Abstract
A method for non-invasive visualization of genetically labelled cells in animal disease models with micron-level resolution would greatly facilitate development of cell-based therapies. Imaging of fluorescent proteins (FPs) using red excitation light in the “optical window” above 600 nm is one potential method for visualizing implanted cells. However, previous efforts to engineer FPs with peak excitation beyond 600 nm have resulted in undesirable reductions in brightness. Here we report three new red-excitable monomeric FPs obtained by structure-guided mutagenesis of mNeptune, previously the brightest monomeric FP when excited beyond 600 nm. Two of these, mNeptune2 and mNeptune2.5, demonstrate improved maturation and brighter fluorescence, while the third, mCardinal, has a red-shifted excitation spectrum without reduction in brightness. We show that mCardinal can be used to non-invasively and longitudinally visualize the differentiation of myoblasts and stem cells into myocytes in living mice with high anatomical detail.
INTRODUCTION
Mammalian tissues are relatively transparent to red wavelengths of light within the “optical window” (600–1000 nm), where hemoglobin and myoglobin absorbances are relatively low (Fig. 1a)1,2. Compared to bluer wavelengths, red light also generates less tissue autofluorescence from excitation of NAD(P)H, flavins, and lipofuscin3. FPs that can be excited by red light are therefore useful as genetically encoded markers for non-invasive deep-tissue imaging in mammals4. Monomeric FPs are particularly desirable, as they can be fused to other protein domains to create reporters of biochemical pathways or cellular states5,6. Monomeric FPs that can be excited in the optical window include conventional autocatalytic far-red FPs7–9, the light-induced far-red fluorescent form of PSmOrange10, and the bacteriophytochrome-based infrared fluorescent protein, IFP1.411.
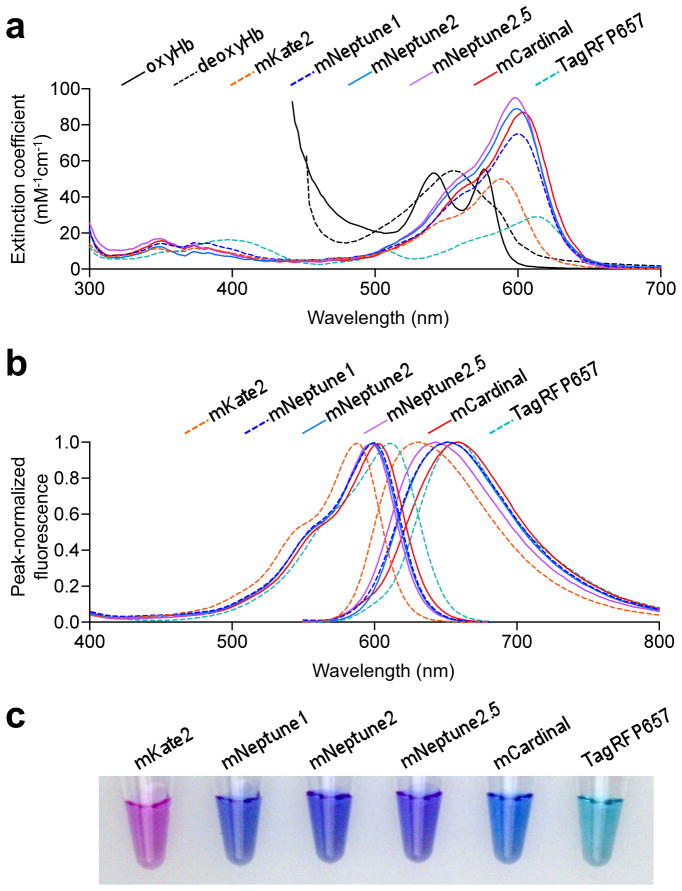
Figure 1.
Spectral characteristics of new far-red FPs. (a) Absorbance spectra of oxygenated hemoglobin (oxyHb), deoxygenated hemoglobin (deoxyHb), and monomeric far-red FPs. Myoglobin spectra are similar to hemoglobin spectra29. (b) Normalized excitation (left) and emission (right) spectra of monomeric far-red FPs. (c) Transmittance of mKate2, mNeptune1, mNeptune2, mNeptune2.5, mCardinal, and TagRFP657 at 1 mg/mL of purified mature protein. Blue transmission of mCardinal is due to efficient absorbance of green and red light.
Of these, the autocatalytic far-red FPs have practical advantages for in vivo imaging. Most of them are brighter than the far-red form of PSmOrange10 or IFP1.411. Furthermore, they can mature in the dark, unlike PSmOrange, and independent of trans-acting factors, unlike IFP1.4. We previously engineered mNeptune, derived from eqFP578 of Entacmaea quadricolor, as a bright autocatalytic FP with peak excitation at 600 nm7. mNeptune has a large Stokes’ shift (50 nm) and intrinsic brightness (ε = 75 mM−1 cm−1, ϕ = 0.23 according to our recent measurements, Table 1) similar to the widely used red FP mCherry. Because subsequent efforts shifting the absorbance peak of FPs beyond 600 nm also resulted in decreased brightness12, mNeptune still generates higher fluorescence contrast over background than other monomeric and oligomeric autocatalytic FPs in living animals when excited with light beyond 620 nm10,13,14. Nevertheless, further red-shifting would be desirable if brightness can be maintained, as wavelengths of light beyond the hemoglobin absorbance tail could then be used for illumination.
Table 1.
Characteristics of far-red and infrared FPs
| mCherryh | mKate 2 | mNeptune 1 | mNeptune 2 | mNeptune 2.5 | mCardinal | TagRFP 657 | E2-Crimsoni | eqFP650i | eqFP670i | IFP1.4j | iRFPk | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Excitation peak (nm) | 587 | 588 | 600 | 599 | 599 | 604 | 611 | 605 | 592 | 605 | 684 | 690 |
| Emission peak (nm) | 610 | 630 | 651 | 651 | 643 | 659 | 659 | 646 | 650 | 670 | 707 | 713 |
| ε at peak (mM−1cm−1)a | 72 | 50 | 75 | 89 | 95 | 87 | 29 | 59 | 65 | 70 | 102 | 105 |
| ϕ totalb | 0.22 | 0.40j | 0.23 | 0.24 | 0.28 | 0.19 | 0.10 | 0.12 | 0.24 | 0.060 | 0.070 | 0.059 |
| BR excited at peakc | 16 | 20 | 17 | 21 | 27 | 17 | 2.9 | 7.1 | 16 | 4.2 | 7.1 | 6.2 |
| ε at 635 nm (mM−1cm−1) | 0.62 | 1.4 | 9.6 | 12 | 11 | 18 | 13 | 19 | 5.9 | 20 | 55 | 52 |
| ϕ > 635 nm | 0.094 | 0.25 | 0.18 | 0.19 | 0.20 | 0.16 | 0.09 | 0.10 | 0.18 | 0.055 | 0.070 | 0.059 |
| BR excited at 635 nmd | 0.058 | 0.35 | 1.9 | 2.2 | 2.2 | 2.8 | 1.2 | 1.9 | 1.1 | 1.1 | 3.9 | 3.1 |
| Maturation half-time (min)e | 40 | 38 | 28 | 27 | 26 | 27 | 88 | 24 | ND | ND | 114 | 168 |
| Maturation efficiencyf | ND | 0.52 | 0.48 | 0.53 | 0.52 | 0.60 | 0.66 | ND | ND | ND | ND | ND |
| Photostability (s)g | 68 | 81 (84j) | 160 | 373 | 506 | 730 | 110 | ND | ND | ND | ND | ND |
| pKa | <4.5 | 6.5 | 5.4 | 6.3 | 5.8 | 5.3 | 5.1 | 4.5 | 5.7 | 4.5 | 4.6 | 4.0 |
| quaternary structure (m = monomer, d = dimer) | m | m | m | m | m | m | m | d | d | d | m | d |
| Red/green fluorescence ratiol | ND | 4.1 | 2.4 | 5.6 | 8.2 | 5.8 | 0.47 | ND | ND | ND | ND | ND |
ε = extinction coefficient, measured as in ref. 30.
ϕ = quantum yield.
Calculated as the product of ε at peak excitation and ϕ in units of mM−1cm−1.
Calculated as the product of ε at 635 nm, emission fraction above 635 nm, and ϕ in units of mM−1cm−1.
Time for fluorescence to obtain half-maximal value after exposure to oxygen.
Functional chromophore concentration divided by total protein concentration.
Predicted time for fluorescence to photobleach by 50% under arc lamp illumination with excitation intensity adjusted to produce 1000 emission photons per molecule per s.
Data from ref. 7.
Data from ref. 14.
Data from ref. 13.
Data from ref. 19.
Ratio of far-red emission peak to green emission peak when excited by 490–510 nm light.
In this study, we applied structure-guided mutagenesis to engineer both brighter and more red-shifted derivatives of mNeptune. The red-shifted variant, mCardinal, can be efficiently imaged in cells and organisms with 633–635 nm laser lines and enables orthogonal imaging with orange-red FPs. We used mCardinal to perform non-invasive visualization of muscle cell regeneration in living mice with high anatomical resolution.
RESULTS
Engineering of far-red fluorescent proteins
First, to improve chromophore maturation and packing, we mutagenized residues surrounding the chromophore. We also mutated Ile-171 in the former cross-dimer interface to reduce hydrophobicity that may be energetically unfavorable for monomer folding. After screening bacterial colonies for chromophore absorbance and maturation by visual inspection, and for fluorescence with 600–620 nm excitation and 630–660 nm emission filters, we obtained one variant with internal mutations A104V and I121L and external mutation I171H (Supplementary Fig. 1). This protein retained the excitation and emission spectra of mNeptune (Fig. 1b, 1c), but was 25% brighter (ε = 89 mM−1 cm−1, ϕ = 0.24). We named it mNeptune2 and designated the original mNeptune as mNeptune1.
We next performed mutagenesis to optimize a hydrogen bond interaction first introduced into FPs in mNeptune and its dimeric variant Neptune. In Neptune, a water molecule donates a hydrogen bond to the carbonyl oxygen in the acylimine portion of the chromophore7. This may have a bathochromic effect by preferentially stabilizing the excited state, in which electron density over the carbonyl oxygen is believed to be increased relative to the ground state. We hypothesized that substituting a genetically encoded hydrogen bond donor for the water molecule could improve brightness by reducing excited state vibrations that could lead to non-radiative decay, and/or cause further red-shifting by increasing the strength or uniformity of the hydrogen bond interaction. Saturation mutagenesis at positions in the beta barrel wall facing the acylimine oxygen (Fig. 2a) yielded one mutant that appeared brighter, mNeptune2 S28H G41N, and another that appeared red-shifted in absorbance, mNeptune2 S28T G41Q. We then mutagenized amino acids interacting with position 28, position 41, or the chromophore phenolate and selected for improved maturation. This yielded mNeptune2 M11T S28H G41N and mNeptune2 S28T G41Q S143T. The first protein, which we designated mNeptune2.5, is 25% brighter than mNeptune2 and 54% brighter than mNeptune1 (ε = 95 mM−1 cm−1, ϕ = 0.28), but is slightly blue-shifted in excitation and emission (peak ex/em, 599/643 nm, Fig. 1b). The second protein is red-shifted in excitation and emission (peak ex/em, 604/659 nm, Fig. 1b) while retaining the brightness of mNeptune1 (ε = 87 mM−1 cm−1, ϕ = 0.19). We named this protein mCardinal for its cardinal-red emission.
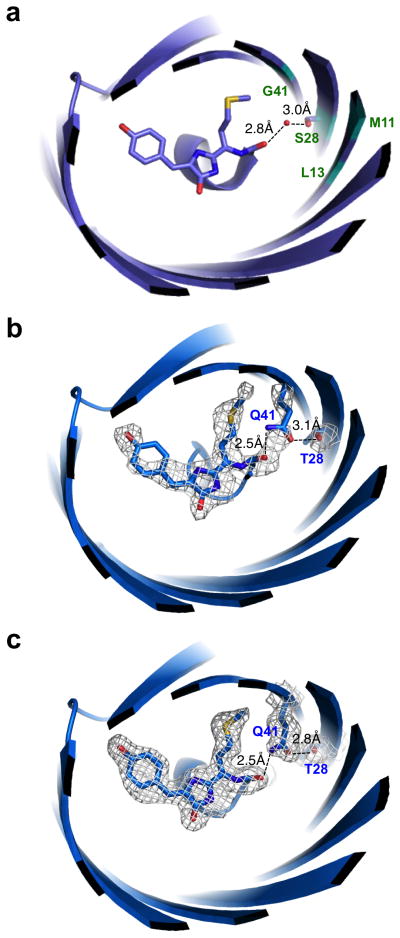
Figure 2.
Structural basis of red-shifting in mCardinal. (a) Structure of Neptune showing the water molecule hydrogen-bonded to the acylimine oxygen of the chromophore. The fluorescent protein is viewed down the axis of the beta-barrel from the direction of the N- and C-termini. Beta-strands and alpha-helices are depicted in cartoon form. The chromophore, the Met63 side chain attached to the chromophore, and the water are depicted in stick representation with carbon colored lavender, nitrogen blue, and oxygen red. Numbers indicate distances between the water oxygen atom and hydrogen-bonding partners. Cyan labels indicate positions mutated to amino acids with side chains capable of donating hydrogen bonds to the acylimine oxygen: Met11, Leu13, Ser28 and Gly41. (b,c) Hydrogen bond interactions between Thr28, Gln41, and the chromophore acylimine in mCardinal (b) or mCardinal-V218E (c). Rendering is of chain A. Distances shown are averages of measurements from chains with temperature factors for Gln41 amide atoms lower than those for the hydrogen-bonding water molecule in the Neptune structure, specifically chains A and B for mCardinal and chains A and C for mCardinal-V218E. Mesh depicts electron density in the Fo-Fc OMIT map contoured to 3.0 sigma within 2.0 Å of the atoms displayed in stick form.
Characterization of far-red fluorescent proteins
To understand the mechanism of red-shifting in mCardinal, we determined its structure at pH 7.5 to 2.2 Å resolution by X-ray crystallography (Supplementary Table 1). The structure revealed that, in place of the water molecule in Neptune (Fig. 2a), the amide group of Gln41 in mCardinal donates a hydrogen bond to the chromophore acylimine oxygen (Fig. 2b). The donor-acceptor distance is shorter in mCardinal compared to Neptune (2.5 vs 2.8 Å), suggesting a stronger hydrogen bond15, which could provide additional stabilization of the excited state in mCardinal compared to Neptune. In addition, the Thr28 hydroxyl group of mCardinal appears to be engaged in a hydrogen bond with the Gln41 amide oxygen, positioning the amide nitrogen for hydrogen bonding to the chromophore acylimine oxygen. In Neptune, Ser28 served a similar role in positioning a water molecule to hydrogen-bond with the chromophore acylimine oxygen. To further confirm these findings, we also crystallized mCardinal with an external V218E mutation (introduced to reduce dimericity) in a different space group and solved its structure at 1.7 Å resolution (Supplementary Table 1). This second structure confirmed the presence of a new shorter hydrogen bond from Gln41 to the chromophore acylimine oxygen, as well a hydrogen bond between Thr28 and Gln41 (Fig. 2c). Thus, crystal structures of mCardinal validate our original design approach of introducing a stronger genetically encoded hydrogen bond to the acylimine oxygen.
mCardinal is more readily excitable at the common laser wavelength of 635 nm than mNeptune1 (ε 18 vs. 9.6 mM−1 cm−1). It also has at least 50% higher quantum yield above 635 nm than the monomeric TagRFP657, dimeric eqFP670, or tetrameric E2-Crimson (Table 1), previously described autofluorescent FPs with redder peak absorbance than mNeptune18,14. Thus mCardinal would be expected to be brighter than all other far-red FPs upon 635 nm excitation, even when collecting emission at infrared wavelengths beyond 700 nm (Supplementary Fig. 2a). In tests with purified proteins, mCardinal was indeed brighter upon excitation by 625–655 nm light than other autofluorescent proteins (Supplementary Fig. 2b). mCardinal was also brighter when excited by light beyond 670 nm (Supplementary Fig. 2b). However, the two-photon cross-section of mCardinal may be smaller than that of mNeptune1, as mCardinal was dimmer at equivalent concentrations when excited at wavelengths from 800 to 1080 nm, the furthest possible with our Ti-Sapphire laser (Supplementary Fig. 3).
We characterized mNeptune2, mNeptune2.5, and mCardinal for chromophore maturation, pH stability, and photostability (Table 1). Compared to mNeptune1, the new FPs exhibit equal or faster red chromophore maturation at 37°C (Supplementary Fig. 4a), higher final maturation efficiency (Table 1), and reduced formation of a green fluorescent side-product that is common to eqFP578 derivatives and is especially high in TagRFP657 (Table 1, Supplementary Fig. 4b). The fluorescence of mCardinal was more resistant to acidic pH than the other mNeptune derivatives (Supplementary Fig. 4c). The new FPs were similar to mNeptune1 in monomeric character (Supplementary Fig. 4d), and improved in photostability under arc lamp illumination in vitro (Supplementary Fig. 5a). In cells under wide-field and laser illumination, mCardinal exhibited photostability between mKate2 and mNeptune1 (Supplementary Fig. 5b,c). All mNeptune-derived FPs exhibited an increase in fluorescence indicating photoactivation prior to photobleaching in vitro and in cells, with the exception of mNeptune1 in vitro.
We further assessed the performance of the new FPs in mammalian cells. We observed that mNeptune2 and mCardinal were approximately 20% and 50% brighter, respectively, than mNeptune1 in HeLa cells, while mNeptune2.5 was 35% dimmer (Supplementary Fig. 6). The difference between mCardinal and mNeptune1 is larger than expected from the spectral properties of purified proteins, indicating higher expression of mCardinal relative to mNeptune1 in HeLa cells. Likewise, the lower brightness of mNeptune2.5 relative to mNeptune1 in cells despite being 40% brighter in vitro suggests lower expression. mNeptune2 and mCardinal perform well in primary neurons (Supplementary Fig. 7a), as well as in primary mouse myoblasts in both proliferating and differentiating states (Supplementary Fig. 7b). A variety of N-terminal and C-terminal fusions to mNeptune2 (Supplementary Fig. 8) or mCardinal (Supplementary Figs. 9 and 10) showed correct subcellular localization and could be imaged with high contrast with 633 nm excitation light.
Performance of far-red fluorescent proteins in vivo
As the performance of mCardinal in all metrics matched or exceeded the other mNeptune variants, we proceeded to test mCardinal in animals. To determine if mCardinal could be used to provide an additional imaging channel beyond orange-red FPs, we created C. elegans worms expressing GFP::Rab-3 in neurons, synaptogyrin::mCardinal in neurons, and the orange-red DsRed in muscles. By epifluorescence microscopy, we observed the expected colocalization of punctate red and green fluorescence in synaptic vesicles apposed to the orange fluorescence of the body wall (Supplementary Fig. 11a), confirming that mCardinal is bright enough to label small subcellular structures and can be visualized orthogonally to orange-red FPs. To test the ability to visualize three different structures, we expressed GFP, mCherry, and mCardinal in primary epithelial cells, muscle cells, and secondary epithelial cells of the vulva, respectively. Each cell type was observed only in its expected channel (Supplementary Fig. 11b), indicating that the low amount of residual green fluorescence in mCardinal does not interfere with GFP imaging and that mCherry and mCardinal can also be visualized orthogonally. mCardinal thus provides another imaging channel that allows separation from common orange or orange-red FPs.
We tested whether mCardinal can be used for extended time-lapse microscopy or fast confocal microscopy. We continuously scanned mCardinal-expressing cells with a 633-nm laser at 60× magnification for over 20 minutes without apparent toxicity (Supplementary Videos 1 and 2). In worms, we were able to visualize mCardinal in pharyngeal muscles during free movement and serotonin-induced pharyngeal contractions by confocal microscopy with a galvonometer-driven 635-nm laser (Supplementary Videos 3 and 4). We observed no toxicity or photobleaching for over 200 frames while tracking the worms. Thus, 633–635 nm excitation light enables extended imaging of mCardinal without toxicity, and mCardinal is bright enough to allow for fast time-lapse confocal imaging in a multi-cellular organism.
Deep-tissue imaging of cellular differentiation
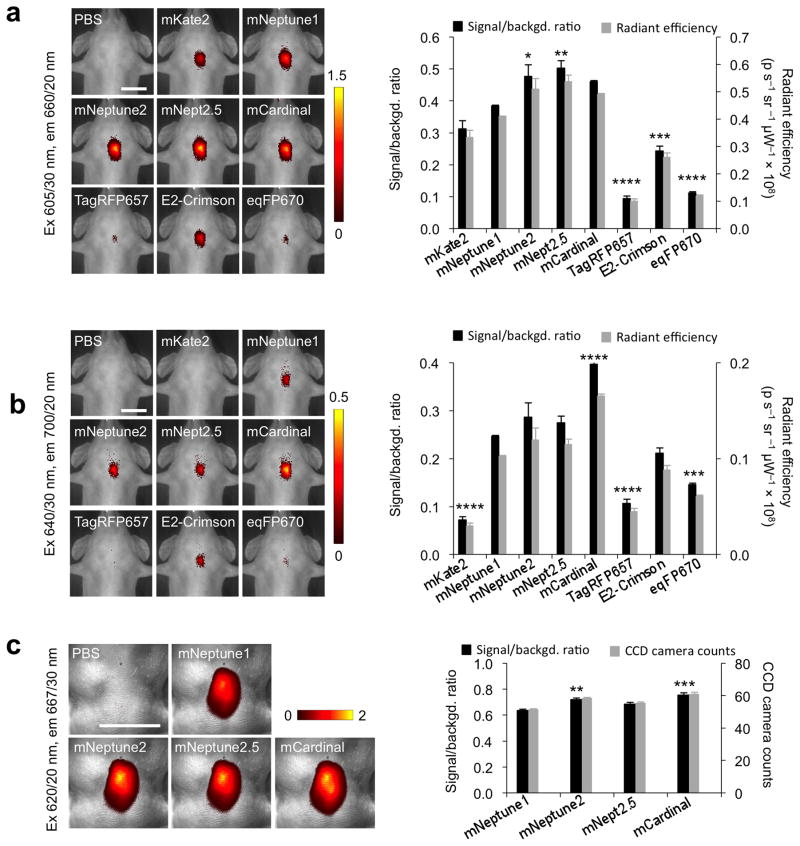
As discussed, a primary motivation for developing far-red FPs is to perform deep-tissue imaging in mammals using excitation wavelengths above 600 nm, which penetrate through hemoglobin-rich tissues more easily than bluer light16. To directly compare the imaging performance of far-red FPs in deep tissue, we first imaged purified far-red proteins placed within the esophagus of an euthanized mouse using a commercial small-animal fluorescence imaging system. Imaged from the ventral side, this location is approximately 7 mm deep. With 605/30 nm excitation light (center wavelength/full width at half-maximum), a range that covers the excitation peaks of all tested FPs, mNeptune2.5 was the brightest (Fig. 3a), consistent with mNeptune2.5 having the highest peak brightness by spectroscopy (Table 1). When using 640/30 nm light, mCardinal was uniquely brighter than all other far-red FPs (Fig. 3b), consistent with its predicted advantage at these redder excitation wavelengths (Table 1). We also compared mCardinal with iRFP, a bacteriophytochrome mutant that binds biliverdin to enhance its infrared fluorescence, for the ability to detect gene expression in the liver following hydrodynamic injection of plasmid DNA. Compared to iRFP excited at 675/30 nm, mCardinal yielded a higher signal/background ratio with 605/30 nm excitation and a similar signal/background ratio with 640/30 nm excitation (Supplementary Fig. 12). In preparation for μm-resolution imaging in mouse tissues, we equipped a fluorescence stereoscope using 620/20 nm excitation filters, selected so that all excitation wavelengths are beyond 600 nm while still efficiently exciting far-red FPs. In this system, mCardinal again yielded the highest brightness among the mNeptune derivatives (Fig. 3c). Thus, mCardinal performs especially well in deep-tissue imaging with wavelengths of light above 600 nm.
Figure 3.
Comparison of far-red FPs for deep-tissue imaging. (a,b,c) Left, representative fluorescence images of equal amounts of purified far-red FPs placed within the esophagus of euthanized mice. Images were acquired with 605/30 nm excitation in an IVIS Spectrum (a), 640/30 nm excitation in an IVIS spectrum (b), or with 620/20 nm excitation on a SZX-12 fluorescence stereomicroscope (c). Pseudocolor scale represents signal-to-background ratio, calculated as (FFP − FPBS)/FPBS, where FFP and FPBS were total intensities measured in a common region of interest encompassing all fluorescence signal in images of FP and PBS, respectively. Scale bars, 1 cm. Right, quantification of signal-to-background ratio and signal intensity subtracted by background in units of 108 photons per second per steradian (p s−1 sr−1 μW−1 × 108) presented as mean ± standard error of the mean (SEM), n = 4 for (a,b) and 5 for (c). Differences are statistically significant by one-way ANOVA (P < 0.0001). Asterisks indicate significant differences by Dunnett’s multiple comparison test versus mNeptune1 (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
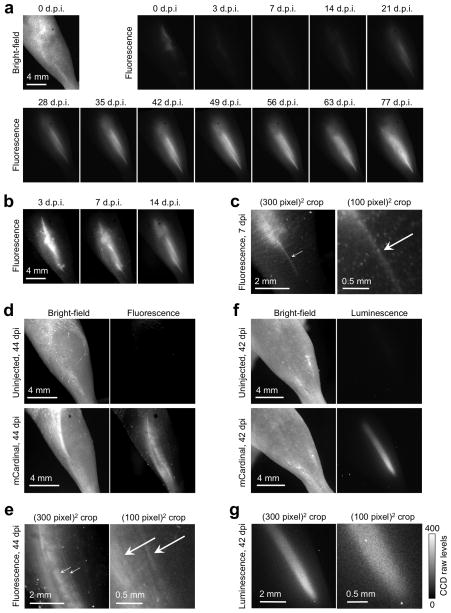
Stem cell therapy for repairing damaged or degenerated muscles is an area of active research. Stem cell performance varies with implantation timing and dosage, as well as cell source and growth conditions ex vivo and in vivo17. Stem cell research could thus benefit from a reliable method for visualizing muscle differentiation non-invasively and with high anatomical detail18. We tested the ability of mCardinal to track the fate of myoblasts in injured tibialis anterior (TA) muscle in living mice, a widely used model for cellular therapy for muscle degenerative diseases19. We created myoblasts stably expressing mCardinal, then injected them into TA muscles injured with notexin to induce muscle fiber degeneration19. We then imaged the legs over many weeks using 620/20 nm light (Fig. 4a–c). Injected cells were visible by mCardinal fluorescence immediately after injection then declined in the first 7 days post-injection (d.p.i.), consistent with previous studies using bioluminescence19. Beginning at 3 d.p.i., thin mCardinal-positive fibers appeared in the leg, gradually increasing in number over the next 46 days and then reaching steady state. Multiple rounds of fluorescence imaging of the same area showed that mCardinal was photostable in vivo (Supplementary Fig. 13). We also tested the ability of mCardinal to image pluripotent muscle stem cells. We injected 1000 stem cells transduced with a mCardinal- and luciferase-expressing lentivirus into TA muscles of SCID mice that had been injured with notexin and 18-Gy irradiation, which enhances engraftment of injected stem cells by incapacitating endogenous myogenic cells19. At 30 d.p.i., notexin was injected again to further expand muscle stem cells, as previously observed19. Strong fluorescence signal was subsequently observed with the morphology of differentiated myofibers throughout a large region of the TA muscle (Fig. 4d,e). Contrast over background from a 1-min exposure was 11 (calculated as the ratio of fluorescence intensity from fibers to autofluorescence intensity from a non-injected leg). Bioluminescence can achieve higher detection sensitivity than fluorescence in animals due to negligible background20; thus we also tested the ability to visualize myofibers derived from stem cells using bioluminescence imaging of luciferase. An 8-min acquisition of bioluminescence signal at a similar pixel sampling resolution (21 μm/pixel vs. 15 for fluorescence) showed only a diffuse signal without structural features (Fig. 4f,g). Thus, fluorescence imaging with mCardinal allows visualization of differentiation and incorporation of stem cell derivatives in situ in living animals with superior detail compared to bioluminescence.
Figure 4.
Non-invasive longitudinal visualization of muscle regeneration in living mice. (a) Tibialis anterior (TA) muscles injected with 1 million myoblasts expressing mCardinal, imaged with a fluorescence stereoscope with 620/20 nm excitation. All images are normalized to the same intensity scale. Series is representative of 5 repeats. (b) Images from 3, 7, and 14 days post-injection (d.p.i.) are shown with intensity scaling tighter than in (a) by a factor of 10, 10, and 5 respectively. (c) Magnified view of the muscle at 7 d.p.i., showing an early regenerating fiber (arrow). The image at right is deliberately enlarged until pixelated to show that this fiber appears just a few pixels wide. (d) Fluorescence signal from TA muscles injected with 1000 muscle stem cells expressing mCardinal. (e) Magnified view of the muscle at 44 d.p.i., showing multiple regenerating fibers (arrows). (f) Bioluminescence imaging at 42 d.p.i. of stem cells shown in (d). A single 8-min bioluminescence image was acquired at the highest possible resolution with no binning. The resulting sampling resolution (21 μm/pixel) is similar to that (15 μm/pixel) of the fluorescence images in (e). (g) Enlargement of the luminescence image in (f) shows the absence of structures resembling myofibers. The right panel is enlarged to show the presence of noise at the level of individual pixels. In a 40-pixel × 30-pixel region containing the brightest pixels, standard deviation was 33 while mean intensity was 290 counts over that of a background region, indicating presence of substantial shot and read noise relative to signal.
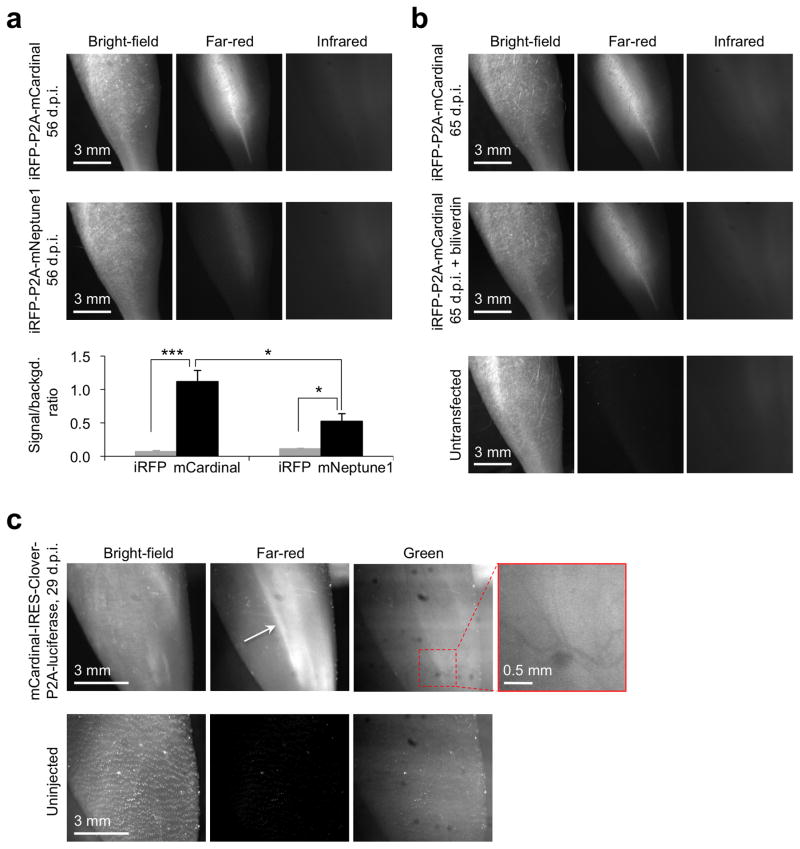
We next assessed whether mCardinal improves non-invasive imaging of myocytes in vivo compared to mNeptune1, iRFP, or Clover, currently the brightest GFP derivative21. We created stable myoblast cell lines coexpressing mCardinal or mNeptune1 with iRFP (Supplementary Fig. 14), injected them into injured TA muscle, then imaged 56 days later with optimized filters (Supplementary Fig. 15). Far-red fluorescence from cells expressing mNeptune1 was dimmer than from cells expressing mCardinal (Fig. 5a). Unexpectedly, with cells coexpressing iRFP and mCardinal, only extremely faint infrared fluorescence from iRFP was visible upon excitation at 680–700 nm light, while far-red fluorescence from mCardinal was easily detectable (Fig. 5a), suggesting that differentiated myocytes may maintain low levels of biliverdin. Intraperitoneal administration of biliverdin did not improve iRFP signal (Fig. 5b), suggesting that myocytes may not take up circulating biliverdin efficiently. We then compared the ability of mCardinal and Clover to image myocyte differentiation in vivo by stably transfecting stem cells with mCardinal and Clover (Supplementary Fig. 16), followed by injection into injured TA muscle. In the green channel, we only observed muscle autofluorescence interrupted by blood vessels, as expected from the absorption of blue and green light by hemoglobin, while mCardinal again gave clear signals (Fig. 5c). These results demonstrate that fluorescence imaging of mCardinal with red excitation light improves non-invasive visualization of cellular differentiation in deep tissues.
Figure 5.
Comparison of mCardinal with mNeptune1, iRFP and Clover GFP for non-invasive visualization of muscle regeneration in living mice. (a) Representative fluorescence images of tibialis anterior (TA) muscles injected with 1 million myoblasts expressing iRFP and mCardinal or iRFP and mNeptune1. Images were taken in the absence of exogenous biliverdin. The bar graph shows contrast over background (mean ± SEM, n = 6 and n = 8 for mNeptune1 and mCardinal, respectively). Asterisks indicate significant differences by unpaired two-tailed Student t test (* P < 0.05 and *** P < 0.001). (b) Representative fluorescence images of TA muscles expressing mCardinal and iRFP in the absence (top panel) and presence (middle panel) of 250 nM biliverdin. The leg injected with untransfected myoblasts (bottom panel) was used as a negative control. (c) Representative fluorescence images of TA muscles injected with 5000 muscle stem cells expressing Clover and mCardinal at 29 d.p.i. The white arrow indicates mCardinal-positive muscle fibers. Shadows of blood vessels (highlighted in the inset) are expected from absorption of green autofluorescence by blood.
DISCUSSION
In fluorescence imaging, the limiting factor in detection is the ratio of specific signal to background autofluorescence, so the concentration and molar brightness of a FP must be sufficiently high to create a signal over background20. Once that condition is achieved, however, shorter integration times are possible with FPs than with luciferase. FPs are capable of emission rates of millions of photons per second per molecule22, orders of magnitude more than the 2.7 photons per second of peak luciferase emission23. In macroscopic applications, illumination intensity is a limiting factor in determining exposure time for fluorescence detection. Using light from an inexpensive 32-W light-emitting diode source passing through a low-numerical aperture lens of a standard variable-magnification dissecting microscope, we were able to image myocytes non-invasively with acquisition times as short as 5 s. These imaging times allow repositioning of the mouse and multiple acquisitions in one session, something not possible with bioluminescence because luciferase signal will decay appreciably over the minutes-long time frame of a single exposure24. We expect that use of higher-power light sources, higher-numerical aperture optics, and stereoscopes optimized for light capture and transmission can reduce acquisition times further to the sub-second range.
Bioluminescence and fluorescence can have complementary roles in tracking cellular differentiation. The greater detection sensitivity of bioluminescence is useful for detecting low numbers of cells, discerning general trends in cell number and location, and screening multiple animals in wide fields of view25. Non-invasive fluorescence imaging with mCardinal can be used to study specific interesting regions at higher magnification, either once or over time to assess how experimental conditions influence their survival or differentiation. In addition to its use in animals, mCardinal should be the FP of choice in microscopy when excitation with 633–635 nm lasers is needed, either for multi-wavelength imaging or to avoid autofluorescence from endogenous compounds3. In particular, mCardinal should improve fluorescence imaging of cells such as hepatocytes, neurons, or retinal pigment epithelial cells that contain high levels of lipofuscin, which is excited efficiently by all visible wavelengths below 620 nm26–28. As a traditional autocatalytic fluorescent protein, mCardinal may provide advantages over phytochrome-biliverdin fluorescent complexes in cells where biliverdin concentrations are low. In summary, with its unique combination of brightness, excitability with red light, and cofactor-independence, mCardinal should be useful in a wide variety of in vivo imaging applications.
ONLINE METHODS
Mutagenesis and screening of libraries
Mutations at specific residues were introduced by overlap-extension PCR. All PCR products were cut and ligated into a constitutive bacterial expression vector pNCS (Allele Biotech) or arabinose-inducible pBAD (Invitrogen). For all library construction methods, chemically competent Escherichia coli strain DH5α (BioExpress) or XL-10 Gold (Invitrogen) were transformed and grown overnight on LB/agar at 37°C and maintained thereafter at room temperature. For each round of mutagenesis, a number of colonies approximately 10-fold higher than the expected library diversity were screened to ensure full coverage. Agar plates were screened for transmitted color by eye and for fluorescence in a light-tight enclosure with a KL2500 fiberoptic light source (Leica) and a ST-8300M cooled CCD camera controlled with CCDOps software (Santa Barbara Instrument Group) on a MacBook Pro running OS 10.6.8 (Apple). 610/20 nm excitation and 645/30 nm emission filters (Chroma) were placed in the light path before the fiberoptic light guide and before the camera, respectively. Bacterial colonies of interest were patched on LB/agar plates and incubated overnight at 37°C. Lysates were extracted with B-PER II (Pierce), and spectra were obtained on a Safire II plate reader (TECAN). DNA sequences of all constructs are available upon request.
Protein production and characterization
Fluorescent proteins with polyhistidine tags were expressed from pNCS vectors in E. coli strain DH5α, purified with cobalt-chelating affinity chromatography (Pierce) and desalted using gel filtration columns (Bio-rad). Excitation and emission spectra of recombinant proteins were measured with a Horiba Jobin Yvon Fluorolog 3 fluorimeter (HORIBA Scientific). Absorbance spectra were obtained on a SafireII plate reader (Tecan). Extinction coefficients were calculated using the base-denaturation method, as previously described for red fluorescent proteins30. Quantum yields were determined by integration of emission curves corrected for detector sensitivity, using mKate2 as a standard (quantum yield 0.4). pH titrations were performed using a series of buffers (1 M HOAc, 1 M NaOAc, 5 M NaCl for pH 3.0–4.5; 1 M NaH2PO4, 1 M Na2HPO4, 5 M NaCl for pH 5–9.0; 100 mM glycine for pH 9.5 and 10). 5 μL of purified protein was diluted in 145 μL buffer with different pH values and the fluorescence brightness was measured.
For maturation experiments, bacteria of E. coli strain TOP10 transformed with pBAD vectors expressing FPs were grown at 37°C in sealed 2 mL tubes filled up with 2.2 mL LB media deoxygenated by bubbling with CO2 gas and supplemented with 4% D-glucose. After 24 h, the culture was spun down, resuspended in supplemented LB media containing 100 μg/mL ampicillin and 0.1% L-arabinose, and cultured for another 3 hours at 30°C in aerobic conditions. Bacteria were then quickly lysed with B-PER II (Pierce) in aerobic conditions and fluorescence of lysates measured as a function of time on the Safire II plate reader.
For semi-native PAGE, 10 μg of purified proteins were loaded without boiling onto 4%–12% Bis-Tris polyacrylamide gels (Invitrogen) and electrophoresed in MOPS electrophoresis buffer. Gels were imaged for fluorescence with 400–500 nm excitation light and a yellow acrylic longpass filter in a BlueView Transilluminator (Vernier).
In vitro photobleaching measurements were performed in aqueous droplets of purified proteins in oil using a Zeiss Axiovert 200M inverted microscope with a 40× 1.2-numerical aperture (NA) water immersion objective (Zeiss), an X-Cite 120-W metal halide lamp (Lumen Dynamics), and a 615/20 nm excitation filter (Chroma). The microscope was operated with Micro-Manager 1.4.2 in MacOS 10.6.8 on a 2.5-GHz Core 2 Duo MacBook Pro computer (Apple). Images were taken every 1 s under continuous illumination. Times were adjusted to produce photon output rates of 1000 per molecule per s as described previously31.
Crystallization and structure determination of mCardinal
Polyhistidine-mCardinal was purified from bacterial lysates with metal affinity resin, then the polyhistidine tag was removed with EKMax Enterokinase and EK-Away Resin (Invitrogen) and the buffer was changed to 20 mM Tris, 20 mM HCl pH 8. The protein was then lysine-methylated and repurified into HEPES-buffered saline pH 7.3 as previously described32. Absorption, excitation, and emission spectra confirmed that lysine-methylation did not affect mCardinal spectral properties. mCardinal was crystallized by the hanging-drop vapor diffusion method by mixing equal volumes of 10 mg/mL protein with of 0.2 M calcium chloride, 10 mM Tris(2-carboxyethyl)phosphine HCl, 0.1 M Tris HCl pH 7.6, and 22–25% PEG 4000. Crystals were cryoprotected in well solution supplemented with 15% ethylene glycol and flash cooled in liquid nitrogen. Diffraction data were collected at beamline 8.2.2 at the Advanced Light Source (Berkeley, US). A 2.2 Å dataset was obtained and processed with XDS33. The structure of mCardinal was solved by molecular replacement using PHASER34 with the structure of Neptune (PDB accession number 3IP2) as the search model. Analysis of the data with PHENIX35 indicated a spacegroup of P2(1) with pseudomerohedral twinning and a refined twin factor of 0.50. Refinement was conducted with PHENIX using least-square twin refinement with twin law h, −k, −l. The model was edited with COOT36. Initial refinement used rigid body, coordinate, and real space refinement with individual atomic displacement parameter refinement and non-crystallography symmetry (NCS) restraints. NCS restraints were omitted in subsequent rounds of refinement, and translation/libration/screw (TLS) refinement was added. The Fo-Fc OMIT map was calculated with PHENIX using the averaged kick approach to minimize model bias37. The OMIT map was contoured to 3.0 sigma with a −30 Å2 temperature factor sharpening factor38.
After purification and tag removal as above, mCardinal-V218E was exchanged into 50 mM Tris (hydroxymethyl)aminomethane, 25 mM NaCl, 4.0 mM TCEP at pH 7.5, then concentrated to 10 mg/mL using an Amicon ultra 10 kDa-cutoff centrifugal unit (Millipore). The protein was mixed in 3:1 ratio with 0.2 M calcium acetate, 0.1 M MES:NaOH pH 6.0, and 20% (w/v) PEG 8000 (condition B6 from MSCG-1 crystallization condition set, Microlytic), then allowed to crystallize for 3 d at 12°C in the dark. The crystals were washed for 20 s in paratone oil for cryoprotection before they were mounted on a MicroLoop (MiTeGen), cryo-cooled in liquid nitrogen, and loaded for remote collection at beamline 12.3.1 at the Advanced Light Source. Diffraction data were processed with XDS. The structure was solved by molecular replacement using MOLREP39 with the structure of Neptune (PDB accession number 3IP2) as the search model. PHENIX was used to automatically build > 95% of the structure. The structure was refined with REFMAC40 and PHENIX using TLS parameters, alternating with manual correction of the model using COOT.
Chains where temperature factors for the Gln41 amide atoms were lower than that of the water oxygen in Neptune (36.9) were used to measure distances between the Gln41 amide and the chromophore acylimine groups and between the Gln41 amide and Thr28 hydroxyl groups.
Fusion gene construction
mCardinal cDNA was PCR amplified with a 5′ primer encoding an AgeI site and a 3′ primer encoding either a BspEI (C1) or NotI (N1) site for C-terminal and N-terminal fusions (with regards to the FP), respectively. The purified and digested PCR products were ligated into similarly digested pEGFP-C1 and pEGFP-N1 cloning vectors to create pmCardinal-C1 and pmCardinal-N1. A cDNA fragment encoding each protein domain was PCR amplified with primers containing the appropriate restriction enzyme sites and ligated into pmCardinal-C1 or pmCardinal-N1. To prepare fusions at the N-terminus of mCardinal, the following digests were performed: human non-muscle α-actinin (GenBank accession number NM_001130005.1, source: Tom Keller, Florida State University, Tallahassee, FL, U.S.A.), EcoRI and NotI; human calnexin (NM_001746.3, Origene), AgeI and NotI; human centromere protein B (NM_001810.5, Alexey Khodjakov, Wadsworth Center, Albany, NY, U.S.A.), BamHI and NotI; rat β-2 connexin-26 (NM_001004099.1, Matthias Falk, Lehigh University, Bethlehem, PA, U.S.A.), EcoRI and BamHI; human keratin 18 (NM_199187.1, Open Biosystems), EcoRI and NotI; yeast Lifeact (IDT DNA), BamHI and NotI; human pyruvate dehydrogenase, (NM_000284, Origene), AgeI and NotI; human peroxisomal membrane protein (NM_018663.1, Origene), NotI and AgeI; human vimentin (NM_003380.3, Origene), NheI and BamHI; chicken paxillin (NM_204984.1, Clare Waterman, NIH, Bethesda, MD, U.S.A), NheI and BamHI; mouse mannosidase II (NM_008549.2, Jennifer Lippincott-Schwartz, NIH), NheI and BamHI; and human histone H1 (NM_008197.3, David Gilbert, Florida State University), BamHI and NheI. To prepare C-terminal fusions to mCardinal, the following digests were performed: human β-actin (NM_001101.3, Clontech), NheI and BglII; human α-tubulin (NM_006082, Clontech), NheI and BglII; human Rab4a (NM_004578.2, Viki Allen, University of Manchester, U.K.), BspEI and BamHI; human lamin B1 (NM_005573.2, George Patterson, NIH), EcoRI and BamHI; human myotilin (NM_006790.1, Origene), AgeI and BspEI; human fibrillarin (NM_001436.3, Evrogen), BglII and BamHI; human tight junction protein ZO1 (NM_003257.1, Origene), AgeI and BspEI; human VE cadherin (NM_001795.3, Origene), BglII and EcoRI; and the 20-amino-acid farnesylation signal from c-Ha-Ras (NM_001130442.1, Clontech), AgeI and BspEI. Full sequences of plasmids are available upon request.
Fluorescence Microscopy
HeLa cells (CCL-2 and S-3, ATCC) were grown in a 50:50 mixture of Dulbecco’s modified Eagle’s medium (DMEM) and Ham’s F12 medium (Sigma-Aldrich) with 15% calf serum (HyClone). NIH 3T3 cells were grown in DMEM with 10% fetal bovine serum (FBS, HyClone). FoLu cells were grown in Eagle’s minimal essential medium with Earle’s balanced salt solution, 1% non-essential amino acids (Sigma-Aldrich), and 10% FBS (HyClone). Cells were transfected with Effectene (Qiagen). Imaging was performed in Delta-T culture chambers (Bioptechs) under a humidified atmosphere of 5% CO2 in air. Single-image microscopy of fusion proteins was performed using either a Nikon Eclipse 80i equipped with a Chroma Cy5 filter set (Ex = 620/60 nm; Em = 700/75 nm; DM = 660 nm LP) or a Zeiss Axiovert equipped with a Semrock Cy5 filter set (Ex = 628/40 nm; Em = 700/75 nm; DM = 660 nm LP). Time-lapse microscopy was performed using an IX81 inverted microscope with a FV1000 confocal scanning unit, a PlanApo 60× 1.4 NA oil objective, a 633 nm helium-neon laser, and a manufacturer-installed Cy5 emission filter (Olympus). For time-lapse imaging of mCardinal-18aa-β-actin, the following settings were used: 2.2× zoom, 12.5 μs pixel dwell time, 400 μm pinhole, 2× Kallman filtering per line, 680 V photomultiplier tube voltage, 645–745 nm emission filter, 18 s interframe interval. For time-lapse imaging of PDHA1-10aa-mCardinal, the following settings were used: 2.2× zoom, 10 μs pixel dwell time, 400 μm pinhole, 2× Kalman filtering per line, 650 V photomultiplier tube voltage, 645–745 nm emission filter, 15 s interframe interval.
Live-cell photobleaching was performed in live HeLa S3 cells expressing far-red FPs fused to human histone H2B under arc lamp and laser illumination. Arc-lamp photobleaching was performed on a TE2000 inverted microscope with a Plan Fluorite 40× 0.85-NA dry objective (Nikon), an X-Cite Exacte 120-W metal halide lamp (Lumen Dynamics), and a Brightline Cy5-4040C-000 filter cube (excitation 628/40 nm; dichroic mirror 660 nm, emission 692/40 nm, Semrock). A 1918-C optical power meter (Newport) measured illumination power at the objective as 19.7 mW. Power moderation was achieved by using neutral density filters contained within the lamp. With a 3.125% neutral density (ND32) filter in place, several regions containing at least 30 nuclei in total were located. The neutral density filter was then removed and the region was photobleached continuously while imaged with a Q-Imaging Retiga EXi camera (Photometrics) with 200-ms exposure times for 6 min for a total of 3100 frames. The raw data was collected using NIS-Elements software (Nikon) and then analyzed with Simple PCI software (Hamamatsu). Laser photobleaching measurements were collected on a FV1000 confocal microscope with a PLAPO 40× 1.0-NA oil-immersion objective (Olympus) and a 543-nm He-Ne laser (Melles Griot) tuned to an output power of 1005 μW, as measured at the objective with a FieldMax II-TO power meter (Coherent). Scan settings were 2× zoom, 600 μm pinhole size, 515 V photomultiplier voltage, 4 μs pixel dwell time. Emission was collected with detector slit settings of 600–700 nm. Several regions containing at least 30 nuclei in total were first located with minimal laser power. The laser power was raised back to 1005 μW and each region was photobleached continuously for ~20 min for a total of 1500 frames. Raw data was collected with the FluoView software (Olympus) and then analyzed with Simple PCI software.
For luciferase-normalized fluorescence intensity measurements, HeLa cells (4 × 106) were lysed in 600 μL of pH 7.2 luciferase reporter buffer (Promega) containing protease inhibitor cocktail (Roche). Fluorescence spectra with three different excitation wavelengths were taken in a 96-well plate in a volume of 150 μL on a plate reader Safire II (Tecan) with corrected excitation and emission. Fluorescence intensities were calculated by integrating the area of the emission spectrum between 660 nm and 800 nm. Bioluminescence measurements were performed in a 96-well plate in a volume of 60 μL (10 μL of lysate and 50 μL of luciferin substrate) on a Veritas microplate luminometer with injectors (Turner Biosystems).
Worm culture and imaging
C. elegans worms were maintained at 20°C on OP50 E. coli-seeded nematode growth medium plates. Transgenic strains were made by micro-injection of plasmid DNA into the gonads of adult hermaphrodites as previously described41. Transgenic line TV13901 (wyEx5674; wyIs251) was generated by injecting a mixture of 10 ng/μL Pmig-13::SNG-1::mCardinal, 5 ng/μL Pmyo-3::DsRed, and 25 ng/μL Podr-1::mRFP into wyIs251 (Pmig-13::GFP::Rab-3) worms. Line TV13928 (wyEx5684; vsIs147; syIs65) was generated by injecting a mixture of 10 ng/μL Pegl-17::mCardinal and 25 ng/μL Podr-1::DsRed into vsIs147 (Punc-103e::mCD8::mCherry); syIs65 (B0034.1::pes-10::GFP) worms. Line TV14404 (wyEx5836) was generated by injecting a mixture of 15 ng/μL Pmyo-2::mCardinal and 25 ng/μL Podr-1::GFP into N2 strain worms. Mixtures were injected at a volume sufficient to visualize gonad filling without causing damage. Podr-1::GFP labels AWC neurons as a positive control.
Three-color imaging was performed in live C. elegans using high NA objectives (40× 1.20-NA water objective for presynaptic vesicles and 100× 1.40-NA oil objective for vulva) on an Axiovert 200M inverted fluorescence microscope (Zeiss). Worms were transferred onto a hydrated 2% agarose pad in M9 solution and immobilized using a mixture of 200 mM 2,3-butanedione monoxime (Sigma-Aldrich, St Louis, MO) and 2.5 mM levamisole (Sigma-Aldrich) in 10 mM Na-HEPES. Fluorescence images were acquired consecutively under three channels: green channel (ex-470/30 nm, DM-510LP nm and em-530/40 nm) for GFP, red channel (ex-545/10 nm, DM-570LP nm and em-590/45 nm) for DsRed or mCherry, and far-red channel (ex-615/30 nm, DM-630LP nm and em-675/50 nm) for mCardinal. Fast time-lapse imaging of freely moving worms or pharyngeal muscle contraction was performed on the FV1000 microscope with a 20× 0.75-NA objective. Worms were transferred onto a hydrated 2% agarose pad in M9 solution with or without the treatment of cyanoacrylate glue, which can partially immobilize the worms. 1 mg/mL serotonin (Sigma-Aldrich) in M9 was added to induce pharyngeal pumping in partially immobilized worms42. Time-series scans, with 256×256 pixels/frame resolution (0.414 μm/pixel), were taken in roundtrip scanning mode with a pixel dwell time of 0.5 μs thereby achieving a frame rate at 125 ms/frame sufficient to resolve free movement and pharyngeal pumping events. A red diode laser was used for excitation (635 nm, 50% power) and emission between 650 nm and 750 nm was collected.
Deep-tissue imaging in euthanized mice
All procedures with mice (esophagus, liver and stem cell imaging) were conducted in accordance with NIH regulations and approved by the Stanford Institutional Animal Care and Use Committee. The minimum number of mice used in this study was calculated as described43. All mice were grouped according to body weight on the day of injection. 10-week-old male nude mice (strain NU/NU) were obtained from the Charles River Laboratories. Mice were fed low-fluorescence purified diet (TD.97184, Harlan) for 1 week. Purified FPs were diluted in PBS to 60 μM, then 4 μL of FPs or PBS was loaded into small glass capillary tubes with 0.8–1.1 mm inner diameter (Kimble Chase, Cat# 34507-99) and centrifuged to the bottom of the tubes at 3000 rpm for 5 min. A mouse was euthanized by an overdose of isoflurane anesthesia, then background fluorescence was allowed to stabilize for 20 min after death, and then the mouse was immobilized for tube placement and imaging. Imaging was performed with an IVIS Spectrum system (Perkin Elmer) with a DW436 back-thinned CCD with 2048×2048 13.5-μm pixels and 95% peak quantum efficiency (Andor), or an SZX12 fluorescence stereoscope with a 1× 0.11-NA PlanApo objective (Olympus), a 35-W 615-nm LED (CoolLED), a 620/20-nm excitation filter, a 667/30-nm emission filter, and a Retiga 4000RV cooled CCD digital camera (QImaging). IVIS Spectrum settings were: field of view 13 cm, aperture f/1, binning 4×4, and exposure time 5 s (605/30 nm excitation) or 10 s (640/30 nm excitation). SZX12 settings were: zoom 7×, binning 2×2, and exposure time 10 s. Each tube in the series was inserted into the esophagus of the same mouse to the same location and imaged, and then the entire set was repeated 4 more times. Quantitative measurements of fluorescence signal (presented as signal/background ratio) were made with Living Image Software 4.3.1 and ImageJ 1.46r. For each FP, a signal-to-background ratio in each channel was calculated as (FFP − FPBS)/FPBS, where FFP and FPBS were total intensities measured in a common region of interest encompassing all fluorescence signal from the images of the FP and PBS, respectively.
Liver transfection and imaging in living mice
6~7 week-old male nude mice of strain J:NU(007850) were obtained from the Jackson Laboratory and fed with low-fluorescence purified diet (TD.97184, Harlan) for a week. Using a hydrodynamic injection protocol, 20 μg of plasmid DNA in 1.8 mL of saline (0.9% NaCl) was injected into the tail vein of the mice within 5–6 seconds. Fluorescence imaging was performed 40 h after injection on the IVIS Spectrum with exposure time 60 s, binning 2×2, aperture f/1, and field of view 22.4 cm. mCardinal was imaged with 605/30 nm excitation and 660/20 nm emission, or 640/30 nm excitation and 700/20 nm emission. iRFP was imaged with 675/30 excitation and 740/20 nm emission. Data analysis was performed as with deep-tissue imaging in euthanized mice above.
Muscle cell transfection and in vivo imaging of muscle cell regeneration
Primary myoblasts and muscle stem cells (MuSCs) were isolated from mice. Briefly, forelimbs and hindlimbs were removed from 8-week-old mice (strain C57/BL6) and bones removed. The muscles were transferred to F10 media and minced into a slurry with razor blades. Muscles were enzymatically dissociated by the addition of 2 mL per g of tissue of a solution of dispase and collagenase. The slurry was maintained at 37°C for 30–45 minutes while triturated every 15 minutes with a 5-mL plastic pipette, then passed through 80 μm nylon. The filtrate was spun at 350× g0 to sediment the dissociated cells, the pellet was resuspended in growth medium, and the suspension was plated on collagen-coated dishes. During the first several passages of the primary cultures, myoblasts were enriched by preplating. Muscle stem cells were prepared by enzymatic digestion of muscle sample, stained with biotin-labeled antibodies (CD45, CD31, CD11b, and Sca1) for magnetic depletion of non-myogenic cells and then with specific antibodies (CD34 and α7-integrin) for myogenic markers. Cells were then purified by two rounds of cell sorting for the (CD45/CD11b/CD31/Sca1)-negative CD34-positive α7-integrin-positive cell population as previously described19.
Constructs expressed in myoblasts were pCMV-piggyBac, MCm-CMV-mCardinal, MCm-CMV-iRFP-P2A-mCardinal, and MCm-CMV-iRFP-P2A-mNeptune1. MCm plasmids are modified minicircle plasmids with piggyBac long terminal repeats. Lentiviral constructs expressed in MuSCs were hLIG-EF1α-mCardinal-IRESPV-luciferase2 and pLL3.7-EF1α-mCardinal-IRESEMCV-Clover-P2A-luciferase2. Full sequences of plasmids are available upon request. One million myoblasts were transfected with 5.3 μg total of minicircle vector and pCMV-piggyBac in a 3:1 molar ratio in Solution V from the Cell Line Optimization Nucleofector Kit (Lonza) with program B-32 on a Nucleofector I electroporation machine (Lonza). Myoblasts were cultured for 10 days, and then the 10% of cells exhibiting the highest far-red fluorescence upon 635 nm excitation were selected by fluorescence activated cell sorting on a FACSAria II (Becton-Dickinson). Myoblasts were further cultured for 7 days, and then injected in injured TA muscles. For MuSC infection, plates were coated with 20 μg/mL retronectin in PBS for 2 h at room temperature, then with 200 μL 2% bovine serum albumim in PBS for 30 min, then washed with PBS. MuSCs and lentivirus were mixed in a 1:10 ratio in media with 5 μg/mL protamine sulfate, then plated and incubated overnight at 37°C. The next day, MuSCs were harvested and injected in injured TA muscles.
All images of muscles were acquired non-invasively on living mice. TA muscles of 4-week-old male mice (SCID Congenic, Charles River Laboratories) were treated with 10 μL of 10 μg/mL notexin (myoblast engraftment) or 18-Gy irradiation plus 20 μL of 10 μg/mL notexin (stem cell engraftment). One day later, 106 myoblasts stably expressing mNeptune1 or mCardinal, or 1000 or 5000 lentivirus-transduced stem cells expressing mCardinal, were suspended in 10 μL of PBS with 2% FBS and 1mM EDTA and slowly injected into each TA muscle. Images were acquired with the SZX12 fluorescence stereoscope equipped with the Retiga 4000RV cooled CCD digital camera. Imaging conditions were: for mCardinal, pE-100 35-W 615-nm LED (CooLED), 620/20 nm excitation filter, 667/30 nm emission filter; for Clover, MME250 250-W metal-halide lamp (Moritex), 475/30 nm excitation filter, 510 nm long-pass emission filter; for iRFP, MME250 250-W metal-halide lamp, 675/20 nm excitation filter, 730/30 nm emission filter. For bioluminescence, after intraperitoneal injection of 100 μL 30 mg/mL D-luciferin in PBS, a single 8-min exposure without binning was acquired on the IVIS Spectrum system.
Data analysis and statistical methods
Images from IVIS Spectrum or non-IVIS systems were analyzed using Living Image 4.2 (Perkin-Elmer) or Fiji (http://fiji.sc/Fiji), respectively. Statistical tests were performed using GraphPad Prism v6.0. Multiple or single comparisons were performed with one-way ANOVA or two-tailed Student’s t tests, respectively, with significance level set at α = 0.05. F-tests were used to determine the equivalence of variance for single comparison, and t-tests assuming equal or unequal variances were selected accordingly. Brown-Forsythe tests were used to determine the equivalence of variance for multiple comparisons, and one-way ANOVA with post-hoc tests of Dunnett (all mNeptune derivatives vs. mNeptune1, Fig. 3) or Tukey (comparison between any two FPs, Supplementary Fig. 12) was performed.
Supplementary Material
Acknowledgments
The authors thank F. Kanokwan for technical support, F. Fang and L. Lisowski for cell sorting, C. Ran and X. Chen for two-photon imaging, F. Zhang for hydrodynamic injection, and S. Classen (Lawrence Berkeley National Laboratory, US) for help with data collection at ALS Beamline 12.3.1. The authors also thank K. Yusa (Wellcome Trust Sanger Institute, UK) for a gift of pCMV-piggyBAC, M. Kay (Stanford University, US) for a gift of minicircle plasmid, N. Deliolanis (Fraunhofer Project Group for Mannheim, Germany) and C. Vinegoni (Harvard University, US) for helpful suggestions on mouse imaging, members of the laboratory of E. Mellins (Stanford University, US) for help with protein purification, and members of the Lin laboratory for advice and assistance. This work was supported by a seed grant from the Center for Biological Imaging at Stanford (J.C.), NIH grant 1R01NS076860-01 (J.C., M.Z.L.), NIH grant T32 HD007249 (R.D.H.), NIH grant 5R01AG020961-08 (S.Y.C., H.M.B.), the Florida State University Research Foundation (P.J.C., M.A.B., M.W.D.), the Chambers Family Foundation and the Pachyonychia Congenita Project (E.G-G., C.H.C), an Irvington Postdoctoral Fellowship from the Cancer Research Institute (J.S.B.), the University of Hawaii (N.J.A., H-L.N.), the Howard Hughes Medical Institute (K.S., K.C.G.), and the Burroughs Wellcome Fund and the Rita Allen Foundation (M.Z.L.).
Footnotes
Accession codes. GenBank accession codes are KJ131553 (mNeptune2), KJ131554 (mNeptune2.5), and KJ131552 (mCardinal). Mammalian expression plasmids are available at Addgene with identification numbers 51309 (mNeptune2), 51310 (mNeptune2.5), and 51311 (mCardinal). PDB accession codes are mCardinal (XXXX) and mCardinal-V218E (4OJ0).
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
J.C. performed protein mutagenesis and characterization, cell imaging, mouse imaging, and data analysis, and co-wrote the paper. S.Y.C. performed myoblast and stem cell purification and transfection. R.D.H. and E.G-G. assisted with animal experiments. P.L. performed worm culture and made transgenic worms. P.J.C., M.B., and M.W.D. performed microscopy of FP fusions. A.J.L. prepared mCardinal proteins for crystallization. J.S.B. and N.J.A. obtained structures of mCardinal and mCardinal-V218E, respectively. H-L.N., K.C.G., M.W.D., C.H.C., K.S., and H.M.B. provided supervision. M.Z.L. performed protein mutagenesis and characterization, analyzed data, co-wrote the paper, provided supervision, and directed the project.
References
- 1.Schenkman KA, Marble DR, Feigl EO, Burns DH. Near-infrared spectroscopic measurement of myoglobin oxygen saturation in the presence of hemoglobin using partial least-squares analysis. Appl Spectrosc. 1999;53:325–331. [Google Scholar]
- 2.Tromberg BJ, et al. Non-invasive in vivo characterization of breast tumors using photon migration spectroscopy. Neoplasia. 2000;2:26–40. doi: 10.1038/sj.neo.7900082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monici M. Cell and tissue autofluorescence research and diagnostic applications. Biotechnol Annu Rev. 2005;11:227–256. doi: 10.1016/S1387-2656(05)11007-2. [DOI] [PubMed] [Google Scholar]
- 4.Deliolanis NC, et al. In vivo tomographic imaging of red-shifted fluorescent proteins. Biomed Opt Express. 2011;2:887–900. doi: 10.1364/BOE.2.000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawai Y, Sato M, Umezawa Y. Single color fluorescent indicators of protein phosphorylation for multicolor imaging of intracellular signal flow dynamics. Anal Chem. 2004;76:6144–6149. doi: 10.1021/ac040037s. [DOI] [PubMed] [Google Scholar]
- 6.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 7.Lin MZ, et al. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem Biol. 2009;16:1169–1179. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morozova KS, et al. Far-red fluorescent protein excitable with red lasers for flow cytometry and superresolution STED nanoscopy. Biophys J. 2010;99:L13–5. doi: 10.1016/j.bpj.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shcherbo D, et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subach OM, et al. A photoswitchable orange-to-far-red fluorescent protein, PSmOrange. Nat Methods. 2011;8:771–777. doi: 10.1038/nmeth.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shu X, et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin MZ. Beyond the rainbow: new fluorescent proteins brighten the infrared scene. Nat Methods. 2011;8:726–728. doi: 10.1038/nmeth.1678. [DOI] [PubMed] [Google Scholar]
- 13.Filonov GS, et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shcherbo D, et al. Near-infrared fluorescent proteins. Nat Methods. 2010;7:827–829. doi: 10.1038/nmeth.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeffrey GA. An introduction to hydrogen bonding. Oxford University Press; New York: 1997. [Google Scholar]
- 16.Rice BW, Cable MD, Nelson MB. In vivo imaging of light-emitting probes. J Biomed Opt. 2001;6:432–440. doi: 10.1117/1.1413210. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert PM, Blau HM. Engineering a stem cell house into a home. Stem Cell Res Ther. 2011;2:3. doi: 10.1186/scrt44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroeder T. Imaging stem-cell-driven regeneration in mammals. Nature. 2008;453:345–351. doi: 10.1038/nature07043. [DOI] [PubMed] [Google Scholar]
- 19.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Contag CH, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- 21.Lam AJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harms GS, Cognet L, Lommerse PH, Blab GA, Schmidt T. Autofluorescent proteins in single-molecule research: applications to live cell imaging microscopy. Biophys J. 2001;80:2396–2408. doi: 10.1016/S0006-3495(01)76209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinde R, Perkins J, Contag CH. Luciferin derivatives for enhanced in vitro and in vivo bioluminescence assays. Biochemistry. 2006;45:11103–11112. doi: 10.1021/bi060475o. [DOI] [PubMed] [Google Scholar]
- 24.Inoue Y, Kiryu S, Watanabe M, Tojo A, Ohtomo K. Timing of imaging after d-luciferin injection affects the longitudinal assessment of tumor growth using in vivo bioluminescence imaging. Int J Biomed Imaging. 2010;2010:471408. doi: 10.1155/2010/471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Condeelis J, Weissleder R. In vivo imaging in cancer. Cold Spring Harb Perspect Biol. 2010;2:a003848. doi: 10.1101/cshperspect.a003848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunk UT, Terman A. Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free Radic Biol Med. 2002;33:611–619. doi: 10.1016/s0891-5849(02)00959-0. [DOI] [PubMed] [Google Scholar]
- 27.Eldred GE, Katz ML. Fluorophores of the human retinal pigment epithelium: separation and spectral characterization. Exp Eye Res. 1988;47:71–86. doi: 10.1016/0014-4835(88)90025-5. [DOI] [PubMed] [Google Scholar]
- 28.Murdaugh LS, et al. Compositional studies of human RPE lipofuscin. J Mass Spectrom. 2010;45:1139–1147. doi: 10.1002/jms.1795. [DOI] [PubMed] [Google Scholar]
- 29.Nighswander-Rempel SP, Kupriyanov VV, Shaw RA. Relative contributions of hemoglobin and myoglobin to near-infrared spectroscopic images of cardiac tissue. Appl Spectrosc. 2005;59:190–193. doi: 10.1366/0003702053085106. [DOI] [PubMed] [Google Scholar]
- 30.Chalfie M, Kain S. Green fluorescent protein : properties, applications, and protocols. Wiley-Interscience; Hoboken, N.J: 2006. [Google Scholar]
- 31.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 32.Walter TS, et al. Lysine methylation as a routine rescue strategy for protein crystallization. Structure. 2006;14:1617–1622. doi: 10.1016/j.str.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Praznikar J, Afonine PV, Guncar G, Adams PD, Turk D. Averaged kick maps: less noise, more signal. and probably less bias. Acta Crystallogr D Biol Crystallogr. 2009;65:921–931. doi: 10.1107/S0907444909021933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bass RB, Strop P, Barclay M, Rees DC. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science. 2002;298:1582–1587. doi: 10.1126/science.1077945. [DOI] [PubMed] [Google Scholar]
- 39.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerr R, et al. Optical imaging of calcium transients in neurons and pharyngeal muscle of C. elegans. Neuron. 2000;26:583–594. doi: 10.1016/s0896-6273(00)81196-4. [DOI] [PubMed] [Google Scholar]
- 43.National Research Council (US) Committee on Guidelines for the Use of Animals in Neuroscience and Behavioral Research. Guidelines for the Care and Use of Mammals in Neuroscience and Behavioral Research. National Academies Press (US); Washington (DC): 2003. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.