Abstract
A total of 310 Salmonella isolates were isolated from 6 broiler farms in Eastern China, serotyped according to the Kauffmann-White classification. All isolates were examined for susceptibility to 17 commonly used antimicrobial agents, representative isolates were examined for resistance genes and class I integrons using PCR technology. Clonality was determined by pulsed-field gel electrophoresis (PFGE). There were two serotypes detected in the 310 Salmonella strains, which included 133 Salmonella enterica serovar Indiana isolates and 177 Salmonella enterica serovar Enteritidis isolates. Antimicrobial sensitivity results showed that the isolates were generally resistant to sulfamethoxazole, ampicillin, tetracycline, doxycycline and trimethoprim, and 95% of the isolates sensitive to amikacin and polymyxin. Among all Salmonella enterica serovar Indiana isolates, 108 (81.2%) possessed the bla TEM, floR, tetA, strA and aac (6')-Ib-cr resistance genes. The detected carriage rate of class 1 integrons was 66.5% (206/310), with 6 strains carrying gene integron cassette dfr17-aadA5. The increasing frequency of multidrug resistance rate in Salmonella was associated with increasing prevalence of int1 genes (rs = 0.938, P = 0.00039). The int1, bla TEM, floR, tetA, strA and aac (6')-Ib-cr positive Salmonella enterica serovar Indiana isolates showed five major patterns as determined by PFGE. Most isolates exhibited the common PFGE patterns found from the chicken farms, suggesting that many multidrug-resistant isolates of Salmonella enterica serovar Indiana prevailed in these sources. Some isolates with similar antimicrobial resistance patterns represented a variety of Salmonella enterica serovar Indiana genotypes, and were derived from a different clone.
Introduction
Salmonella is a leading cause of microbial food poisoning cases and salmonellosis is one of the most important bacterial diseases in chicken farms, leading to large numbers of chicken death and significant loss to the poultry industry [1]. The widespread use of antimicrobial agents in food-animal production has contributed to the decreased susceptibility of Salmonella to antibiotics, which can be transmitted to humans through food of animal origin [2]. In recent years, the rapid emergence of multidrug resistance bacteria caused by the wide application of antibiotics in clinical practices has caused great difficulties in medical treatment [3]. The integron gene cassette system is one of the main reasons for the rapid development of multidrug resistance in Salmonella [4]. The same gene cassette found in different genotype strains indicates that it can spread in clinical strains through horizontal transfer. Moreover, integrons can propagate vertically [5]–[6]. The existence and flexible transmission of integrons was proven suitable for the spread of drug resistant genes and the acceleration of multidrug resistance [7]. In the current study, the drug sensitivity of Salmonella strains isolated in chicken farms from Eastern China in 2009 was tested, and the prevalent class 1 integrons and integron-carrying gene cassette in connection with resistance in the strains was studied. We assessed the relationship between multidrug resistance of Salmonella and class 1 integrons by statistical analyses.
Materials and Methods
Salmonella isolation, identification and serotyping
A total of 1024 strains were collected in 2008–2009 from 6 broiler farms in 3 regions of Eastern China. Fecal samples were taken in cloaca with disposable sterile swabs from two chicken farms in Lin Yi (280 samples), one chicken farm in Zou Ping (190 samples) and three chicken farms in Yan Tai (554 samples). Dr. Yuqing Liu (Institute of Animal Science and Veterinary Medicine Shandong Academy of Agricultural Sciences, China) responsible for contact takes the strains. The strains were collected according to acquisition guidelines. The field studies did not involve endangered or protected species.
After the samples incubated in sterile Selenite Cystine Broth (SC) at 37°C for 24 h, all of the samples incubated were grown on chromogenic medium for Salmonella (CHROM agar, Paris, France) at 37°C for 24–48 h. Only one colony per plate was picked for further study, and purple-colored colonies on the culture plates were regarded as presumptive Salmonella colonies. Suspected colonies were isolated and grown on nutrient agar (Dibco), and then identified by transferring to tubes with triple sugar iron agar, indole-lysin motility semisolid agar, Voges Proskauer semisolid media, urease test broth, and Simmons citrate agar. The isolates examined with these media were simultaneously investigated for presence of the invA gene with the polymerase chain reaction (PCR).
Salmonella isolates were serotyped based on slide agglutination for O and H antigens according to Kauffmann-White [8], and using the same antisera produced at the National Institute of Biological Sciences in Beijing.
Antimicrobial sensitivity
Minimum inhibitory concentrations (MICs) of the Salmonella isolates were determined by broth micro dilution according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [9]–[10]. Escherichia coli ATCC 25922 was used as the quality control strain. Seventeen antimicrobials were tested (MIC break point Sensitivity/Resistance µg/mL): namely, ampicillin (≤8/≥32), amoxicillin-clavulanic acid (≤4/≥16), cefazolin (≤8/≥32), ceftiofur (≤2/≥8), tetracycline (≤4/≥16), doxycycline (≤4/≥16), chloramphenicol (≤8/≥32), florfenicol (≤4/≥16), kanamycin (≤16/≥64), gentamicin (≤4/≥16), sulfisoxazole (≤256/≥512), trimethoprim (≤38/≥76), enrofloxacin (≤0.25/≥2), norfloxacin (≤4/≥16), ciprofloxacin (≤0.06/≥2), amikacin (≤16/≥64) and polymyxin (≤2/≥4).
PCR amplification and DNA sequencing of resistance genes and integrase genes
According to the MIC data, multidrug-resistant genes of 133 Salmonella enterica serovar Indiana and 177 Salmonella enterica serovar Enteritidis were analyzed by PCR. Antimicrobial resistance genes were detected for ampicillin-resistant isolates (bla TEM and bla PSE); chloramphenicol/florfenicol-resistant isolates (catA1, catA2, catA3, cmlA and floR); tetracycline-resistant isolates (tetA, tetB, tetC, tetD and tetG), streptomycin-resistant isolates (aadA, strA and strB), fluoroquinolone-resistant isolates (qnrA, qnrB, qnrS, aac(6′)-Ib-cr and qepA), and integrase genes (int1, int2 and int3) [11]. The DNA sequences obtained were compared with those in GenBank using the BLAST program.
Pulsed field gel electrophoresis (PFGE)
Chromosomal DNA of 104 Salmonella enterica serovar Indiana isolates carrying the int1, bla TEM, floR, tetA, strA, and aac(6’)-Ib-cr genes were digested with the restriction enzyme XbaI and then subjected to PFGE analysis according to the Pulse Net Standardized Laboratory Protocol (U.S. Centers for Disease Control and Prevention, Atlanta, GA) using the CHEF MAPPER System (Bio-Rad Laboratories, Hercules, CA). The gels were run at 6.0 V/cm with an initial and final switch time of 2.16 s and 54.17 s at an angle of 120 degrees and 14°C for 18 h. The Salmonellaser. Braenderup H9812 standard served as size markers. Cluster analysis of pulsotypes was carried out using Dice's coefficient in UPGMA with InfoQuest FP Software/Version 4.5 (Bio-Rad). [12]
Statistical analysis
The relation between multidrug resistance rate and prevalence of int1 genes was assessed by calculating simple linear regression and the corresponding P value. All statistical analyses were done using SPSS software (SPSS, Chicago, IL).
Results
Strain identification and serotyping
A total of two serotypes were identified among 310 strains of Salmonella, accounting for 133 strains of Salmonella enterica serovar Indiana and 177 strains of Salmonella enterica serovar Enteritidis. No other serotypes were identified in all Salmonella isolates.
Antimicrobial sensitivity and multidrug resistance
As shown in Table 1, the resistance rates of the Salmonella enterica serovar Indiana isolates were 100% to sulfamethoxazole; above 90% to ampicillin, florfenicol, tetracycline, doxycycline, kanamycin, gentamicin and trimethoprim; above 80% to amoxicillin/clavulanic acid, cefazolin, ceftiofurand chloramphenicol; over 60% to enrofloxacin, norfloxacin and ciprofloxacin; and about 5% to amikacin and polymyxin E. Antimicrobial sensitivity showed that the Salmonella enterica serovar Indiana strains were multidrug resistant and only sensitive to amikacin and polymyxin. The isolates of Salmonella enterica serovar Enteritidis were resistant to ampicillin, tetracycline, doxycycline and trimethoprim above 60%, and were sensitive to all other tested drugs.
Table 1. Number and antimicrobial resistance profiles of resistant Salmonella strains within each serogroup.
| Sources (no. of isolates) | Serovar (no. of isolates) | Percentage of resistance (no. of isolates) | ||||||||||||||||
| AMP | AMC | CEF | XNL | CHL | FFN | TET | DOX | KAN | GEN | AMI | SUL | TMP | ENR | NOR | CIP | POL | ||
| Lin Yi (119) | Indiana (5) | 100 (5) | 100 (5) | 60 (3) | 60 (3) | 100 (5) | 60 (3) | 100 (5) | 100 (5) | 60 (3) | 60 (3) | 0 | 100 (5) | 100 (5) | 60 (3) | 80 (4) | 40 (2) | 0 |
| Enteritidis (114) | 78.1 (89) | 42.1 (48) | 5.3 (6) | 6.1 (7) | 5.3 (6) | 0.9 (1) | 73.7 (84) | 73.7 (84) | 9.6 (11) | 1.8 (2) | 0 | 100(114) | 74.6 (85) | 0 | 2.7 (3) | 0.9 (1) | 0.9 (1) | |
| Zou Ping (51) | Indiana (28) | 96.4 (27) | 82.1 (23) | 92.9 (26) | 85.7 (24) | 75 (21) | 96.4 (27) | 89.3 (25) | 92.9 (26) | 82.1 (23) | 85.7 (24) | 17.9(5) | 100 (28) | 96.4 (27) | 53.6(15) | 71.4 (20) | 53.6(15) | 7.1 (2) |
| Enteritidis (23) | 69.6 (16) | 17.4 (4) | 17.4 (4) | 13.0 (3) | 4.3 (1) | 0 | 52.2 (12) | 43.5 (10) | 4.3 (1) | 4.3(1) | 4.3(1) | 100 (23) | 60.9 (14) | 0 | 4.3(1) | 13.0 (3) | 0 | |
| Yan Tai (140) | Indiana (100) | 98 (98) | 89 (89) | 88 (88) | 87 (87) | 87 (87) | 91 (91) | 100 (100) | 100 (100) | 94 (94) | 96 (96) | 1 (1) | 100(100) | 100 (100) | 69 (69) | 81 (81) | 62 (62) | 5 (5) |
| Enteritidis (40) | 62.5 (25) | 17.5 (7) | 5 (2) | 5 (2) | 0 | 0 | 55 (22) | 47.5 (19) | 2.5(1) | 12.5(5) | 2.5(1) | 100 (40) | 30 (12) | 0 | 2.5 (1) | 2.5 (1) | 12.5(5) | |
| Total (310) | Indiana (133) | 97.7 (130) | 887.9 (117) | 87.9 (117) | 85.7(114) | 84.9(113) | 90.9(121) | 97.7(130) | 98.5(131) | 90.2(120) | 92.5(123) | 4.5(6) | 100(133) | 99.2(132) | 65.4(87) | 78.9(105) | 59.4(79) | 5.3 (7) |
| Enteritidis (177) | 73 (130) | 33.3 (59) | 6.8 (12) | 6.8 (12) | 3.9 (7) | 0.6 (1) | 66.7(118) | 63.8(113) | 7.3(13) | 4.5(8) | 1.1(2) | 100(177) | 62.7(111) | 0 | 2.8(5) | 2.8 (5) | 3.4 (6) | |
AMP, ampicillin; AMC, amoxicillin/clavulanic acid; CEF, cefalotin; XNL, ceftiofur; CHL, chloramphenicol; FFN, florfenicol; TET, tetracycline; DOX, doxycycline; KAN, kanamycin; GEN, gentamicin; AMI, amikacin; SUL, sulfamethoxazole; TMP, trimethoprim; ENR, enrofloxacin; NOR, norfloxacin; CIP, ciprofloxacin; POL, polymyxin.
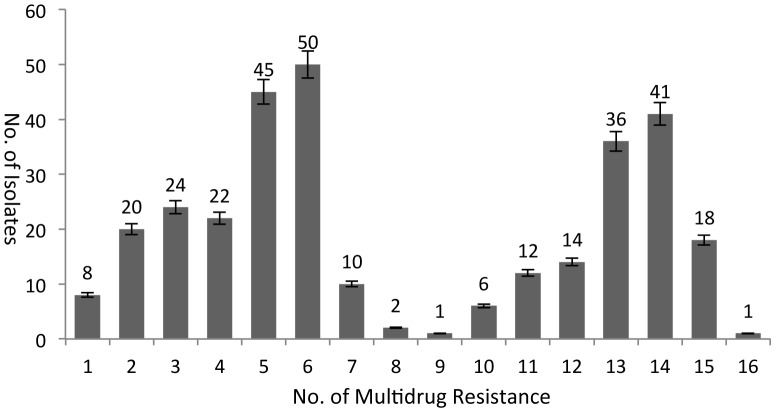
The 310 strains of Salmonella were strongly resistant to 17 antibiotics generally used in clinical practice (Figure 1). The most common Salmonella strains were resistant to 5 and 6 drugs of 44 and 50 strains, respectively, accounting for 30.6% of the total, with most represented by Salmonella enterica serovar Enteritidis. The next most common was Salmonella with 13 and 14 drug resistance of 36 and 41 strains, respectively, accounting for 24.8% of the total, with most represented by Salmonella enterica serovar Indiana, whose drug resistance of the two serotypes showed double peaks. A total of 74 (23.8%) Salmonella strains were below 4 drug resistance, 45 (14.5%) Salmonella strains had 7 to 12 drug resistance, 18 (5.8%) Salmonella strains had 15 drug resistance, and one Salmonella strain had 16 drug resistance. Salmonella with 15 and 16 drug resistances were Salmonella enterica serovar Indiana. No strain of Salmonella showed zero drug resistance. The above results showed that Salmonella with above 10 drug resistance were mainly Salmonella enterica serovar Indiana. A total of 282 (90.9%) Salmonella strains were resistant to more than 3 kind of drug, demonstrating its multidrug resistance.
Figure 1. No. of multidrug resistance to 17 drugs in 310 Salmonella isolates.
The 310 strains of Salmonella were strongly resistant to 17 antibiotics generally used in clinical practice. The most common Salmonella strains were resistant to 5 and 6 drugs of 44 and 50 strains. The next most common was Salmonella with 13 and 14 drug resistance of 36 and 41 strains.
PCR amplification and DNA sequencing resistance genes
As shown in Table 2, bla TEM, catA1, floR, tetA, strA and aac(6′)-Ib-cr were the most prevalent resistance genes and were present in Salmonella enterica serovar Indiana isolates from all chicken farm sources. The bla TEM gene was prevalent in Salmonella enterica serovar Indiana (78.2%) and Salmonella enterica serovar Enteritidis (62.4%) isolates from the three sources. The floR, tetA and strA genes were highly prevalent in Salmonella enterica serovar Indiana isolates, and were detected in 92% of the isolates from the three chicken farms. The aac (6′)-Ib-cr gene was highly prevalent in Salmonella enterica serovar Indiana isolates, and was present in 95 (95%) of the 100 isolates from Yan Tai chicken farm, 25 (89.3%) of the 28 isolates from ZouPing chicken farm, and 3 (60%) of the 5 isolates from Lin Yi chicken farm. Among all Salmonella enterica serovar Indiana isolates, 108 (81.2%) possessed the bla TEM, floR, tetA, strA and aac(6')-Ib-cr resistance genes, which were the most frequently observed combination in this study.
Table 2. Distribution of antimicrobial resistance genes and integrase genes among Salmonella.
| Sourcce (no. of isolates) | Serovar (no. of isolates) | Percentage (no.) of isolates carrying resistance gens and integrase gens | ||||||||
| bla TEM | catA1 | floR | clmA | tetA | strA | aac(6′)-Ib-cr | int1 | Multidrug resistance rate (no. of isolates) | ||
| Lin Yi (119) | Indiana (5) | 80 (4) | 40 (2) | 60 (3) | 0 | 60(3) | 60(3) | 60(3) | 80 (4) | 100 (5) |
| Enteritidis (114) | 69.6 (80) | 4.3 (5) | 5.2 (6) | 1.7(2) | 3.5(4) | 2.6(3) | 0 | 57 (65) | 88.6 (101) | |
| Zou Ping (51) | Indiana (28) | 89.3 (25) | 89.3 (25) | 100 (28) | 0 | 96.4(27) | 96.4(27) | 89.3(25) | 96.4 (27) | 100 (28) |
| Enteritidis (23) | 56.5 (13) | 8.7 (2) | 4.4 (1) | 0 | 0 | 0 | 0 | 43.5 (10) | 82.6 (19) | |
| Yan Tai (140) | Indiana (100) | 75 (75) | 77 (77) | 97(97) | 7(7) | 94(94) | 93(93) | 95(95) | 85 (85) | 96 (96) |
| Enteritidis (40) | 45 (18) | 0 | 0 | 2.5(1) | 0 | 0 | 0 | 37.5 (15) | 82.5 (33) | |
| Total (310) | Indiana (133) | 78.2 (104) | 78.2 (104) | 96.2(128) | 5.3(7) | 93.2(124) | 92.5(123) | 92.5(123) | 87.2 (116) | 97 (129) |
| Enteritidis (177) | 62.4 (111) | 3.9 (7) | 3.9(7) | 1.7(3) | 2.2(4) | 1.7(3) | 0 | 50.8 (90) | 86.4 (153) | |
Prevalence of class I integrons
Among the 310 Salmonella strains, 206 were amplified to int1 fragments of about 856 bp with a positive rate of 66.5%. The int1 positive rate was 87.2% (116) for the 133 Salmonella enterica serovar Indiana isolates and 50.8% (90) for the 177 Salmonella enterica serovar Enteritidis isolates. The increasing frequency of multidrug resistance rate in Salmonella was associated with increasing prevalence of int1 genes (r2 = 0.938, P = 0.00039). One 1515 bp amplicon was obtained from the variable region of the class 1 integrons by PCR. Sequencing and nucleotide sequence analysis of gene cassettes deposited in GenBank showed that this amplicon contained dfr17 and aadA5 with a detection rate of 2.91% (6/206). The drug resistant gene cassette dfr17-aadA5 was correlated with drug resistance to trimethoprim and aminoglycosides, respectively.
Pulsed-field gel electrophoresis
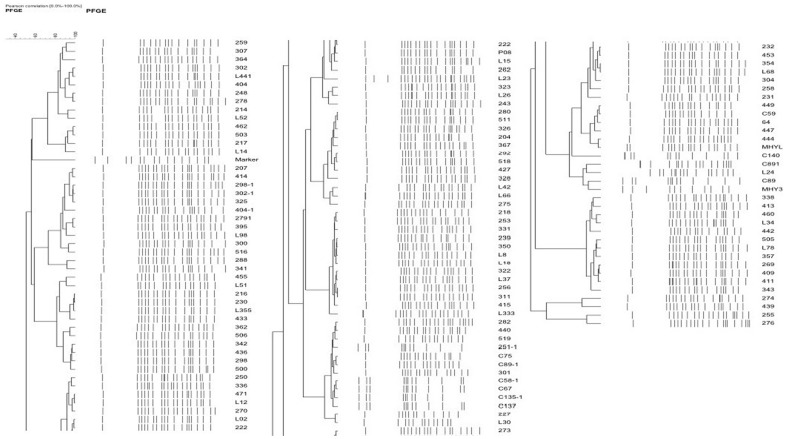
The int1, bla TEM, floR, tetA, strA and aac (6') -Ib-cr positive Salmonella enterica serovar Indiana isolates showed five major patterns as determined by PFGE (Figure 2). Most isolates had showed the common PFGE patterns in all 6 chicken farms, suggesting that many multidrug resistant Salmonella enterica serovar Indiana isolates prevailed in the three sources. Some were not derived from a specific clone, but represented a variety of different genotypes.
Figure 2. PFGE pattern of 104 Salmonella enterica serovars Indiana.
Chromosomal DNA of 104 Salmonella enterica serovar Indiana isolates carrying the int1, bla TEM, floR, tetA, strA, and aac(6′)-Ib-cr genes were digested with the restriction enzyme XbaI and then subjected to PFGE analysis. The results showed five major patterns as determined by PFGE.
Discussion
Human and animal Salmonella involves numerous serotypes, which exhibit certain correlation in distribution, host, separation time, and source. For example, research in Ireland from 1998 to 2003 showed that Salmonella enterica serovar Typhimurium was mainly isolated from cattle and swine, with the isolation rate decreasing year by year, and Salmonella enterica serovar Kentucky was mainly isolated from poultry [13]. A variety of serotypes were found in 137 strains of isolated food borne Salmonella in provinces and cities of China [14], in which the seven serotypes of Salmonella enterica serovar Derby, Salmonella enterica serovar Agona, Salmonella enterica serovar Enteritidis, Salmonella enterica serovar Reading, Salmonella enterica serovar Anatis, Salmonella enterica serovar Chester and Salmonella enterica serovar Typhimurium accounted for 80% of the isolates.
In this study, the multidrug resistance of 310 Salmonella strains to 17 antibiotics commonly used in clinical practice was examined. The investigation on drug resistance of 51 strains of human Salmonella enterica serovar Typhimurium conducted by Biendo [15] showed that multidrug resistance of the strains was 98%, with more than 90% of isolates resistant to sulfonamides, ampicillin, streptomycin and tetracycline, and sensitive to amikacin and cephalosporins. Analysis of the drug resistance of animal and human Salmonella enterica serovar Typhimurium isolated by Graziani [16] showed that 64% of strains were resistant to more than four drugs, the frequently seen drug resistance spectrum was ACSSuT type, and most strains were resistant to sulfamethoxazole. Recently, the emergence of MDR Salmonella enterica serovar Typhimurium, Salmonella enterica serovar Paratyphi and Salmonella enterica serovar Agona suggests that this multidrug-resistant phenotype may emerge in other Salmonella enterica serovar Enteritidis serotypes [17]–[18]. The existence of multidrug-resistant geneson Salmonella enterica serovar Indiana was not found. While the Salmonella enterica serovar Indiana isolated in this study was not only wide spread in the farms, but also showed characteristics of multidrug resistance. These strains had high resistance not only to streptomycin and tetracycline, but also to chloramphenicol, fluoroquinolones and cephalosporins.
Integrons are a proposed spreading mechanism of drug resistance and are widely distributed in nature [19]. Integrons have been found in human clinical isolates, avian, livestock, and animal strains, as well as in bacteria in soil and aquatic ecosystems [20]. Integrons are a movable genetic element; they can move the gene cassette by capturing and shearing mode, on the other hand, integrons were found on transposon, plasmid and other movable genetic elements, which enable their transportation and dissemination of resistance genes. Integrons can integrate drug resistance genes of almost all antimicrobial agents such as aminoglycosides, β-lactam, chloramphenicols, sulfonamides, trimethoprim, macrolides and rifampin [21]. In this study, the prevalent rate of class I integrons in Salmonella was 64.9% in the 310 Salmonella strains, which was consistent with previous findings of 59%–75% [22]. The detection rate of the class I integrons was 87.2% in the 133 Salmonella enterica serovar Indiana strains, and 48.3% in the 178 Salmonella enterica serovar Enteritidis strains, with the former slightly higher than and the latter similar to the previous report [23]. The positive rates of class I integrons in the Salmonella enterica serovar Indiana and Salmonella enterica serovar Enteritidis strains with multidrug resistance (above 5-fold resistance) were 92.5% and 92.6%, respectively, demonstrating the close relationship between multidrug resistance of strains and the prevalence of class I integrons, which was consistent with earlier research [24]–[25]. The class I integron-carrying resistance gene cassettes of Salmonella were mainly two gene families encoding trimethoprim and aminoglycosides (dfr17 and aadA5), as observed in other studies [26]–[27].
After the isolation of drug resistant Salmonella from different regions of Eastern China and analysis of the molecular epidemiology of integrons and drug resistance genes, we found that Salmonella was strongly resistant to antibiotics commonly used in clinical practice, and the carrying rate of integrons and drug resistance genes was positively correlated with drug resistance phenotype. This provides a scientific basis for guiding the rational clinical use of antibiotics, and for preventing and controlling the spread of drug-resistant bacteria and drug resistant genes.
PFGE analysis suggested that most pan-resistant isolates were distinctly different based on their macro-restriction patterns, which suggests that the multidrug-resistant Salmonella isolates carrying the int1, bla TEM, floR, tetA, strA and aac(6′)-Ib-cr genes were not derived from a specific clone, but represented a wide variety of genotypes.
In conclusion, 310 Salmonella strains were isolated from several broiler chicken farms in Eastern China, and manifested themselves in two serotypes. A total of 282 (90.9%) Salmonella strains were presenting multidrug resistance. The detected carriage rate of class 1 integrons was 66.5%, with 6 strains carrying gene integron cassette dfr17-aadA5. The increasing frequency of multidrug resistance rate in Salmonella was associated with increasing prevalence of int1 genes.
Acknowledgments
We are grateful to Dr. Yu-Qing Liu (Institute of Animal Science and Veterinary Medicine Shandong Academy of Agricultural Sciences, China) who kindly provided assistance in the isolation of the Salmonella.
Ethical approval: Not required.
Funding Statement
This study was supported by the National Natural Science Foundation of China (31201949), the National Natural Science Foundation of China (31172362); it was also supported by the Scientific Research Improvement Foundation of Beijing University of Agriculture (GJB2012003). The funders had no role in study design,data collection and analysis,decision to publish,or preparation of the manuscript.
References
- 1. Rostagno MH, Wesley IV, Trampel DW (2006) Salmonella Prevalence in Market-Age Turkeys On-Farm and at Slaughter. Poultry Sci 85: 1838–1842. [DOI] [PubMed] [Google Scholar]
- 2. Threlfall EJ (2002) Antimicrobial drug resistance in Salmonella: problems and perspectives in food and water borne infections. FEMS Microbiol 26: 141–148. [DOI] [PubMed] [Google Scholar]
- 3. Shan X, Huang M (2010) Multidrug resistance of bacteria and integron gene cassette system. Med Recapitulate 16: 354–356. [Google Scholar]
- 4. Hu XH, Li GM (2009) Advances in the relationship between integrons and drug resistance of bacteria. World Notes on Antibiotics 30: 255–263. [Google Scholar]
- 5. Boucher Y, Labbate M, Koenig JE (2007) Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol 15: 301–309. [DOI] [PubMed] [Google Scholar]
- 6. Nemergut DR, Martin AP, Schmidt SK (2004) Integron diversity in heavy-metal-contaminated mine tailings and inferences about integron evolution. Appl Environ Microbiol 70: 1160–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen L, Wu CM, Shen JZ (2008) Detection of antimicrobial resistance and class 1 integrons among Salmonella isolates from animals. Chin J Vet Med 44: 6–9. [Google Scholar]
- 8.Popoff MY, Minor L (1992) Antigenic Formulas of the Salmonella Serovars, 6th revision. WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur. [Google Scholar]
- 9.[CLSI] Clinical and Laboratory Standards Institute, 2007. Performance Standards for Antimicrobial Susceptibility Testing: Seventeenth Informational Supplement. CLSI document M100-S17. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- 10.[CLSI] Clinical and Laboratory Standards Institute, 2008. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals; Approved Standard-Third Edition. Informational Supplement. CLSI document M31-A3. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- 11. Yan L, Cong MW, Guo JW (2011) Prevalence of Antimicrobial Resistance among Salmonella Isolates from Chicken in China. Fooodborne Pathog Dis 8: 45–53. [DOI] [PubMed] [Google Scholar]
- 12. Xia LN, Li L, Wu CM (2010) A Survey of Plasmid-Mediated Fluoroquinolone Resistance Genes from Escherichia coli Isolates and Their Dissemination in Shandong, China. Fooodborne Pathog Dis 7: 207–215. [DOI] [PubMed] [Google Scholar]
- 13. O'Hare C, Doran G, Delappe N (2004) Antimicrobial resistance and phage types of human and non-human Salmonella enterica isolates in Ireland, 1998–2003. Commun Dis Public Health 7: 193–199. [PubMed] [Google Scholar]
- 14. Wang MQ, Ran L, Wang ZT (2004) Study on national active monitoring for food borne pathogens and antimicrobial resistance in China 2001. J Hyg Res 33: 49–52. [PubMed] [Google Scholar]
- 15. Biendo M, Laurans G, Thomas D (2005) Molecular characterization and mechanisms of resistance of multidrug-resistant human Salmonella enterica serovar Typhimurium isolated in Amiens (France). Int J Antimicrob Agents 26: 219–229. [DOI] [PubMed] [Google Scholar]
- 16. Graziani G, Busani L, Dionisi AM (2008) Antimicrobial resistance in Salmonella enterica serovar Typhimurium from human and animal sources in Italy. Vet Microbiol 128: 414–418. [DOI] [PubMed] [Google Scholar]
- 17. Boyd DA, Peters GA, Cloeckaert A (2001) Complete nucleotide sequence of a 43-kilobase genomic island associated with the multi-drug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol 183: 5725–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meunier D, Boyd D, Mulvey MR (2002) Salmonella enterica serotype Typhimurium DT (2002) 104 antibiotic resistance genomic island I in serotype ParatyphiB. Emerg Infect Dis 8: 430–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Idrees M, Mussarat U, Badshah Y, Qadir M, Bokhari H (2011) Prevalence of antimicrobial resistance and integrons in Escherichia Coli from Punjab, Pakistan. Braz J Microbiol 42: 462–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kennedy FC, Ifeoma CA, Didacus CE, Sergio S, Vanesa E, et al. (2010) Antimicrobial resistance, integrons and plasmid replicon typing in multi-resistant clinical Escherichia coli strains from Enugu State, Nigeria. J Basic Microb 50: 1–7. [DOI] [PubMed] [Google Scholar]
- 21. Peters ED, L-van MA, Box AT (2001) Novel gene cassettes and integrons. Antimicrob Agents Chemother 45: 2961–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fluit AC, Schmitz FJ, Eur J (1999) Class 1 integrons gene cassettes mobility and epidemiology. Clin Microbiol Infect Dis 18: 761–770. [DOI] [PubMed] [Google Scholar]
- 23. Fan W, Hamilton T, Webster SS (2007) Multiplex real-time SYBR Green Ι PCR assay for detection of tetracycline efflux genes of Gram-negative bacteria. Mol Cell Probes 21: 245–256. [DOI] [PubMed] [Google Scholar]
- 24. Shi L, Chen X, Xiao ZH (2007) Detection and characterization of integron structure in clinical bacteria isolates. Chinese J Antibiot 32: 951–953. [Google Scholar]
- 25. Molla B, Miko A, Pries K (2007) Class 1 integron and resistance gene cassettes among multidrug resistant Salmonella serovars isolated from slaughter animals and foods of animal origin in Ethiopia. Acta Trop 103: 142–149. [DOI] [PubMed] [Google Scholar]
- 26. Kang HY, Jeong YS, Oh JY (2005) Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J Antimicrob Chemother 55: 639–644. [DOI] [PubMed] [Google Scholar]
- 27. Shu CH, Tsai HC, Jen CP (2006) Characterisation of antimicrobial resistance patterns and class 1 integrons among Escherichia coli and Salmonella enteric serovar Choleraesuis strains isolated from humans and swine in Taiwan. Int J Antimicrob Agents 27: 383–391. [DOI] [PubMed] [Google Scholar]