Abstract
Exploratory (i.e., voxelwise) spatial methods are commonly used in neuroimaging to identify areas that show an effect when a region-of-interest (ROI) analysis cannot be performed because no strong a priori anatomical hypothesis exists. However, noise at a single voxel is much higher than noise in a ROI making noise management critical to successful exploratory analysis. This work explores how preprocessing choices affect the bias and variability of voxelwise kinetic modeling analysis of brain positron emission tomography (PET) data. These choices include the use of volume- or cortical surface-based smoothing, level of smoothing, use of voxelwise partial volume correction (PVC), and PVC masking threshold. PVC was implemented using the Muller-Gartner method with the masking out of voxels with low gray matter (GM) partial volume fraction. Dynamic PET scans of an antagonist serotonin-4 receptor radioligand ([11C]SB2307145) were collected on sixteen healthy subjects using a Siemens HRRT PET scanner. Kinetic modeling was used to compute maps of non-displaceable binding potential (BPND) after preprocessing. The results showed a complicated interaction between smoothing, PVC, and masking on BPND estimates. Volume-based smoothing resulted in large bias and intersubject variance because it smears signal across tissue types. In some cases, PVC with volume smoothing paradoxically caused the estimated BPND to be less than when no PVC was used at all. When applied in the absence of PVC, cortical surface-based smoothing resulted in dramatically less bias and the least variance of the methods tested for smoothing levels 5mm and higher. When used in combination with PVC, surface-based smoothing minimized the bias without significantly increasing the variance. Surface-based smoothing resulted in 2-4 times less intersubject variance than when volume smoothing was used. This translates into more than 4 times fewer subjects needed in a group analysis to achieve similarly powered statistical tests. Surface-based smoothing has less bias and variance because it respects cortical geometry by smoothing the PET data only along the cortical ribbon and so does not contaminate the GM signal with that of white matter and cerebrospinal fluid. The use of surface-based analysis in PET should result in substantial improvements in the reliability and detectability of effects in exploratory PET analysis, with or without PVC.
Introduction
Exploratory spatial methods are used in neuroimaging to find areas that show an effect of diagnosis, demographics, treatment, etc., where no strong a priori anatomical hypothesis exists. To do this, a parametric map of some neuroimaging measure is acquired for each subject. This map is then transformed into a common space where subjects can be compared independently at each voxel. Based on this test, each voxel is assigned a statistic to create a statistical parametric map (SPM). The effect under study can be declared significant only after correcting the SPM for multiple spatial comparisons, usually by creating clusters of contiguous voxels whose statistic exceeds a threshold (Friston et al., 1993). These clusters need not have well-defined anatomical boundaries and so might not be found with region-of-interest (ROI) analysis. Exploratory analysis has been applied extensively in PET neuroimaging (e.g., Haahr et al., 2012b;Park et al., 2006; Kochunov et al., 2009; Protas et al., 2010; Becker et al., 2011; Kraus et al., 2012; see also references in Table 1).
Table 1.
A sample of studies that perform exploratory voxelwise analysis of PET or SPECT data with MG PVC as implemented in this study.
| Reference | FWHM | Threshold | Tracer | Scanner | PSF |
|---|---|---|---|---|---|
| Matsuda et al., 2002 | 12mm | 35% | 99mTc-ECD | Multispect3 | 9mm |
| R Matsuda et al., 2003 | 12mm | 35% | 99mTc-ECD | Multispect3 | 9mm |
| Chetelat et al., 2003 | 14mm | ? | FDG | ECAT Exact HR+ | 12mm |
| Yanase et al., 2005 | 9mm | 35% | FDG | GE Advance | 6mm |
| Inoue et al., 2005 | 12mm | 25% | 99mTc-ECD | SPECT-2000H | 7mm |
| Haltia et al., 2007 | 5mm | ?b | 11C-Raclopride | GE Advance | ? |
| R Samuraki et al., 2007 | 12mm | 35% | FDG | GE Advance | 6mm |
| Mevel et al., 2007 | 14mm | 25%a | FDG | ECAT Exact HR+ | 12mm |
| Hurlemann et al., 2008 | 10mm | 50%a | 18F-Altanserin | ECAT Exact HR+ | 6mm |
| Chetelat et al., 2008 | 14mm | 20% | FDG | ECAT Exact HR+ | 12mm |
| Van Laere et al., 2008 | 10mm | ?a | 18F-MK-9470 | HR+ | ? |
| Kalpouzos et al., 2009a | 14mm | 50%a | FDG | ECAT Exact HR+ | 12mm |
| Kalpouzos et al., 2009b | 14mm | 50%a | FDG | ECAT Exact HR+ | 12mm |
| Bourgeat et al., 2010 | 12mm | ?a | 11C-PIB | Allegro | ? |
| Curiati et al., 2011 | 12mm | ?b | FDG | Biograph-16 | 4mm |
| Uchida et al., 2011 | 8mm | ?a | 18F-Setoperone | ECAT HRRT | 3 mm |
The current study used a Siemens ECAT HRRT scanner with PSF=4mm. FWHM is the applied smoothing level.
Thresholding performed after smoothing; this makes the threshold essentially 0% for purposes of comparison with this study. All others performed thresholding before smoothing.
Used method from Quarantelli et al., 2004 which thresholds based on tissue type with maximum PVF which makes the threshold voxel-dependent in the range of 33% to 50%.
These studies report reduction in FDG or 99mTc-ECD uptake after PVC. A question mark indicates that a parameter was not given.
A disadvantage with exploratory analysis is that the measurement at a single voxel is often quite noisy which reduces the statistical power and makes it difficult to find clusters. To compensate, spatial smoothing is widely applied in exploratory analysis (Worsley et al., 1996). Spatial smoothing is the process of replacing the value at a voxel by a distance-weighted average of neighboring voxels. If the signal is more similar over the neighborhood than the noise, then the averaging process yields a boost in the signal-to-noise ratio (SNR). Spatial smoothing can profoundly affect the results of an exploratory spatial analysis by increasing the statistical power at individual voxels (Strother et al., 2004). The weight of a neighbor is determined by the distance to the center voxel and choice of full-width/half-maximum (FWHM) of the Gaussian weighting kernel. In volumetric smoothing, the neighborhood is defined in three-dimensional space, encompassing all voxels within a surrounding sphere irrespective of whether a given voxel within that sphere is of the same tissue type as the central voxel. For example, if the central voxel is within cortical gray matter (GM), then voxels within the smoothing kernel may be white matter (WM), cerebrospinal fluid (CSF), subcortical GM, or cortical GM from a neighboring gyrus. In contrast, surface-based smoothing defines a neighborhood to be only along the cortical surface (i.e., within the “cortical ribbon”) with distances computed along the ribbon. This prevents blurring effects of neighboring WM, CSF, and subcortical GM with cortical GM and also prevents blurring between two cortical areas that are close in Euclidian space but far apart along the surface (e.g., precentral and superior temporal gyri). Thus, surface-based smoothing should be used instead of volume-based smoothing for the analysis of cortical structures.
PET imaging is susceptible to partial volume effects (PVE) for reasons involving the finite size of detector crystals, detector principles, traveling distance before annihilation, Poisson count statistics, and reconstruction methods. The reconstructed image can be approximated by assuming that the true underlying PET image has been volume-smoothed with a Gaussian of a known FWHM (the point spread function of the scanner). This implicit smoothing is distinct from the explicit smoothing performed in an exploratory analysis as mentioned above. The PVE causes the radiotracer signal to spill over between tissue types. Typically this results in an underestimation of radiotracer concentration in GM. The amount of underestimation at a location depends on the volume of GM near that area. When studying diseases where GM volume is changing (such as Alzheimer's disease), PVEs can create uncertainty as to whether a change in radiotracer concentration is due to a change in tissue uptake or simply a change in GM volume (Thomas et al., 2012). Either can change the measured radiotracer concentration.
Post-reconstruction methods have been developed to correct for PVEs (PVC) on a voxel-wise basis given a coregistered tissue segmentation from CT or MRI and the FWHM of the PET point spread function. The most common of these is the Muller-Gartner (MG) method (Muller-Gartner et al., 1992) but others have also been proposed (Meltzer et al., 1990; Meltzer et al., 1996; Thomas et al., 2012). The principle is that the PET signal spilling into one tissue type from an adjacent tissue type is estimated and subtracted and, subsequently, each voxel is divided by the partial volume fraction (PVF) for its tissue type. The resulting image is then transformed to common space, spatially smoothed, and compared across subject; some examples of studies that have taken this approach are shown in Table 1.
Evaluation of voxelwise PVC performance has been limited to how accurately the PET signal can be recovered inside of a ROI (Meltzer et al., 1990; Muller-Gartner et al., 1992; Yanase et al., 2005; Thomas et al., 2012; Hutton et al., 2013). The performance of PVC has not been evaluated after the explicit spatial smoothing operation ubiquitously performed in exploratory analysis. This is a critical omission because MG PVC can cause noise amplification (Rousset et al., 2007) due to the division by the PVF, a number always less than 1 (sometimes much less). If the spill-in subtraction is inaccurate, then this inaccuracy will also be amplified. The more distant a voxel is from GM, the smaller the GM PVF and the more the noise amplification. When the data are volume-smoothed, problematic voxels (i.e., those with low PVF) will be smoothed into, and in so doing contaminate, the high PVF voxels.
PET time series data are often analyzed using kinetic or graphical models to determine the binding potential (BPND) of a radioligand (Logan et al., 1990; Ichise et al., 1996; Lammertsma et al., 1996; Lammertsma and Hume, 1996; Ichise et al., 2003). Ichise et al., 2003 demonstrated that BPND estimates computed from kinetic models are subject to noise-dependent bias. This means that as the noise level increases, not only does the variance of the estimated BPND increase, but it systematically deviates from the true underlying BPND. The noise-dependent bias can be expected to be much worse in an exploratory analysis where the voxel-wise noise level is much greater than that in a ROI. If the noise level is different across subjects, then the bias will increase the intersubject variability. If the noise level is different across groups, then noise-dependent bias can contribute to inter-group bias. This emphasizes the need to properly manage time series noise in exploratory kinetic modeling applications.
To summarize, (1) exploratory analysis is needed to find effects for which there is no spatial a priori hypothesis, (2) spatial smoothing is needed in exploratory analysis to improve voxel-wise SNR, and (3) volume-based spatial smoothing can interact with MG PVC to cause an increase in noise and bias. As a result, exploratory analysis with PVC of brain PET data is problematic. The purpose of this study is to document the noise and bias amplification that results from the interaction of MG-style PVC and volume-based smoothing and to show that these problems are greatly reduced when cortical surface-based smoothing is used instead. We also demonstrate that the bias and noise properties of surface-based smoothing are superior to that of volume-based even when PVC is not used. The demonstration uses a kinetic modeling application, which is particularly sensitive to noise and so has the most to gain from the new surface-based technique.
Methods
Participants
Sixteen healthy male participants (age: mean ± s.d. = 25.9 ± 3.85, range = 20 – 35 years) were recruited. These data sets have been used in two other studies. However, one was a ROI-based analysis (Fisher et al., 2012) and the second focused on specific binding only within hippocampus (Haahr et al., 2012a). The protocol was approved by the Ethics Committee of Copenhagen and Frederiksberg, Denmark, and all subjects gave written informed consent.
PET Acquisition and Preprocessing
The serotonin 5-HT4 PET radioligand [11C]SB207145 was synthesized as described previously (Marner et al., 2009). An intravenous bolus injection of [11C]SB207145 (injected radioactivity: mean ± s.d. = 589 ± 31 MBq, range = 489 - 608 MBq) was given over 20 seconds. Immediately following injection, a 2-hour dynamic emission scan including 38 time frames (6×5, 10×15, 4×30, 5×120, 5×300, and 8×600 seconds) was acquired using a Siemens ECAT HRRT scanner operating in 3D-acquisition mode with an approximate in-plane resolution of 2 mm and point spread function (PSF) of 4mm. Dynamic PET images were reconstructed using a 3D-OSEM-PSF (Sureau et al., 2008). Single-subject PET frames were motion corrected to frame 26 using AIR 5.2.5 (Woods et al., 1992). The aligned maps of time activity curves (TACs) were used as input to the pipelines described below.
Structural MRI and Anatomical Analysis
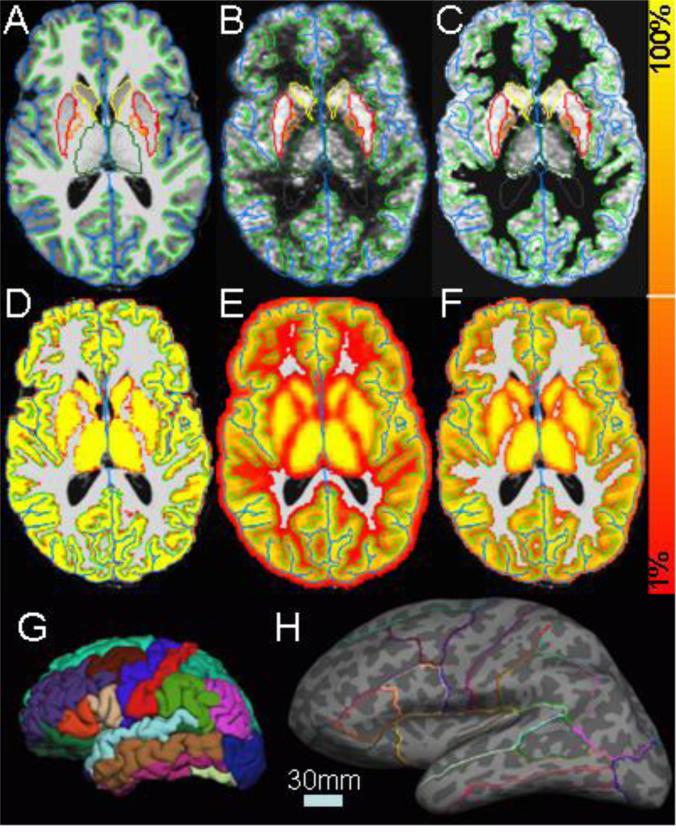
A 3D T1-weighted MPRAGE scan (TE/TR/TI = 3.04/1550/800 ms; bandwidth=170 Hz/Px; echo spacing= 7.7 ms; flip angle= 9°; 1mm3 voxel size) was acquired for each subject using a Siemens Trio 3T MR scanner. All MRI scans were corrected for spatial distortions due to gradient non-linearity (Jovicich et al., 2006) so that they were distortion-free relative to the PET. All subjects were analyzed in FreeSurfer (FS, www.surfer.nmr.mgh.harvard.edu, version 5.3) to provide detailed anatomical information customized for each subject (Dale et al., 1999; Fischl et al., 1999a; Fischl et al., 1999b; Fischl and Dale, 2000; Segonne et al., 2004; Segonne et al., 2007). An example FS analysis is shown in Figure 1A, 1G, and 1H. In Figure 1A; one can see the green and blue lines that surround cortex. These are the “white” and “pial” surfaces, respectively. Smoothness constraints allow the surfaces to cut through voxels and provide subvoxel segmentation accuracy. While rendered in a 2D slice, these surfaces are actually 2D manifolds embedded in 3D space. The 2D-in-3D pial surface is shown in Figure 1G. The folded surface can be unfolded (“inflated”) to see inside the sulci as shown in Figure 1H. FreeSurfer also segments seven subcortical GM structures per hemisphere (four of which are shown Figure 1A) as well as ventricles, cerebral WM, cerebellum GM and WM, and brainstem (Fischl et al., 2002). The cortical surface is further segmented into gyral regions as shown in Figures 1G and 1H. A total of 82 segmentations (including left and right hemispheres) were used in this study (Fischl et al., 2004; Desikan et al., 2006). FS provides excellent MRI segmentation accuracy, something that has been shown to be important in PET PVC (Quarantelli et al., 2004). The cortical surfaces of each subject are further registered to a standard surface atlas space (similar to Talairach but on the surface) in which comparisons across subject can be made (Fischl et al., 1999b).
Figure 1.
A. T1 MRI with white surface (light green), pial surface (blue), putamen (red), pallidum (orange), caudate (yellow), and thalamus (green). Note that insular cortex and putamen are very close to each other. B. PET image summed over time and co-registered with the MRI (gray scale intensity is raw summed PET value). C. Same as B after MG PVC and 20% masking. D. Gray matter partial volume fraction superimposed on the MRI. The color scale to the right has a minimum of 1% and a maximum of 100%. E. Same as C but smoothed by 4mm (the PSF of the scanner) and thresholded at 1%. F. Same as D but thresholded at 20%. G. Pial surface with surface segmentations. H. Inflated surface with segmentation outlines. Dark gray areas are sulci; light gray areas are gyri. The light blue scale bar shows the length of 30mm in both G and H.
Surface Smoothing
The 2D-in-3D surface model consists of a dense mesh of triangles, the edges of which are about 1mm in length. The point where neighboring triangles meet is called a vertex1. Each vertex can be assigned a value such as cortical thickness or PET intensity or segmentation label. Surface smoothing is then accomplished by averaging this value among nearby vertices (Hagler et al., 2007). This type of smoothing is constrained to be only within cortical GM, so WM, CSF, and subcortical GM never get smoothed into cortical GM. Like volume smoothing, surface smoothing is quantified by its FWHM. Several surface-based PET studies have been performed with voxelwise PVC (Park et al., 2006; Kochunov et al., 2009; Protas et al., 2010; Becker et al., 2011); however, they did not use MG PVC, did not perform a systematic comparison between volume and surface smoothing at different smoothing levels, and did not perform surface-based kinetic modeling.
Multimodal Integration
For each subject, mean PET uptake images were computed across all time points in the TAC. This was aligned to the MRI using Boundary-based Registration (BBR, Greve and Fischl, 2009) using a 6 degree of freedom linear transform. BBR is very robust with respect to image artifacts and so an ideal candidate for PET-MR registration. The registration accuracy was assessed visually for each subject, a typical example of which is shown in Figure 1B. Accurate registration has been shown to be important in PET PVC (Quarantelli et al., 2004). Once the registration was established, the FreeSurfer segmentations were mapped into the PET space. In addition, the segmented MRIs were used to create partial volume fraction maps for WM and GM in the PET space (Figure 1D). These were volume smoothed by the PSF of the HRRT (4mm FWHM; Figure 1E) in preparation for performing PVC. The registration also allowed for the PET data to be sampled onto the cortical surface. The PET data were always sampled half way between the white and pial surfaces (i.e., in the middle of the cortical ribbon). Varnas et al., 2003 found that cortical layers I and II have higher BPND than deeper layers, so sampling in the middle captures the average. Given that cortex is only 3mm thick on average and the voxels were 1.2mm, there is not sufficient resolution to claim that a particular layer of cortex is being sampled. Sampling at the halfway point is a way to assure that the sample comes from a point that is least affected by PVE
Spatial Processing Pipelines
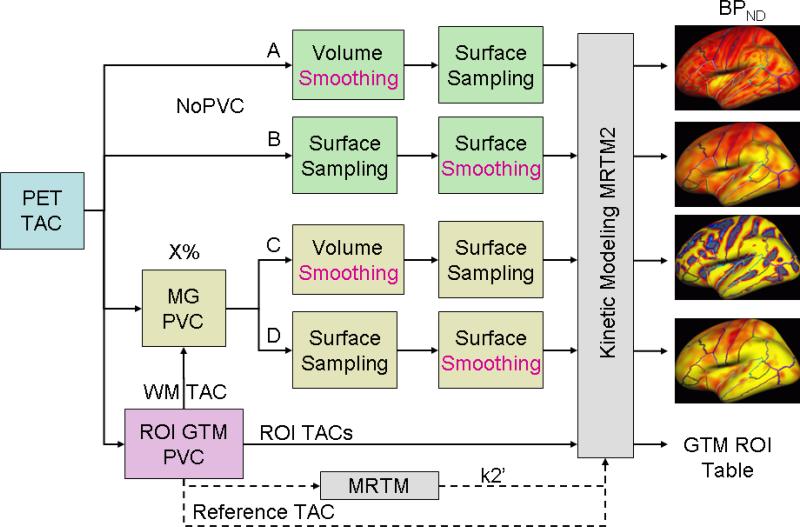
An overview of the pipelines under study is shown in Figure 2. There are four major manipulations of the pipeline based on 1) whether PVC was used or not and 2) whether smoothing was performed in the volume or on the surface. In each case the raw PET TAC served as the input. In the Surface Smoothing pipelines, the PET TAC2 was sampled onto the subject's native surface and then transformed to the FreeSurfer standard surface space after which it was surface-smoothed. In pipelines with Volume Smoothing, the TACs were sampled onto the surface after smoothing in the volume. The result of each pipeline is a set of TACs sampled onto the standard surface space. In PET exploratory analysis, the 3-D FWHM of applied smoothing generally ranges from 5mm-14mm (Table 1). In this study, the smoothing level was varied from 0mm to 15mm. At a smoothing level of 0, the volume- and surface-based streams are identical. Comparisons between the volume- and surface-based pipelines were always done in the standard surface space to assure that differences are only due to either PVC or type of smoothing.
Figure 2.
Overview of processing streams under test. A: Volume smoothing without PVC (NoPVC+VolSm). B. Surface smoothing without PVC (NoPVC+SurfSm). C. Volume Smoothing after PVC (PVC+VolSm). D. Surface smoothing after PVC (PVC+SurfSm). The GTM stream is a ROI-based analysis. X% refers to the GM PVF masking threshold. The Reference TAC comes from the cerebellum.
ROI-based Partial Volume Effect Correction
Though we are interested in testing voxelwise analysis, a ROI-based PVC method was also implemented for two reasons: (1) the ROI method supplies information needed for voxelwise analysis (namely the and mean WM TAC), and (2) it provides a natural way to assess the effect of smoothing on voxelwise analysis. The method used, the geometric transfer matrix (GTM; Rousset et al., 1998), assumes each ROI to have a constant but unknown mean value. A forward linear model is constructed that relates all unknown means, the spatial definitions of all ROIs, the PSF, and the observed PET intensities. This model is then inverted to solve for the unknown means while simultaneously removing spill-in and compensating for spill-out without suffering from noise amplification present in MG. The GTM was implemented using all 82 ROIs, which covered the entire brain (i.e., cortical and subcortical, GM, WM, and CSF). The output is a set of 82 TACs.
Voxel-based Partial Volume Effect Correction
The MG algorithm was implemented as described in the original paper (Muller-Gartner et al., 1992) with several modifications introduced in later papers. MG assumes that the true PET signal in WM is the same across all WM voxels. This value is obtained from the GTM (making this the “modified” MG method (Quarantelli et al., 2004)) and then multiplied by the WM PVF to get a “synthetic” WM image which is then subtracted from the raw PET to remove the contribution of WM from all voxels. The voxels in this image are then divided by the GM PVF to correct for the reduction in GM intensity due to the scanner PSF. Researchers performing exploratory analysis have modified MG further by thresholding the corrected image based upon the amount of GM in a voxel (Rousset et al., 2007; see also Table 1 for references). Typical thresholds are in the range of 20%-50% (Table 1). In this study, we varied the threshold from 1% to 50%. The TACs in voxels that did not meet this criterion were set to zero. Figures 1E and 1F show the masks for 1% and 20%, thresholding. Figure 1C shows the uptake image from Figure 1B after MG correction and 20% masking but without any smoothing. In addition, any voxel whose TAC sum was less than zero was set to 0, which can happen when the synthetic WM TAC is too large. The MG TAC images were then processed in the volume- and surface-based streams (Figure 2) to provide TAC maps for the kinetic modeling.
Kinetic Modeling (KM)
KM was performed using the two-stage Multilinear Reference Tissue Model (MRTM2; Ichise et al., 2003). In the first stage, the global washout rate constant from the reference region, , was computed from high-binding regions (putamen, caudate, and pallidum for 5-HT4) using MRTM (Ichise et al., 1996) with cerebellar GM as the reference. Once the was established, the rate constant for transfer from target tissue to plasma (k2) and the apparent rate constant (k2a) were estimated in the second stage from the TAC of each ROI/voxel by solving for k2 and k2a in the following formula:
| (1) |
where Cj(t) is the TAC of the target ROI/voxel j, and Cr(t) is the reference TAC. The non-displaceable binding potential was computed as . For pipelines A-D in Figure 2, the result is a map of binding potentials for each subject in the standard surface space. The output of the GTM pipeline is a set of BPND values for each ROI and subject. All pipelines use the same cerebellar reference TAC and the same .
Performance Criteria
We evaluated the performance of the four pipelines in anticipation of their use in an exploratory map-based analysis using two performance criteria: bias and variance (Thomas et al., 2012; Hutton et al., 2013). Bias indicates how much the value at a voxel systematically varies from the “true” value at that voxel as a percentage of the “true” value3. We do not have the true value at each voxel, but we do have the value of a ROI from the GTM. To compute the bias, we average the voxelwise BPND over a ROI and compare it to the BPND from the GTM. In addition, we performed a noiseless simulation in which the value at a voxel in a ROI was set to the group average GTM BPND for that ROI afterwhich the map was smoothed by the 4mm PSF. Simulated BPND maps were generated for each subject and analyzed in each pipeline afterwhich the bias was computed as above comparing the group mean ROI value against the known simulation value.
Variance indicates how much the results tend to be spread around the average (without regard to any bias). We use the coefficient of variation (CoV) as our metric of variance. The CoV was computed as follows. The mean and standard deviation of the BPND across subject was computed at each voxel. The CoV was computed as the voxel standard deviation divided by the voxel mean. The CoV was then averaged across all voxels in the ROI to give an estimate of the amount of total cross-subject variability at each voxel in the ROI. This CoV metric is meant to measure the average intersubject variability at a voxel inside the ROI. It is not a measure of intervoxel variability. The CoV is the inverse of the effect size and so can be used in a power analysis. Simulations were not used to evaluate variance because of the difficulties in simulating both within and between-subject noise.
For both the bias and variance metrics, averages for a ROI were computed using a robust mean in which the highest and lowest 5% of the data within a ROI were excluded (Thomas et al., 2012; Hutton et al., 2013). Note that while we are reporting ROI-based metrics, the bias and variance measures are meant to reflect performance at a voxel level and so applicable to exploratory analysis.
Results
Table 2 summarizes the group mean GTM ROI results for a subset of ROIs and provides a comparison with other studies. Group mean (+/−s.d.) BPND values for each cortical ROI and each processing stream are given in Supplementary Table S1. Since the GTM produces one value for each ROI, it should always result in variance reduction due to averaging over the ROI. However, the matrix inversion can result in noise amplification if there are many ROIs or if some ROIs are very small. This was not an issue for any of the 82 ROIs in any subject. The worst case was the nucleus accumbens in which the variance was reduced by a factor of 95. For cortex, the reduction ranged from 600 to 2000. Thus all ROIs had very stable, robust mean estimates making the GTM a stable benchmark against which the voxelwise results can be compared. The intersubject standard deviations in Table 2 confirm that the GTM is not much noisier than non-PVC methods.
Table 2.
Distribution of 5-HT4 BPND throughout the brain as found in this and other studies.
| ROI | GTM | Simple Mean | Fisher | Marner | Varnas |
|---|---|---|---|---|---|
| Caudate | 5.41+/-0.63 | 3.30+/-0.41 | 2.97+/-0.83 | 2.62+/-0.33 | 32.0 |
| Lentiform | 3.82+/-0.50 | 2.83+/-0.29 | 3.03+/-0.74 | 2.60+/-0.22 | 29.0 |
| Parietal Cortex | 1.51+/-0.13 | 0.89+/-0.09 | N/A | 0.47+/-0.08 | N/A |
| Temporal Cortex | 1.43+/-0.14 | 0.92+/-0.10 | N/A | 0.62+/-0.08 | 22.0 |
| Posterior Cingulate | 1.30+/-0.11 | 0.92+/-0.09 | N/A | 0.59+/-0.10 | N/A |
| Anterior Cingulate | 1.28+/-0.14 | 0.93+/-0.11 | N/A | 0.59+/-0.09 | N/A |
| Prefrontal Cortex | 1.26+/-0.13 | 0.79+/-0.10 | N/A | 0.42+/-0.06 | 15.0 |
| Hippocampus | 1.22+/-0.09 | 0.91+/-0.07 | 0.95+/-0.20 | 0.91+/-0.13 | 23.5 |
| Sensorimotor Cortex | 1.17+/-0.11 | 0.68+/-0.08 | N/A | 0.27+/-0.06 | N/A |
| Amygdala | 1.16+/-0.17 | 0.96+/-0.12 | 0.85+/-0.18 | 0.87+/-0.14 | 18.0 |
| Occipital Cortex | 1.10+/-0.09 | 0.65+/-0.06 | N/A | 0.38+/-0.06 | 11.9 |
| Insula | 1.10+/-0.11 | 0.86+/-0.08 | N/A | 0.77+/-0.10 | 16.0 |
| Thalamus | 0.73+/-0.12 | 0.63+/-0.09 | N/A | 0.54+/-0.07 | 11.1 |
“GTM” is for this study where the 5-HT4 BPND was computed from the GTM. “Simple Mean” is for this study where the 5-HT4 BPND was computed from a simple mean of the TACs in the ROI (and so more consistent with Marner and Fisher). “Fisher” refers to the 5-HT4 BPND as found in a PET study by Fisher et al., 2012 using an overlapping data set. “Marner” refers to the 5-HT4 BPND as found in a PET study by Marner et al., 2010. “Varnas” is the approximate number of binding sites found by Varnas et al., 2003 in the postmortem human brain using autoradiography. +/- indicates cross-subject standard deviation. N/A: not available. The ROIs are sorted in descending GTM BPND order. ROI definitions are approximate across the studies.
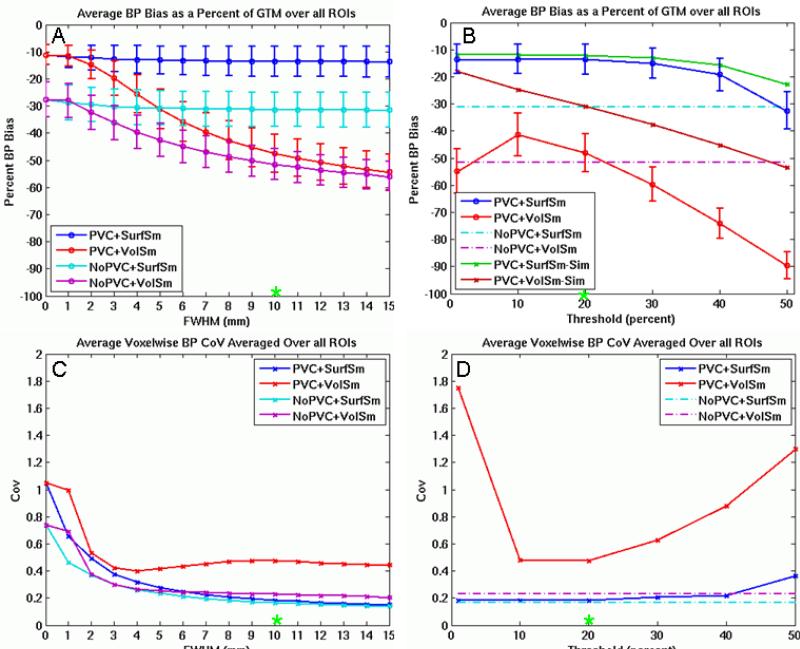
Figure 3 shows a summary of the main results for this study. Figure 3A shows the bias for each of the four pipelines as a function of smoothing level at a fixed threshold of 20%. Figure 3B shows the bias as a function of threshold at a fixed smoothing level of 10mm and includes the noiseless simulation results. Each point in Figures 3A and 3B represents the average percent bias across subjects. The average percent bias of a subject is the average of the absolute biases across all ROIs within that subject. The absolute bias could reflect negative bias (i.e., the ROI value is less than the GTM value) or positive bias (i.e., the ROI value is greater than the GTM value). The absolute bias is given a negative sign to reflect that the vast majority have negative bias (though see Figure 4D for an example of positive bias). While the BPND will vary across ROI, it is appropriate to average the bias metric because it is normalized to the GTM BPND in each ROI thus making the bias in each ROI commensurate. The error bars are the standard deviation of the bias across subject after averaging over the ROI which is why they change little as the smoothing level is changed in Figure 3A. Thus, these error bars represent the ROI-wise, not voxelwise, variance and are not closely related to the CoV in Figures 3C and 3D, which does change with FWHM. The two plots on the bottom row (3C and 3D) show how the CoV compares across the pipelines as a function of smoothing and threshold. Each data point represents the average CoV across all ROIs. The CoV already represents the variance across subjects, so cross-subject error bars are not possible. Note that non-PVC pipelines do not have a threshold parameter so they are represented by flat dashed lines in Figures 3B and 3D.
Figure 3.
Effect of volume (3D) and surface (2D) smoothing and thresholding on bias and variance. A. Bias of each pipeline as a function of smoothing level at a threshold of 20%. B. Bias of the PVC pipelines as a function of threshold at a FWHM of 10mm (including simulations). C. Average voxelwise CoV for each pipeline as a function of smoothing level at a threshold of 20%. D. Average voxelwise CoV for each pipeline as a function of threshold at a FWHM of 10mm. The green asterices (*) indicate the same data points. Error bars reflect standard deviation across subjects, which is not directly related to the CoV.
Figure 4.
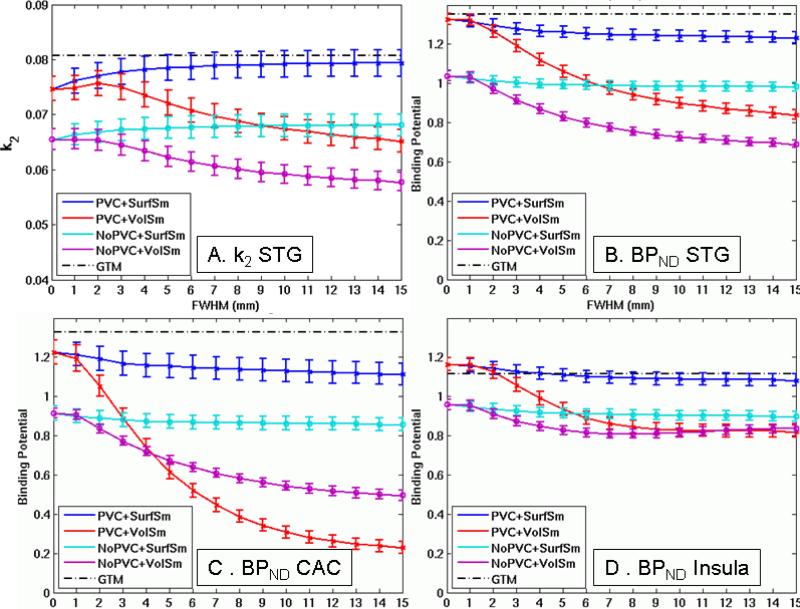
Effect of smoothing on k2 and BPND in three representative ROIs. A. k2 (min−1) for Superior temporal gyrus (STG). B. BPND for STG. C. BPND for Caudal anterior cingulate (CAC). D. BPND for Insula. Error bars reflect standard error of the ROI mean across subjects (not related to the voxelwise CoV measure).
Figure 4A shows the effect of FWHM on the k2 KM rate parameter for superior temporal gyrus (STG). Figures 4B-D show the effect of FWHM on BPND for three representative ROIs. Each point is the robust average of the BPND values across the given ROI for the given pipeline, and the error bars are standard deviation across subject (again not necessarily related to CoV). All plots used a threshold of 20%. The horizontal black line is the GTM result (the “ideal” value).
Figure 5 shows the effects of smoothing and threshold on the group average voxelwise BPND maps, with and without PVC, projected onto the lateral and medial surfaces of the inflated left hemisphere. These maps represent the data from which the plots in Figures 3A, 3B and 4 were generated. Figure 6 shows the effects of threshold on the group average voxelwise BPND maps (FWHM=10mm). These maps represent the data from which the plots in Figure 3B were generated. One always expects BPND to be greater than or equal to zero (red/yellow); however, many of the maps have negative (blue) BPND for reasons explained below.
Figure 5.
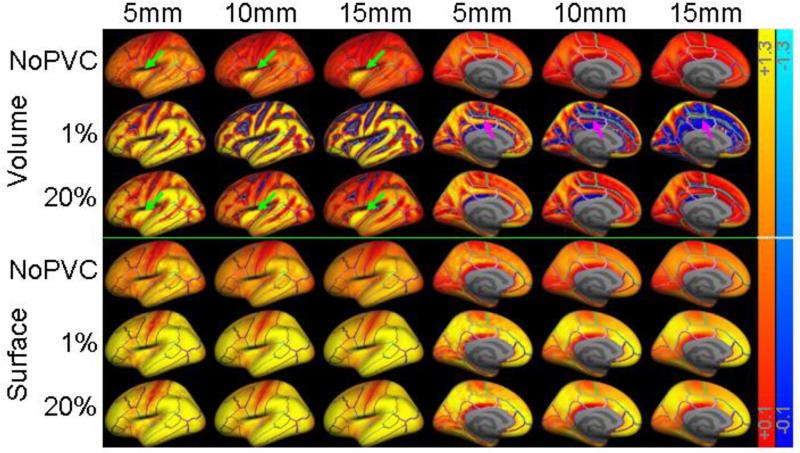
Binding potential maps as a function of volume and surface smoothing level. Voxels are transparent (i.e., gray) if they are not in cortex or their BPND value is in the range −0.1 to + 0.1. The color scales go from +/−0.1 to +/−1.3 (warm colors are positive BPND; blue are negative). The first row is the volume smoothing method without PVC. The next two rows are the volume method with PVC and thresholds of 1% and 20%. The next three rows are the same for surface-based smoothing. The first three columns are of the lateral view of left hemisphere; the second three columns are of the medial view. Green arrows point to activity from the high binding putamen “leaking” into cortex. Magenta arrows point to increases in negative BPND in the medial wall.
Figure 6.
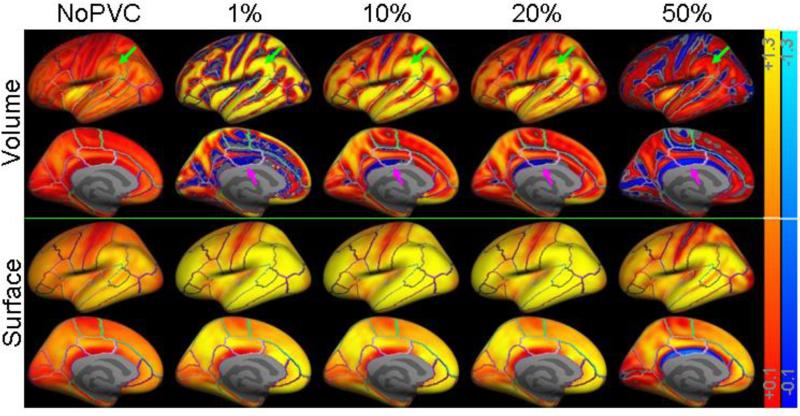
Binding potential maps as a function of threshold (at volume or surface smoothing of FWHM=10mm). The color scales go from +/−0.1 to +/−1.3 (warm colors are positive BPND; blue are negative). The first two rows are the lateral and medial views of the results for the volume smoothing method. The next two rows are that for the surface-based smoothing method. Green arrows point to a location where BPND drops with threshold eventually becoming less than when no PVC is applied. Magenta arrows point to a location with a large negative BPND at low threshold which decreases at moderate threshold then becomes larger at high thresholds.
Discussion
Kinetic Modeling
Kinetic modeling of [11C]SB2307145 has been extensively studied in Marner et al., 2009 and Marner et al., 2010 using data from the (lower resolution) GE Advance scanner, using the SRTM instead of MRTM2, defining ROIs based on Quarantelli et al., 2004 instead of FreeSurfer, and not using PVC. Fisher et al., 2012 used the same processing as Marner et al., 2010 with a superset of the data in this study. Neither Fisher et al., 2012 nor Marner et al., 2010 used PVC, so, for comparison, we analyzed the ROI data using a simple mean of the TACs. Table 2 shows that the “Simple Mean” results of the current study are very close (within 1 s.d.) to that of Fisher et al., 2012 despite very different processing; the standard deviation is always less for the current study. Our results are also very close to those of Marner et al., 2010 in ROIs less susceptible to PVE (eg, caudate, lentiform, hippocampus, amygdale, thalamus). For cortical ROIs, our BPND values are substantially larger than that of Marner et al., 2010. This is because Marner et al., 2010 used a lower resolution scanner and cortical ROIs are much more susceptible to PVE than subcortical ROIs. The GTM results are always greater than the others because the GTM uses PVC. This comparison shows that the kinetic modeling performs as well as and is consistent with that of Marner et al., 2010 and Fisher et al., 2012. Table 2 also shows that the GTM BPND rank order generally agrees with the number of binding sites in the post mortem autoradiography study of Varnas et al., 2003.
No Partial Volume Correction (NoPVC)
As expected the bias is generally higher without PVC than the equivalent PVC pipeline (but not always, see below). As smoothing increases, the bias also increases (Figures 3A, 3B, and 4), but the increase is much more for volume smoothing (NoPVC+VolSm) than for surface smoothing (NoPVC+SurfSm). The reason for this is that NoPVC+VolSm uses a 3D smoothing kernel which indiscriminately smoothes in signal from neighboring WM and CSF tissue around cortex (as well as true signal from adjacent cortical GM). Smoothing in of signal from neighboring WM and CSF will artificially reduce estimated binding potential values within GM because any [11C]SB207145 binding in WM and CSF reflects non-specific binding. In effect, volume-smoothing without PVC is the same as collecting data on a lower resolution scanner with a larger PSF.
The surfaced-based analysis without PVC has similar bias as the volume-based analysis at low smoothing levels (they are the exact same analysis at FWHM=0). However, at higher smoothing levels, the bias of NoPVC+SurfSm is virtually unchanged. This is because smoothing along the surface does not dilute the GM TAC with that from WM and CSF but only smoothes in signal from adjacent cortex. Though there is variation in BPND of the 5-HT4 receptor across cortex, adjacent regions tend to have similar BPND (Varnas et al., 2003; Marner et al., 2010), so smoothing along cortex tends to have a small effect. Figure 1H shows that even a 15mm kernel is much smaller than the cortical structures involved.
Volume-based smoothing has another undesirable effect in that it can smooth subcortical GM into cortex or vice versa. In this data, this effect is evident in insula and putamen. Putamen is a high binding area that is very close to insula (only a few mm separate them; see Figure 1A). Thus, volume smoothing will cause signal from putamen to be smoothed into insula. This can be clearly seen in Figure 5. As volume smoothing is increased, the high BPND spot in insula gets bigger (green arrows). In Figure 4D, one can see that the NoPVC+VolSm estimates level out at a much lower FWHM than the other ROIs and actually increases slightly as FWHM is increased. This increase is evidence of contamination of the insula from putamen. The same phenomenon was observed in the simulations (not shown). Again, the surface-based result does not suffer from this effect. In Figure 4D, one can also see that the MG PVC results are actually higher than the GTM at low smoothing levels (i.e., positive bias). This is because the GTM appropriately removes the spill-over between insula and putamen whereas MG PVC cannot correct for spillover between two GM structures.
PVC and Volumetric Smoothing
In general, MG PVC resulted in larger BPND and less bias with respect to the GTM BPND, both expected results. While this is generally true, it is not always the case, and conditions where PVC actually reduces BPND relative to NoPVC are important and informative. For example, Figure 3B shows that the bias in PVC+VolSm (red line) is worse than that of NoPVC+VolSm (dashed magenta line) when the threshold exceeds 25%. Even at a threshold of 20%, the PVC+VolSm of the caudal anterior cingulate (Figure 4C) is more biased than NoPVC+VolSm when smoothing level exceeds 4mm. The reason for this is due to an interaction between the volume smoothing and the PVC. The MG method simulates the WM TAC at each point in the brain, subtracts this TAC from the raw PET, then divides each voxel by the GM PVF, and then masks the TAC based on GM PVF. Violations of the MG model can manifest themselves in several ways. The subtraction of the WM from the total PET can result in negative TACs (and so negative BPND). These negatives are then amplified by division by the GM PVF. This is most likely to happen in putative areas of WM away from GM. Volume smoothing then smoothes these negatives into the GM thereby reducing signal and possibly creating more bias than if no PVC was done at all. One can clearly see negative BPND (blue in the medial wall pointed to by magenta arrows in Figure 5) at 1% threshold and that the negative area gets larger as smoothing level is increased. Note that the direction of the effect depends upon whether the synthetic WM TAC at a voxel is over or underestimated. Underestimation will give rise to a positive bias.
This problem at low GM PVF voxels led to modifications to the MG method in which voxels with low amounts of GM were masked out (i.e., replaced with zeros). One must choose a threshold to define which voxels have “low PVF”; thresholds used by other authors range from 20%-50% (Table 1). The choice of threshold is important. At a threshold of 1%, the PVC+VolSm is slightly more biased than NoPVC+VolSm (Figure 3B). At thresholds of 10-20%, PVC+VolSm has less bias, but beyond 25% PVC actually creates more bias than not doing PVC at all. Figure 6 shows that this is caused by two effects. At 1% there are many areas in the volume analysis that have high BPND (bright yellow; e.g., area pointed to by the second green arrow). As the threshold increases, the estimated BPND decreases (becomes darker red) and, at 50%, are darker than when no PVC is applied at all (i.e., area pointed to by the last green arrow darker than that of the first green arrow). One can also see that there are large areas of negative BPND (blue; e.g., area pointed to by first magenta arrow) at 1% probably caused by WM oversubtraction as described above. These negatives cause the ROI average to drop even when the ROI has areas of high positive BPND. At 10% and 20%, one can see that the negative regions recede as negative TACs are masked out (second and third magenta arrows). At 50%, negative regions reappear (forth magenta arrow) because there are many zero-valued TACs that get smoothed into the unmasked TACs. This causes the unmasked TACs to have a value less than that of the reference region, which forces the BPND to be negative. Thus some of the bias is due to an interaction with kinetic modeling, and some would be present even if kinetic modeling were not performed.
PVC and Surface Smoothing
These problems are greatly reduced when MG PVC is used in the context of the surface-based analysis. Figure 3A shows that increasing the FWHM has very little effect on the bias. Figure 3B shows that changing the threshold has very little effect as well. The reason for this is that the surface sampling selects points that are only well within cortical gray matter to begin with. This tends to exclude data points where MG PVC has the most noise amplification or where masking creates zeros. The surface smoothing then only smoothes among these good data points and excludes WM and CSF. Problems arise only at the highest thresholds where cortical voxels themselves start to be masked out.
Effects on k2
the k2 parameter is the rate constant for transfer from target tissue to plasma. It is strongly affected by the various preprocessing options as indicated by Figure 4A for STG. In general, all k2 values were less than that of the GTM. Surface smoothing causes k2 estimates to increase (i.e., become less biased) while volume smoothing causes them to decrease (i.e., more biased). PVC caused k2 to be less biased in most ROIs with CAC again being an exception. The reasons for this behavior mirror those for the behavior of BPND discussed above.
Voxelwise Intersubject Variability
The different pipelines in Figure 2 reflect different ways to prepare single-subject data in for cross-subject analysis. While the choice of pipeline obviously affects the within-subject variance, it also affects between-subject variance because spatial smoothing creates an interaction with the individual anatomy. Thus, the variance metric should be sensitive to both within- and between-subject variance (i.e., the mixed effects variance). For this reason, we have chosen a variance metric (the CoV) that reflects total cross-subject variance. Smoothing can reduce this variation in two ways (Worsley et al., 1996). First, it can reduce the within subject temporal noise by averaging nearby TACs. This reduces the noise in the MRTM2 parameter estimates and so reduces noise in the final BPND measurement (this also reduces noise-dependent bias). Second, it can improve the effective intersubject alignment in cases where intersubject alignment is off by the FWHM. Figure 3C shows that the CoV generally decreases with FWHM for all pipelines (at a threshold of 20%). Given the problems with volume smoothing mentioned above, one might be tempted not to smooth at all. However, one can see that without smoothing (volume or surface) the CoV is 2-3 times greater than when moderate amounts of smoothing are used. Similar noise reductions are seen in fMRI with smoothing (Strother et al., 2004). Smoothing is generally advantageous; however, if an effect is small relative to the smoothing kernel, there is a risk that the smoothing will washout the effect.
Regardless of FWHM, surface smoothing without PVC (NoPVC+SurfSm) has the best CoV performance. This is because noise from WM and CSF does not get smoothed into cortex and there is no noise amplification due to PVC. Note that, while NoPVC+SurfSm has the best CoV performance, it does not have the best bias performance (Figure 3A). The CoV metric used in this study is independent of the bias metric. For example, if all subjects had the same negative BPND value at a voxel, the bias would be high but the CoV would be zero. So it is possible that different pipelines or manipulation to a pipeline parameter can have different effects on bias and CoV, i.e., there is a bias-variance tradeoff. The CoV performance of volume smoothing without PVC (NoPVC+VolSm) is only a little worse than the surface-based methods at FWHM > 7mm. While WM and CSF voxels contaminate the GM voxels, the noise in them has not been amplified by PVC and there are no zeros getting smoothed in.
PVC with volume smoothing (PVC+VolSm) has the worst noise performance. As volume smoothing FWHM increases, more voxels from adjacent WM and CSF are smoothed into GM. These voxels may be noisy because of noise amplification or because the WM signal has not been effectively removed or because they have been replaced by zeros. Whatever the source, the intersubject variability will increase because the effect depends upon the individual subject's anatomy.
Notably, at a 20% threshold and FWHM of 10mm, the CoV for PVC+VolSm is more than twice that of PVC with surface smoothing (Fig 3C/D). This means that a study analyzed with PVC+VolSm would require roughly four times more subjects than if it were analyzed using PVC with surface smoothing to attain comparable statistical power. At a threshold of 40%, the CoV for PVC+VolSm was four times worse, meaning that sixteen times more subjects would be needed to attain comparable power.
Simulation Results
Bias of the noiseless simulation results matched the real data quite well for the surface-based analysis but underestimated the bias for the volume-based analysis (Figure 3B). The likely reason for this is that noise causes additional bias in the BPND estimates (Ichise et al., 2003) that is not reflected in the noiseless simulation. While there is noise in the surface-based analysis, the volume-based analysis suffers from a high degree of noise amplification due to the already documented interaction between volume smoothing and MGPVC.
Limitations
These results were derived from [11C]SB2307145 data analyzed with a one-compartment MRTM2; however, we fully expect the results to generalize to other radiotracers and kinetic models. The current study was performed on a molecular imaging application. While more traditional PET imaging studies (eg, FDG) are less sensitive to noise because KM is not used, they will still be susceptible to bias and noise amplification and so should benefit from surface-based analysis. For example, two of the FDG studies in Table 1 actually report decreases in PET signal in GM after PVC, a phenomenon associated with shortcomings in the volume-based method in this study. The other studies in Table 1 did not test for decreases caused by PVC.
Only healthy young subjects were studied. However, this methodology is not only appropriate for analyses of elderly subjects with substantial cortical atrophy, it may even be necessary. FreeSurfer will accurately model the cortical surface even in cases of extreme atrophy (e.g., see Salat et al., 2004 and Dickerson et al., 2009). Such accurate segmentation is critical to removing the PVE. Similar surface-based methods have been applied extensively in fMRI analysis of such populations (e.g., Dickerson et al., 2004; Dickerson et al., 2005).
Surface-based methods do not allow for the analysis of subcortical structures. Subcortical structures are better suited to be analyzed using a volume-based approach, which employs volume-based smoothing, perhaps constrained to avoid smoothing across subcortical GM and WM boundaries. This implies separate processing streams for cortical and subcortical spaces. The details of such a pipeline are beyond the scope of this manuscript. However, we point out that one has already been implemented in fMRI4 (e.g., Schadwinkel and Gutschalk, 2011; Holzel et al., 2013). While we do not foresee a case where a surface-based analysis is not advantageous, surface-based analysis is technically and computationally more challenging than its volume-based counterpart because: it requires an 8-16 hour FreeSurfer analysis for each subject; the PET must be sampled to and analyzed on the surface; and the user must deal with unfamiliar surface formats and visualization.
The voxelwise PVC method used in this paper reflects a common way in which voxelwise PVC has been applied over the last decade (Table 1). More recent post-reconstruction PVC methods (e.g., Hutton et al., 2013) may work better with volume-based smoothing by reducing noise amplification and replacing masked out values with something more reasonable than zero, but it is our opinion that smoothing in a manner that recognizes the actual geometry and anatomy (i.e., surface-smoothing) of the brain is inherently sensible and more likely to avoid smoothing-induced noise and bias regardless of the PVC method used. Surface-based smoothing will improve with these new methods just as it does MG. Note that we are not in a position to recommend the GTM PVC over MG PVC because the GTM PVC cannot be used for voxelwise analysis.
Conclusions
This study explored how PVC and smoothing choices made when preprocessing PET data affect the performance of exploratory analysis of cortical regions. Without PVC, volumetric smoothing increased the BPND bias by reducing signal in gray matter. The bias was much less when surface smoothing was employed. The use of MG PVC with volume smoothing reduced bias at some smoothing levels but increased it at others and often caused a dramatic increase in variance due to noise amplification and masking. Surface-based smoothing resulted in substantially less bias without an increase in variance. When used with PVC, surface smoothing resulted in the standard deviation decreasing by 2-4 times compared to the equivalent volume-smoothed analysis. This translates into at least 4-16 times fewer subjects needed in a group analysis to achieve the same significance. Surface-based smoothing has less bias and variance because it respects cortical geometry by smoothing the PET data only along the cortical ribbon and so does not contaminate the GM signal with that of white matter and cerebrospinal fluid. Thus, the use of surface-based analysis in PET should result in improvements in the reliability and detectability of effects in exploratory PET analysis, with or without PVC.
Supplementary Material
This is a study of the effects of preprocessing on exploratory PET analysis
Volumetric 3D smoothing of PET data exacerbates the partial volume effect
Volumetric smoothing and partial volume correction combine to increase variability
Cortical surface-based smoothing does not exacerbate the partial volume effect
Surface-based kinetic analysis reduces intersubject variance by a factor of 4.
Acknowledgements
We would like to thank M. Haahr, G. Thomsen, C. Jensen, S. Larsen, A. Dyssegaard, K. Christiansen, L. Freyr for their assistance in scheduling and data collection at both the MR and PET centers. We would like to gratefully acknowledge The John and Birthe Meyer Foundation for the donation of the Cyclotron and PET-scanner. We would like to thank the Danish Research Centre for Magnetic Resonance for the MRI resources. This study was funded by a center grant to Cimbi from the Lundbeck Foundation. Support for this research was provided in part by the National Institutes of Health grants 5R01EB006758-04, 5R01NS052585-05, 5R21NS072652-02, P41-RR14075, R01RR16594-01A1, and by the Capital Region of Denmark
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
A vertex is a data point in a spatial location, so it can be thought of as a voxel. Below we use “voxel” as a single term to indicate either a voxel from a volume-based analysis or a vertex from a surface-based analysis.
I.e., the radioactivity corrected for decay.
Some researchers (Erlandsson, et al, 2012, Thomas, et al, 2013, Hutton, et al, 2013) use the recovery coefficient (RC) to measure bias. The RC and bias are related through the equation RC = (Bias/100 +1).
The FreeSurfer Functional Analysis Steam (FSFAST) surfer.nmr.mgh.harvard.edu/fswiki/FsFast
References
- Becker JA, Hedden T, Carmasin J, Maye J, Rentz DM, Putcha D, Fischl B, Greve DN, Marshall GA, Salloway S, Marks D, Buckner RL, Sperling RA, Johnson KA. Amyloid-beta associated cortical thinning in clinically normal elderly. Ann Neurol. 2011;69:1032–1042. doi: 10.1002/ana.22333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeat P, Chetelat G, Villemagne VL, Fripp J, Raniga P, Pike K, Acosta O, Szoeke C, Ourselin S, Ames D, Ellis KA, Martins RN, Masters CL, Rowe CC, Salvado O. Beta-amyloid burden in the temporal neocortex is related to hippocampal atrophy in elderly subjects without dementia. Neurology. 2010;74:121–127. doi: 10.1212/WNL.0b013e3181c918b5. [DOI] [PubMed] [Google Scholar]
- Chetelat G, Desgranges B, de la Sayette V, Viader F, Berkouk K, Landeau B, Lalevee C, Le Doze F, Dupuy B, Hannequin D, Baron JC, Eustache F. Dissociating atrophy and hypometabolism impact on episodic memory in mild cognitive impairment. Brain. 2003;126:1955–1967. doi: 10.1093/brain/awg196. [DOI] [PubMed] [Google Scholar]
- Chetelat G, Desgranges B, Landeau B, Mezenge F, Poline JB, de la Sayette V, Viader F, Eustache F, Baron JC. Direct voxel-based comparison between grey matter hypometabolism and atrophy in Alzheimer's disease. Brain. 2008;131:60–71. doi: 10.1093/brain/awm288. [DOI] [PubMed] [Google Scholar]
- Curiati PK, Tamashiro-Duran JH, Duran FL, Buchpiguel CA, Squarzoni P, Romano DC, Vallada H, Menezes PR, Scazufca M, Busatto GF, Alves TC. Age-related metabolic profiles in cognitively healthy elders: results from a voxel-based [18F]fluorodeoxyglucose-positron-emission tomography study with partial volume effects correction. AJNR Am J Neuroradiol. 2011;32:560–565. doi: 10.3174/ajnr.A2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical Surface-Based Analysis I: Segmentation and Surface Reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Dickerson B, Salat D, Bates J, Atiya M, Killiany R, Greve D, Dale A, Stern C, Blacker D, Albert MS, RA S. Medial temporal lobe function and structure in mild cognitive impairment. Annals of Neurology. 2004;56:27–35. doi: 10.1002/ana.20163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Bakkour A, Salat DH, Feczko E, Pacheco J, Greve DN, Grodstein F, Wright CI, Blacker D, Rosas HD, Sperling RA, Atri A, Growdon JH, Hyman BT, Morris JC, Fischl B, Buckner RL. The cortical signature of Alzheimer's disease: regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb Cortex. 2009;19:497–510. doi: 10.1093/cercor/bhn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Salat DH, Greve DN, Chua EF, Rand-Giovannetti E, Rentz DM, Bertram L, Mullin K, Tanzi RE, Blacker D, Albert MS, Sperling RA. Increased hippocampal activation in mild cognitive impairment compared to normal aging and AD. Neurology. 2005;65:404–411. doi: 10.1212/01.wnl.0000171450.97464.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proceedings of the National Academy of Sciences. 2000;97:11044–11049. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Albert M, Dieterich M, Haselgrove C, Kouwe A.v.d., Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999a;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Human Brain Mapping. 1999b;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM. Automatically parcellating the human cerebral cortex. Cereb Cortex. 2004;14:11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Fisher PM, Holst KK, Mc Mahon B, Haahr ME, Madsen K, Gillings N, Baare WF, Jensen PS, Knudsen GM. 5-HTTLPR status predictive of neocortical 5-HT4 binding assessed with [(11)C]SB207145 PET in humans. Neuroimage. 2012;62:130–136. doi: 10.1016/j.neuroimage.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Worsley KJ, Frackowiak RSJ, Mazziotta JC, Evans AC. Assessing the significance of focal activations using their spatial extent. Hman Brain Mapping. 1993;1:210–220. doi: 10.1002/hbm.460010306. [DOI] [PubMed] [Google Scholar]
- Greve D, Fischl B. Accurate and robust brain image alignment using boundary-based registration. Neuroimage. 2009;48:63–72. doi: 10.1016/j.neuroimage.2009.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haahr ME, Fisher P, Holst K, Madsen K, Jensen CG, Marner L, Lehel S, Baare W, Knudsen G, Hasselbalch S. The 5-HT(4) receptor levels in hippocampus correlates inversely with memory test performance in humans. Hum Brain Mapp. 2012a doi: 10.1002/hbm.22123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haahr ME, Rasmussen PM, Madsen K, Marner L, Ratner C, Gillings N, Baare WF, Knudsen GM. Obesity is associated with high serotonin 4 receptor availability in the brain reward circuitry. Neuroimage. 2012b;61:884–888. doi: 10.1016/j.neuroimage.2012.03.050. [DOI] [PubMed] [Google Scholar]
- Hagler DJ, Jr., Riecke L, Sereno MI. Parietal and superior frontal visuospatial maps activated by pointing and saccades. Neuroimage. 2007;35:1562–1577. doi: 10.1016/j.neuroimage.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltia LT, Rinne JO, Merisaari H, Maguire RP, Savontaus E, Helin S, Nagren K, Kaasinen V. Effects of intravenous glucose on dopaminergic function in the human brain in vivo. Synapse. 2007;61:748–756. doi: 10.1002/syn.20418. [DOI] [PubMed] [Google Scholar]
- Holzel BK, Hoge EA, Greve DN, Gard T, Cresswell JD, Brown KW, Lazar SW. Neural mechanisms of symptom improvements in generalized anxiety disorder following mindfulness training. Neuroimage: Clinical. 2013 doi: 10.1016/j.nicl.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlemann R, Matusch A, Kuhn KU, Berning J, Elmenhorst D, Winz O, Kolsch H, Zilles K, Wagner M, Maier W, Bauer A. 5-HT2A receptor density is decreased in the at-risk mental state. Psychopharmacology (Berl) 2008;195:579–590. doi: 10.1007/s00213-007-0921-x. [DOI] [PubMed] [Google Scholar]
- Hutton BF, Thomas BA, Erlandsson K, Bousse A, Reilhac-Laborde A, Kazantsev D, Pedemonte S, Vunckx K, Arridge S, Ourselin S. What approach to brain partial volume correction is best for PET/MRI? Nuclear Instruments and Methods in Physics Research A. 2013:29–33. [Google Scholar]
- Ichise M, Ballinger JR, Golan H, Vines D, Luong A, Tsai S, Kung HF. Noninvasive quantification of dopamine D2 receptors with iodine-123-IBF SPECT. J Nucl Med. 1996;37:513–520. [PubMed] [Google Scholar]
- Ichise M, Liow JS, Lu JQ, Takano A, Model K, Toyama H, Suhara T, Suzuki K, Innis RB, Carson RE. Linearized reference tissue parametric imaging methods: application to [11C]DASB positron emission tomography studies of the serotonin transporter in human brain. J Cereb Blood Flow Metab. 2003;23:1096–1112. doi: 10.1097/01.WCB.0000085441.37552.CA. [DOI] [PubMed] [Google Scholar]
- Inoue K, Ito H, Goto R, Nakagawa M, Kinomura S, Sato T, Sato K, Fukuda H. Apparent CBF decrease with normal aging due to partial volume effects: MR-based partial volume correction on CBF SPECT. Ann Nucl Med. 2005;19:283–290. doi: 10.1007/BF02984620. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, Fischl B, Dale A. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage. 2006;30:436–443. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Kalpouzos G, Chetelat G, Baron JC, Landeau B, Mevel K, Godeau C, Barre L, Constans JM, Viader F, Eustache F, Desgranges B. Voxel-based mapping of brain gray matter volume and glucose metabolism profiles in normal aging. Neurobiol Aging. 2009a;30:112–124. doi: 10.1016/j.neurobiolaging.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Kalpouzos G, Chetelat G, Landeau B, Clochon P, Viader F, Eustache F, Desgranges B. Structural and metabolic correlates of episodic memory in relation to the depth of encoding in normal aging. J Cogn Neurosci. 2009b;21:372–389. doi: 10.1162/jocn.2008.21027. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Ramage AE, Lancaster JL, Robin DA, Narayana S, Coyle T, Royall DR, Fox P. Loss of cerebral white matter structural integrity tracks the gray matter metabolic decline in normal aging. Neuroimage. 2009;45:17–28. doi: 10.1016/j.neuroimage.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus C, Hahn A, Savli M, Kranz GS, Baldinger P, Hoflich A, Spindelegger C, Ungersboeck J, Haeusler D, Mitterhauser M, Windischberger C, Wadsak W, Kasper S, Lanzenberger R. Serotonin-1A receptor binding is positively associated with gray matter volume -- a multimodal neuroimaging study combining PET and structural MRI. Neuroimage. 2012;63:1091–1098. doi: 10.1016/j.neuroimage.2012.07.035. [DOI] [PubMed] [Google Scholar]
- Lammertsma AA, Bench CJ, Hume SP, Osman S, Gunn K, Brooks DJ, Frackowiak RS. Comparison of methods for analysis of clinical [11C]raclopride studies. J Cereb Blood Flow Metab. 1996;16:42–52. doi: 10.1097/00004647-199601000-00005. [DOI] [PubMed] [Google Scholar]
- Lammertsma AA, Hume SP. Simplified reference tissue model for PET receptor studies. Neuroimage. 1996;4:153–158. doi: 10.1006/nimg.1996.0066. [DOI] [PubMed] [Google Scholar]
- Logan J, Fowler JS, Volkow ND, Wolf AP, Dewey SL, Schlyer DJ, MacGregor RR, Hitzemann R, Bendriem B, Gatley SJ, et al. Graphical analysis of reversible radioligand binding from time-activity measurements applied to [N-11C-methyl]-(−)-cocaine PET studies in human subjects. J Cereb Blood Flow Metab. 1990;10:740–747. doi: 10.1038/jcbfm.1990.127. [DOI] [PubMed] [Google Scholar]
- Marner L, Gillings N, Comley RA, Baare WF, Rabiner EA, Wilson AA, Houle S, Hasselbalch SG, Svarer C, Gunn RN, Laruelle M, Knudsen GM. Kinetic modeling of 11C-SB207145 binding to 5-HT4 receptors in the human brain in vivo. J Nucl Med. 2009;50:900–908. doi: 10.2967/jnumed.108.058552. [DOI] [PubMed] [Google Scholar]
- Marner L, Gillings N, Madsen K, Erritzoe D, Baare WF, Svarer C, Hasselbalch SG, Knudsen GM. Brain imaging of serotonin 4 receptors in humans with [11C]SB207145-PET. Neuroimage. 2010;50:855–861. doi: 10.1016/j.neuroimage.2010.01.054. [DOI] [PubMed] [Google Scholar]
- Matsuda H, Kanetaka H, Ohnishi T, Asada T, Imabayashi E, Nakano S, Katoh A, Tanaka F. Brain SPET abnormalities in Alzheimer's disease before and after atrophy correction. Eur J Nucl Med Mol Imaging. 2002;29:1502–1505. doi: 10.1007/s00259-002-0930-2. [DOI] [PubMed] [Google Scholar]
- Matsuda H, Ohnishi T, Asada T, Li ZJ, Kanetaka H, Imabayashi E, Tanaka F, Nakano S. Correction for partial-volume effects on brain perfusion SPECT in healthy men. J Nucl Med. 2003;44:1243–1252. [PubMed] [Google Scholar]
- Meltzer CC, Leal JP, Mayberg HS, Wagner HN, Jr., Frost JJ. Correction of PET data for partial volume effects in human cerebral cortex by MR imaging. J Comput Assist Tomogr. 1990;14:561–570. doi: 10.1097/00004728-199007000-00011. [DOI] [PubMed] [Google Scholar]
- Meltzer CC, Zubieta JK, Links JM, Brakeman P, Stumpf MJ, Frost JJ. MR-based correction of brain PET measurements for heterogeneous gray matter radioactivity distribution. J Cereb Blood Flow Metab. 1996;16:650–658. doi: 10.1097/00004647-199607000-00016. [DOI] [PubMed] [Google Scholar]
- Mevel K, Desgranges B, Baron JC, Landeau B, De la Sayette V, Viader F, Eustache F, Chetelat G. Detecting hippocampal hypometabolism in Mild Cognitive Impairment using automatic voxel-based approaches. Neuroimage. 2007;37:18–25. doi: 10.1016/j.neuroimage.2007.04.048. [DOI] [PubMed] [Google Scholar]
- Muller-Gartner HW, Links JM, Prince JL, Bryan RN, McVeigh E, Leal JP, Davatzikos C, Frost JJ. Measurement of radiotracer concentration in brain gray matter using positron emission tomography: MRI-based correction for partial volume effects. J Cereb Blood Flow Metab. 1992;12:571–583. doi: 10.1038/jcbfm.1992.81. [DOI] [PubMed] [Google Scholar]
- Park HJ, Lee JD, Chun JW, Seok JH, Yun M, Oh MK, Kim JJ. Cortical surface-based analysis of 18F-FDG PET: measured metabolic abnormalities in schizophrenia are affected by cortical structural abnormalities. Neuroimage. 2006;31:1434–1444. doi: 10.1016/j.neuroimage.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Protas HD, Huang SC, Kepe V, Hayashi K, Klunder A, Braskie MN, Ercoli L, Bookheimer S, Thompson PM, Small GW, Barrio JR. FDDNP binding using MR derived cortical surface maps. Neuroimage. 2010;49:240–248. doi: 10.1016/j.neuroimage.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarantelli M, Berkouk K, Prinster A, Landeau B, Svarer C, Balkay L, Alfano B, Brunetti A, Baron JC, Salvatore M. Integrated software for the analysis of brain PET/SPECT studies with partial-volume-effect correction. J Nucl Med. 2004;45:192–201. [PubMed] [Google Scholar]
- Rousset O, Rahmim A, Alavi A, Zaidi H. Partial volume correction strategies in PET. PET Clinics. 2007;2:235–249. doi: 10.1016/j.cpet.2007.10.005. [DOI] [PubMed] [Google Scholar]
- Rousset OG, Ma Y, Evans AC. Correction for partial volume effects in PET: principle and validation. J Nucl Med. 1998;39:904–911. [PubMed] [Google Scholar]
- Salat DH, Buckner RL, Snyder AZ, Greve DN, Desikan RS, Busa E, Morris JC, Dale AM, Fischl B. Thinning of the cerebral cortex in aging. Cereb Cortex. 2004;14:721–730. doi: 10.1093/cercor/bhh032. [DOI] [PubMed] [Google Scholar]
- Samuraki M, Matsunari I, Chen WP, Yajima K, Yanase D, Fujikawa A, Takeda N, Nishimura S, Matsuda H, Yamada M. Partial volume effect-corrected FDG PET and grey matter volume loss in patients with mild Alzheimer's disease. Eur J Nucl Med Mol Imaging. 2007;34:1658–1669. doi: 10.1007/s00259-007-0454-x. [DOI] [PubMed] [Google Scholar]
- Schadwinkel S, Gutschalk A. Transient bold activity locked to perceptual reversals of auditory streaming in human auditory cortex and inferior colliculus. J Neurophysiol. 2011;105:1977–1983. doi: 10.1152/jn.00461.2010. [DOI] [PubMed] [Google Scholar]
- Segonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK, Fischl B. A hybrid approach to the skull stripping problem in MRI. Neuroimage. 2004;22:1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Segonne F, Pacheco J, Fischl B. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Trans Med Imaging. 2007;26:518–529. doi: 10.1109/TMI.2006.887364. [DOI] [PubMed] [Google Scholar]
- Strother s., La Conte S, Hansen LK, Anderson J, Zhang J, Pulapura S, Rottenberg DA. Optimizing the fMRI data-processing pipeline using prediction and reproducibility performance metrics: I. A preliminary group analysis. Neuroimage. 2004;23:S196–S207. doi: 10.1016/j.neuroimage.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Sureau FC, Reader AJ, Comtat C, Leroy C, Ribeiro MJ, Buvat I, Trebossen R. Impact of image-space resolution modeling for studies with the high-resolution research tomograph. J Nucl Med. 2008;49:1000–1008. doi: 10.2967/jnumed.107.045351. [DOI] [PubMed] [Google Scholar]
- Thomas BA, Erlandsson K, Modat M, Thurfjell L, Vandenberghe R, Ourselin S, Hutton BF. The importance of appropriate partial volume correction for PET quantification in Alzheimer's disease. Eur J Nucl Med Mol Imaging. 2012;38:1104–1119. doi: 10.1007/s00259-011-1745-9. [DOI] [PubMed] [Google Scholar]
- Uchida H, Chow TW, Mamo DC, Kapur S, Mulsant BH, Houle S, Pollock BG, Graff- Guerrero A. Effects of aging on 5-HT(2A) R binding: a HRRT PET study with and without partial volume corrections. Int J Geriatr Psychiatry. 2011;26:1300–1308. doi: 10.1002/gps.2682. [DOI] [PubMed] [Google Scholar]
- Van Laere K, Goffin K, Casteels C, Dupont P, Mortelmans L, de Hoon J, Bormans G. Gender-dependent increases with healthy aging of the human cerebral cannabinoid-type 1 receptor binding using [(18)F]MK-9470 PET. Neuroimage. 2008;39:1533–1541. doi: 10.1016/j.neuroimage.2007.10.053. [DOI] [PubMed] [Google Scholar]
- Varnas K, Halldin C, Pike VW, Hall H. Distribution of 5-HT4 receptors in the postmortem human brain--an autoradiographic study using [125I]SB 207710. Eur Neuropsychopharmacol. 2003;13:228–234. doi: 10.1016/s0924-977x(03)00009-9. [DOI] [PubMed] [Google Scholar]
- Woods RP, Cherry SR, Mazziotta JC. Rapid Automated Algorithm for Aligning and Reslicing PET Images. J of Computer Assisted Tomography. 1992;16:620–633. doi: 10.1097/00004728-199207000-00024. [DOI] [PubMed] [Google Scholar]
- Worsley KJ, Marrett S, Neelin P, Evans AC. Searching Scale Space for Activation in PET Images. Human Brain Mapping. 1996;4:74–90. doi: 10.1002/(SICI)1097-0193(1996)4:1<74::AID-HBM5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Yanase D, Matsunari I, Yajima K, Chen W, Fujikawa A, Nishimura S, Matsuda H, Yamada M. Brain FDG PET study of normal aging in Japanese: effect of atrophy correction. Eur J Nucl Med Mol Imaging. 2005;32:794–805. doi: 10.1007/s00259-005-1767-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.