Abstract
A high-quality NMR solution structure is presented for protein hMcl-1(171–327) which comprises residues 171–327 of the human anti-apoptotic protein Mcl-1 (hMcl-1). Since this construct contains the three Bcl-2 homology (BH) sequence motifs which participate in forming a binding site for inhibitors of hMcl-1, it is deemed to be crucial for structure-based design of novel anti-cancer drugs blocking the Mcl1 related anti-apoptotic pathway. While the coordinates of an NMR solution structure for a corresponding construct of the mouse homologue (mMcl-1) are publicly available, our structure is the first atomic resolution structure reported for the ‘apo form’ of the human protein. Comparison of the two structures reveals that hMcl-1(171–327) exhibits a somewhat wider ligand/inhibitor binding groove as well as a different charge distribution within the BH3 binding groove. These findings strongly suggest that the availability of the human structure is of critical importance to support future design of cancer drugs.
Introduction
The malfunctioning of cellular apoptosis [1] is a major hallmark of cancer. The regulation of apoptosis depends on the family of Bcl-2 proteins which contain one or several Bcl-2 homology (BH) sequence motifs. Based on their function and the similarity of their respective BH sequence motifs, these proteins can be grouped into three classes [2],[3]: (i) multi-domain pro-apoptotic proteins such as Bax and Bak, (ii) anti-apoptotic (i.e., pro-survival) proteins such as Mcl-1, Bcl-1, Bcl-xL, Bcl-w and Bfl-1/A1, all of which exhibit a similar architecture as Bax and Bak, and (iii) several pro-apoptotic proteins comprising only a single BH3 sequence motif such as Bid, Bad, Bim, Puma, Noxa, Hrk, Bmf, and Nbk/Bik (‘BH3-only’ proteins). The BH3 motif of class (iii) proteins forms an amphipathic α-helix which interacts specifically with a hydrophobic pocket formed in both pro-apoptotic class (i), and anti-apoptotic class (ii) proteins with participation of their respective BH motifs [2],[3]. Inhibition of the resulting protein-protein complex formation offers a promising strategy to treat cancer. For example, the small molecule Bcl-2 antagonist ABT-737 [4] inhibits anti-apoptotic class (ii) proteins Bcl-xL, Bcl-w and Bcl-1, and a congener [5] that can be orally administered is currently in clinical trials.
The anti-apoptotic, pro-survival 350-residue protein Mcl-1 (‘myeloid cell leukemia-1’) [2] is primarily anchored in the outer mitochondrial membrane by a C-terminal trans-membrane domain and contains three BH sequence domains: BH3 (residues 209–223), BH1 (252–272) and BH2 (residues 304–319) [2]. Mcl-1 inhibits death receptor-induced apoptosis by selectively binding to truncated Bid (tBid) [6] and can sequester endogenous Bak to block Bak-mediated cell death. Moreover, Mcl-1 interacts with several BH3-only proteins (Bim, Bid and Puma, Noxa and Bak). Hence, Mcl-1 plays an early role in response to signals directing either cell survival or cell death [2] and has been shown to be up-regulated in numerous malignant tumors. Approaches abrogating the Mcl-1’s anti-aptototic function either by reducing its abundance or by inactivating its functional BH3-binding groove show great promise for the cancer treatment [2],[4],[6],[7]. Here we present the high-quality NMR solution structure of polypeptide segment 171–327 of human Mcl-1 (hMcl-1) which comprises the three BH motifs deemed to be crucial for structure based drug design.
Results and Discussion
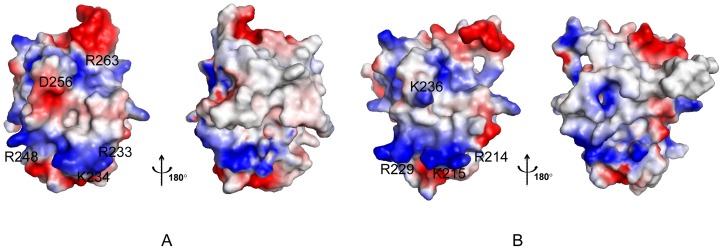
A high-quality NMR structure of hMcl-1(171–327) was obtained (Table 1) and the coordinates were deposited in the PDB [8] (accession code 2mhs). The structure comprises seven α-helices α1-α7 (residues 173–191, 204–235, 240–253, 262–280, 284–301, 303–308 and 311–319) arranged to form the characteristic ‘Bcl-2 core’ structure [9] (Figure 1). The helices are locally and globally well-defined, while the C-terminus (residues 320–327) and the loops connecting, respectively, helices α1 and α2, helices α3 and α4, and helices α4 and α5 are flexibly disordered. The central helix α4 is surrounded by the other six helices, with α1, α2, α3 and α5 packed around one side, and α6 and α7 packed against its N-terminus. Helices α2, α3, α4 and α7 participate in forming the BH3 binding groove. The electrostatic protein surface potential is positive at both ends of the BH3 binding groove (due to the presence of Arg 233, Lys 234, Arg 248 and Arg 263) and negative at the side of helix α3 side (due to Asp 256) (Figure 2). This shows that the charge distribution in the BH3 binding groove of hMcl-1(171–327) differs distinctly from other anti-apoptotic proteins [10].
Table 1. Statistics of hMcl-1(171–327) NMR Structure.
| Completeness of stereo-specific assignments [%]a | |
| αCH2 of Gly | 55 (6/11) |
| βCH2 | 38 (27/71) |
| Val and Leu methyl groups | 100 (27/27) |
| Conformationally restricting distance constraints | |
| Intra-residue [i = j] | 1052 |
| Sequential [|i-j| = 1] | 1062 |
| Medium range [1 < |i-j| < 5] | 1197 |
| Long range [|i-j| ≥ 5] | 1058 |
| Total | 4369 |
| Didedral angle constraints | |
| φ | 113 |
| ψ | 113 |
| Number of constraints per residue (170–327) | 29.1 |
| Number of long range constraints per residue (170–327) | 6.7 |
| CYANA target function [Å2] | 0.88±0.12 |
| Number of distance constraints violations per CYANA conformer | |
| 0.2–0.5 Å | 0 |
| > 0.5 Å | 0 |
| Number of dihedral-angle constraint violations per CYANA conformer | |
| > 5° | 0 |
| Average rmsd to the mean CNS coordinates [Å] | |
| Α-helices,b backbone heavy atoms N, Cα, C’ | 0.42±0.05 |
| Α-helices,b all heavy atoms | 0.88±0.07 |
| Residues 172–312, backbone heavy atoms N, Cα, C’ | 0.65±0.13 |
| all residues, all heavy atoms | 1.05±0.10 |
| PROCHECK [38] G-factors raw score | |
| (φ and ψ/all dihedral angles)c | 0.34/0.22 |
| PROCHECK [38] G-factor Z - score | |
| (φ and ψ/all dihedral angles)c | 1.65/1.30 |
| MOLPROBITY[39] clash score (raw/Z - score)c | 20.88/−2.06 |
| AutoQF R/P/F/DP scores [40] (%) | 96/97/96/81 |
| Ramachandran plot summaryc | |
| most favorable regions | 92.7 |
| additionally allowed regions | 7.3 |
| generously allowed regions | 0.0 |
| disallowed regions | 0.0 |
Related to pairs with non-degenerate chemical shift.
Regular secondary element: α-helical residues 173–191, 204–235, 240–253, 262–280, 284–301, 303–308 and 311–319.
Ordered residues: 172–192,194–198, 204–235, 238–255, 262–321 with dihedral angle order parameters S(φ) and S(ψ) > 0.90. Z-scores were computed relative to corresponding structure quality measures for high resolution X-ray crystal structures [42].
Figure 1. NMR structure of hMcl-1(171–327).
(A) Backbone of the 20 CYANA conformers representing the solution structure of hMcl-1(171–327) after superposition of backbone N, Cα and C’ atoms of the α-helices for minimal rmsd. The three BH sequence motifs are colored in green (BH3), red (BH1) and blue (BH2), respectively. (B) Ribbon drawing of the lowest energy conformer of hMcl-1(171–327). α-helices α1-α7 are labeled and colored differently, and the N- and C-termini are labeled as “N’ ” and “C’ ”. The figures were generated using the programs MOLMOL [36] and PYMOL [37].
Figure 2. Electrostatic surface potentials.
(A) For human hMcl-1(171–327) in the orientation shown in Figure 1 (left) and after rotation by 180° about the vertical axis (right). Surface colors (blue for positively charged; red for negatively charged) indicated the electrostatic potential calculated by using PYMOL [37] and its default vacuum electrostatics protocol. (B) Same as in (A) but for mouse mMcl-1(152–308).
Including our hMcl-1(171–327) structure, twenty atomic resolution structures containing different Mcl-1 constructs are currently deposited in the PDB. In addition to the two ‘apo’ proteins hMcl-1(171–327) and mouse mMcl-1(152–308) [10] [PDB accession code 1wsx, 89% sequence identity with the human protein], the structures for nineteen protein-ligand complexes were deposited (Table 2) [9],[11]–[18]. Clearly, the large number of available structures reflects the outstanding interest in Mcl-1 as a target for the development of new cancer drugs. Superposition of the α-helices reveals, as expected, close structural similarity for all Mcl-1 proteins structures (Figure 3): the root mean square deviation (rmsd) values range from 1.05 to 1.54 Å relative to hMcl1-1(171–327) (Table 2). However, comparison of the two apo protein structures of hMcl-1(171–327) and mMcl-1(152–308) with the complex structures shows that the binding pocket is widened upon complex formation (Table 2): the distances between the Cα-atoms of residues His 224 in helix α2 (His 205 in mMcl-1) and His 252 (His 233 in mMcl-1) at the C-terminus of helix α3 are, respectively, ∼16 Å and ∼14 Å in hMcl-1(171–327) and mMcl-1(152–308), and ∼18–21 Å in the complexes.
Table 2. Rmsd values for comparison of the NMR structure of hMcl-1(171–327) with the structures of mouse mMcl-1(152–308) and Mcl-1complexes.a .
| Mcl-1 structures | 172–193, 203–321b | α1–α7c | 209–321b | α2–α4d | α2–α7d | dCA224,252 d |
| mMcl-1e | 1.60±0.09 | 1.52±0.06 | 1.61±0.10 | 1.21±0.07 | 1.53±0.06 | 13.2–14.6 |
| hMcl-1-hBimf | 1.76±0.10 | 1.41±0.06 | 1.87±0.11 | 1.52±0.09 | 1.53±0.09 | 19.9 |
| rMcl-1-hMcl-1-mNoxaBg | 1.46±0.12 | 1.05±0.08 | 1.53±0.14 | 1.00±0.10 | 1.08±0.09 | 19.9 |
| mMcl-1-mNoxaA h | 1.52±0.11 | 1.16±0.07 | 1.59±0.13 | 1.11±0.11 | 1.18±0.08 | 18.8–20.2 |
| mMcl-1-mPumai | 1.57±0.09 | 1.30±0.08 | 1.59±0.11 | 1.24±0.11 | 1.30±0.09 | 18.3–19.9 |
| mMcl-1-mNoxaBj | 1.46±0.09 | 1.13±0.05 | 1.53±0.11 | 1.12±0.08 | 1.18±0.06 | 18.3–19.6 |
| rMcl-1-hMcl-1-hBimk | 1.75±0.09 | 1.44±0.06 | 1.86±0.11 | 1.58±0.09 | 1.56±0.07 | 19.9 |
| rMcl-1-hMcl-1-hBim(L62A, F78A) l | 1.80±0.09 | 1.44±0.06 | 1.90±0.11 | 1.53±0.08 | 1.55±0.07 | 19.8 |
| hMcl1-hBidBH3J | 1.84±0.15 | 1.54±0.09 | 1.57±0.20 | 1.30±0.15 | 1.30±0.12 | 20.8–21.4 |
| hMcl1-hBIMBH3k | 1.38±0.16 | 1.06±0.07 | 1.43±0.18 | 0.93±0.10 | 1.09±0.09 | 19.4 |
| hMcl1-BimL12Yl | 1.75±0.13 | 1.50±0.08 | 1.85±0.15 | 1.48±0.12 | 1.59±0.09 | 20.2 |
| hMcl1-BimBH3 2dAm | 1.75±0.13 | 1.50±0.08 | 1.84±0.14 | 1.46±0.12 | 1.57±0.09 | 19.7 |
| hMcl1-BimBH3 F4aEn | 1.75±0.13 | 1.47±0.08 | 1.84±0.15 | 1.44±0.08 | 1.55±0.08 | 19.9 |
| hMcl1 -B7o | 1.73±0.14 | 1.46±0.08 | 1.83±0.11 | 1.42±0.11 | 1.54±0.08 | 19.6–19.9 |
| hMcl1–hMcl1BH3p | 1.47±0.16 | 1.21±0.09 | 1.48±0.18 | 1.08±0.15 | 1.24±0.11 | 20.3 |
| hMcl1-BaxBH3q | 1.69±0.12 | 1.46±0.08 | 1.79±0.13 | 1.43±0.12 | 1.54±0.09 | 19.4 |
| mMcl1-NoxaBH3r | 1.43±0.16 | 1.20±0.10 | 1.48±0.17 | 1.10±0.13 | 1.23±0.11 | 19.1 |
| hMcl1-compound53s | 1.45±0.12 | 1.22±0.08 | 1.48±0.14 | 1.19±0.10 | 1.28±0.08 | 18.7 |
| hMcl1-compound60t | 1.45±0.13 | 1.22±0.08 | 1.47±0.12 | 1.12±0.09 | 1.25±0.08 | 17.9–19.6 |
| hMcl1-BH3u | 1.51±0.16 | 1.22±0.09 | 1.57±0.18 | 1.06±0.12 | 1.27±0.10 | 20.3 |
Average pairwise rmsd values (Å) were calculated for backbone heavy atoms N, Cα, and C’ between the 20 conformers of Mcl-1(171–327) and corresponding polypeptide segments in the other structures. The distances dCA (in Å) between the Cα-atoms of residues His 224 in helix α2 (His 205 in mMcl-1) and His 252 (His 233 in mMcl-1) at the C-terminus of helix α3 are provided as a measure for the width of the BH3 binding groove.
Residue numbers are for hMcl-1(171–327); residues 194–202 were excluded since one structure (2nl9k) does not contain the corresponding residues; residues 172–193 and 203–321 correspond to residues 153–174 and 184–302 in mMcl-1, and residues 209–321 correspond to residues 190–302 in mMcl-1.
Helices α1–α7 in hMcl-1 comprise residues 173–191, 204–235, 240–253, 262–280, 284–301, 303–308 and 311–319; the corresponding residues in mMcl-1 are: 155–172, 185–216, 221–234, 243–261, 265–282, 284–289 and 292–300.
Helices α2–α7 in hMcl-1 and residues 204–208 (numbers in hMcl-1) were excluded
Mouse mMcl-1(152–308), PDB accession code 1wsx (the mean NMR coordinates were used) [10].
Human hMcl-1 complexed with human hBim BH3, 2pqk [11].
Chimiric rat-human rMcl-1(171–208)hMcl-1(209–327) complexed with mouse mNoxaB BH3, 2nla [9].
Mouse mMcl-1 complexed with mouse mNoxaA BH3, 2rod [12].
Mouse mMcl-1 complexed with mouse mPuma BH3, 2roc [12].
Mouse mMcl-1 complexed with mouse mNoxaB BH3, 2jm6 [9].
Chimiric rat-human Mcl-1 complexed with human hBim BH3, 2nl9 [9].
Chimiric rat-human Mcl-1 complexed with human hBim (L62A, F68A), 3d7v [13].
Human hMcl1 complexed with human Bid BH3, 2kbw [15].
Human hMcl-1 complexed with human Bim BH3 mutant I2dY, 3kj0 [11].
Human hMcl-1 complexed with human BimL12Y, 3io9 [16].
Human hMcl1 complexed with human Bim BH3 mutant I2dA, 3kj1 [11].
Human hMcl1 complexed with human Bim BH3 mutant F4aE, 3kj2 [11].
Human hMcl-1 complexed with Mcl1 specific selected peptide B7, 3kz0 [41].
Human hMcl-1 complexed with human Mcl1 BH3, 3mk8 [17].
Human hMcl1 complexed with human Bax BH3, 3pk1.
Mouse mMcl-1 complexed with mouse Noxa BH3, 4g35 [18].
Human hMcl-1 complexed with 6-chloro-3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1H-indole-2-carboxylic acid, 4hw2 [14].
Human hMcl-1 complexed with 6-chloro-3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1H-indole-2-carboxylic acid, 4hw3 [14].
Human hMcl-1 complexed with human Mcl1 BH3, 4hw4 [14].
Figure 3. Superposition of selected Mcl-1 structures.
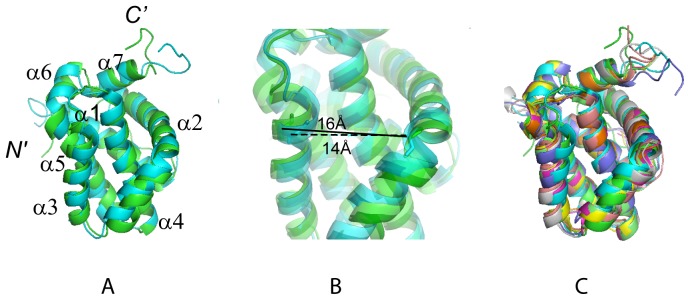
(A) Structures of hMcl-1(171–327) (green) and mMcl-1(152–308) (cyan, PDB accession code 1wsx) after superposition of the backbone N, Cα and C’ atoms of the α-helices for minimal rmsd. (B) Ribbon drawing (zoomed into (A)) showing the different binding groove widths of human (green) and mouse (cyan) protein. The distances between the Cα-atoms of residues His 224 in helix α2 (His 205 in mMcl-1) and His 252 (His 233 in mMcl-1) at the C-terminus of helix α3 are highlighted: ∼16 Å in hMcl-1(171–327) and ∼14 Å mMcl-1(152–308) (C) Superposition as in (A) of hMcl-1(171–327) (green) and mMcl-1(152–308) (cyan, 1wsx), and six selected Mcl-1 complex structures (see also Table 2): human Mcl-1 complexed with Bim BH3 (magenta, 2nla); chimeric rat-human rMcl-1(171–208)hMcl-1(209–327) complexed with mouse mNoxaB BH3 (yellow, 2rod); mouse mMcl-1(152–308) complexed with mouse NoxaA BH3 (pink, 2roc); mouse mMcl-1(152–308) complexed with mouse Puma BH3 (grey, 2jm6); mouse mMcl-1(152–308) complexed with mouse NoxaB BH3 (purple, 2rod); chimeric rat-human mMcl-1(171–208)hMcl-1(209–327) complexed with human Bim BH3 (orange, 2nl9); chimeric rat-human mMcl-1(171–208)hMcl-1(209–327) complexed with human Bim BH3(L62A, F68A) (light green, 3d7v).The figures were prepared with the programs MOLMOL [36] and PYMOL [37].
The fact that the human apo protein exhibits a somewhat wider binding groove than the mouse homologue (Table 2) can be, at least partially, ascribed to the side chain of Leu 246 in the human protein which is not buried as deeply as the corresponding Phe side chain in the mouse protein. Furthermore, when comparing the human and the mouse protein, differences are observed for the charge distributions in the BH3-binding groove (Figure 2): the human protein is negatively charged on the side of helix α3, while the corresponding surface of mouse protein is positively charged. This difference arises from Ser 255 corresponding to Lys 236 in the mouse protein. Remarkably, hMcl-1(171–327) is structurally more similar to the hMcl1(171–327)-hBim BH3 complex (Figure 3) than to apo mMcl-1(152–308) (Table 2).
Taken together, structural comparisons show that, in spite of the 89% sequence identity between human and mouse protein, the availability of the human hMcl-1(171–327) structure can be expected to be of critical importance for supporting future design of cancer drugs.
Materials and Methods
NMR Sample Preparation
Preliminary studies showed that hMcl-1(171–327) (UniProtKB/Swiss-Prot ID Q07820/MCL1_HUMAN) is not stable in solution. However, the mutant Cys 286 → Ser is stable for several weeks at concentrations ∼0.7 mM, and both wild-type and mutant bind the Bim-BH3 peptide with the same affinity (Kd ∼ 60 pM) in a Biacore assay. Hence, we solved the NMR structure of hMcl-1(171–327) Cys 286 → Ser referred to as hMcl-1(171–327) in this publication.
hMcl-1(171–327) was cloned, expressed, refolded and purified following standard protocols to produce a uniformly 13C, 15N-labeled protein sample [19]. Briefly, the gene was cloned into a pET21d (Novagen) derivative yielding plasmid pSR482-21.1. The resulting construct contains seven nonnative residues at the C-terminus (LHHHHHH) to facilitate protein purification. Escherichia Coli BL21 (DE3) pMGK cells, a codon enhanced strain, were transformed with pMcl1-21.1, and cultured in MJ9 minimal medium [20] containing (15NH4)2SO4 and U-13C-glucose as sole nitrogen and carbon sources. Double-washed inclusion bodies containing hMcl-1(171–327) were solubilized in 8 M buffered guanidine hydrochloride (VWR) containing 5 mM DTT, slowly diluted into nine volumes of 20 mM Tris-HCl, 250 mM NaCl, 0.5 M urea, 10% glycerol, pH 7.4, and refolded within 72 hours at 4°C. hMcl-1(171–327) was purified using a Talon affinity resin (Clontech) applied to a HiTrap SP High Performance column (GE Healthcare). The final yield of purified U-13C, 15N protein (> 98% homogeneous by SDS-PAGE; 20.3 kDa by MALDI-TOF mass spectrometry) was ∼25 mg/L. In addition, a U-15N and 5% biosynthetically directed fractionally 13C-labeled sample [21] was generated for stereo-specific assignment of isopropyl methyl groups. NMR samples were prepared at 0.7 mM protein concentration. An isotropic overall rotational correlation time of ∼10 ns was inferred from 15N spin relaxation times indicating that hMcl-1(171–327) is monomeric in solution.
NMR spectroscopy
NMR spectra were recorded at 25°C. Five G-matrix Fourier transform (GFT) NMR experiments [22],[23] and a simultaneous 3D 15N/13Caliphatic/13Caromatic-resolved NOESY [24],[25] spectrum (mixing time 60 ms; measurement time: 48 hours) were acquired on a Varian INOVA 750 MHz spectrometer equipped with a conventional probe. 2D constant-time [13C,1H]-HSQC spectra (18 hours) were recorded for the 5% biosynthetically directed fractionally 13C-labeled sample on a Varian INOVA 600 MHz spectrometer equipped with a cryogenic probe as was described [21],[26]. Spectra were processed and analyzed using the programs NMRPipe [27] and XEASY [28].
Sequence specific backbone (HN, Hα, N, Cα) and Hβ/Cβ resonance assignments were obtained by using (4,3)D HNNC αβ C α (63 hours)/(4,3)D C αβ C α(CO)NHN (62 hours), and (4,3)D H αβ C αβ (CO)NHN (69 hours) [23] along with the program AUTOASSIGN [29]. More peripheral side chain chemical shifts were assigned with aliphatic (4,3)D HCCH (87 hours) [23] and 3D 15N/13Caliphatic/13Caromatic-resolved [1H,1H]-NOESY [24],[25]. Overall, assignments were obtained for 96% of backbone and 1Hβ/13Cβ resonances and for 93% of the side chain resonances which are assignable with the NMR experiments listed above (excluding the N-terminal NH3 +, Pro 15N, 13C’ preceding prolyl residues, Lys NH3 +, Arg NH2, OH, side chain 13C’ and aromatic 13Cγ). Furthermore, 100%/100% of Val and Leu isopropyl moieties with non-degenerate proton chemical shifts were stereo-specifically assigned (Table 1). Chemical shifts were deposited in the BioMagResBank [30] (accession code 19654). 1H–1H upper distance limit constraints for structure calculations were obtained from NOESY (Table 1). In addition, backbone dihedral angle constraints were derived from chemical shifts using the program TALOS [31] for residues located in well-defined secondary structure elements (Table 1). The programs CYANA [32],[33] and AUTOSTRUCTURE [34] were used in parallel to assign long-range NOEs [24]. The final structure calculations were performed using CYANA followed by explicit water bath refinement using the program CNS [35].
Acknowledgments
We thank Dr. Janet Cheetham for the valuable suggestion to consider the C286S mutant of hMcl-1(171–327) for NMR structure determination.
Funding Statement
This study was funded by Amgen Inc. (www.amgen.com). LP, KA, HY and JL are employees of Amgen Inc. and Amgen Inc. played a role in study design, data collection and analysis, decision to publish and preparation of the manuscript.
References
- 1. Vogler M, Dinsdale D, Dyer MJ, Cohen GM (2009) Bcl-2 inhibitors: small molecules with a big impact on cancer therapy. Cell Death Differ 16: 360–367. [DOI] [PubMed] [Google Scholar]
- 2. Warr MR, Shore GC (2008) Unique biology of Mcl-1: therapeutic opportunities in cancer. Curr Mol Med 8: 138–147. [DOI] [PubMed] [Google Scholar]
- 3. Gelinas C, White E (2005) BH3-only proteins in control: specificity regulates MCL-1 and BAK-mediated apoptosis. Genes Dev 19: 1263–1268. [DOI] [PubMed] [Google Scholar]
- 4. Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, et al. (2005) An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435: 677–681. [DOI] [PubMed] [Google Scholar]
- 5. Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, et al. (2008) ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res 68: 3421–3428. [DOI] [PubMed] [Google Scholar]
- 6. Simmons MJ, Fan G, Zong WX, Degenhardt K, White E, et al. (2008) Bfl-1/A1 functions, similar to Mcl-1, as a selective tBid and Bak antagonist. Oncogene 27: 1421–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kang MH, Reynolds CP (2009) Bcl-2 inhibitors: targeting mitochondrial apoptotic pathways in cancer therapy. Clin Cancer Res 15: 1126–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. (2000) The Protein Data Bank. Nucleic Acids Res 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Czabotar PE, Lee EF, van Delft MF, Day CL, Smith BJ, et al. (2007) Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc Natl Acad Sci U S A 104: 6217–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Day CL, Chen L, Richardson SJ, Harrison PJ, Huang DC, et al. (2005) Solution structure of prosurvival Mcl-1 and characterization of its binding by proapoptotic BH3-only ligands. J Biol Chem 280: 4738–4744. [DOI] [PubMed] [Google Scholar]
- 11. Fire E, Gulla SV, Grant RA, Keating AE (2010) Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes. Protein Sci 19: 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Day CL, Smits C, Fan FC, Lee EF, Fairlie WD, et al. (2008) Structure of the BH3 domains from the p53-inducible BH3-only proteins Noxa and Puma in complex with Mcl-1. J Mol Biol 380: 958–971. [DOI] [PubMed] [Google Scholar]
- 13. Lee EF, Czabotar PE, van Delft MF, Michalak EM, Boyle MJ, et al. (2008) A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation. J Cell Biol 180: 341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Friberg A, Vigil D, Zhao B, Daniels RN, Burke JP, et al. Discovery of potent myeloid cell leukemia 1 (Mcl-1) inhibitors using fragment-based methods and structure-based design. J Med Chem 56: 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Q, Moldoveanu T, Sprules T, Matta-Camacho E, Mansur-Azzam N, et al. (2010) Apoptotic regulation by MCL-1 through heterodimerization. J Biol Chem 285: 19615–19624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee EF, Czabotar PE, Yang H, Sleebs BE, Lessene G, et al. (2009) Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands. J Biol Chem 284: 30508–30517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stewart ML, Fire E, Keating AE, Walensky LD (2010) The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol 6: 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muppidi A, Doi K, Edwardraja S, Drake EJ, Gulick AM, et al. (2012) Rational design of proteolytically stable, cell-permeable peptide-based selective Mcl-1 inhibitors. J Am Chem Soc 134: 14734–14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Acton TB, Gunsalus KC, Xiao R, Ma LC, Aramini J, et al. (2005) Robotic cloning and Protein Production Platform of the Northeast Structural Genomics Consortium. Methods Enzymol 394: 210–243. [DOI] [PubMed] [Google Scholar]
- 20. Jansson M, Li YC, Jendeberg L, Anderson S, Montelione GT, et al. (1996) High-level production of uniformly N-15- and C-13-enriched fusion proteins in Escherichia coli. J Biomol NMR 7: 131–141. [DOI] [PubMed] [Google Scholar]
- 21. Neri D, Szyperski T, Otting G, Senn H, Wuthrich K (1989) Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28: 7510–7516. [DOI] [PubMed] [Google Scholar]
- 22. Kim S, Szyperski T (2003) GFT NMR, a new approach to rapidly obtain precise high-dimensional NMR spectral information. J Am Chem Soc 125: 1385–1393. [DOI] [PubMed] [Google Scholar]
- 23. Atreya HS, Szyperski T (2004) G-matrix Fourier transform NMR spectroscopy for complete protein resonance assignment. Proc Natl Acad Sci U S A 101: 9642–9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu GH, Shen Y, Atreya HS, Parish D, Shao Y, et al. (2005) NMR data collection and analysis protocol for high-throughput protein structure determination. Proc Natl Acad Sci U S A 102: 10487–10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shen Y, Atreya HS, Liu GH, Szyperski T (2005) G-matrix Fourier transform NOESY-based protocol for high-quality protein structure determination. J Am Chem Soc 127: 9085–9099. [DOI] [PubMed] [Google Scholar]
- 26. Penhoat CH, Li Z, Atreya HS, Kim S, Yee A, et al. (2005) NMR solution structure of Thermotoga maritima protein TM1509 reveals a Zn-metalloprotease-like tertiary structure. J Struct Funct Genomics 6: 51–62. [DOI] [PubMed] [Google Scholar]
- 27. Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, et al. (1995) Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. J Biomol NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- 28. Bartels C, Xia TH, Billeter M, Guntert P, Wuthrich K (1995) The Program Xeasy for Computer-Supported Nmr Spectral-Analysis of Biological Macromolecules. J Biomol NMR 6: 1–10. [DOI] [PubMed] [Google Scholar]
- 29. Moseley HNB, Monleon D, Montelione GT (2001) Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Meth Enzymology 339: 91–108. [DOI] [PubMed] [Google Scholar]
- 30. Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, et al. (2008) BioMagResBank. Nucleic Acids Res 36: D402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13: 289–302. [DOI] [PubMed] [Google Scholar]
- 32. Guntert P, Mumenthaler C, Wuthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Bio 273: 283–298. [DOI] [PubMed] [Google Scholar]
- 33. Herrmann T, Guntert P, Wuthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Bio 319: 209–227. [DOI] [PubMed] [Google Scholar]
- 34. Huang YPJ, Moseley HNB, Baran MC, Arrowsmith C, Powers R, et al. (2005) An integrated platform for automated analysis of protein NMR structures. Meth Enzymology 394: 210–243. [DOI] [PubMed] [Google Scholar]
- 35. Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography 54: 905–921. [DOI] [PubMed] [Google Scholar]
- 36. Koradi R, Billeter M, Wuthrich K (1996) MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph 14: 51–55. [DOI] [PubMed] [Google Scholar]
- 37.Delano WL (2002) PyMOL molecular graphics system. Available: http://www.pymol.org
- 38. Laskowski RA, Macarthur MW, Moss DS, Thornton JM (1993) Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr 26: 283–291. [Google Scholar]
- 39. Word JM, Bateman RC, Presley BK, Lovell SC, Richardson DC (2000) Exploring steric constraints on protein mutations using MAGE/PROBE. Pro Sci 9: 2251–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang YJ, Powers R, Montelione GT (2005) Protein NMR recall, precision, and F-measure scores (RPF scores): Structure quality assessment measures based on information retrieval statistics. J Am Chem Soc 127: 1665–1674. [DOI] [PubMed] [Google Scholar]
- 41. Dutta S, Gulla S, Chen TS, Fire E, Grant RA, et al. (2010) Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL. J Mol Biol 398: 747–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bhattacharya A, Tejero R, Montelione GT (2007) Evaluating protein structures determined by structural genomics consortia. Proteins 66, 778–795. [DOI] [PubMed] [Google Scholar]