Abstract
The Ribosomal Database Project (RDP) grew out of Carl Woese’s vision of how rRNA comparative methods could transform biology. First at the University of Illinois Urbana-Champaign, and later at Michigan State University’s Center for Microbial Ecology, the project has grown from a few hundred to several million rRNA gene sequences. In the years since Woese started the RDP, publications describing the database and related tools have been cited over 11 000 times in journals spanning a wide range of disciplines, while the RDP website is accessed by 10 000 researchers in over 20 000 analysis sessions each month. This article describes the history of RDP’s development over the last two decades.
Keywords: rRNA, phylogeny, microbial ecology, microbiology, database, Woese
Beginning Days of RDP
Carl Woese and his colleague Gary Olsen realized that making the rRNA sequence data they were using to derive phylogenetic relationships between organisms available to the research community would help stimulate more such research. In 1989 they obtained initial funding from the National Science Foundation to help make these resources available and on 5 January 1992, the first version of the Ribosomal Database Project (RDP) was released.1 This first version was available via an FTP site hosted by Argonne National Laboratory, an early indication of the importance to multiple government agencies of the types of science enabled by RDP. This release consisted of 471 hand-aligned 16S rRNA sequences mostly generated by Woese’s own research group, plus the “AE2” alignment editor to enable researchers to add their locally generated sequences to this alignment. By Release 3 in August 1993, the number of bacterial 16S rRNA sequences had increased to 1379,2 a great scientific achievement, but one that was taking more and more time to properly curate using the tools available at the time.
November 1993 Phone Call
In November 1993 I (Tiedje) received a message that I was to call Carl Woese, and that he wanted to talk about the future of RDP. I was on sabbatical at the University of Hawai’i with Dave Karl. I phoned him back and his point was this: he wanted to plan to get RDP located at a place where the ongoing science meshed with RDP so that RDP’s development could be guided by science needs, and he thought microbial ecology would be the driver of ribosomal sequence data use. We, being the Center for Microbial Ecology (CME), were in our fourth year of funding by the National Science Foundation as a Science and Technology Center, one of the first class with 11 such centers. This new NSF program was widely publicized and, hence, to Carl and others, it was seen that microbial ecology had legs and more solid prospects for stable funding. The NSF funding was for 11 years, and database funding even at that time was difficult to obtain, so Carl also hinted at the fact that stable funding would be needed to grow RDP. In hindsight, Carl was right in foreseeing microbial ecology as the lead dog for RDP and that the funding had to be a part of the strategy.
A perhaps interesting sidelight of my November phone call from Hawai’i is that Carl’s brother-in-law was an executive of Hawaiian Airlines and that Carl was familiar with the islands, having visited his relatives on several occasions. This led to our discussing whether the young age and geographic isolation of Hawai’i might be seen in ecological and evolutionary patterns of microbes as has been so famous for a number of higher organisms. His opinion reflected his focus, that Hawai’i soil microbes would be no different than those anywhere (in a similar habitat), but he did acknowledge that his opinion was based on the SSU rRNA gene. Perhaps this fortuitous Hawai’i link got the Michigan State University–University of Illinois relationship off on a comfortable footing.
At the time of Carl’s phone call, RDP had 1687 aligned sequences, the database consisted of flat files, and he personally curated every sequence, even changing bases in conserved regions that he knew must be sequencing errors (something that would never be done later without documenting the change). While the quality of the database was very high for the knowledge at the time, the mode of development could never keep up with data growth and RDP was lagging farther and farther behind. A criticism voiced at the time was that RDP was behind and that Carl was not releasing important data that would help the community gain a broader perspective of microbial phylogeny. It was clear, however, that both the database structure and curation model had to change for RDP to meet the user community’s expectations.
The CME Scientific Advisory Committee was very supportive of the importance of CME investing in RDP as a community service, something appropriate for an STC Center to do. Carl came in person to our 1994 site visit, something very impressive to us since at that time he was not traveling except in exceptional situations (Fig. 1).
Figure 1. Picture cropped from group photo taken at the 1994 CME site visit. From left to right: Rich Lenski, Jim Tiedje, Larry Forney, and Carl Woese.
Joint Development Phase, and Move to MSU
One critical need to alleviate the curatorial problem was to move the RDP from its flat-file curatorial model to a modern database schema modeled to handle the unique features of sequence alignments and associated annotation. In 1995, NSF awarded joint funding for Gary Olsen at UIUC and Sakti Pramanik in MSU’s Computer Science Department to develop a database schema for RDP and to migrate RDP to a robust database system. The ObjectStore commercial object-oriented database management system (DBMS) was chosen for this implementation. Although in hindsight it seems an unusual choice, at the time, the interfaces or “glue” necessary for interfacing between object-orientated languages and relational databases were still primitive. Also, mapping of sequence data and associated metadata to an object-oriented database was considered more natural by many. For example, Lincoln Stein and colleagues at the Whitehead/MIT Genome Center developed a large genome-mapping database using ObjectStore.3
One of the strengths of ObjectStore was the use of C++ objects to model data, making data access very fast. This was also its main weakness, as developing a database in C++ is more difficult and much more bug-prone than relational database development using SQL. One of us (Cole) became involved as a postdoctoral associate helping with the biological aspects of schema development, while a computer science student was recruited to do the actual database development. When the student quit before coding started, Cole took over both coding and schema development, enduring steep learning curves in both database design and C++ programming. The resulting database was used for several years, only being phased out in favor of a new relational database schema when the amount of data overwhelmed the limitations of the ObjectStore platform on the hardware of the time.
With DBMS migration underway, official support of the RDP by the Center for Microbial Ecology at MSU started December 1997 and the RDP officially moved to MSU with the release of a new website on 31 July 1998.4 During the first year of operation at MSU, this new website served over 15 000 distinct hosts from over 40 countries and averaged over 26 M bytes of data transferred per day, a significant volume for the time and indicative of the rapid adoption of rRNA comparative analysis in many branches of microbiology. Two of the RDP staff from UIUC moved to MSU, Bonnie Maidak to handle curation and Niels Larsen to handle website and tool development. Their participation was crucial for a smooth transition and for the first phase of continuing development at MSU. Also in 1998, the US DOE Office of Biological and Environmental Research (BER) began its funding of RDP, in recognition of the critical role of rRNA data to that agency’s mission as well as for many branches of science. Funding of RDP by DOE BER has continued to the present day and has been crucial in development of the RDP and, through its use, to microbial ecology globally.
This was a time of rapid development of new methods, both bench and informatics, for rRNA analysis. The ARB tool for sequence analysis was being developed by Oliver Strunk in the laboratory of Wolfgang Ludwig in Munich (Strunk and Ludwig; 1997; ARB, a software environment for sequence data; retrieved from http://download.arb-home.de/documentation/arb.pdf). The CME hosted Oliver for an extended visit, and one of the outcomes was a modified version of ARB able to directly load sequence and trees from RDP’s ObjectStore database. This early collaboration is indicative of the collegial relationship that has continued to exist between those researchers developing rRNA resources.
Further Development at MSU
Along with the increased volume of rRNA data and usage of the website came the need for improved curatorial infrastructure to handle this increase. In 2002, NSF joined DOE with a five-year award of joint funding along with a contribution by NIH to improve infrastructure and allow the RDP to become up-to-date in cataloging the increasing volume of 16S rRNA sequence data.
Two major bottlenecks in sequence annotation needed rethinking before RDP could become up-to-date with the rapid increase in rRNA sequences. First, it was clear that the number of sequences had become too large for hand aligning. The problem was that a good rRNA alignment requires attention to both primary sequence similarity and RNA secondary structure. In June 2000, RDP 8 was released with 16 277 aligned and annotated prokaryotic sequences, but with a backlog of 30 322 additional sequences awaiting alignment and classification. To solve this, we turned to RNAcad5 the first practical implementation of a stochastic context-free grammar-based automated aligner capable of aligning full-length rRNA gene sequences. (Interestingly, development of RNAcad was also funded by DOE.) This class of aligners is able to take into account the conserved rRNA secondary structure. After training on a small hand alignment, alignment of new sequences requires no manual tuning. The second bottleneck was phylogenetic placement of new sequences to provide order to the collection. Phylogenetic tree construction just did not reliably scale to the numbers of rRNA sequences available. Out of desperation, we developed an early version of what eventually became the RDP Classifier.6 With this tool we could rapidly assign most sequences into a new phylogenetically informed taxonomy,7 reserving the more time-consuming analysis to sequences representing novel lineages. With these new tools we were able to rapidly progress through the backlog and released RDP 9 in September 2002 with 50,055 aligned and annotated sequences.8 Because we were unsure how this “radical” new automated alignment and taxonomic assignment methodology would be accepted by our user community, we provided both the final hand-aligned release (RDP 8.1) and the new Release 9 in parallel for two years, by which time there were 101 632 aligned and annotated sequences in RDP 9.21, before completely committing to the new automated methods. These methods have stood the test of time. Alignment methods continued to improve with the adoption of the Infernal SCFG aligner in 2008,9 and there have been several updates to the RDP Classifier, but since Release 9.0 there have been 92 incremental sequence updates and the current release (11.1) contains 2 872 266 aligned and annotated sequences.10
Ribosomal Resources Beyond RDP
Carl’s original vision had included more than just rRNA sequence data. He had envisioned the need for a central resource for all types of ribosome-related information. Due to the limitations of funding this never happened, but important resources exist covering other areas of ribosomal data. In particular, the Comparative RNA website contains secondary-structure information for rRNA and other structural RNAs11 and is a resource critical to RDP’s alignment strategy. With the advent of next-gen sequencing, the RDP made a decision to leave curation of high-throughput short rRNA gene segments to the primary nucleic-acid databases (INSDC), as it is difficult to use these to define new lineages due to their short length and relatively low accuracy.
Carl’s vision on the importance of ribosomal data led to cousins of RDP: the ARB/SILVA project was started by Wolfgang Ludwig at the Technical University Munich to support his phylogenetic research;12,13 GreenGenes was developed at Lawrence Berkeley National Laboratory as an outgrowth of the Phylochip development by Gary Anderson and colleagues;14 and EzTaxon was developed by Jongsik Chun at Seoul National University initially for identification of validly named organisms and with a system for adding new unnamed taxa.15 All of these separately developed databases have strong followings in the microbial community but all have worked together on common issues. In fact, these three, along with RDP, applied to US DOE BER in 2010 for joint funding to better harmonize the nomenclature between the databases. The proposal was not funded and it is interesting to note that a major deficiency was that we had not included the excellent taxonomy group at NCBI in the proposal. That had been a tactical decision on our part, but reviewer criticism demonstrated that we had misjudged the DOE program’s commitment to good science across agencies.
These projects continue to cooperate on many issues important to the research community. For example, scientists from RDP, GreenGenes, and ARB/Silva are all active in the Genomic Standards Consortium16 and work together to develop metadata standards important for users of rRNA data.
Impact
The number of sequences maintained by RDP has increased over 6000-fold since Carl Woese first released RDP 1 in 1992 (Fig. 2), while the RDP website now hosts over 22 000 analysis sessions by more than 10 000 researchers each month. In 2007, we polled our users as to their field of research (Fig. 3). Since the first published article describing the RDP in 1991,17 16 additional articles have been published describing RDP. These articles have been cited over 11 000 times in journals covering many areas of research such as phylogeny, bioinformatics, dairy science, environmental microbiology, fermentation, bioengineering, gastroenterology, veterinary medicine, and AIDS to name just a few (Fig. 4).
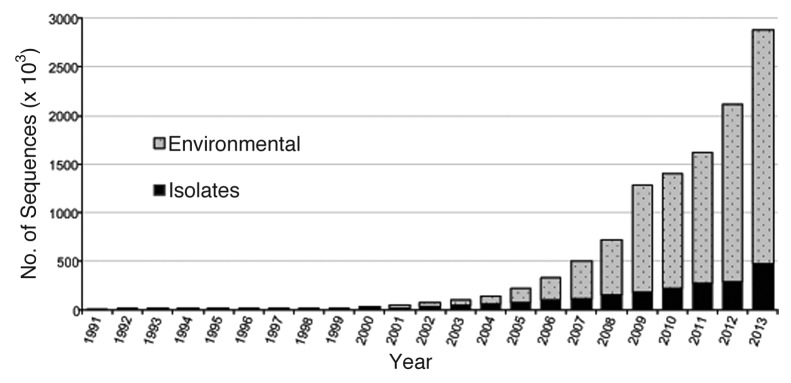
Figure 2. Growth in rRNA sequences maintained by RDP. Those coming from cultures (isolates) and from environment are delineated.
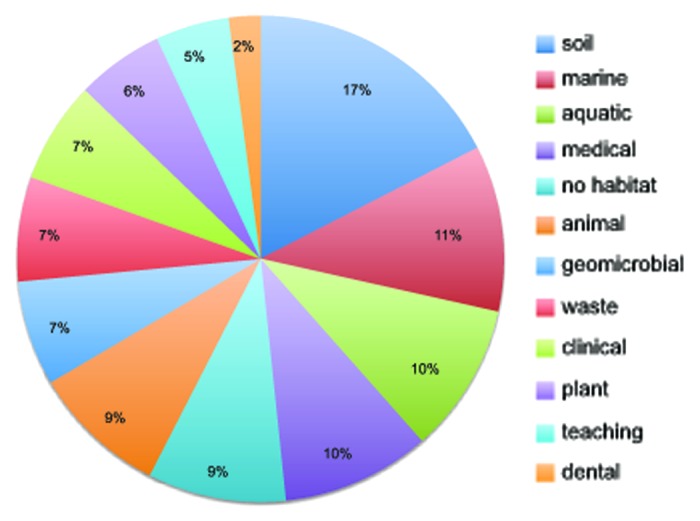
Figure 3. The broad use of RDP is shown by responses to a 2007 user survey of their field of microbiology.

Figure 4. Number of citations to the 16 publications describing RDP, by year of citation.
In the future, as more genomic data accumulates, as techniques such as single-cell sequencing become high-throughput, and as metagenomic coverage and assembly improves, it seems likely that single gene analysis will be supplanted by more comprehensive data from these newer techniques. But the knowledge gained using the rRNA-based methods pioneered by Carl Woese and his collaborators will form the backbone to be enriched by newer techniques. The insights gained into phylogenetics and the conventions developed for describing uncultured clades, for example, will be used well beyond the rRNA era.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Our continued development and the operation of RDP has been primarily supported by the U.S. Department of Energy Office of Science, Office of Biological and Environmental Research grants DE-FG02-99ER62848 and DE-FG02-98ER62678. Additional funding for RDP and related microbial informatics has come from: the U.S. Department of Energy Office of Science, through the Great Lakes Bioenergy Research Center (DE-FC02-07ER64494), DOE GTL:Genomics and Carbon Cycling Programs (DE-SC0004601), NSF (DBI-0328255), NIEHS (P42-ES004911), and NIH HMP (UH3 DK083993). We also thank the primary RDP staff for their extraordinary contributions and extra effort over the last decade: Qiong Wang, Benli Chai, Jordan Fish, Ryan Farris, Paul Saxman, Chuck Parker, Siddique Kulam, and Donna McGarrell.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/28306
References
- 1.Olsen GJ, Overbeek R, Larsen N, Marsh TL, McCaughey MJ, Maciukenas MA, Kuan WM, Macke TJ, Xing Y, Woese CR. The Ribosomal Database Project. Nucleic Acids Res. 1992;20(Suppl):2199–200. doi: 10.1093/nar/20.suppl.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maidak BL, Larsen N, McCaughey MJ, Overbeek R, Olsen GJ, Fogel K, Blandy J, Woese CR. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–7. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rozen S, Stein L, Goodman N. LabBase: A Database to Manage Laboratory Data in a Large-Scale Genome-Mapping Project. Proceedings IEEE Computers in Medicine and Biology; 1995; 702 p. [Google Scholar]
- 4.Maidak BL, Cole JR, Parker CT, Jr., Garrity GM, Larsen N, Li B, Lilburn TG, McCaughey MJ, Olsen GJ, Overbeek R, et al. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–3. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown MPS. Small subunit ribosomal RNA modeling using stochastic context-free grammar. Proceedings of the Eighth International Conference on Intelligent Systems for Molecular Biology (ISMB 2000); 2000; San Diego, CA, USA; 57 p. [PubMed] [Google Scholar]
- 6.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garrity GM, Winters M, Kuo AW. Searles DB. Taxonomic Outline of the Prokaryotes. Bergey’s Manual of Systematic Bacteriology, 2nd ed. New York: Springer-Verlag; 2002. [Google Scholar]
- 8.Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, et al. Ribosomal Database Project The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003;31:442–3. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–42. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Müller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chun J, Lee J-H, Jung Y, Kim M, Kim S, Kim BK, Lim Y-W. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol. 2007;57:2259–61. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- 16.Yilmaz P, Kottman R, Field D, Knight R, Cole JR, Amaral-Zettler L, Gilbert JA, Karsch-Mizrachi I, Johnston A, Cochrane G, et al. The “Minimum Information about a MARKer gene Sequence” (MIMARKS) checklist: Capturing contextualcontextual data about marker gene sequences and introducing MIxS, a unified standard for sequence checklist development including environmental data. Nat Biotechnol. 2011;29:415–20. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsen GJ, Larsen N, Woese CR, Woese CR. The ribosomal RNA database project. Nucleic Acids Res. 1991;19(Suppl):2017–21. doi: 10.1093/nar/19.suppl.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]



