Abstract
Gene single nucleotide polymorphisms (SNPs) have been extensively studied in association with development and prognosis of various malignancies. However, the potential role of genetic polymorphisms of cancer stem cell (CSC) marker genes with respect to cancer risk has not been examined. We conducted a case-control study involving a total of 1000 subjects (500 lung cancer patients and 500 age-matched cancer-free controls) from northeastern China. Lung cancer risk was analyzed in a logistic regression model in association with genotypes of four lung CSC marker genes (CD133, ALDH1, Musashi-1, and EpCAM). Using univariate analysis, the Musashi-1 rs2522137 GG genotype was found to be associated with a higher incidence of lung cancer compared with the TT genotype. No significant associations were observed for gene variants of CD133, ALDH1, or EpCAM. In multivariate analysis, Musashi-1 rs2522137 was still significantly associated with lung cancer when environmental and lifestyle factors were incorporated in the model, including lower BMI; family history of cancer; prior diagnosis of chronic obstructive pulmonary disease, pneumonia, or pulmonary tuberculosis; occupational exposure to pesticide; occupational exposure to gasoline or diesel fuel; heavier smoking; and exposure to heavy cooking emissions. The value of the area under the receiver-operating characteristic (ROC) curve (AUC) was 0.7686. To our knowledge, this is the first report to show an association between a Musashi-1 genotype and lung cancer risk. Further, the prediction model in this study may be useful in determining individuals with high risk of lung cancer.
Introduction
Lung cancer is one of the most commonly diagnosed malignancies and the leading cause of cancer-related death in the world [1]. Cigarette smoking is considered as an important risk factor for lung cancer. However, only 10–15% of smokers develop lung cancer, suggesting that individual variation in genetic susceptibility to lung cancer in the general population may play a role. Cancer stem cells (CSCs) are a small minority of cells in a heterogeneous tumor population that drives tumor growth and have been associated with resistance to chemo- and radiation-therapies [2]–[4]. It has been demonstrated that lung CSCs play an important role in tumor initiation [5], [6]. CDCs also share some similarities with normal stem cells, including self-renewal and differentiation, in addition to their potent tumor-driving capability [7]–[10]. CSCs are characterized by expression of particular molecular markers that play an important role in promoting stem cell self-renewal and maintenance [11].
Aberrant expression of CSC markers is associated with the initiation and development of lung cancer, including cluster of differentiation 133 (CD133), Musashi RNA-binding protein 1 (Musashi-1), aldehyde dehydrogenase 1 (ALDH1), epithelial cell adhesion molecule (EpCAM), B-cell-specific moloney murine leukemia virus integration site 1 (Bmi-1), Octarner binding factor 4 (OCT-4), and Glycine dehydrogenase (GLDC) [12]–[20]. CD133, initially described as a surface antigen specific for human hematopoietic stem cells [21], [22], is now being used as an isolation marker of CSCs from lung cancer [23]. Musashi-1, an RNA-binding protein, is expressed in various epithelial stem cells and plays an important role in regulating the maintenance and differentiation of stem/precursor cells [24]–[26]. Musashi-1 is over-expressed in several tumor tissues, including lung cancer [17], gliomas [27], intestinal adenomas [28], [29] and hepatomas [30], suggesting a correlation with oncogenic development. ALDH1 is widely regarded as a surface marker of CSCs in lung cancer [31]–[33]. ALDH positive lung cancer stem-like cells have longer telomeres than the non-CSC cells [34]. EpCAM, a type I transmembrane glycoprotein of ∼40 kDa, is overexpressed in a variety of epithelial tumors, including lung cancer [18]–[20]. EpCAM is involved in intercellular adhesion and interacts with E-cadherin to induce cell adhesion [35]. Overexpression of EpCAM is linked directly to stimulation of the cell cycle and proliferation by upregulating c-myc and cyclin A/E [36]. Inhibition of EpCAM by small inhibitory RNA diminishes cell proliferation, migration and invasiveness [37].
Together, these observations have correlated aberrant function of CSC marker molecules with cellular hallmarks of cancer: hyperproliferation and metastatic behaviors. While SNPs have been extensively studied for their association with the risk and prognosis of cancers, little is known about the potential role of SNPs in CSC marker genes with relation to cancer. In this study, we examined the association of lung cancer risk in a Chinese population with polymorphisms of the well-established CSC marker genes CD133, ALDH1, Musashi-1 and EpCAM. A forecasting model was constructed using CSC marker SNPs and epidemiologic factors; the results provide a novel method to predict individuals at increased risk of developing lung cancer.
Methods
Study population
We conducted a hospital-based, case-control study involving a total of 1000 subjects from northeastern China (Changchun City, Jilin province). All subjects were local residents of Han descent, consisting of 500 patients clinically diagnosed with lung cancer and 500 cancer-free controls. Patients had histologically-confirmed primary lung cancer without previous cancer history, did not receive radiotherapy, chemotherapy or other anti-cancer therapy. Controls were randomly selected normal individuals receiving routine physical examinations in the same hospital. Case matching was performed based on age, gender and place of residence. The study was approved by the Ethics Committee of the First Hospital of Jilin Medical University, and conducted according to the Declaration of Helsinki Principles. All subjects were provided written informed consent.
Diagnostic criteria and Data Collection
A standardized interview was conducted by trained interviewers in the hospital or at the homes of participating individuals. Information regarding socio-demographic details, medical history, family history, lifestyle history, and cancer diagnosis was recorded. Risk factor information and peripheral blood lymphocytes were collected at the time of diagnosis for cancer patients or on the day of interview for controls.
CSC marker gene polymorphism selection
We used a candidate gene approach [38]–[40] to select SNPs for this study. Four well-established CSC marker genes (CD133, ALDH1, Musashi-1 and EpCAM) were selected in the study design. Expression of these four proteins had been reported as a marker to identify lung CSCs [41], [42].
Three predefined criteria were used for CSC SNP selection: (a) minor allele frequency (MAF) ≥5% in the HapMap CHB population; (b) SNPinfo website (http://snpinfo.niehs.nih.gov) for candidate CSC gene SNP selection, and (c) publications showing clinical correlations with cancer risk/outcome or recurrence. Using these criteria, five CSC candidate SNPs were chosen in our model analysis: Rs2286455 in the CD133 gene, rs1342024 and rs13959 in the ALDH1 gene, rs2522137 in the Musashi-1 gene, and rs17036526 in the EpCAM gene (Table 1). Based upon literature information, we excluded polymorphisms previously implicated in COPD or lung cancer. Additionally, we did not select SNPs in genes encoding proteins involved in pathways of cell-cycle control, oxidant response, apoptosis and airways inflammation. Finally, we avoided SNPs known to have either functional effects on in vitro assays, or were non-synonymous or in regulatory regions.
Table 1. Single nucleotide polymorphisms in cancer stem cell marker genes.
| Gene | SNP | Base exchange | Gene location | Reference |
| CD133 | rs2286455 | G>A | Splice Site | nd |
| ALDH1A1 | rs1342024 | G>C | Upstream | [54] |
| rs13959 | C>T | Synonymous coding | nd | |
| MSI-1 | rs2522137 | T>G | 3′ UTR | nd |
| EpCAM | rs17036526 | G>C | Splice site | nd |
UTR: untranslated region; nd: no data.
Genotyping and quality control
Genomic DNA was isolated from peripheral blood lymphocytes. MassArray (Sequenom, San Diego, CA) was used to genotype CSC markers using allele specific MALDI–TOF mass spectrometry. Primers and multiplex reactions were designed using the RealSNP.com Website. Concordance among the 3 genomic control DNA samples present in duplicate was 100%. Of the SNPs with genotyping data, the sample call rates were more than 95%.
Statistical analysis
The Hardy-Weinberg equilibrium (HWE) was tested by a best fit chi-square (χ2) test that compared expected genotype frequencies with observed genotype frequencies in cancer-free controls. The model was also used to determine the presence of significant differences in genotype and allele distribution as well as SNP frequency between clinically diagnosed lung cancer and controls. A logistic regression model was used to identify independent risk factors for lung cancer. The forward stepwise likelihood ratio method was employed to screen variables in model selection, where the cut-off for variables in the model was 0.05 and the cut-off for variables outside of model was 0.10; an optimal model with minimum akaike information criterion was selected. All categorical variables were set as dummy variables, and the first category of each variable was selected as baseline. The classification ability of the model was evaluated using the area under the receiver operating characteristic (ROC) curve (AUC), and the optimal operating point (OPP) was given afterwards. All analyses were conducted using SPSS v19.0 software (SPSS, Inc., Chicago, IL, USA). All P-values were two-sided, and P-values <0.05 were considered statistically significant.
Results
Distribution of genotype and its characteristics in cancer and control populations
We recruited 500 cases of lung cancer and 500 cancer free controls between 2010 and 2012. Table 2 shows the distribution and frequency of study-specific risk factors between cancer patients and controls.
Table 2. Distribution of CSC marker genotypes and characteristics of the case group and the healthy control group.
| Characteristic | Case group | Control group | |
| (n = 500) | (n = 500 ) | ||
| rs2286455 | AA | 46 (9.2%) | 37 (7.4%) |
| GG | 266 (53.2%) | 262 (52.4%) | |
| GA | 188 (37.6%) | 201 (40.2%) | |
| rs1342024 | GG | 97 (19.4%) | 92 (18.4%) |
| CC | 164 (32.8%) | 166 (33.2%) | |
| GC | 239 (47.8%) | 242 (48.4%) | |
| rs13959 | TT | 89 (17.8%) | 99 (19.8%) |
| CC | 176 (35.2%) | 159 (31.8%) | |
| CT | 235 (47.0%) | 242 (48.4%) | |
| rs2522137 | TT | 219 (43.8%) | 219 (43.8%) |
| GG | 67 (13.4%) | 39 (7.8%) | |
| GT | 214 (42.8%) | 242 (48.4%) | |
| rs17036526 | GG | 224 (44.8%) | 233 (46.6%) |
| CC | 55 (11.0%) | 65 (13.0%) | |
| CG | 221 (44.2%) | 202 (40.4%) | |
| Gender | male | 305 (61%) | 302 (60.4%) |
| female | 195 (39%) | 198 (39.6%) | |
| Age | <30 | 2 (0.4%) | 5 (1.0%) |
| 30–39 | 14 (2.8%) | 16 (3.2%) | |
| 40–49 | 64 (12.8%) | 70 (14.0%) | |
| 50–59 | 176 (35.2%) | 196 (39.2%) | |
| 60–69 | 174 (34.8%) | 148 (19.7%) | |
| ≥70 | 70 (14.0%) | 65 (13.0%) | |
| Education | Junior high school and lower | 318 (63.6%) | 130 (26.0%) |
| High school | 97 (19.4%) | 144 (28.8%) | |
| Greater than high school | 85 (17.0%) | 226 (45.2%) | |
| Smoking | Pack years | 14.25 (0–36.0) | 0.0 (0.0–6.9) |
| exposure to | Absent | 398 (79.6%) | 473 (94.6%) |
| Pesticide | Present | 102 (20.4%) | 27 (5.4%) |
| exposure to | Absent | 487 (97.4%) | 496 (99.2%) |
| Gasoline/diesel | Present | 13 (2.6%) | 4 (0.8%) |
| exposure to Ink | Absent | 493 (98.6%) | 497 (99.4%) |
| Present | 7 (1.4%) | 3 (0.6%) | |
| Cooking emissions | Absent | 244 (48.8%) | 250 (50.0%) |
| (Total dish-years) | ≤50 | 149 (29.8%)) | 152 (30.4%) |
| 51–100 | 61 (12.2%) | 80 (16.0%) | |
| 101–150 | 46 (9.2%) | 18 (3.6%) | |
| Pneumonia | History Absent | 477 (95.4%) | 490 (98.0%) |
| History Present | 23 (4.6%) | 10 (2.0%) | |
| COPD | History Absent | 449 (89.8%) | 489 (97.8%) |
| History Present | 51 (10.2%) | 11 (2.2%) | |
| Pulmonary | History Absent | 470 (94.0%) | 486 (97.2%) |
| tuberculosis | History Present | 30 (6.0%) | 14 (2.8%) |
| Bronchial asthma | History Absent | 488 (97.6%) | 495 (99.0%) |
| History Present | 12 (2.4%) | 5 (1.0%) | |
| Family history | History Absent | 330 (66.0%) | 397 (79.4%) |
| of cancer | History Present | 170 (34.0%) | 103 (20.6%) |
| BMI | <18.5 | 49 (9.8%) | 15 (3.0%) |
| (kg/m2) | 18.5–24 | 302 (60.4%) | 230 (46.0%) |
| ≥24 | 149 (29.8%) | 255 (51.0%) | |
Association of CSC marker gene SNPs with lung cancer risk in univariate analysis
We first evaluated lung cancer risk using univariate analysis. Among CSC marker gene SNPs selected, the Musashi-1 rs2522137 GG genotype had a tendency toward a higher incidence of lung cancer than the rs2522137 GG genotype in both recessive model (P = 0.004) and additive model. However, no significant differences were noticed for SNPs in other CSC marker genes in dominant, recessive, additive, or multiplicative models ( Table 3 ).
Table 3. Association of SNPs with lung cancer risk in univariate analysis.
| Genotype | Univariate OR (95%CI) | P value | |
| rs17036526 | |||
| Recessive model | CC + GC | 1 | 0.568 |
| GG | 0.930 (0.725–1.193) | ||
| Dominant model | CC | 1 | 0.331 |
| GG + GC | 1.209 (0.825–1.773) | ||
| Additive model | CC | 1 | |
| GC | 1.293 (0.861–1.942) | 0.216 | |
| GG | 1.136 (0.759–1.700) | 0.535 | |
| Multiplicative model | G allele | 1.004 (0.837–1.205) | 0.963 |
| rs2522137 | |||
| Dominant model | TT | 1 | 1.000 |
| GG + TG | 1.000 (0.779–1.284) | ||
| Recessive model | TT + TG | 1 | 0.004 |
| GG | 1.829 (1.207–2.733) | ||
| Additive model | TT | 1 | |
| TG | 0.884 (0.680–1.150) | 0.359 | |
| GG | 1.718 (1.110–2.6591) | 0.015 | |
| Multiplicativemodel | G allele | 1.138 (0.942–1.374) | 0.179 |
| rs2286455 | |||
| Recessive model | GG + GA | 1 | 0.303 |
| AA | 1.268 (0.807–1.992) | ||
| Dominant model | GG | 1 | 0.800 |
| AA + GA | 0.968 (0.755–1.241) | ||
| Additive model | GG | 1 | |
| GA | 0.921 (0.709–1.197) | 0.540 | |
| AA | 1.225 (0.769–1.950) | 0.393 | |
| Multiplicative model | A allele | 0.963 (0.803–1.153) | 0.806 |
| rs1342024 | |||
| Recessive model | CC + CG | 1 | 0.686 |
| GG | 1.067 (0.778–1.465) | ||
| Dominant model | CC | 1 | 0.893 |
| GG + CG | 1.018 (0.782–1.325) | ||
| Additive model | CC | 1 | |
| CG | 1.000 (0.755–1.323) | 0.998 | |
| GG | 1.067 (0.746–1.526) | 0.722 | |
| Multiplicative model | G allele | 1.028 (0.863–1.226) | 0.754 |
| rs13959 | |||
| Recessive model | TT + TC | 1 | 0.255 |
| CC | 1.165 (0.896–1.515) | ||
| Dominant model | TT | 1 | 0.418 |
| CC + TC | 1.140 (0.830–1.566) | ||
| Additive model | TT | 1 | |
| TC | 1.080 (0.770–1.514) | 0.655 | |
| CC | 1.231 (0.861–1.761) | 0.254 | |
| Multiplicative model | C allele | 1.114 (0.935–1.327) | 0.228 |
Association of SNPs with lung cancer risk in multivariate analysis
Next, we evaluated independent risk factors of lung cancer using multivariate analysis. By incorporating environmental and lifestyle parameters, we found that in the recessive model, Musashi-1 rs2522137 was still significantly associated with lung cancer. These environmental and lifestyle parameters included lower BMI, family history of cancer, prior diagnosis of COPD, pneumonia or pulmonary tuberculosis, occupational exposure to pesticide, occupational exposure to gasoline or diesel, heavier smoking, and exposure to heavier cooking emission (Table 4). These data suggest that the Musashi-1 rs2522137 GG genotype is a significant genetic risk factor for lung cancer.
Table 4. Multivariate risk model with adjusted odds ratios and 95% confidence intervals.
| Risk factors | Exp (B) | 95% C.I. | P value |
| Occupational exposure to pesticide | |||
| Absence | 1.00 | Reference | 0.000 |
| Presence | 3.390 | (2.093–5.493) | |
| exposure to gasoline/diesel: | |||
| Absence | 1.00 | Reference | 0.012 |
| Presence | 4.653 | (1.402–15.448) | |
| Smoking | 1.032 | (1.024–1.041) | 0.000 |
| Cooking emission(Total dish-years) | 0.001 | ||
| ≤50 | 1.00 | Reference | |
| 51–100 | 1.304 | (0.934–1.819) | 0.119 |
| 101–150 | 0.941 | (0.608–1.457) | 0.785 |
| >150 | 3.375 | (1.779–6.402) | 0.000 |
| COPD: | |||
| History absence | 1.00 | Reference | 0.000 |
| History presence | 3.775 | (1.809–7.878) | |
| Pneumonia: | |||
| History presence | 1.00 | Reference | 0.021 |
| History absence | 0.369 | (0.158–0.860) | |
| Pulmonary tuberculosis: | |||
| History presence | 1.00 | Reference | 0.022 |
| History absence | 0.428 | (0.207–0.884) | |
| Family history of cancer: | |||
| History absence | 1.00 | Reference | 0.000 |
| History presence | 1.848 | (1.338–2.553) | |
| BMI(kg/m2): | 0.000 | ||
| <18.5 | 1.00 | Reference | |
| 18.5–24 | 0.370 | (0. 192–0. 714) | 0.003 |
| ≥24 | 0.168 | (0.086–0. 328) | 0.000 |
| rs2522137: | |||
| TT+TG | 1.00 | Reference | |
| GG | 1.926 | (1.209–3.070) | 0.006 |
COPD: chronic obstructive pulmonary disease
ROC analysis
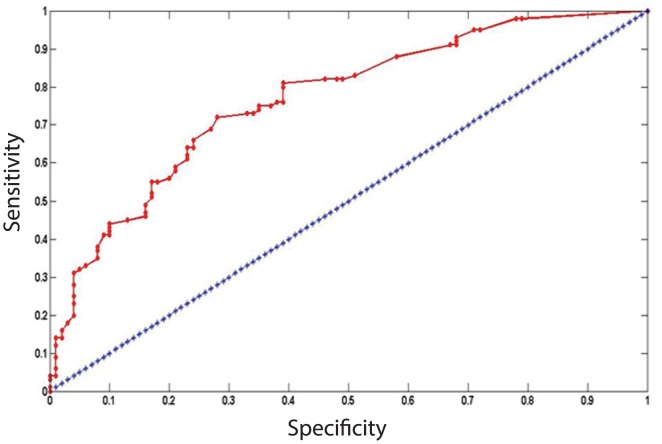
The classification ability of the multivariate model was further evaluated using the area under ROC curve (AUC) and the optimal operating point (OPP). Figure 1 shows the ROC curve derived from our model; AUC was calculated as 0.7686. Furthermore, the OPP was obtained when the cutoff was set at 0.47. The estimated false positive rates, true positive rates, and Youden index were determined to be 0.28, 0.72, and 0.44, respectively.
Figure 1. ROC plots for lung cancer risk prediction model.
The ROC AUC was 0.7686. The straight line represented the ROC curve expected by chance alone.
Correlation between SNPs and lung cancer type
Finally, we looked for correlations between CSC SNPs and lung cancer type (squamous cell, adenocarcinoma, small cell) along with age at onset and gender of lung cancer. We did not observe statistically significant differences between these CSC SNPs and age or gender at the onset of lung cancer ( Tables 5 ). In the pathology-stratified analysis, however, CD133 SNP rs2286455 was significantly correlated with lung cancer type (P = 0.048) ( Table 6 ). No differences were observed between the remaining SNPs being considered with lung cancer type.
Table 5. Association of SNPs with gender and age from lung cancer patients.
| Characteristic | Gender | P value | Age (yrs.) | P value | ||
| M* | F* | |||||
| rs2286455 | AA | 29 | 17 | 0.184 | 58.87±10.18 | reference |
| GG | 171 | 95 | 57.07±11.34 | 0.257 | ||
| GA | 105 | 83 | 59.02±9.22 | 0.872 | ||
| rs1342024 | CC | 99 | 65 | 0.806 | 58.85±9.95 | reference |
| GG | 62 | 35 | 59.86±9.95 | 0.432 | ||
| GC | 144 | 95 | 58.25±9.93 | 0.550 | ||
| rs13959 | CC | 103 | 73 | 0.574 | 59.36±9.78 | reference |
| TT | 58 | 31 | 60.15±9.84 | 0.541 | ||
| CT | 144 | 91 | 57.79±10.03 | 0.112 | ||
| rs2522137 | GG | 39 | 28 | 0.877 | 58.27±10.52 | reference |
| TT | 135 | 84 | 58.07±10.15 | 0.885 | ||
| GT | 131 | 83 | 59.62±9.50 | 0.331 | ||
| rs17036526 | CC | 31 | 24 | 0.740 | 59.80±9.90 | reference |
| GG | 139 | 85 | 58.90±9.08 | 0.547 | ||
| CG | 135 | 86 | 58.36±10.76 | 0.338 | ||
*M, male; F, female.
Table 6. Association of SNPs with histology types from lung cancer patients.
| Characteristic | Histology types | P value | ||||
| SQ* | AD* | SC* | OC* | |||
| rs2286455 | AA | 78 | 93 | 70 | 25 | 0.048 |
| GG | 10 | 16 | 18 | 2 | ||
| GA | 53 | 67 | 38 | 30 | ||
| rs1342024 | CC | 41 | 59 | 43 | 21 | 0.640 |
| GG | 30 | 39 | 20 | 8 | ||
| GC | 70 | 78 | 63 | 28 | ||
| rs13959 | CC | 49 | 64 | 42 | 21 | 0.324 |
| TT | 23 | 39 | 22 | 5 | ||
| CT | 69 | 73 | 62 | 31 | ||
| rs2522137 | GG | 21 | 25 | 18 | 3 | 0.361 |
| TT | 65 | 69 | 59 | 26 | ||
| GT | 55 | 82 | 49 | 28 | ||
| rs17036526 | CC | 15 | 19 | 13 | 8 | 0.780 |
| GG | 70 | 76 | 52 | 26 | ||
| CG | 56 | 81 | 61 | 23 | ||
* SQ: squamous cell; AD, adenocarcinoma; SC, small cell; OC, other carcinomas.
Discussion
Single nucleotide polymorphisms (SNPs) have been extensively examined in practically all cancer types in an effort to identify inherited cancer susceptibility genes and their interaction with environmental factors. Cancer stem cells (CSCs) play an important role in tumor initiation, metastases, and recurrence. We have examined the potential correlation between SNPs present in CSCs and the likelihood of lung cancer. This is particularly important since SNPs in CSC-directing genes could provide a genetic link to cancers that are particularly challenging to treat. While typical cancer therapies may eliminate most of the tumor mass, a small population of CSCs with the potential to repopulate the tumor may remain [5]. It is generally accepted that CSCs are characterized by the unique expression of cell surface molecules called CSC marker genes. CSC markers play an important part in the maintenance of self-renewal and resistance to apoptosis pathway activation in these cells. In this study, we obtained information to support the hypothesis that clinical outcome in lung cancer patients may be influenced by genetic variants of CSC marker genes.
The potential impact of CSC marker gene polymorphisms on lung cancer susceptibility has not been previously explored. In this study we took the advantage of a hypothesis-driven candidate gene approach [38]–[40] to identify potentially functional SNPs associated with histologically validated lung cancer. In contrast to genome-wide association (GWA) and quantitative trait locus (QTL) approaches, the candidate gene approach is economical and has rather high statistical power [38]. We focused on four CSC marker genes that have been used to isolate CSCs: CD133, ALDH1, EpCAM, and Musashi-1 [41], [42]. Using the candidate gene approach, we selected a panel of SNPs in these CSC gene loci from SNP websites and peer-reviewed literature. SNPs identified to have high allele frequency were genotyped in 500 lung cancer cases along with 500 age-matched controls. Our results have identified the Musashi-1 variant as an independent risk factor for lung cancer. It is also interesting to note that the Musashi-1 rs2522137 genotype was still associated with lung cancer risk in a multivariate regression model that considered several environmental and lifestyle factors. Taken together, this study provides the first evidence to correlate the Musashi-1 rs2522137 SNP variant with lung cancer.
Currently, we know very little about the detailed molecular mechanisms by which Musashi-1 rs2522137 polymorphisms might contribute to lung cancer development. Musashi-1 is an evolutionarily conserved RNA-binding protein that has profound implications in cellular processes, such as stem cell maintenance, nervous system development, and tumorigenesis. Musashi-1 is highly expressed in many cancers, whereas in normal tissues, its expression is restricted only to stem cells. It is now clear that this RNA-binding protein is involved in cell asymmetric division and is required for the maintenance of stem cell identity [43]–[45]. Interestingly, Musashi-1 mRNA transcript contains an 1811-bp long 3′-untranslated region (3′-UTR). The 3′-UTR of mRNA transcripts usually buries the target site for regulatory microRNA (miRNA). SNPs in the 3′-UTRs have been shown to have functional effects on control of mRNA stability and/or translational efficiency through the regulation of miRNA. The binding of miRNAs to the 3′-UTRs may play an important overall role in gene expression.
The 3′-UTR of mature Musashi-1 mRNA is potentially targeted by several tumor suppressor miRNAs, including miR-34a, -101, -128, -137 and -138 [46]. In addition, the Musashi-1 mRNA 3′-UTR contains several AU- and U-rich sequences that are targeted by an evolutionarily conserved RNA-binding protein HuR [47]. HuR is a member of the Hu/ELAV (embryonic lethal abnormal vision) family, which is highly expressed in tumor tissue and enhances tumorigenesis by interacting with a subset of mRNAs that encode proteins in the regulation of cell proliferation, cell survival, angiogenesis, invasion, and metastasis [48], [49]. Using the SNPinfo website (http://snpinfo.niehs.nih.gov/), we found that the Musashi-1 3′-UTR also buries potential target sites for miRNAs hsa-miR-1275, hsa-miR-1285, hsa-miR-483-5p, hsa-miR-486-3p, hsa-miR-612, and hsa-miR-625. It is worthwhile noting that Musashi-1 rs2522137 is located within these miRNA and HuR binding sites. Future studies are needed to delineate whether rs2522137 variants may affect the binding of these regulatory miRNAs and HuR protein. Presumably, the Musashi-1 rs2522137 GG variant may interfere with the binding of miRNAs and HuR factor, thus increasing the stability of Musashi-1 mRNA. If true, this mechanism could provide the basis for the Musashi-1 rs2522137 variant to maintain self-renewing lung cancer stem cells.
SNPs represent inherited genetic variations that occur during the lifetime of an individual. It is well known that non-genetic risk factors, such as age, history of lung disease, and smoking history are also very important and can be combined to develop risk-based models of cancer. Robert et al [50], [51] have suggested that SNPs need to be combined with other risk variables to identify individuals who are most susceptible to developing lung cancer. Similarly, the Liverpool Lung Project risk model improves its predictive capability of lung cancer by adding a marker SNP (rs663048) in the SEZ6L gene [52], [53]. In this study, we identified a correlation between lung cancer and a specific SNP within a CSC marker gene. Environmental and lifestyle factors included in this analysis, such as occupational exposure to pesticide, occupational exposure to gasoline/diesel prior diagnosis of pulmonary tuberculosis, and cooking emission, provide a similar correlation relative to an age-matched control population.
In summary, this study revealed a significantly increased risk of lung cancer for the CSC marker Musashi-1 rs2522137-GG compared with -TT and -TG SNPs in a Chinese population. The ROC AUC of our model was 0.7686, indicating the potential to identify high-risk individuals in the Chinese population by focusing on information that can be readily obtained in the primary care setting. Finally, this lung cancer risk prediction model discriminated between high- and low-risk individuals. Further studies are needed in larger cohorts of unselected cases and controls to further validate and extent these initial observations.
Acknowledgments
We thank Li Deng for technical assistance in sample collection and preparation as well as Lina Jin and Hua He for their help in data and statistical analysis.
Funding Statement
This work was supported by the National Natural Science Foundation of China grants (81071920, 81372835), Jilin Provincial Science and Technology Department Grant (201201023), the Key Clinical Project of the Ministry of Health of the People's Republic of China Grant (2001133), National Institutes of Health grant (1R43CA103553-01), California Institute of Regenerative Medicine (CIRM) grant (RT2-01942), Jilin International Collaboration Grant (20120720), the National Natural Science Foundation of China grant (81272294), and the grant of Key Project of Chinese Ministry of Education 311015). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Siegel R, Xu J, Ward E (2010) Cancer statistics, 2010. CA Cancer J Clin 60: 277–300. [DOI] [PubMed] [Google Scholar]
- 2. Soltanian S, Matin MM (2011) Cancer stem cells and cancer therapy. Tumour Biol 32: 425–440. [DOI] [PubMed] [Google Scholar]
- 3. Kemper K, Grandela C, Medema JP (2010) Molecular identification and targeting of colorectal cancer stem cells. Oncotarget 1: 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Keysar SB, Jimeno A (2010) More than markers: biological significance of cancer stem cell-defining molecules. Mol Cancer Ther 9: 2450–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Reya T, Morrison SJ, Clarke MF, Weissman IL (2001) Stem cells, cancer, and cancer stem cells. Nature 414: 105–111. [DOI] [PubMed] [Google Scholar]
- 6. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF (2003) Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 100: 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Clarke MF (2005) A self-renewal assay for cancer stem cells. Cancer Chemother Pharmacol 56 Suppl 164–68. [DOI] [PubMed] [Google Scholar]
- 8. Soltysova A, Altanerova V, Altaner C (2005) Cancer stem cells. Neoplasma 52: 435–440. [PubMed] [Google Scholar]
- 9. Ratajczak MZ (2005) Cancer stem cells—normal stem cells "Jedi" that went over to the "dark side". Folia Histochem Cytobiol 43: 175–181. [PubMed] [Google Scholar]
- 10. Sales KM, Winslet MC, Seifalian AM (2007) Stem cells and cancer: an overview. Stem Cell Rev 3: 249–255. [DOI] [PubMed] [Google Scholar]
- 11. Marhaba R, Klingbeil P, Nuebel T, Nazarenko I, Buechler MW, et al. (2008) CD44 and EpCAM: cancer-initiating cell markers. Curr Mol Med 8: 784–804. [DOI] [PubMed] [Google Scholar]
- 12. Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, et al. (2012) Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 148: 259–272. [DOI] [PubMed] [Google Scholar]
- 13. Meng X, Wang Y, Zheng X, Liu C, Su B, et al. (2012) shRNA-mediated knockdown of Bmi-1 inhibit lung adenocarcinoma cell migration and metastasis. Lung Cancer 77: 24–30. [DOI] [PubMed] [Google Scholar]
- 14. Shien K, Toyooka S, Ichimura K, Soh J, Furukawa M, et al. (2012) Prognostic impact of cancer stem cell-related markers in non-small cell lung cancer patients treated with induction chemoradiotherapy. Lung Cancer 77: 162–167. [DOI] [PubMed] [Google Scholar]
- 15. Kimura M, Takenobu H, Akita N, Nakazawa A, Ochiai H, et al. (2011) Bmi1 regulates cell fate via tumor suppressor WWOX repression in small-cell lung cancer cells. Cancer Sci 102: 983–990. [DOI] [PubMed] [Google Scholar]
- 16. Barr MP, Gray SG, Hoffmann AC, Hilger RA, Thomale J, et al. (2013) Generation and characterisation of cisplatin-resistant non-small cell lung cancer cell lines displaying a stem-like signature. PLoS One 8: e54193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moreira AL, Gonen M, Rekhtman N, Downey RJ (2010) Progenitor stem cell marker expression by pulmonary carcinomas. Mod Pathol 23: 889–895. [DOI] [PubMed] [Google Scholar]
- 18. Went P, Vasei M, Bubendorf L, Terracciano L, Tornillo L, et al. (2006) Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. Br J Cancer 94: 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim Y, Kim HS, Cui ZY, Lee HS, Ahn JS, et al. (2009) Clinicopathological implications of EpCAM expression in adenocarcinoma of the lung. Anticancer Res 29: 1817–1822. [PubMed] [Google Scholar]
- 20. van der Gun BT, Melchers LJ, Ruiters MH, de Leij LF, McLaughlin PM, et al. (2010) EpCAM in carcinogenesis: the good, the bad or the ugly. Carcinogenesis 31: 1913–1921. [DOI] [PubMed] [Google Scholar]
- 21. Miraglia S, Godfrey W, Yin AH, Atkins K, Warnke R, et al. (1997) A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood 90: 5013–5021. [PubMed] [Google Scholar]
- 22. Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, et al. (1997) AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood 90: 5002–5012. [PubMed] [Google Scholar]
- 23. Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, et al. (2008) Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ 15: 504–514. [DOI] [PubMed] [Google Scholar]
- 24. Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, et al. (2005) Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res 306: 349–356. [DOI] [PubMed] [Google Scholar]
- 25. Okano H, Imai T, Okabe M (2002) Musashi: a translational regulator of cell fate. J Cell Sci 115: 1355–1359. [DOI] [PubMed] [Google Scholar]
- 26. Kaneko Y, Sakakibara S, Imai T, Suzuki A, Nakamura Y, et al. (2000) Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev Neurosci 22: 139–153. [DOI] [PubMed] [Google Scholar]
- 27. Kanemura Y, Mori K, Sakakibara S, Fujikawa H, Hayashi H, et al. (2001) Musashi1, an evolutionarily conserved neural RNA-binding protein, is a versatile marker of human glioma cells in determining their cellular origin, malignancy, and proliferative activity. Differentiation 68: 141–152. [DOI] [PubMed] [Google Scholar]
- 28. Wang T, Ong CW, Shi J, Srivastava S, Yan B, et al. (2011) Sequential expression of putative stem cell markers in gastric carcinogenesis. Br J Cancer 105: 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fan LF, Dong WG, Jiang CQ, Xia D, Liao F, et al. (2010) Expression of putative stem cell genes Musashi-1 and beta1-integrin in human colorectal adenomas and adenocarcinomas. Int J Colorectal Dis 25: 17–23. [DOI] [PubMed] [Google Scholar]
- 30. Shu HJ, Saito T, Watanabe H, Ito JI, Takeda H, et al. (2002) Expression of the Musashi1 gene encoding the RNA-binding protein in human hepatoma cell lines. Biochem Biophys Res Commun 293: 150–154. [DOI] [PubMed] [Google Scholar]
- 31. Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, et al. (2007) ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1: 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, et al. (2009) Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res 7: 330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liang D, Shi Y (2012) Aldehyde dehydrogenase-1 is a specific marker for stem cells in human lung adenocarcinoma. Med Oncol 29: 633–639. [DOI] [PubMed] [Google Scholar]
- 34. Serrano D, Bleau AM, Fernandez-Garcia I, Fernandez-Marcelo T, Iniesta P, et al. (2011) Inhibition of telomerase activity preferentially targets aldehyde dehydrogenase-positive cancer stem-like cells in lung cancer. Mol Cancer 10: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Litvinov SV, Balzar M, Winter MJ, Bakker HA, Briaire-de Bruijn IH, et al. (1997) Epithelial cell adhesion molecule (Ep-CAM) modulates cell-cell interactions mediated by classic cadherins. J Cell Biol 139: 1337–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Munz M, Kieu C, Mack B, Schmitt B, Zeidler R, et al. (2004) The carcinoma-associated antigen EpCAM upregulates c-myc and induces cell proliferation. Oncogene 23: 5748–5758. [DOI] [PubMed] [Google Scholar]
- 37. Osta WA, Chen Y, Mikhitarian K, Mitas M, Salem M, et al. (2004) EpCAM is overexpressed in breast cancer and is a potential target for breast cancer gene therapy. Cancer Res 64: 5818–5824. [DOI] [PubMed] [Google Scholar]
- 38. Amos W, Driscoll E, Hoffman JI (2011) Candidate genes versus genome-wide associations: which are better for detecting genetic susceptibility to infectious disease? Proc Biol Sci 278: 1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clark AG (2004) The role of haplotypes in candidate gene studies. Genet Epidemiol 27: 321–333. [DOI] [PubMed] [Google Scholar]
- 40. Leng S, Stidley CA, Liu Y, Edlund CK, Willink RP, et al. (2012) Genetic determinants for promoter hypermethylation in the lungs of smokers: a candidate gene-based study. Cancer Res 72: 707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eramo A, Haas TL, De Maria R (2010) Lung cancer stem cells: tools and targets to fight lung cancer. Oncogene 29: 4625–4635. [DOI] [PubMed] [Google Scholar]
- 42. Koren A, Motaln H, Cufer T (2013) Lung cancer stem cells: a biological and clinical perspective. Cell Oncol (Dordr) 36: 265–275. [DOI] [PubMed] [Google Scholar]
- 43. Siddall NA, McLaughlin EA, Marriner NL, Hime GR (2006) The RNA-binding protein Musashi is required intrinsically to maintain stem cell identity. Proc Natl Acad Sci U S A 103: 8402–8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sakakibara S, Imai T, Hamaguchi K, Okabe M, Aruga J, et al. (1996) Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev Biol 176: 230–242. [DOI] [PubMed] [Google Scholar]
- 45. Nishimura S, Wakabayashi N, Toyoda K, Kashima K, Mitsufuji S (2003) Expression of Musashi-1 in human normal colon crypt cells: a possible stem cell marker of human colon epithelium. Dig Dis Sci 48: 1523–1529. [DOI] [PubMed] [Google Scholar]
- 46. Vo DT, Qiao M, Smith AD, Burns SC, Brenner AJ, et al. (2011) The oncogenic RNA-binding protein Musashi1 is regulated by tumor suppressor miRNAs. RNA Biol 8: 817–828. [DOI] [PubMed] [Google Scholar]
- 47. Vo DT, Abdelmohsen K, Martindale JL, Qiao M, Tominaga K, et al. (2012) The oncogenic RNA-binding protein Musashi1 is regulated by HuR via mRNA translation and stability in glioblastoma cells. Mol Cancer Res 10: 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barreau C, Paillard L, Osborne HB (2005) AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res 33: 7138–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Abdelmohsen K, Gorospe M (2010) Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA 1: 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Young RP, Hopkins RJ, Hay BA, Epton MJ, Mills GD, et al. (2009) Lung cancer susceptibility model based on age, family history and genetic variants. PLOS One 4: e5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Young RP, Hopkins RJ, Whittington CF, Hay BA, Epton MJ, et al. (2011) Individual and cumulative effects of GWAS susceptibility loci in lung cancer: associations after sub-phenotyping for COPD. PLOS One 6: e16476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cassidy A, Myles JP, van Tongeren M, Page RD, Liloglou T, et al. (2008) The LLP risk model: an individual risk prediction model for lung cancer. Br J Cancer 98: 270–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Raji OY, Agbaje OF, Duffy SW, Cassidy A, Field JK (2010) Incorporation of a genetic factor into an epidemiologic model for prediction of individual risk of lung cancer: the Liverpool Lung Project. Cancer Prev Res (Phila) 3: 664–669. [DOI] [PubMed] [Google Scholar]
- 54. Gerger A, Zhang W, Yang D, Bohanes P, Ning Y, et al. (2011) Common cancer stem cell gene variants predict colon cancer recurrence. Clin Cancer Res 17: 6934–6943. [DOI] [PubMed] [Google Scholar]