind & Smith, 1992). While IF2-2/3 may exert an effect that aggravates transcription-replication conflicts, these same effects may be beneficial when replication forks are more likely to collide directly with DNA lesions. Effects such as the inhibition of fork regression or assembly of replisomes without additional accessory proteins may enable more efficient resumption of DNA synthesis under these conditions. This may ensure completion of replication forks and entry into a maintenance mode that preserves genome integrity.
No matter what the underlying mechanism for the differences between IF2-1 and IF2-2/3, each has considerable impact for cell survival depending on whether the growth rate is high or low, indicating differential functions for the IF2 forms in preserving genome integrity by a process adaptable to environmental and physiological conditions.
Experimental Procedures
Bacterial strains
All strains are derivatives of E. coli K-12 and are listed in Table S1. Strain constructions and verification were mostly carried out as previously described (Madison et al., 2012). Sequencing of dnaC was routinely conducted with mutants potentially defective in replication restart to ensure that no suppressor mutations had arisen. PCR primers GTTGCGATTTGCCGATTTCGGCAGGTCTGGTCCCT and CCCTTTCCTCAAACCGCTATCATATGTAGATACAG were used to verify knockout of relA. Upon introduction of a relA knockout allele, the resulting strain was subjected to the SMG growth test, which verifies the inability of a relA mutant to grow in minimal media containing serine, methionine, and glycine (Uzan & Danchin, 1976). GCTATTGCTGAAGGTCGTCGTTAATCACAAAGCGG and GCATTTCGCAGATGCGTGCATAACGTGTTGGGTTC were used to verify knockout of spoT, and the former oligo was also used to sequence spoT to verify spoT202 and spoT203 alleles. The varying ppGpp levels promoted by the various relA and spoT alleles were further verified by the inverse linear relationship between growth rates and ppGpp levels (Sarubbi et al., 1988). Double relA spoT knockout mutants were routinely streaked on Davis minimal media (Difco) supplemented with thiamine and any amino acid(s) specified by other auxotrophic markers to ensure no suppressor mutations that allow growth on this media had arisen (Xiao et al., 1991). Suppressor mutants (M+) were isolated by selecting on this medium. For each independent isolation, a suppressor-free ppGpp0 colony was used to inoculate 5 ml LB, which was grown up by shaking overnight at 37°C. The culture was passaged 3 times or more in 5 ml LB at 37°C before selection for M+ mutants.
Plasmids
The pSPCnusAinfB plasmids (simply referred to as pSPCinfB in the main text), which bear various infB alleles, were constructed as previously described (Madison et al., 2012). Briefly, these have the nusA infB operon inserted into the NsiI-BamHI site of pBAD43, a relatively low copy number plasmid with a pSC101 origin (Stewart et al., 1999). Sequences required for arabinose-based gene expression in pBAD43 have been replaced by the operon and are absent from all pSPCinfB plasmids. All plasmid-bearing strains have a knockout of the natural infB site (del(infB)1::tet), and where indicated, a second infB allele was introduced as a nusA infB operon inserted into the flgG gene (flgG::<nusAinfB>).
Plasmid pTYB12-S-IF2-2 was constructed by cutting out the S-tagged IF2-2 (S-IF2-2) coding sequence from pBAD24-S-IF2-2 (North et al., 2007) with NdeI and SpeI, and the DNA fragment was ligated into the NdeI-SpeI site of pTYB12 (New England Biolabs).
Proteins
All proteins were purified as previously described (North et al., 2007). S-IF2-2 was expressed in intein-tagged form from pTYB12-S-IF2-2 and was purified by the same procedure that we used to purify untagged IF2-2 from intein-tagged form (North et al., 2007).
Bacteriophage Mu DNA replication in vitro
The in vitro transposition reaction was performed in two stages: strand transfer and DNA replication. Reaction conditions for each have been previously described (North et al., 2007, North & Nakai, 2005). Purified proteins used in the second stage include ClpX, IF2-2, and a set of purified replication proteins (gyrase A and B subunits, SSB, primase, β subunit of pol III holoenzyme, pol III*, DNA ligase, DNA pol I, PriA, PriC, DnaT, and the DnaB-DnaC complex). In reactions containing GDP, GTP, or ppGpp, 50 ng purified S-IF2-2 was assembled on ice with GDP, GTP, or ppGpp in 5 μl strand transfer reaction buffer for a final concentration of 1 mM in 50-μl replication reaction mixtures. This 5-μl mixture of S-IF2-2 and guanine nucleotides were assembled just before starting the replication reaction, being allowed to sit on ice for a uniform 5 min before use. Quantified levels of Mu DNA replication are the average of six determinations with error expressed as ± SD.
ppGpp was a generous gift from Dr. Michael Cashel (National Institutes of Health). Experiments were also repeated using ppGpp purchased from TriLink Biotechnologies.
S-IF2-2 binding assay
Binding of S-IF2-2 to the Mu STC was assayed by a two-stage procedure. In stage one, STC-DF nucleoprotein complexes were formed in a strand transfer reaction mixture (50 μl) containing 40 μg/ml pGG215 donor DNA, 80 μg/ml pACYC184 target DNA, 20 μg/ml MuA, 12 μg/ml MuB, and 7.6 μg/ml HU, 24 μg/ml ClpX protein, and 40 U/ml MRFα-DF; all other components were identical to the standard strand transfer reaction mixture (North & Nakai, 2005), which includes 2 mM ATP and an ATP-regenerating system. The reaction mixture was incubated at 37°C for 30 min. The resulting complex (STC-DF) was purified from unbound proteins on a 1 ml Bio-Gel A-15m column equilibrated in 25 mM Hepes (K+), pH 7.5, 12 mM magnesium acetate, 5 mM dithiothreitol (DTT), 50 μg/ml BSA, and 50 mM potassium chloride at room temperature. The DNA-containing fractions in the void volume (three peak fractions of 40 μl each) were pooled. For the second stage, a 5 μl mixture that includes purified S-IF2-2 (50 ng) and GDP, GTP, or ppGpp (for a final concentration of 1 mM in the final binding reaction mixture) was assembled in reaction buffer on ice (such mixtures were on ice for a uniform 5 min before the next step). It was then added to 45 μl of the isolated STC-DF. The mixture was incubated at 37°C for 30 min, and the resulting complexes were purified from unbound proteins on a 1 ml Bio-Gel A-15m column equilibrated in 25 mM Hepes(K+), pH 7.5, 12 mM magnesium acetate, 5 mM DTT, 50 mM potassium chloride at room temperature. The DNA-containing fractions in the void volume (three peak fractions of 40 μl each) were pooled and resolved on a 12.5% SDS-polyacrylamide gel for Western blot analysis, using the S-tag HRP LumiBlot kit (Novagen) as previously described (North et al., 2007).
Other methods
Size ranges of bacterial cells grown in LB and Mu plating efficiency on various strains were measured as previously described (Madison et al., 2012). All plating efficiency is expressed relative to the number of plaques obtained on a lawn of GTN932 (arbitrarily set to 100%).
All results are the average of at least 3 determinations with the error expressed as ± SD unless otherwise indicated. Viability of cells during growth in MMS and NFZ were measured by plating on LB plates containing 6 mM MMS and 10 μM NFZ, respectively. MMS plates were used within 6 hours of preparation and NFZ plates within 24 hours. UV irradiation at 254 nm and all other procedures are as previously described (Madison et al., 2012).
Supplementary Material
Acknowledgments
We thank Mona R. Abdelmeguid for assistance in strain constructions and Elliott Crooke for critically reading the paper. This investigation was supported by NIH grant R01 GM49649 (to H.N.).
References
- Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37:3475–3492. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharoglu Z, Lestini R, Duigou S, Michel B. RNA polymerase mutations that facilitate replication progression in the rep uvrD recF mutant lacking two accessory replicative helicases. Mol Microbiol. 2010;77:324–336. doi: 10.1111/j.1365-2958.2010.07208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubakri H, de Septenville AL, Viguera E, Michel B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 2010;29:145–157. doi: 10.1038/emboj.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas T, Laalami S, Richarme G. Chaperone properties of bacterial elongation factor EF-G and initiation factor IF2. J Biol Chem. 2000;275:855–860. doi: 10.1074/jbc.275.2.855. [DOI] [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VJ, Vinella D. The stringent response. In: Neidhardt FC, editor. Escherichia coli and Salmonella: cellular and molecular biology. Washington, DC: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- Cenatiempo Y, Deville F, Dondon J, Grunberg-Manago M, Sacerdot C, Hershey JW, Hansen HF, Petersen HU, Clark BF, Kjeldgaard M, et al. The protein synthesis initiation factor 2 G-domain. Study of a functionally active C-terminal 65-kilodalton fragment of IF2 from Escherichia coli. Biochemistry. 1987;26:5070–5076. doi: 10.1021/bi00390a028. [DOI] [PubMed] [Google Scholar]
- Chaconas G, Harshey RM. Transposition of phage Mu DNA. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, D.C: ASM Press; 2002. pp. 384–402. [Google Scholar]
- Cohen SE, Lewis CA, Mooney RA, Kohanski MA, Collins JJ, Landick R, Walker GC. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc Natl Acad Sci USA. 2010;107:15517–15522. doi: 10.1073/pnas.1005203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Olsson CL, Hershey JW, Grunberg-Manago M, Nomura M. Feedback regulation of rRNA synthesis in Escherichia coli. Requirement for initiation factor IF2. J Mol Biol. 1987;198:383–392. doi: 10.1016/0022-2836(87)90288-9. [DOI] [PubMed] [Google Scholar]
- De Septenville AL, Duigou S, Boubakri H, Michel B. Replication fork reversal after replication-transcription collision. PLoS Genet. 2012;8:e1002622. doi: 10.1371/journal.pgen.1002622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferullo DJ, Lovett ST. The stringent response and cell cycle arrest in Escherichia coli. PLoS Genet. 2008;4:e1000300. doi: 10.1371/journal.pgen.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores MJ, Ehrlich SD, Michel B. Primosome assembly requirement for replication restart in the Escherichia coli holDG10 replication mutant. Mol Microbiol. 2002;44:783–792. doi: 10.1046/j.1365-2958.2002.02913.x. [DOI] [PubMed] [Google Scholar]
- Giuliodori AM, Brandi A, Gualerzi CO, Pon CL. Preferential translation of cold-shock mRNAs during cold adaptation. RNA. 2004;10:265–276. doi: 10.1261/rna.5164904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Halpern E, Trisler P. Role of sulA and sulB in filamentation by lon mutants of Escherichia coli K-12. J Bacteriol. 1981;148:265–273. doi: 10.1128/jb.148.1.265-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grompone G, Ehrlich SD, Michel B. Replication restart in gyrB Escherichia coli mutants. Mol Microbiol. 2003;48:845–854. doi: 10.1046/j.1365-2958.2003.03480.x. [DOI] [PubMed] [Google Scholar]
- Guy CP, Atkinson J, Gupta MK, Mahdi AA, Gwynn EJ, Rudolph CJ, Moon PB, van Knippenberg IC, Cadman CJ, Dillingham MS, Lloyd RG, McGlynn P. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol Cell. 2009;36:654–666. doi: 10.1016/j.molcel.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Marians KJ. The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Mol Cell. 2005a;17:733–743. doi: 10.1016/j.molcel.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ. Unwinding of the Nascent Lagging Strand by Rep and PriA Enables the Direct Restart of Stalled Replication Forks. J Biol Chem. 2005b;280:34143–34151. doi: 10.1074/jbc.M507224200. [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006a;439:557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006b;7:932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- Howe JG, Hershey JW. Initiation factor and ribosome levels are coordinately controlled in Escherichia coli growing at different rates. J Biol Chem. 1983;258:1954–1959. [PubMed] [Google Scholar]
- Hubert M, Nyengaard NR, Shazand K, Mortensen KK, Lassen SF, Grunberg-Manago M, Sperling-Petersen HU. Tandem translation of Bacillus subtilis initiation factor IF2 in E. coli. FEBS Lett. 1992;312:132–138. doi: 10.1016/0014-5793(92)80920-c. [DOI] [PubMed] [Google Scholar]
- Huisman O, D’Ari R, Gottesman S. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc Natl Acad Sci USA. 1984;81:4490–4494. doi: 10.1073/pnas.81.14.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiani C, Langston LD, Yurieva O, Goodman MF, O’Donnell M. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc Natl Acad Sci USA. 2009;106:6031–6038. doi: 10.1073/pnas.0901403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiani C, Patel M, Goodman MF, O’Donnell ME. RecA acts as a switch to regulate polymerase occupancy in a moving replication fork. Proc Natl Acad Sci USA. 2013;110:5410–5415. doi: 10.1073/pnas.1303301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JM, Nakai H. The phiX174-type primosome promotes replisome assembly at the site of recombination in bacteriophage Mu transposition. EMBO J. 1997;16:6886–6895. doi: 10.1093/emboj/16.22.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JM, Nakai H. Duplex opening by primosome protein PriA for replisome assembly on a recombination intermediate. J Mol Biol. 1999;289:503–515. doi: 10.1006/jmbi.1999.2783. [DOI] [PubMed] [Google Scholar]
- Kanjee U, Ogata K, Houry WA. Direct binding targets of the stringent response alarmone (p)ppGpp. Mol Microbiol. 2012;85:1029–1043. doi: 10.1111/j.1365-2958.2012.08177.x. [DOI] [PubMed] [Google Scholar]
- Laursen BS, de ASSA, Hedegaard J, Moreno JM, Mortensen KK, Sperling-Petersen HU. Structural requirements of the mRNA for intracistronic translation initiation of the enterobacterial infB gene. Genes Cells. 2002;7:901–910. doi: 10.1046/j.1365-2443.2002.00571.x. [DOI] [PubMed] [Google Scholar]
- Lee EH, Kornberg A. Replication deficiencies in priA mutants of Escherichia coli lacking the primosomal replication n’ protein. Proc Natl Acad Sci USA. 1991;88:3029–3032. doi: 10.1073/pnas.88.8.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Autret S, Seror SJ. A checkpoint involving RTP, the replication terminator protein, arrests replication downstream of the origin during the Stringent Response in Bacillus subtilis. Mol Microbiol. 1995;15:287–295. doi: 10.1111/j.1365-2958.1995.tb02243.x. [DOI] [PubMed] [Google Scholar]
- Levine A, Vannier F, Dehbi M, Henckes G, Seror SJ. The stringent response blocks DNA replication outside the ori region in Bacillus subtilis and at the origin in Escherichia coli. J Mol Biol. 1991;219:605–613. doi: 10.1016/0022-2836(91)90657-r. [DOI] [PubMed] [Google Scholar]
- Liu J, Nurse P, Marians KJ. The ordered assembly of the phiX174-type primosome. III. PriB facilitates complex formation between PriA and DnaT. J Biol Chem. 1996;271:15656–15661. doi: 10.1074/jbc.271.26.15656. [DOI] [PubMed] [Google Scholar]
- Madison KE, Abdelmeguid MR, Jones-Foster EN, Nakai H. A new role for translation initiation factor 2 in maintaining genome integrity. PLoS Genet. 2012;8:e1002648. doi: 10.1371/journal.pgen.1002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdi AA, Briggs GS, Lloyd RG. Modulation of DNA damage tolerance in Escherichia coli recG and ruv strains by mutations affecting PriB, the ribosome and RNA polymerase. Mol Microbiol. 2012;86:675–691. doi: 10.1111/mmi.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool JD, Ford CC, Sandler SJ. A dnaT mutant with phenotypes similar to those of a priA2::kan mutant in Escherichia coli K-12. Genetics. 2004;167:569–578. doi: 10.1534/genetics.103.025296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P, Al-Deib AA, Liu J, Marians KJ, Lloyd RG. The DNA replication protein PriA and the recombination protein RecG bind D-loops. J Mol Biol. 1997;270:212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 2013;41:6175–6189. doi: 10.1093/nar/gkt302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrikh H, Zhang Y, Grossman AD, Wang JD. Replication-transcription conflicts in bacteria. Nature reviews Microbiology. 2012;10:449–458. doi: 10.1038/nrmicro2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B, Grompone G, Florès MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc Natl Acad Sci USA. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milon P, Tischenko E, Tomsic J, Caserta E, Folkers G, La Teana A, Rodnina MV, Pon CL, Boelens R, Gualerzi CO. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc Natl Acad Sci USA. 2006;103:13962–13967. doi: 10.1073/pnas.0606384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy H, Cashel M. Isolation of RNA polymerase suppressors of a (p)ppGpp deficiency. Methods Enzymol. 2003;371:596–601. doi: 10.1016/S0076-6879(03)71044-1. [DOI] [PubMed] [Google Scholar]
- North SH, Kirtland SE, Nakai H. Translation factor IF2 at the interface of transposition and replication by the PriA-PriC pathway. Mol Microbiol. 2007;66:1566–1578. doi: 10.1111/j.1365-2958.2007.06022.x. [DOI] [PubMed] [Google Scholar]
- North SH, Nakai H. Host factors that promote transpososome disassembly and the PriA-PriC pathway for restart primosome assembly. Mol Microbiol. 2005;56:1601–1616. doi: 10.1111/j.1365-2958.2005.04639.x. [DOI] [PubMed] [Google Scholar]
- Nurse P, Zavitz KH, Marians KJ. Inactivation of the Escherichia coli PriA DNA replication protein induces the SOS response. J Bacteriol. 1991;173:6686–6693. doi: 10.1128/jb.173.21.6686-6693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyengaard NR, Mortensen KK, Lassen SF, Hershey JW, Sperling-Petersen HU. Tandem translation of E. coli initiation factor IF2 beta: purification and characterization in vitro of two active forms. Biochem Biophys Res Commun. 1991;181:1572–1579. doi: 10.1016/0006-291x(91)92118-4. [DOI] [PubMed] [Google Scholar]
- Nystrom T. Growth versus maintenance: a trade-off dictated by RNA polymerase availability and sigma factor competition? Mol Microbiol. 2004;54:855–862. doi: 10.1111/j.1365-2958.2004.04342.x. [DOI] [PubMed] [Google Scholar]
- Ona KR, Courcelle CT, Courcelle J. Nucleotide excision repair is a predominant mechanism for processing nitrofurazone-induced DNA damage in Escherichia coli. J Bacteriol. 2009;191:4959–4965. doi: 10.1128/JB.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Sacerdot C, Vachon G, Laalami S, Morel-Deville F, Cenatiempo Y, Grunberg-Manago M. Both forms of translational initiation factor IF2 (alpha and beta) are required for maximal growth of Escherichia coli. Evidence for two translational initiation codons for IF2 beta. J Mol Biol. 1992;225:67–80. doi: 10.1016/0022-2836(92)91026-l. [DOI] [PubMed] [Google Scholar]
- Sandler SJ. Multiple genetic pathways for restarting DNA replication forks in Escherichia coli K-12. Genetics. 2000;155:487–497. doi: 10.1093/genetics/155.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler SJ, Marians KJ, Zavitz KH, Coutu J, Parent MA, Clark AJ. DnaC mutations suppress defects in DNA replication and recombination functions in priB and priC double mutants in E. coli K-12. Mol Microbiol. 1999;34:91–101. doi: 10.1046/j.1365-2958.1999.01576.x. [DOI] [PubMed] [Google Scholar]
- Sandler SJ, McCool JD, Do TT, Johansen RU. PriA mutations that affect PriA-PriC function during replication restart. Mol Microbiol. 2001;41:697–704. doi: 10.1046/j.1365-2958.2001.02547.x. [DOI] [PubMed] [Google Scholar]
- Sarubbi E, Rudd KE, Cashel M. Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol Gen Genet. 1988;213:214–222. doi: 10.1007/BF00339584. [DOI] [PubMed] [Google Scholar]
- Sassanfar M, Roberts JW. Nature of the SOS-inducing signal in Escherichia coli: The involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- Schreiber G, Ron EZ, Glaser G. ppGpp-mediated regulation of DNA replication and cell division in Escherichia coli. Curr Microbiol. 1995;30:27–32. doi: 10.1007/BF00294520. [DOI] [PubMed] [Google Scholar]
- Selby CPC, Sancar AA. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- Sikora A, Mielecki D, Chojnacka A, Nieminuszczy J, Wrzesinski M, Grzesiuk E. Lethal and mutagenic properties of MMS-generated DNA lesions in Escherichia coli cells deficient in BER and AlkB-directed DNA repair. Mutagenesis. 2010;25:139–147. doi: 10.1093/mutage/gep052. [DOI] [PubMed] [Google Scholar]
- Slominska M, Neubauer P, Wegrzyn G. Regulation of bacteriophage lambda development by guanosine 5′-diphosphate-3′-diphosphate. Virology. 1999;262:431–441. doi: 10.1006/viro.1999.9907. [DOI] [PubMed] [Google Scholar]
- Stewart EJ, Katzen F, Beckwith J. Six conserved cysteines of the membrane protein DsbD are required for the transfer of electrons from the cytoplasm to the periplasm of Escherichia coli. EMBO J. 1999;18:5963–5971. doi: 10.1093/emboj/18.21.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehranchi AK, Blankschien MD, Zhang Y, Halliday JA, Srivatsan A, Peng J, Herman C, Wang JD. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell. 2010;141:595–605. doi: 10.1016/j.cell.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell. 2005;19:247–258. doi: 10.1016/j.molcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Travers AA, Debenham PG, Pongs O. Translation initiation factor 2 alters transcriptional selectivity of Escherichia coli ribonucleic acid polymerase. Biochemistry. 1980;19:1651–1656. doi: 10.1021/bi00549a020. [DOI] [PubMed] [Google Scholar]
- Uzan M, Danchin A. A rapid test for the relA mutation in E. coli. Biochem Biophys Res Commun. 1976;69:751–758. doi: 10.1016/0006-291x(76)90939-6. [DOI] [PubMed] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- Yeeles JT, Marians KJ. The Escherichia coli replisome is inherently DNA damage tolerant. Science. 2011;334:235–238. doi: 10.1126/science.1209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harbor perspectives in biology. 2013;5:a012815. doi: 10.1101/cshperspect.a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavitz KH, Marians KJ. ATPase-deficient mutants of the Escherichia coli DNA replication protein PriA are capable of catalyzing the assembly of active primosomes. J Biol Chem. 1992;267:6933–6940. [PubMed] [Google Scholar]
- Zyskind JW, Smith DW. DNA replication, the bacterial cell cycle, and cell growth. Cell. 1992;69:5–8. doi: 10.1016/0092-8674(92)90112-p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.