Abstract
In a series of conceptual articles published around the millennium, Carl Woese emphasized that evolution of cells is the central problem of evolutionary biology, that the three-domain ribosomal tree of life is an essential framework for reconstructing cellular evolution, and that the evolutionary dynamics of functionally distinct cellular systems are fundamentally different, with the information processing systems “crystallizing” earlier than operational systems. The advances of evolutionary genomics over the last decade vindicate major aspects of Woese’s vision. Despite the observations of pervasive horizontal gene transfer among bacteria and archaea, the ribosomal tree of life comes across as a central statistical trend in the “forest” of phylogenetic trees of individual genes, and hence, an appropriate scaffold for evolutionary reconstruction. The evolutionary stability of information processing systems, primarily translation, becomes ever more striking with the accumulation of comparative genomic data indicating that nearly allof the few universal genes encode translation system components. Woese’s view on the fundamental distinctions between the three domains of cellular life also withstand the test of comparative genomics, although his non-acceptance of symbiogenetic scenarios for the origin of eukaryotes might not. Above all, Woese’s key prediction that understanding evolution of microbes will be the core of the new evolutionary biology appears to be materializing.
Keywords: Darwinian threshold, cellular evolution, domains of life, evolutionary transitions, horizontal gene transfer, progenote
Carl Woese’s Concept of Cellular Evolution
There seems to be something peculiar, almost paradoxical, about the legacy of truly great scientists. Their crown achievements can be so overwhelmingly momentous that they almost completely overshadow other works which in themselves would have been more than sufficient for an outstanding career in science. Carl Woese is most famous as the founder of molecular phylogenetics, the creator of the ribosomal Tree of Life, and the discoverer of the Archaea.1-3 These are indeed some of the defining events in biology over the last half-century. But Woese’s legacy goes far beyond these indisputable feats and encompasses the deepest questions of evolutionary biology, the very nature of the evolutionary process, and hence, the nature of life. And in that capacity his work is not textbook matter but a living part of an intense, at times, strained debate.
In this brief discussion, I would like to focus primarily on a series of four articles that Carl Woese published around the turn of the millennium, between 1998 and 2002, and broadly centered on the origin and evolution of cells.4-7 In the course of the discussion, however, I will find it necessary to turn both to some of Woese’s earlier work and to several subsequent papers that fulfilled parts of the research program outlined in the “millennial series.” The articles in this series are highly unusual in format and style, and are difficult to classify under any standard rubric of scientific publication. Although at least the first of these articles4 has been hailed as a new theory of evolution and Woese himself refers to this work as a “genetic annealing model for the universal ancestor of all extant life,” the paper that is all text, without a single formula, plot, or schematic, hardly fits the standard perception of a theory or a model. It is both less and more than a model in the regular sense. Less—because Woese’s analysis is qualitative rather than quantitative, general rather than specific, and does not strive to make concrete predictions (hence not a falsifiable hypothesis sensu Popper). More—thanks to the same generality that allows, not for a single model that is doomed to be over-specified and hence wrong, but for a diverse family of models all of which would fit the general framework outlined by Woese. So this and the other three papers in the series are not exactly original research. Neither are these papers reviews, “opinions,” or “hypotheses.” They might be classified as essays but such a “light” definition does not give justice to the true gravitasof these papers, which together present a coherent, broad vision of the nature of biological evolution. If one is hard pressed to define the genre of these papers, perhaps the old-fashioned “treatise” or “tract” would fit best; these indeed read like a series of short treatises on interlocked major subjects of evolutionary biology.
Let us try to list, in the briefest possible form, the key propositions of Woese’s evolutionary vision that he denoted the genetic annealing model in the first of the treatises4 (Fig. 1).
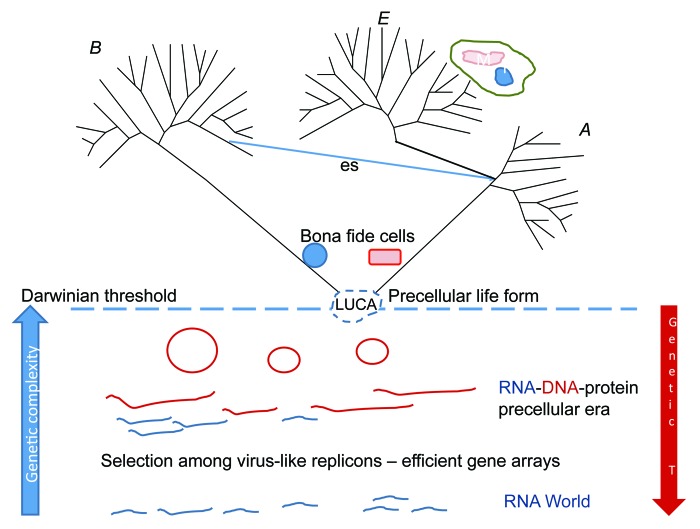
Figure 1. Scenario of cellular evolution according to Woese, with additions and modifications. A, archaea; B, bacteria; E, eukaryotes; es, endosymbiosis that is considered to have occurred by engulfment of an α-proteobacterium by an archaeon of the TACK superphylum (see text); N, nucleus, M, mitochondrion; T, “temperature” (sensu Woese, i.e., intensity of genetic exchanges); LUCA, Last Universal Common (Cellular) Ancestor that is envisaged as a pre-cellular life form with a primitive, possibly porous membrane.
(1) The Universal Ancestor (UA) was not a modern-type cellular organism but rather a community of progenotes (a much earlier concept of Woese8), primitive entities with imprecise, “statistical” translation, and multiple, small genomic segments, conceivably present in multiple copies in each (proto)cell.
(2) The protocells divided through the simplest imaginable mechanism, namely physical pinching of the membrane vesicles.
(3) The UA was characterized by extremely high “genetic temperature,” i.e., high rate of change represented by both mutational processes and horizontal gene transfer (HGT).
(4) Different functional systems “crystallized,” i.e., became largely refractory to HGT, asynchronously, with the translation system “crystallizing” first.
These seem to be the principal theses from which important corollaries follow:
•The UA was not an “organism” and not even, in the regular sense, a community of organisms. Here is Woese’s striking wording: “The universal ancestor is not an entity, not a thing. It is a process characteristic of a particular evolutionary stage...”
•The (ribosomal) Tree of Life was not an “organismal tree” at the time of the UA but subsequently became one, once the crystallization of the major cellular systems was (largely) complete. To use another quote: “…communal ancestor has a physical history but not a genealogical one.”4
Woese developed the themes of the “Universal Ancestor” article in the subsequent papers of the series. The next paper on “Interpreting the universal phylogenetic tree” capitalizes on the simple but non-trivial and powerful idea that emergence of biological complexity is contingent on vertical evolution, or more precisely, coordinated, coherent evolution of coevolving gene ensembles. Remarkably, this is in effect the complexity hypothesis of Lake and colleagues9 in reverse: Lake and co-workers presented evidence of reduced HGT for genes encoding components of multi-subunit complexes, whereas Woese postulated that curtailment of HGT itself was a condition of the evolution of complex cellular organization. The difference is not trivial, not only because reversals of true statements are generally not guaranteed to be true, neither in formal logic nor in real life, but for the more specific reason of causation reversal. Under the complexity hypothesis, fine-tuned complexes evolve, gradually making HGT of the genes encoding their components increasingly deleterious. Conversely, Woese’s scenario holds that, for appreciable complexity to evolve, “genetic temperature” has to drop first, thus allowing coherent evolution of the componentry of complex systems. The two views can be reconciled by postulating concomitant complexification and “genetic cooling” (Fig. 1).
In this second paper, Woese also, to my knowledge, for the first time, links HGT and the universality of the genetic code, a long-standing, fundamental evolutionary enigma and the first major direction of Woese’s research on evolution10: “Horizontal gene transfer selectively maintains the universality of the genetic code (regardless of how it became established in the first place) because the code is an evolutionary lingua franca required for an essential “genetic commerce” among lineages”.6 This theme was further developed in the later, joint work of Woese with Vetsigian and Goldenfeld, in which the feasibility of selection for a universal genetic code as an “innovation-sharing protocol” is supported by detailed mathematical modeling.11 The principle of sharing novelty as the mainstream route of evolution is likely to be quite general. As elegantly formulated by Woese, “only global invention arising in a diverse collection of primitive entities is capable of providing the requisite novelty.”6
The principal message of Woese’s second tract seems to be that “the universal phylogenetic tree based on rRNA is a valid representation of organismal genealogy.”6 Moreover, this tree is perceived as a major evolutionary trend that “transcends the era of modern cells; its deepest branchings extend back in time to an era when cellular entities were considerably more primitive than cells are today.” According to Woese, the initial bifurcation of the universal, three-domain tree of life, at which the bacterial domain diverged from the common ancestor of archaea and eukaryotes, corresponds to the stage of “genetic cooling” when the cohesiveness between the evolution of different components becomes sufficient for the existence of defined organismal lineages.
The third treatise in the series directly addresses the problem of cellular evolution, arguably the paramount problem of evolutionary biology (“the greatest of evolutionary problems” according to Woese), and the central theme of the “millennial series” of treatises.5 Woese’s key proposition is that the root of the universal tree, i.e., the Last Universal Common Ancestor (LUCA; specified here more precisely than the UA, which was initially treated as the universal ancestor not directly linked to the tree4), could have been an entity that was different in kind from any modern cells. In other words, in part, cellular evolution is postulated to have occurred within the span of the universal tree, and hence, should be tractable through comparative genomic approaches. Woese further equates the LUCA with a “Darwinian Threshold,” a major evolutionary transition that involves cohesion of cellular lineages, i.e., the onset of speciation or simply origin of species (hence Woese’s name for this transition).5,12 Much of the discussion revolves around the fundamental importance of the universal ribosomal tree and its spirited defense against claims that the tree has been “uprooted” by the discovery of extensive horizontal gene transfer (HGT) in the microbial world.13-15 To Woese, the ribosomal tree remains the essential framework of cellular evolution (strikingly, this 2002 treatise reproduces the single illustration of the 2000 paper, which is a rough sketch of the ribosomal tree; the abstract tree that is the only illustration in Darwin’s book12 inevitably comes to mind). Woese vehemently rejects the anti-tree argument, and notably with it, the symbiogenesis scenarios for the origin of eukaryotes:16,17 to him, the three domains of cellular life discerned in the ribosomal tree represent the fundamental truth of evolution, not to be compromised by postulating that one of the domains evolved through a symbiosis between fully developed cells of the other two domains.
The theme of the three domains of life reemerges in the fourth article of the series7 that I mention here out of the chronological order due to the special circumstances of its appearance that make this article something of an appendix to the three main treatises in the series, yet an integral part of it. The article is a spirited refutation of a paper by a great evolutionary biologist of a different, classical school, Ernst Mayr, in which he proposes abandoning the three-domain classification of life forms for the more traditional prokaryote–eukaryote dichotomy.18 Woese’s point in his rebuttal is simple but powerful: paraphrasing Darwin, he states that “Our classifications will come to be, as far as they can be so made, “representations of the evolutionary course.”7 The word “course” appears important here, for the implication that it is a central evolutionary trend not necessarily all the details of the universal ribosomal tree (let alone of the evolution of other genes) that are to be reflected in the global classification. That general course of evolution, according to Woese (and the ribosomal tree), firmly establishes the three-domain classification as opposed to Mayr’s two empires.
The View From 2013
The four single author papers of Carl Woese discussed above, with their dates symmetrically framing the beginning of the new millennium, present a coherent vision of biological evolution (at least, its early, crucial stages that involved the origin of cells and differentiation of the domains of cellular life forms) by perhaps the most important architect of the new (after molecular biology) revolution in life sciences.19 Such a vision is important by definition, so I believe it is not only interesting but genuinely instructive to take stock of the status of Woese’s ideas 15 y after the appearance of the first article in the series and 11 y past the last article. Given the inherent generality and depth of Woese’s concepts, their fate is bound to go beyond simple “right or wrong”: some of his thoughts turn out to be prescient in ways Woese himself hardly could anticipate, whereas others fail, also in an unexpected manner. In what follows, I trace some of the key new developments that extend and transform Woese’s ideas.
The Decline and Renaissance of “Tree Thinking”
Without second-guessing the motivations behind Woese’s conceptual treatises, it is safe to note that the universal ribosomal tree, the discovery of Woese’s life, is at the center of his vision of evolution. Naturally, he carefully explores the meaning and implications of this tree, and critically scrutinizes the arguments that have been harnessed against its status as the “Tree of Life” (TOL). There were, indeed, good reasons to be concerned over the fate of the ribosomal TOL in the genomic era. Only a decade after the TOL was firmly established and applied to resolve the major groups of bacteria, archaea, and eukaryotes,20 it was challenged by the first results of phylogenomics, which indicated that the tree topologies for most genes did not agree with the topology of the TOL.13-15,21 Although some of these discrepancies certainly stemmed from inaccuracies of phylogenetic methods, the early results of comparative genomics made it abundantly clear that the incongruence of tree topologies was due primarily to HGT, which has emerged as a major genome-wide phenomenon, at least in the evolution of bacteria and archaea.14,22 In particular, strong evidence of massive gene transfer between hyperthermophilic archaea and bacteria has been obtained.23,24 This fundamental dichotomy—the universality and stability of the ribosomal TOL vs the diversity of tree topologies of other genes—produced a strong tension and considerable controversy in phylogenomics, and more generally in evolutionary biology.
The discovery of extensive HGT triggered a concerted attack on “tree thinking” in evolutionary biology.13-15,21,25 Under this view, the TOL has no merit as a depiction of the course of evolution, a tool of evolutionary reconstruction or even a metaphor.26,27 As long as the TOL is uprooted, the very distinction between vertical and horizontal evolutionary processes becomes moot, and the appropriate depiction of evolution would have to be a network of gene exchanges rather than a tree.28-30 Furthermore, it has been proposed that the appearance of a tree-like relationship between organisms could come solely from a gradient of gene exchanges such that groups of organisms that most extensively intermix their gene pools would come across as “monophyletic.”31
These conceptual developments steeped in the data of comparative genomics certainly did not succeed in banishing the TOL from the practice of genomics and microbiology: it continued to be routinely used for purposes of classification and taxonomy. Moreover, considerable effort was being undertaken to refine the TOL, in particular, with regard to resolving deep branches through phylogenetic analysis of multiple universal genes whose evolutionary history was thought to be more or less free of HGT. Such effort typically involves construction of a phylogenetic tree from concatenated alignments of genes encoding ribosomal proteins and other components of the translation machinery.32 Perhaps, the most poignant episode of the TOL saga is the debate around a well-publicized attempt by Bork and colleagues to develop an automated pipeline for buildinga “highly resolved tree of life” by constructing a phylogeny of 31 universal genes that primarily encode translation system components.33 This new genomic TOL was immediately dubbed the “Tree of one percent” by Dagan and Martin, who eloquently argued that the history of one percent of the genes, even if accurately reconstructed (which is far from being guaranteed), at best represents just that, the history of 1% of the genetic material rather than the history of the respective organisms.34 Hence, the already familiar call for replacing a tree with a network as the major framework of evolutionary study.
Taking a step back and considering the problem in the conceptual plane, it is impossible to deny that a tree is a natural representation of evolution of an individual gene inasmuch as the frequency of recombination within a gene is substantially lower than the frequency of intergenic recombination.35 This is certainly the case for genes from organisms that are sufficiently divergent to rule out homologous recombination. The pertinent question with regard to the status of the TOL, then, is whether the “forest” of individual gene trees encompasses any order (i.e., congruence of tree topologies), and if so, which trees best reflect that order. My colleagues and I compared the topologies of approximately 7000 trees of the most commonly occurring bacterial and archaeal genes.36 We found that, despite the widespread inconsistency of tree topologies in this “forest of life,” the trees of approximately 100 nearly universal genes, almost all of them encoding translation system components, showed distinct and special properties. These trees were not only, to a large extent, topologically congruent among themselves, but also displayed highly significant topological similarity to more than half of the entire set of the analyzed trees. Thus, this group of nearly universal trees represented a unique, statistically significant central trend in the phylogenetic forest—a strong signal of vertical evolution. A quantitative estimate of the tree-like and net-like contributions to the evolution of archaea and bacteria showed that although evolutionary relationships between bacterial and archaeal genomes are shaped primarily by horizontal gene transfers, the central tree-like trend embodied in the evolution of the nearly universal genes is the strongest specific phylogenomic signal.37 The results of these comprehensive phylogenomic studies are strikingly compatible with Woese’s vision: evolution of the translation system indeed seems to represent a central tree-like trend with which evolution of other cellular systems conforms in varying degrees, depending on the extent of “crystallization.” This vertical trend is the best candidate for a TOL that has to be re-conceptualized as a “statistical tree of life” but remains the best available framework for evolutionary reconstruction.38 Although Woese did not consider such reinterpretation of the TOL, it seems to be fully within the spirit of his treatise, which emphasizes the dynamics of genomes, especially at the early stages of evolution, along with the stability of the translation system.
Asynchronous Crystallization of Cellular Systems: A General Guiding Principle of Cellular Evolution?
“Asynchronous crystallization” is perhaps the most original and productive aspect of Woese’s vision of cellular evolution. The results of comparative genomics bring this concept into an ever-sharpening focus. As already pointed out above, there are very few genes that are conserved (i.e., represented by readily detectable orthologs) in all cellular life forms. Altogether, this small, nearly universal set consists of approximately 100 RNA and protein-coding genes, nearly all of which are involved in translation.39 The divide between this exclusive set and the rest of the gene universe is quite sharp as the great majority of the other genes are relatively rare (Fig. 2).
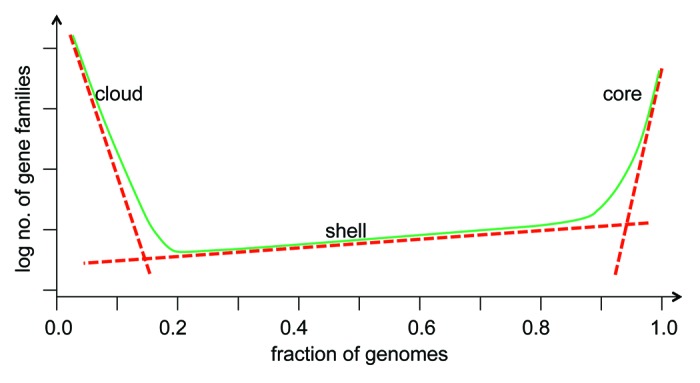
Figure 2. The universal core and the dispersed periphery of the gene universe. The plot is an idealized gene frequency distribution: the green curve shows the number of gene families that are represented in a given fraction of archaeal and bacterial genomes. The three parts of the distribution, the nearly universal core, the moderately conserved “shell,” and the “cloud” of rare genes are approximated by three different exponential functions (red lines). Modified with permission from reference 78.
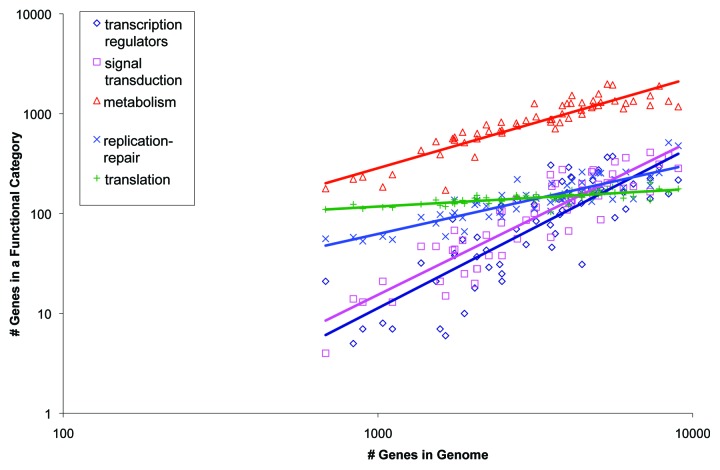
The special position of the translation system (and to a lesser extent, other information processing systems, namely transcription and replication) in all organisms becomes apparent not only through evolutionary reconstructions but also in direct analysis of the gene composition of modern life forms. Functional classes of genes scale differently with the total number of genes in a genome that, at a coarse grain approximation, converge to three distinct exponents: 0 for the information processing, 1 for metabolic enzymes and transporters, and 2 for genes involved in regulation and signal transduction (Fig. 3).40-42 These distinct scaling laws are (nearly) universal among bacteria and archaea, i.e., are reproduced with good precision in diverse groups of microbes43 In other words, the set of genes encoding the components of information processing systems, primarily translation, remains nearly constant, independent of the genome size, in contrast to the dynamic evolution of the genes encoding operational functions. Thus, Woese’s concept of asynchronous crystallization of cellular systems seems to capture a central principle of the evolution of life (as in an even more general sense does Schroedinger’s “aperiodic crystal”44). Conceivably, a stable, “crystalline” information processing system is a necessary condition for the very existence of elaborate, multifunctional supramolecular machines that are even the simplest cells.
Figure 3. Scaling of different functional classes of bacterial and archaeal genes with the total number of genes in a genome. Modified with permission from reference 41.
The Importance of Diverse Gene Pools: From Genomics to Pangenomics
In accord with Woese’s evolutionary vision, the dynamic evolution of operational systems is complementary to the stability of the translation apparatus. Analysis of the rapidly growing database of complete genomes of diverse archaea and bacteria yields ample evidence in support of this complementarity and suggests a broader distinction between the core genome that is common to all isolates of a given species (the difficulties of defining species in bacteria and archaea notwithstanding) and the isolate-specific accessory genome.41,45,46 For most bacteria and archaea, even closely related isolates (as judged, for example, by near identity of rRNA sequences) often possess widely different repertoires of accessory genes, clearly due to an interplay betweenextensive gene loss and HGT. Thus, the gene complement of a species is adequately described not by genome that shows substantial between-isolate variation but by pangenome, i.e., the union of the genes that are accessible to the organisms of the given species.45 The current sampling of microbial genome diversity is insufficient to obtain a reliable pangenome size distribution but it is clear that many pangenomes are quite large, at least an order of magnitude bigger than typical genomes, so that genome sequencing of numerous isolates does not approach saturation of accessory gene repertoires.46
There is increasing evidence that microbial genome diversity not only translates into vast pangenomes but represents an essential aspect of the life of microbial communities (microbiomes), providing both for cooperation between microbes, e.g., through diverse “public goods” supplied by specific metabolic pathways, and for antagonistic relationships, e.g., those mediated by antibiotics.47-49 Thus, the dynamic evolution of the “genomic periphery” (the genes outside of the stable cores) emphasized by Woese as a condition for early cellular evolution remains a perpetually essential aspect of microbial evolution and global ecology.
The Domains of Life and Origin of Eukaryotes
Woese perceived the three-domain structure of the TOL as a fundamental division of life forms that under his vision emerged as the final step in the evolution of the basic cellular organization. This firm belief in the “crystallization” of the three domains from primitive cell-like entities fueled Woese’s aversion to symbiotic scenarios for the origin of eukaryotes.6 In my view, this position ignores the plain fact that the ribosomal TOL, all its importance notwithstanding (see above), reflects primarily the evolution of information processing systems, above all, translation, but not the entire course of cellular evolution. The core genome of eukaryotes is a chimera that consists of approximately one-third genes of archaeal descent that largely encode components of information processing systems and about two-thirds genes of bacterial origin, which encode mostly operational functions.50-52 This chimeric genetic complement seems to be best compatible with the scenario in which the symbiosis between an archaeon and an α-proteobacterium (the mitochondrial ancestor) triggered eukaryogenesis. Strong arguments have been presented that a large cell of typical eukaryotic dimensions simply cannot function without a highly efficient energy-transforming system that can be provided only by endosymbionts,53 and plausible scenarios have been proposed for the origin of the complex internal organization of the eukaryotic cell, including the nucleus, in the wake of the archaeo-bacterial symbiosis.54,55 Furthermore, there is growing evidence that the genes of archaeal provenance in eukaryotes evolved from a distinct branch of archaea, possibly the so-called TACK (Thaumarchaeota-Aigarchaeota-Crenarchaeota-Korarchaeota) superphylum,56-58 rather than from a common ancestor with all extant Archaea as implied by Woese’s three-domain model (Fig. 1).
Given the above observations and argument, but perhaps even more importantly, because all extant eukaryotes appear to possess mitochondria or their degraded derivatives,59,60 the symbiogenetic scenarios for the origin of eukaryotes now seem to hold the upper hand over the original three-domain scenario. This does not in any way undermine the fundamental importance of the three-domain structure of the ribosomal TOL, which indeed appears to be an accurate reflection of the history of cellular information processing systems. Remarkably, despite all the advances of genomics and metagenomics, so far there are no convincing indications that any additional domains of cellular life exist. Thus, under the symbiogenetic scenario for the origin of eukaryotes, there are two basic cell types, archaeal and bacterial, which differ in many fundamental features, above all, the structure and biogenesis of the cell membranes and the organization and origin of the DNA replication machinery.61,62 These major differences between archaeal and bacterial cells give credence to Woese’s idea that the last common ancestor of modern cells (LUCA) lacked a fully established cellular organization and at least in some respects was a primitive entity compared with modern cells.4-6,63 Specific evolutionary scenarios along these lines have been proposed under some of which the LUCA was a pre-cellular ensemble of semi-autonomous genetic elements that replicated inside networks of inorganic compartments64-66 whereas others postulate a cell-like LUCA with primitive membranes.67,68
Darwinian Threshold and the Agency of Selection
Woese aligned the Darwinian threshold, the origin of species, with the divergence of the three cellular types (Fig. 1). There seems to be some lack of clarity here with respect to the agency of selection at early stages of evolution antedating LUCA. The divergence of the cellular domains—as discussed above, more likely two than three—might correspond to the origin of cellular species. However, competition between and selection of distinct replicating entities is essential for evolution from the outset, thus, a different type of “organisms” (molecular species) subject to selection must have preceded the cellular life forms. Conceivably, such primitive units of evolution could have been represented by small, virus-like replicons that populated abiogenic lipid vesicles or inorganic compartments and were subject to selection for replication efficiency.65,69 In the course of evolution, such small replicons would accrete to form larger genomes, those that carried favorable combinations of genes attaining selective advantage. Once such growing replicons reached the level of complexity sufficient for the formation of cells, the major evolution transition to cellular life forms (reproducers sensu Maynard Smith and Szathmary70) associated with viral parasites would occur—Woese’s Darwinian threshold would be crossed.
The Progenote: Back to the RNA World
The central idea of the progenote concept was originally proposed by Woese in the context of the discussion of the origin of the universal genetic code. In his early prescient work, Woese proposed that the modern, unambiguous code evolved from a primitive, ambiguous one that provided for the synthesis of “statistical” polypeptides whose sequence was determined by the sequence of the cognate gene sequences with only limited accuracy.10,71 Although the principal features of the translation system that provide for high efficiency and accuracy of protein synthesis are fixed in all extant life forms, it is unimaginable that the translation system emerged in this advanced form.72 Thus, some type of progenote stage appears to be an almost inevitable evolutionary intermediate. Woese’s hypothesis is more specific in proposing that the LUCA was actually a progenote. However, this does not appear to be a realistic possibility given the advanced stage of protein evolution that already was reached by the time the LUCA existed, according to comparative-genomic reconstructions.39 In particular, the catalytic domains of class I aminoacyl-tRNA synthetases, universal, essential enzymes of the translation system that are readily traced to LUCA, represent only a small twig in the evolutionary tree of the Rossmann-fold of nucleotide-binding domains that evolved only after the diversification of several major branches of these domains.73 Fully compatible observations were made on the evolution of GTPases where the essential, universal translation factors represent a late development.74 Evolution of numerous, diversified protein domains would not have been possible with a “statistical” translation system, hence the non-trivial conclusion that high efficiency and fidelity of protein synthesis were already achieved with a primarily RNA-based translation system.75 Thus, Woese’s progenote apparently belongs at a very early stage of evolution, within the hypothetical primordial RNA World. In contrast, the LUCA, regardless of its other characteristics, possessed a much more advanced translation system that was required for the advent of an extensive repertoire of protein domain that can be traced to this era in the evolution of life.
The Microbial World and Evolution of Life
One of the major themes,to which Carl Woese repeatedly turned in his conceptual articles, is the crucial importance of microbes for the maturation of new evolutionary biology, free from the tight constraints of the Modern Synthesis.76 In my view, he was absolutely correct in this perspective on the evolution of life, which even now might not be fully appreciated by the mainstream evolutionary biology. Thanks in a major part to the efforts of Woese and his school, we now have a solid phylogenetic scaffold against which evolution of cellular systems and biological phenomena can be reconstructed. Such reconstructions reveal a picture of evolution that is incomparably richer, more complex, and multidimensional than the architects of the Modern Synthesis envisaged when they integrated Darwinian principles with population genetics in the middle of the 20th century. Indeed, comparative genomics of bacteria and archaea, followed up by molecular biological and evolutionary experiments, revealed a panoply of novel evolutionary phenomena.77,78 The foremost among these are: (1) pervasive HGT, in large part mediated by viruses and plasmids, which shapes the pangenomes of archaea and bacteria, and demands reconceptualization (but not abandonment) of the Tree of Life, (2) Lamarckian-type inheritance that appears to be critical for antivirus defense and other forms of adaptation in prokaryotes, (3) evolution of evolvability, i.e., dedicated mechanisms for evolution such as vehicles for HGT and stress-induced mutagenesis systems. In addition, we now realize the paramount importance of the non-cellular part of the microbial world, namely viruses and related selfish genetic elements. Phylogenomics and metagenomics have revealed enormous genetic and molecular diversity and extremely high abundance of viruses and related selfish genetic elements that come across as the dominant biological entities on earth. Furthermore, the perennial arms race between viruses and their cellular hosts is one of the defining factors of evolution. Microbial phylogenomics adds new dimensions to the fundamental picture of evolution, demonstrating that the traditional concepts based on the study of the evolution of multicellular animals and plants represent only the proverbial tip of the enormous iceberg of life’s history. I cannot think of a scientist that made a greater contribution to this new era of evolutionary biology than Carl Woese.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
E.V.K. is supported by intramural funds of the US Department of Health and Human Resources (to the National Library of Medicine, NIH).
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/27673
References
- 1.Sapp J, Fox GE. The singular quest for a universal tree of life. Microbiol Mol Biol Rev. 2013;77:541–50. doi: 10.1128/MMBR.00038-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albers SV, Forterre P, Prangishvili D, Schleper C. The legacy of Carl Woese and Wolfram Zillig: from phylogeny to landmark discoveries. Nat Rev Microbiol. 2013;11:713–9. doi: 10.1038/nrmicro3124. [DOI] [PubMed] [Google Scholar]
- 3.Doolittle WF. Carl R. Woese (1928–2012) Curr Biol. 2013;23:R183–5. doi: 10.1016/j.cub.2013.01.057. [DOI] [PubMed] [Google Scholar]
- 4.Woese C. The universal ancestor. Proc Natl Acad Sci U S A. 1998;95:6854–9. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woese CR. On the evolution of cells. Proc Natl Acad Sci U S A. 2002;99:8742–7. doi: 10.1073/pnas.132266999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woese CR. Interpreting the universal phylogenetic tree. Proc Natl Acad Sci U S A. 2000;97:8392–6. doi: 10.1073/pnas.97.15.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woese CR. Default taxonomy: Ernst Mayr’s view of the microbial world. Proc Natl Acad Sci U S A. 1998;95:11043–6. doi: 10.1073/pnas.95.19.11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woese CR, Fox GE. The concept of cellular evolution. J Mol Evol. 1977;10:1–6. doi: 10.1007/BF01796132. [DOI] [PubMed] [Google Scholar]
- 9.Jain R, Rivera MC, Lake JA. Horizontal gene transfer among genomes: the complexity hypothesis. Proc Natl Acad Sci U S A. 1999;96:3801–6. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woese C. The Genetic Code. (Harper & Row, 1967). [Google Scholar]
- 11.Vetsigian K, Woese C, Goldenfeld N. Collective evolution and the genetic code. Proc Natl Acad Sci U S A. 2006;103:10696–701. doi: 10.1073/pnas.0603780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darwin C. On the origin of species. 1859. [Google Scholar]
- 13.Pennisi E. Is it time to uproot the tree of life? Science. 1999;284:1305–7. doi: 10.1126/science.284.5418.1305. [DOI] [PubMed] [Google Scholar]
- 14.Doolittle WF. Lateral genomics. Trends Cell Biol. 1999;9:M5–8. doi: 10.1016/S0962-8924(99)01664-5. [DOI] [PubMed] [Google Scholar]
- 15.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–9. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 16.Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:255–74. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 17.Margulis L. Origin of eukaryotic cells. (Yale University Press, 1970). [Google Scholar]
- 18.Mayr E. Two empires or three? Proc Natl Acad Sci U S A. 1998;95:9720–3. doi: 10.1073/pnas.95.17.9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldenfeld N, Woese C. Biology’s next revolution. Nature. 2007;445:369. doi: 10.1038/445369a. [DOI] [PubMed] [Google Scholar]
- 20.Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–71. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doolittle WF. Uprooting the tree of life. Sci Am. 2000;282:90–5. doi: 10.1038/scientificamerican0200-90. [DOI] [PubMed] [Google Scholar]
- 22.Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu Rev Microbiol. 2001;55:709–42. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aravind L, Tatusov RL, Wolf YI, Walker DR, Koonin EV. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 1998;14:442–4. doi: 10.1016/S0168-9525(98)01553-4. [DOI] [PubMed] [Google Scholar]
- 24.Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–9. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 25.O’Malley MA, Boucher Y. Paradigm change in evolutionary microbiology. Stud Hist Philos Biol Biomed Sci. 2005;36:183–208. doi: 10.1016/j.shpsc.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Doolittle WF, Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc Natl Acad Sci U S A. 2007;104:2043–9. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bapteste E, Susko E, Leigh J, MacLeod D, Charlebois RL, Doolittle WF. Do orthologous gene phylogenies really support tree-thinking? BMC Evol Biol. 2005;5:33. doi: 10.1186/1471-2148-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci U S A. 2008;105:10039–44. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–87. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 30.Dagan T. Phylogenomic networks. Trends Microbiol. 2011;19:483–91. doi: 10.1016/j.tim.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–38. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 32.Wolf YI, Rogozin IB, Grishin NV, Koonin EV. Genome trees and the tree of life. Trends Genet. 2002;18:472–9. doi: 10.1016/S0168-9525(02)02744-0. [DOI] [PubMed] [Google Scholar]
- 33.Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–7. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 34.Dagan T, Martin W. The tree of one percent. Genome Biol. 2006;7:118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koonin EV, Wolf YI. The fundamental units, processes and patterns of evolution, and the tree of life conundrum. Biol Direct. 2009;4:33. doi: 10.1186/1745-6150-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puigbò P, Wolf YI, Koonin EV. Search for a ‘Tree of Life’ in the thicket of the phylogenetic forest. J Biol. 2009;8:59. doi: 10.1186/jbiol159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Puigbò P, Wolf YI, Koonin EV. The tree and net components of prokaryote evolution. Genome Biol Evol. 2010;2:745–56. doi: 10.1093/gbe/evq062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Malley MA, Koonin EV. How stands the Tree of Life a century and a half after The Origin? Biol Direct. 2011;6:32. doi: 10.1186/1745-6150-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koonin EV. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol. 2003;1:127–36. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- 40.van Nimwegen E. Scaling laws in the functional content of genomes. Trends Genet. 2003;19:479–84. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- 41.Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grilli J, Bassetti B, Maslov S, Cosentino Lagomarsino M. Joint scaling laws in functional and evolutionary categories in prokaryotic genomes. Nucleic Acids Res. 2012;40:530–40. doi: 10.1093/nar/gkr711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molina N, van Nimwegen E. Scaling laws in functional genome content across prokaryotic clades and lifestyles. Trends Genet. 2009;25:243–7. doi: 10.1016/j.tig.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 44.Schroedinger E. What is Life?: With Mind and Matter and Autobiographical Sketches. Cambridge University Press. [Google Scholar]
- 45.Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008;11:472–7. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 46.Lapierre P, Gogarten JP. Estimating the size of the bacterial pan-genome. Trends Genet. 2009;25:107–10. doi: 10.1016/j.tig.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 47.Morris JJ, Lenski RE, Zinser ER. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio. 2012;3:3. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sachs JL, Hollowell AC. The origins of cooperative bacterial communities. MBio. 2012;3:3. doi: 10.1128/mBio.00099-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boon E, Meehan CJ, Whidden C, Wong DH, Langille MG, Beiko RG. Interactions in the microbiome: communities of organisms and communities of genes. FEMS Microbiol Rev. 2014;38:90–118. doi: 10.1111/1574-6976.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Esser C, Ahmadinejad N, Wiegand C, Rotte C, Sebastiani F, Gelius-Dietrich G, Henze K, Kretschmann E, Richly E, Leister D, et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 2004;21:1643–60. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- 51.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–30. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 52.Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929–34. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 53.Koonin EV. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010;11:209. doi: 10.1186/gb-2010-11-5-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–5. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 55.López-García P, Moreira D. Selective forces for the origin of the eukaryotic nucleus. Bioessays. 2006;28:525–33. doi: 10.1002/bies.20413. [DOI] [PubMed] [Google Scholar]
- 56.Guy L, Ettema TJ. The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 2011;19:580–7. doi: 10.1016/j.tim.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Williams TA, Foster PG, Nye TM, Cox CJ, Embley TM. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc Biol Sci. 2012;279:4870–9. doi: 10.1098/rspb.2012.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martijn J, Ettema TJ. From archaeon to eukaryote: the evolutionary dark ages of the eukaryotic cell. Biochem Soc Trans. 2013;41:451–7. doi: 10.1042/BST20120292. [DOI] [PubMed] [Google Scholar]
- 59.van der Giezen M. Hydrogenosomes and mitosomes: conservation and evolution of functions. J Eukaryot Microbiol. 2009;56:221–31. doi: 10.1111/j.1550-7408.2009.00407.x. [DOI] [PubMed] [Google Scholar]
- 60.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4:a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leipe DD, Aravind L, Koonin EV. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peretó J, López-García P, Moreira D. Ancestral lipid biosynthesis and early membrane evolution. Trends Biochem Sci. 2004;29:469–77. doi: 10.1016/j.tibs.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 63.Koonin EV. On the origin of cells and viruses: primordial virus world scenario. Ann N Y Acad Sci. 2009;1178:47–64. doi: 10.1111/j.1749-6632.2009.04992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin W, Russell MJ. On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Philos Trans R Soc Lond B Biol Sci. 2003;358:59–83, discussion 83-5. doi: 10.1098/rstb.2002.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koonin EV, Martin W. On the origin of genomes and cells within inorganic compartments. Trends Genet. 2005;21:647–54. doi: 10.1016/j.tig.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martin W, Baross J, Kelley D, Russell MJ. Hydrothermal vents and the origin of life. Nat Rev Microbiol. 2008;6:805–14. doi: 10.1038/nrmicro1991. [DOI] [PubMed] [Google Scholar]
- 67.Mulkidjanian AY, Bychkov AY, Dibrova DV, Galperin MY, Koonin EV. Origin of first cells at terrestrial, anoxic geothermal fields. Proc Natl Acad Sci U S A. 2012;109:E821–30. doi: 10.1073/pnas.1117774109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mulkidjanian AY, Galperin MY, Koonin EV. Co-evolution of primordial membranes and membrane proteins. Trends Biochem Sci. 2009;34:206–15. doi: 10.1016/j.tibs.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koonin EV, Senkevich TG, Dolja VV. The ancient Virus World and evolution of cells. Biol Direct. 2006;1:29. doi: 10.1186/1745-6150-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maynard Smith J, Szathmary E. The Major Transitions in Evolution. Oxford University Press. 1997. [Google Scholar]
- 71.Woese CR. On the evolution of the genetic code. Proc Natl Acad Sci U S A. 1965;54:1546–52. doi: 10.1073/pnas.54.6.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolf YI, Koonin EV. On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization. Biol Direct. 2007;2:14. doi: 10.1186/1745-6150-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aravind L, Anantharaman V, Koonin EV. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA. Proteins. 2002;48:1–14. doi: 10.1002/prot.10064. [DOI] [PubMed] [Google Scholar]
- 74.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 75.Aravind L, Mazumder R, Vasudevan S, Koonin EV. Trends in protein evolution inferred from sequence and structure analysis. Curr Opin Struct Biol. 2002;12:392–9. doi: 10.1016/S0959-440X(02)00334-2. [DOI] [PubMed] [Google Scholar]
- 76.Woese CR, Goldenfeld N. How the microbial world saved evolution from the scylla of molecular biology and the charybdis of the modern synthesis. Microbiol Mol Biol Rev. 2009;73:14–21. doi: 10.1128/MMBR.00002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koonin EV. The Logic of Chance: The Nature and Origin of Biological Evolution. FT Press. 2011. [Google Scholar]
- 78.Koonin EV, Wolf YI. Evolution of microbes and viruses: a paradigm shift in evolutionary biology? Front Cell Infect Microbiol. 2012;2:119. doi: 10.3389/fcimb.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]