Abstract
Autophagy is a protein and organelle degradation pathway that is involved in diverse diseases including cancer. Recent evidence suggests that autophagy is a cell survival mechanism in tumor cells and that its inhibition especially in combination with other therapy could be beneficial but it remains unclear if all cancer cells behave the same way when autophagy is inhibited. We inhibited autophagy in a panel of breast cancer cell lines and found that some of them are dependent on autophagy for survival even in nutrient rich conditions without any additional stress while others need autophagy only when stressed. Survival under unstressed conditions is due to cell type specific autophagy regulation of STAT3 activity and this phenotype is enriched in triple negative cell lines. This autophagy-dependency affects response to therapy because autophagy inhibition reduced tumor growth in vivo in autophagy-dependent but not in autophagy-independent breast tumors while combination treatment with autophagy inhibitors and other agent was preferentially synergistic in autophagy-dependent cells. These results imply that autophagy-dependence represents a tumor cell specific characteristic where autophagy inhibition will be more effective. Moreover, our results suggest that autophagy inhibition might be a potential therapeutic strategy for triple negative breast cancers, which currently lack an effective targeted treatment.
Keywords: autophagy, breast cancer, STAT3
Introduction
Autophagy is a long-lived protein and organelle degradation pathway in which cytoplasmic material is engulfed in a double membrane structure known as the autophagosome and later delivered to the lysosome for degradation. Macroautophagy (referred herein as autophagy) is thought to be the major type of autophagy.(1) It has been extensively studied in recent years, increasing our understanding of the major constituents of the autophagic pathway, encoded by the ATG genes, and unraveling many of its roles in homeostasis and development. Most importantly, defects in the autophagic pathway have been involved in diverse diseases, including cancer. (1–4)
Under normal conditions, basal autophagy has been proposed to function as a tumor suppressor mechanism by reducing oxidative stress, inflammation and genome instability.(2) However, autophagy has also been suggested to act as a survival mechanism in established tumors. It is well known that in cells under stress - including starvation, growth factor deprivation, hypoxia, radiation and chemotherapy - autophagy is up-regulated to recycle cytoplasmic components and provide the cell with amino acids and ATP to support essential metabolic pathways and protein synthesis.(2) This is critical in tumor cells, which are constantly exposed to both metabolic stress via hypoxia, inadequate glucose supply and increased energetic demands of rapidly proliferating cells as well as proteotoxic stress induced by high levels of genomic instability found in cancers. Although this autophagy requirement could be a generally important mechanism of survival in tumor cells, recent evidence suggests that certain tumors are “autophagy addicted.” In this regard, RAS-transformation is known to induce high levels of basal autophagy in cancer cell lines; autophagy is required for efficient RAS-induced tumorigenesis, and many, but not all, RAS-transformed cell lines are highly dependent on autophagy. (5–7)
Despite implementation of prevention programs and novel therapeutic strategies, breast cancer remains the second leading cause of cancer death among women in the United States.(8) One of the biggest recognized barriers to progress in prevention, diagnosis and treatment is the clinical and genetic heterogeneity of the disease.(9) In this regard, gene expression analyses have led to the definition of five molecular “intrinsic” subtypes of breast cancer (Luminal A, Luminal B, HER2-enriched, basal-like and claudin-low), which have differences in incidence, survival and response to treatment.(10) Basal-like and claudin-low tumors comprise the majority of triple negative breast cancers (TNBC),(10) a subgroup of tumors that do not express clinically significant levels of estrogen receptor (ER), progesterone receptor (PgR) and HER2 and thus cannot be treated with endocrine or anti-HER2 targeted therapies. They include 10–24% of invasive breast cancers and have a worse prognosis when compared to other tumor subtypes. Importantly, although patients with TNBC do benefit from chemotherapy, there are no known targeted agents for this type of cancer and after therapy, they tend to relapse with distant metastases, resulting in a worse overall survival (11) and underscoring the need to develop better, less toxic treatment approaches.
Among many other distinctive characteristics, TNBC cells are known to have high levels of activation of the JAK-STAT pathway.(12) STATs (signal transducers and activators of transcription) are transcription factors that are activated in the cytoplasm by tyrosine phosphorylation in response to cytokine receptor activation (IFN or IL-6) and their associated Janus kinase (JAK) or to growth factor receptor signaling (EGF, PDGF), either directly or through recruitment of associated proteins.(13) Cytoplasmic kinases such as SRC and ABL1 can also phosphorylate and activate STATs.(14) Phosphorylated STATs dimerize and translocate to the nucleus activating transcription. Ligand-dependent activation of STATs is associated with cellular differentiation and growth regulation. Conversely, their constitutive activation is associated with tumorigenesis by inducing the transcription of genes that promote cell cycle progression, prevention of apoptosis (BCL2L1, CCND1 and MYC), cancer inflammation, and inhibition of anti-tumor immunity.(14–16)
We found that autophagy inhibition is much more important in some breast lines than others and that cancer cell lines with constitutively high levels of STAT3 activity were more sensitive to autophagy inhibition than those with low levels of basal STAT3 activity. The most autophagy-dependent lines require autophagy to survive even under conditions with no added metabolic stress and are enriched in the basal-like and claudin-low subtypes of breast cancer.(10) These effects were due to autophagy-dependent STAT3 activation and were also associated with increased ability to synergize when autophagy was inhibited in combination with another anti-cancer drug. Our results suggest that autophagy inhibition might prove beneficial especially for “basal-like” and “claudin-low” breast cancers, which mostly constitute the TNBC subgroup and that currently lack effective treatment strategies.
Materials and Methods
Cell Culture
All cell lines were acquired from the University of Colorado Tissue Culture Core in 2012 and were authenticated by allele testing (date of authentication is as follows: MCF10A, MCF7, T47D, HCC1937, MDAMB468, MDAMB231: October, 2011; MCF12A, BT549: May, 2012). After acquisition, cells were grown, frozen and each time they were thawed, they were not used for longer than 6 months. Cell culture media used is shown in Supplementary Experimental Procedures. STAT3C plasmid and its empty vector (Addgene plasmids 24983 and 14883) were a gift from Linzhao Cheng (17) and David Baltimore (18), respectively. MDAMB231 and MDAMB468 cells were transduced with lentiviruses expressing these plasmids and sorted for a high green fluorescent population.
Autophagy shRNA library
We designed a lentiviral shRNA library with 629 shRNAs targeting autophagy proteins from the autophagic core machinery, proteins described in the autophagy interactome (19) and apoptosis-related proteins. A pool of shRNA-expressing plasmids from the TRC library (pLKO.1 vector) was prepared at the Functional Genomics Core of the University of Colorado Cancer Center. 3–5 shRNAs per gene target were used. Lentiviruses were generated according to protocols from the RNAi Consortium (http://www.broadinstitute.org/rnai/trc/lib) and cells were transduced at a MOI of 0.3 and selected with puromycin. Post-infection samples were collected 18 h after the addition of virus in quintuplicate. Another sample of transduced cells was re-plated with puromycin and allowed to grow for 15 days after which the growth sample was collected in quintuplicate. Samples were barcoded as described previously (20) and analyzed in an Illumina Genome Analyzer IIX.
Functional genetic screening data were analyzed using our in-house bio-informatics pipeline,(20–24) and as described in Supplemental Materials and Methods.
shRNA Lentiviral Transduction
Lentiviruses were prepared according to protocols published at the RNAi Consortium webpage (http://www.broadinstitute.org/rnai/trc/lib). Viruses (containing either non-silencing, ATG5 TRCN0000151474, ATG7 TRCN0000007587, BECN1 TRCN0000033549, STAT3 TRCN0000020840 or STAT3 TRCN0000020842 pLKO.1 vectors) were added to achieve an MOI of 5–10. Cells were selected with puromycin according to a dose-response curve and with the concentrations shown on Supplementary Experimental Procedures for 2–3 days, then tripsinized and used for experiments.
Viability assays
For long-term clonogenic assays, cells were plated in 12 well plates and allowed to grow for 8–10 days. They were fixed and stained with crystal violet (BD). Stain was solubilized and absorbance was measured at 540 nm. For proliferation curves, cells were counted at 3, 6, 8 or 10 days after plating. Trypan blue negative cells were considered viable.
For MTS assays, cells were plated in 96 well plates and allowed to grow for 2 to 6 days. Cells were treated with MTS reagent (Promega) according to manufacturer’s instructions. For cell death assessment, media was collected and evaluated for lactate dehydrogenase activity with LDH cytoscan cytotoxicity kit (G-Biosciences) according to manufacturer’s instructions. For time lapse movies, cells were plated at equal densities and imaged for 48 h. For Propidium Iodide (PI) staining, cells transduced with the different shRNAs were plated, allowed to grow for 48 h, stained with PI and imaged.
Protein Isolation and Western Blots
2–3 days after plating, cells were lysed with RIPA buffer containing protease inhibitor (Roche) and Halt phosphatase inhibitor cocktail (Thermo Scientific). Protein was quantitated using Bradford reagent. Antibodies used are listed in Supplemental Materials and Methods. Western Blot figures show a portion of the membranes containing the corresponding bands cropped for clarity.
Animal Studies
All animal studies were performed in accordance with the Colorado State University’s Animal Care and Use Committee. Female Nude nu/nu mice were purchased from the National Cancer Institute (Frederick, MD) and challenged with 5 × 106 MDAMB231 cells. Female nude nu/nu mice were first ovariectomized and received a subcutaneous, 60 day release, 0.25mg estradiol implant (Innovative Research). Mice were then challenged, 7 days later, with 5 × 106 MCF7 cells. 100 μL of cells in 50% serum free media and 50% Matrigel (BD Biosciences) were injected into the 4th mammary fat pad. Upon reaching a tumor volume of 100 mm3, mice received either 60 mg/kg chloroquine diphosphate salt or 0.9% saline given by intraperitoneal injection, once daily for the duration of the study. Study was followed until tumors reached 4 times initial volume (TV*4) (25).
Statistical Analysis
Statistical differences were determined using a two-sample equal variance Student’s t-Test. Log rank analysis for the Kaplan-Meier curves was done in Prism v5.0a. Three MCF7 xenografts did not quite reach 4 times initial volume (TV*4), so time to reach TV*4 was extrapolated using calculated tumor doubling time. Combination index was calculated using CompuSyn (ComboSyn, Inc.).
A more detailed description of the Materials and Methods section was included in Supplemental Materials and Methods.
Results
Autophagy-focused shRNA lentiviral library
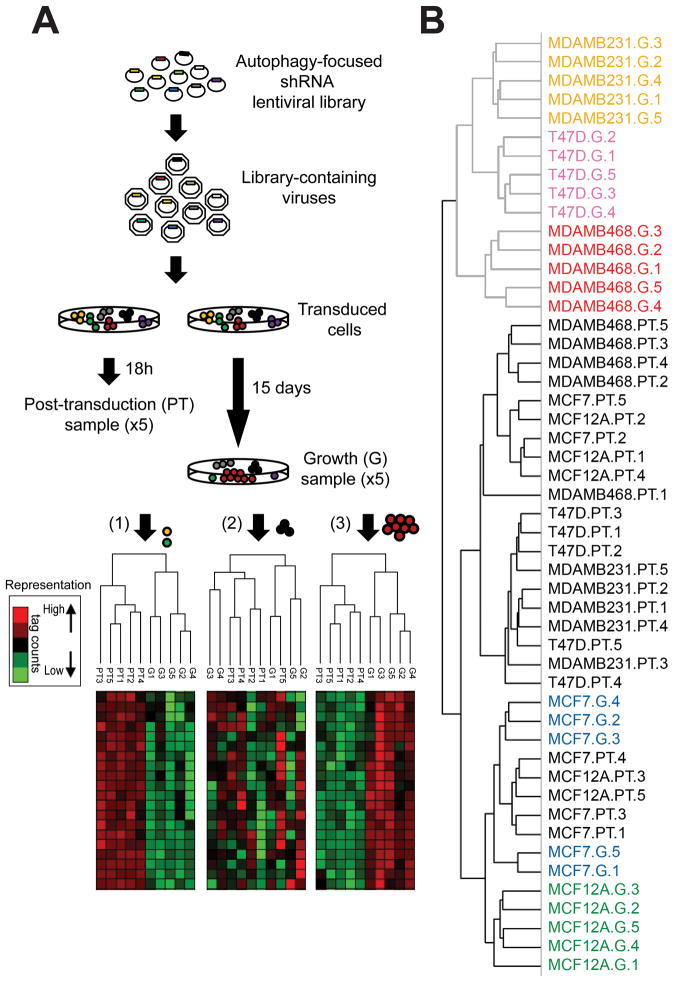
We designed an autophagy-specific shRNA library, targeting the core components of the autophagic pathway (ATG genes) as well as some of their interacting proteins (19) and used this library to evaluate “autophagy dependence” in a panel of breast cancer cell lines. Since functions independent from autophagy have been described for ATG genes,(26–28) we reasoned that if autophagy is important for tumor cell survival and proliferation to different extents in different cell lines, this would be manifested by differential selection for or against shRNAs that target the autophagy pathway during cell growth, leading to overall differences in the pattern of shRNA representation of the library in different breast cell populations, and not to changes in the shRNA of a particular ATG protein. On the other hand, if autophagy is not important for the growth or survival of cells, shRNA representation after growth selection should be similar to the starting population. Because the approach examines large numbers of shRNAs whose only similarity is their connection to autophagy, differences in the overall representation of the whole library should represent the importance of autophagy itself rather than any other function for individual autophagy regulators (Figure 1A). We used a non-transformed cell line (MCF12A), 2 luminal (MCF7, T47D), and two triple negative cell lines, one basal (MDAMB468) and one claudin-low (MDAMB231), according to the classification by Prat, A., et al.(10) One DNA sample was collected 18 h after transduction (post-transduction, PT) and another sample collected 15 days following cell growth in complete medium (G), both in quintuplicate. Samples were barcoded and analyzed by next generation sequencing as described previously.(20) Hierarchical clustering of the shRNA readouts produced two main clusters (Figure 1B). The first cluster contained all the PT samples combined with the MCF7 and MCF12A growth samples, indicating a similar shRNA representation among these conditions. The second cluster included MDAMB231, T47D and MDAMB468 growth samples, suggesting differences in shRNA representation between this group of samples and the other cluster, but a similar representation among them. Those shRNAs “lost” in the G sample but present in the PT represent proteins essential for growth, since their knockdown causes decreased proliferation or cell death and a lack of representation of the shRNA in the growth sample (as in Figure 1A(1)). One condition cluster in the heat map showed shRNAs that had less reads in growth samples of MDAMB231, T47D and MDAMB468 cell lines when compared to the PT cluster (Supplementary Figure S1). This cluster included mostly shRNAs that target autophagy at the nucleation and expansion steps, indicating that MDAMB231, T47D and MDAMB468 had a decreased proliferation or death induction when transduced with these shRNAs, suggesting these cells were sensitive to autophagy inhibition. On the other hand, MCF7 and MCF12A cell lines clustered with the post-transduction samples with no significant selection for or against the shRNAs during the 15 day growth period, indicating they proliferate normally despite having shRNAs that target the autophagic pathway. These results suggest that breast cancer cell lines behave differently when autophagy is modulated and that some breast cancer cell lines are dependent on autophagy for proliferation or survival while others are much less dependent on it. A complete list of the shRNAs in the library and their representation per cell line is included in Supplementary Table 1.
Figure 1. Autophagy-focused shRNA library.
A) Experimental outline. Cells were transduced with a shRNA library containing autophagy-focused shRNAs. After transduction, Post-transduction samples, (PT) were collected. Growth samples (G) were collected after 15 days in culture, both in quintuplicate. Hypothetical heat maps for the three possible outcomes are shown in the figure, showing different representations of shRNA tag counts. If most shRNAs targeted a process important for cell survival (yellow cell), or proliferation (green cell), they would be under-represented in the growth sample when compared with post-transduction shRNAs and the overall representation of shRNAs would look as in (1). The dendogram would produce two different clusters with quintuplicates grouping together. If shRNAs in this cluster represent shRNAs that target the autophagic pathway, then autophagy would be important for proliferation/ survival in these cells. If the shRNAs had no effect on proliferation/survival (black cells), the heatmap would look as in (2) and samples would cluster without a clear distinction between PT and G. If this cluster was enriched in shRNAs targeting the autophagic pathway it would mean that autophagy has no effect on growth/proliferation. Finally, if the shRNAs gave a proliferative advantage (red cells), they would be over-represented in the G sample when compared with the PT and representation would look as in (3). The dendogram would produce two well defined groups representing post-transduction and growth samples. If these were shRNAs inhibiting autophagy, it would mean that autophagy decreases proliferation or induces cell death in these cells. B) Hierarchical clustering of breast cancer cell lines transduced with an autophagy-focused shRNA library produced two main clusters. PT samples from all cell lines are labeled in black. First cluster included all the post-transduction samples as well as MCF7 and MCF12A growth samples. Second cluster included MDAMB231, T47D and MDAMB468 growth samples, showing a differential response to autophagy manipulation among breast cancer cell lines.
Breast cancer cell lines have a differential sensitivity to autophagy inhibition
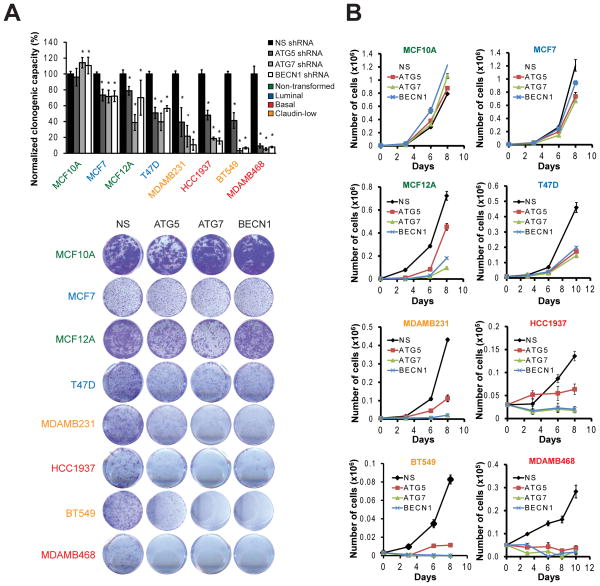
We validated our shRNA library results with individual shRNAs targeting the core autophagy proteins ATG5, ATG7 and BECN1 that were each independently validated as inhibiting autophagy. Figure 2A shows a panel of breast cell lines organized in increasing sensitivity to autophagy inhibition in complete medium when measured by a clonogenic survival assay. These cell lines were differentially affected in their growth / survival when autophagy was inhibited. MCF10A cells were the least affected by autophagy inhibition while MDAMB468 were the most affected. To test how inhibition of autophagy was affecting cell number, we performed a growth curve (Figure 2B). MCF10A cells were resistant to autophagy inhibition; in fact, they proliferated faster. Although there was a decrease in cell number in MCF7, MCF12A and T47D cells with autophagy inhibition, the fact that cell number continued to increase over time in the knockdown cells suggested that their proliferation rate was only decreased. The most affected cell lines were MDAMB231, HCC1937, BT549 and MDAMB468 (Figure 2A, B), where autophagy inhibition greatly decreased proliferation, or completely inhibited it. Importantly, the three shRNAs showed a similar response in each cell line. These results, together with the autophagy library screen, suggest that inhibition of the autophagic pathway and not an unspecific effect of the inhibition of a certain ATG gene differentially affects breast cancer cell proliferation or survival.
Figure 2. Breast cancer cell lines have a differential sensitivity to autophagy inhibition.
A panel of breast cancer cell lines including 2 non-transformed (MCF10A, MCF12A), two luminal (MCF7, T47D), and four TNBC cell lines (two basal, HCC1937 and MDAMB468, and two claudin low, MDAMB231 and BT549) were transduced with shRNAs against ATG5, ATG7 or BECN1 and tested for clonogenic proliferation 8 days after plating. In A, cell lines were ordered according to their increasing sensitivity to autophagy inhibition as measured by crystal violet quantitation. Sample clonogenic images are shown. Alternatively, in B, live (trypan blue negative) cells were counted at different time points after plating and graphed in a proliferation curve. Graph in A shows mean ± SE of three independent experiments performed in triplicate. * = different from its respective non-silencing control; p < 0.01. In B, graphs show one representative experiment performed in triplicate.
Analysis of clonogenic experiments and growth curves from all the cell lines indicated that TNBC cell lines (basal and claudin-low) were the most sensitive to autophagy inhibition (Figure 2A, B). Conversely, in non-transformed and luminal cells, proliferation was induced (MCF10A) or was decreased to a much lesser extent (MCF12A, MCF7 and T47D) than TNBC cell lines, suggesting a delay in cell proliferation rather than cell death. The shRNAs were effective at knocking down their target proteins in all cell lines, showing markedly decreased starvation-induced autophagy as measured by LC3-II accumulation when autophagic flux was blocked (Supplementary Figure S2).
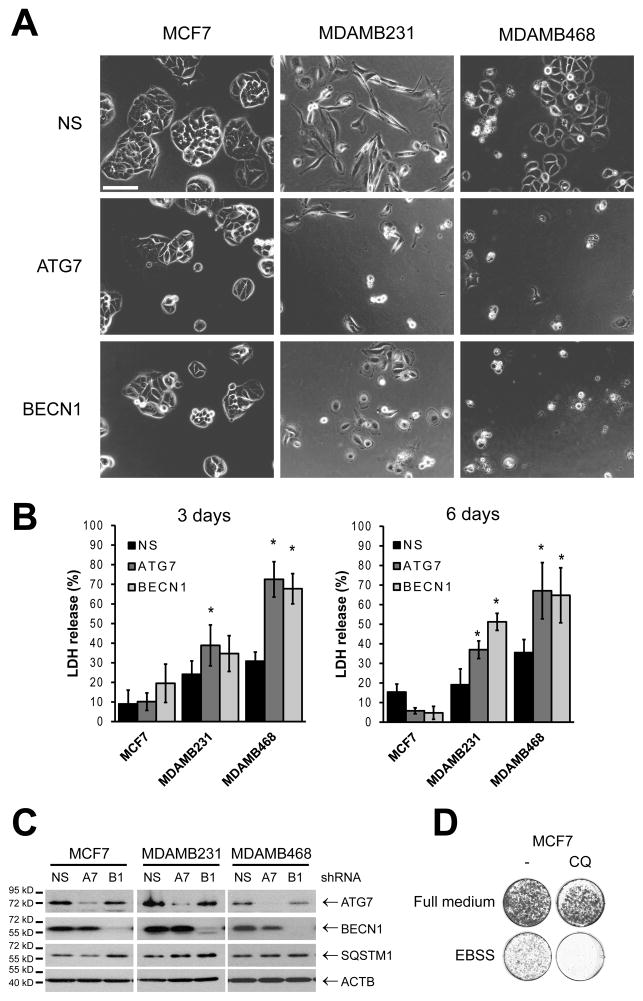
Decreased clonogenic growth could be caused by decreased proliferation or cell death. To test these possibilities, we chose a luminal line with only a modest effect of autophagy inhibition (MCF7) and two TNBC cell lines (MDAMB231 and MDAMB468) where autophagy inhibition had a significant effect and imaged them by time-lapse microscopy after transduction with non-silencing (NS), ATG7 or BECN1 shRNAs (Figure 3A and Supplementary Movies 1–6). The same number of cells were plated for all conditions and Figure 3A shows the cells after 48 h. ATG7 and BECN1 knockdown resulted in a decreased number of cells when compared to the non-silencing controls in the three cell lines. MCF7 cells after ATG7 or BECN1 knockdown looked normal and only proliferated at a somewhat slower rate compared with the non-silencing cells, resulting in smaller colonies. Conversely, MDAMB231 cells showed significantly decreased proliferation when compared to the non-silencing controls and some cells died. The most affected cells were MDAMB468s, where ATG7 and BECN1 knockdown cells did not proliferate and most of these cells died during the 48 h period. In agreement with these observations, MCF7 cells did not show an increase in LDH release in a 6 day period when autophagy was inhibited (Figure 3B). MDAMB231 cells showed increased LDH release after 6 days and MDAMB468 cells showed a maximum LDH release since day 3. All three cell lines had a similar knockdown when measured by Western Blot (Figure 3C). Similar results were found in a propidium iodide (PI) staining experiment to measure cell death, where the highest increase in PI staining was seen on the MDAMB468 cells in which autophagy had been inhibited (Supplementary Figure S3).
Figure 3. Autophagy inhibition induces cell death in some cell lines but not others.
MCF7, MDAMB231 and MDAMB468 cells were transduced with lentiviruses expressing non-silencing, ATG7 or BECN1 shRNAs, plated and evaluated for cell death in a time-lapse movie (A and Supplementary Movies). Frames show a representative image taken 48h after plating; bar represents 200 μM. B). Cell death was also evaluated by LDH release. In C, cell lysates were evaluated for protein knockdown (NS, non-silencing; A7, ATG7; B1, BECN1 shRNAs). D) MCF7 cells were starved in EBSS for 24 h +/− 10 μM chloroquine (CQ). Pictures show a representative image taken 5 days after the removal of the starvation medium. Graphs in B show mean ± SE of three independent experiments performed in triplicate. * = different from its respective non-silencing control (p < 0.05).
Since autophagy inhibition did not induce cell death in MCF7 cells, we tested the functionality of the autophagic pathway in these cells by starving them with EBSS, a standard treatment for autophagy induction. Pharmacological inhibition of autophagy with chloroquine treatment (Figure 3D) or inhibition of autophagy with ATG7 or BECN1 shRNAs (Supplementary Figure S3B) greatly decreased cell viability in a clonogenic assay suggesting that MCF7 cells need autophagy for survival but only under stressed conditions.
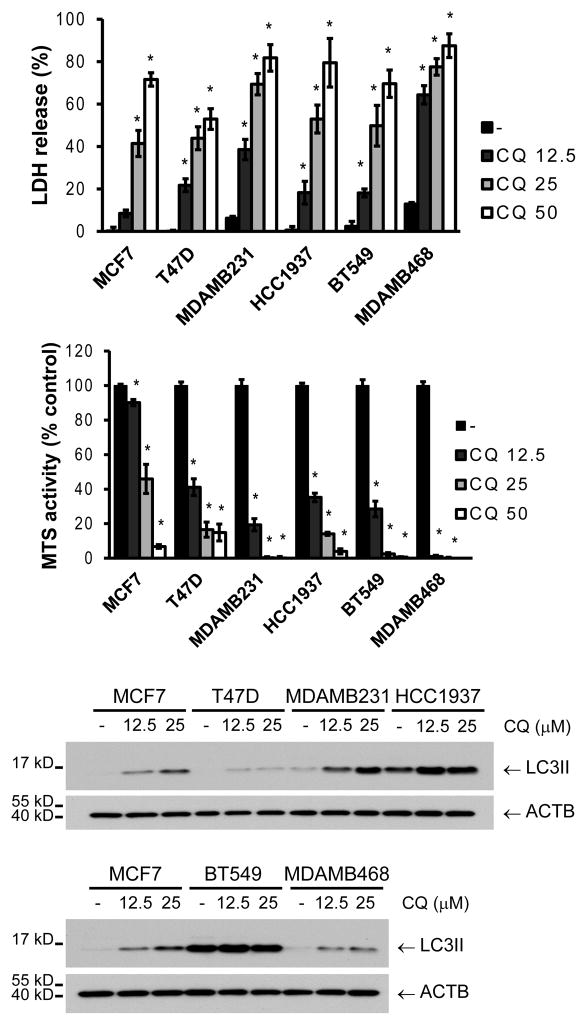
Chloroquine treatment had a similar effect as ATG7 or BECN1 knockdown when cells were grown in full medium (Figure 4). A differential sensitivity to chloroquine was found among all the breast cancer cell lines, with the TNBC cells again being the most sensitive. An increase in LC3II was observed in all the cell lines with chloroquine treatment (Figure 4). However, this increase in LC3II did not correlate with sensitivity to autophagy inhibition, suggesting that differences in basal autophagy among the cell lines are not the cause of autophagy dependence. Together, these data show that breast cancer cell lines differ greatly in their dependency on autophagy, with some lines barely affected when autophagy is inhibited, some showing only relatively some growth inhibition, and some dying. The most autophagy-dependent cells tended to be the triple negatives, which require autophagy for their survival even under conditions of no added stress.
Figure 4. Breast cancer cell lines have a differential sensitivity to pharmacological inhibition of autophagy.
Cells were treated with different concentrations of chloroquine (CQ, μM) and evaluated for death/survival by LDH release or MTS reagent after 48 h of treatment (A). LC3II accumulation was evaluated after a 4 h CQ treatment in all cell lines (B). MCF7 cells were included in both gels for comparison. Graphs show mean ± SE of three independent experiments performed in triplicate. * = different from its respective untreated control (p < 0.05).
Autophagy regulates STAT3 phosphorylation and cell survival
Autophagy has recently been linked to JAK/STAT pathway activation (29–31) and some breast cancer cell lines are known to have high levels of constitutively activated STAT3, particularly those belonging to the TNBC subgroup.(12, 32) Importantly, basal and claudin-low subtypes of breast cancer comprise most (70–80%) of TNBC (10) and the basal and claudin-low cell lines used in this study are known to be triple negative.(33)
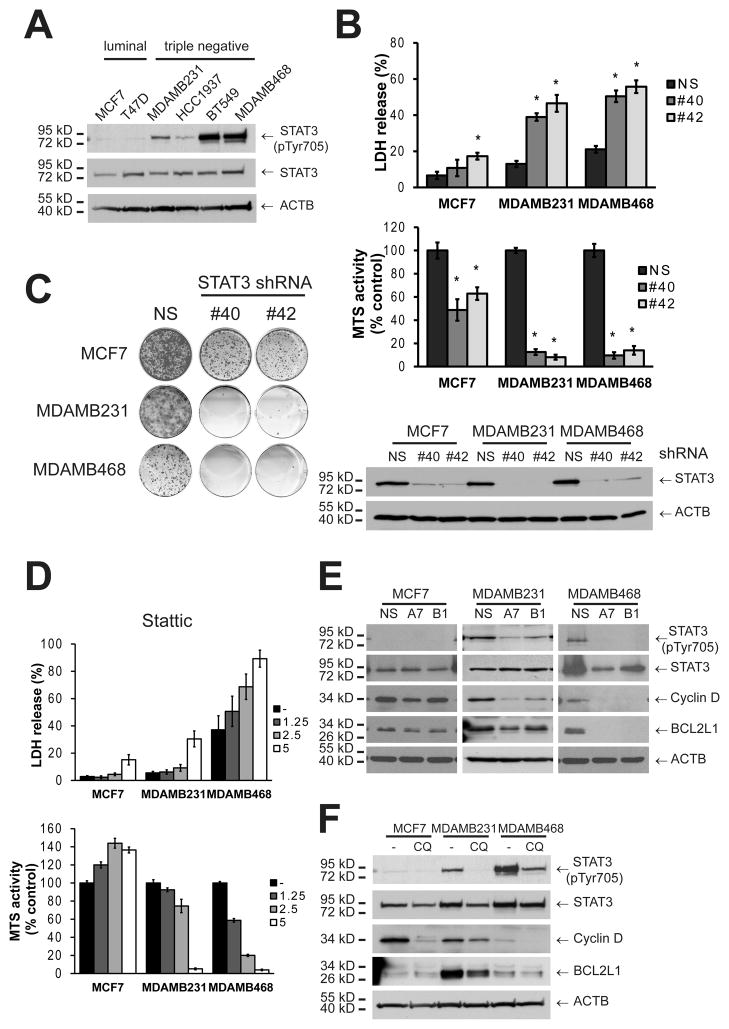
In agreement with previous reports,(12) TNBC cell lines (MDAMB231, HCC1937, BT549, and MDAMB468) showed substantially higher levels of activated STAT3 by Tyr phosphorylation (Figure 5A) than the luminal cell lines. These cells were determined to be STAT3-dependent, since genetic inhibition of STAT3 by gene knockdown showed a significant increase in LDH release as well as decreased MTS activity (Figure 5B) and clonogenic growth (Figure 5C) in both MDAMB231 and MDAMB468. Although MCF7 cells showed a decrease in MTS activity and clonogenic growth, STAT3 knockdown did not induce a substantial amount of LDH release, suggesting that proliferation was impaired but that MCF7 cells are not STAT3 dependent. Moreover, pharmacological inhibition of STAT3 with Stattic, a STAT3 small molecule-inhibitor,(34) killed breast cancer cell lines in a similar manner and with the same relative specificity to autophagy inhibition by chloroquine or ATG gene knockdown (Figure 5D and Supplementary Figure S4A).
Figure 5. TNBC cell lines have constitutively activated JAK/STAT pathway and autophagy regulates their STAT3 phosphorylation status.
A) A panel of breast cancer cell lines was tested for STAT3 Tyr-phosphorylation by Western Blot. B, C) MCF7, MDAMB231 and MDAMB468 cells were tested for STAT3 dependence by transduction with STAT3 (S3) shRNAs (#40 and #42) and evaluated for death/ survival with (B) an LDH release or an MTS assay, or (C) with a clonogenic growth assay. In B, cells were collected 48 h post-transduction and evaluated for STAT3 knockdown. In D, the same cell lines were tested for their sensitivity to pharmacological inhibition of STAT3 with Stattic treatment at the indicated concentrations (72h, μM). In E and F, autophagy was inhibited by transduction with a shRNA for ATG7 (A7) or BECN1 (B1) or treated with chloroquine (CQ), and STAT3 phosphorylation was evaluated by Western Blot either (E) 48h after plating, or (F) 24 h after CQ treatment. In F, CQ concentration was as follows: MCF7 30 μM, MDAMB231 25 μM and MDAMB468 10 μM; which were the highest concentrations that did not kill a significant amount of cells at this time point and if anything, underestimates the effect of CQ treatment on MDAMB231 and MDAMB468 cells. Graphs in B and D show mean ± SE of three independent experiments performed in triplicate. * = different from its respective non-silencing control (p < 0.01).
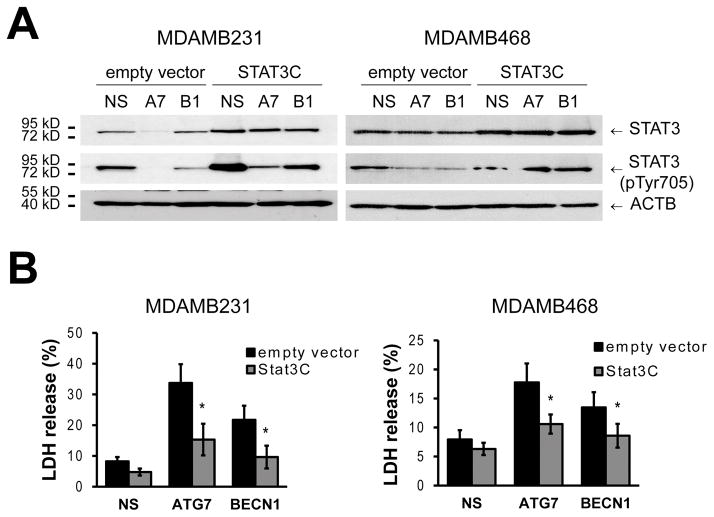
Tyrosine-phosphorylated STAT3 could not be detected in MCF7 cells in basal conditions (Figure 5A) or upon autophagy inhibition (Figure 5E). On the other hand, ATG gene knockdown decreased tyrosine-phosphorylated STAT3 in both the MDAMB231 and MDAMB468 cells and led to a reduction in the levels of transcriptional targets of STAT3 like Cyclin D and BCL2L1 (also known as BCL-XL, Figure 5E). Similar results were observed in the other triple-negative cell lines (HCC1937 and BT549, Supplementary Figure S4B). Pharmacological inhibition of autophagy by chloroquine treatment (Figure 5F) also decreased phosphorylation of STAT3 in MDAMB231 and MDAMB468 cell lines. Together, these data suggest that autophagy inhibition decreases STAT3 phosphorylation in TNBC breast cancer cell lines and that this reduction in STAT3 activity induces cell death. To test this, we overexpressed a constitutively active form of STAT3 (STAT3C), which partially restored phospho-Tyr STAT3 levels (Figure 6A) and decreased death induced by autophagy inhibition in both MDAMB231 and MDAMB468 cell lines (Figure 6B).
Figure 6. Constitutively active STAT3 decreases cell death induced by autophagy inhibition.
MDAMB231 and MDAMB468 cells were transduced with a lentivirus expressing a constitutively active form of STAT3 (STAT3C). A) As shown by Western analysis, STAT3C increased p-Tyr STAT3 levels after autophagy inhibition with ATG7 (A7) or BECN1 (B1) shRNAs. B) Cells were then evaluated for death with an LDH assay 3 days (MDAMB231) or 1 day (MDAMB468) after plating. Graphs show mean ± SE of three independent experiments performed in triplicate. * = different to its empty vector control, p < 0.05.
Chloroquine increases the median survival of MDAMB231 mouse xenografts
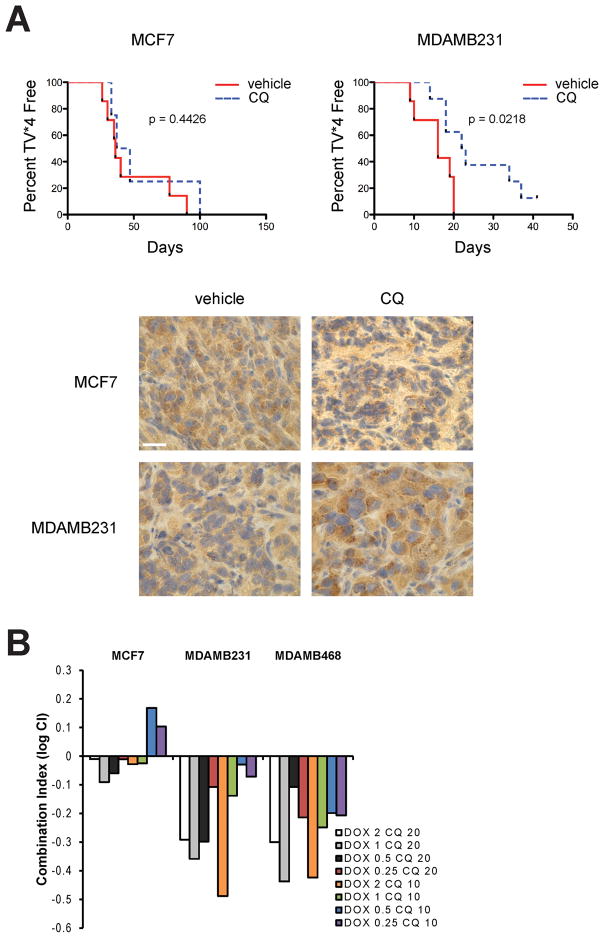
In order to evaluate the effect of autophagy inhibition in vivo, we performed mouse xenograft studies with the MCF7 and MDAMB231 cell lines. Tumor growth was followed until tumors reached 4 times initial volume (TV*4) as suggested by Teicher et al (Teicher 2002). Chloroquine treatment significantly delayed the median time to reach TV*4 of mice injected with MDAMB231 cells and not in those having MCF7 xenografts (Figure 7A). LC3 staining revealed more LC3 positive puncta in the chloroquine treated tumors when compared to their respective vehicle-treated controls, indicating inhibition of autophagy in both MCF7 and MDAMB231 chloroquine-treated tumors. These results suggest that autophagy inhibition has differential effects on breast cancer tumors in vivo and that TNBC tumors similar to the MDAMB231 xenografts are more likely to benefit from autophagy inhibition even in the absence of treatment with other agents.
Figure 7. Autophagy inhibition with chloroquine increases the median event free survival of MDAMB231 but not MCF7 breast tumor xenografts and synergizes with chemotherapy primarily in autophagy dependent cells.
A) Female nu/nu mice were injected with 5 × 106 MDAMB231 or MCF7 cells into the 4th mammary fat pad. Upon reaching a tumor volume of 118 mm3 in MCF7 and 126 mm3 in MDAMB231, mice were treated with chloroquine (60 mg/kg/day) or 0.9% saline, intraperitoneally. Data is presented as time to reach 4-times initial volume (TV*4). Log-rank (Mantel-Cox) analysis of the curves shows significant difference for the MDAMB231 tumors and no significant difference for the MCF7 tumors. Tumor sections were stained for LC3 and pictures show a representative image. Bar represents 200 μM. B) MCF7, MDAMB231 and MDAMB468 cell lines were treated with doxorubicin (DOX) at different doses (μM) in combination with CQ (μM). Combination index (CI) was determined where log CI < 0 is synergistic, CI = 0 additive and CI > 0 is antagonistic. Graph shows one representative experiment of two independent experiments.
Autophagy inhibition with chloroquine synergizes with chemotherapy primarily in autophagy dependent cells
Since MDAMB231 and MDAMB468 cells were the most affected by autophagy inhibition, we hypothesized that chloroquine treatment would synergize with chemotherapy in these cells. Figure 7B shows the combination index for treatment with doxorubicin and chloroquine at different concentrations. MDAMB231 and MDAMB468 cells showed the highest synergy (log CI < 0), while the combined treatment in MCF7 cells was only weakly synergistic in most concentrations tested and even antagonistic in others (log CI > 0).
Discussion
Autophagy is frequently increased in tumors, particularly in areas of low oxygen and blood supply, where it serves as a survival mechanism that helps the cells survive periods of metabolic stress.(35) Although autophagy could, in principle, serve as a general survival mechanism in all tumor cells, recent studies suggest that certain types of cancers are particularly sensitive to autophagy inhibition. In this regard, pancreatic and RAS transformed cancers seem to have high energetic requirements that are supported by autophagic mitigation of ROS,(6) and autophagic maintenance of mitochondrial (5) and glucose metabolism.(7) However, even with RAS-driven cancers, the role of autophagy is context dependent since a recent study suggests that autophagy inhibition prevents Kras driven tumor development when p53 is wild type but promotes tumorigenesis and progression when p53 is deleted.(36)
In this work, we studied autophagy dependence in a panel of breast cancer cell lines representing different intrinsic subtypes of the disease. Importantly, breast cancer has not previously been considered particularly addicted to autophagy. Only two previous studies have suggested that autophagy could be a therapeutic target in breast cancer: one using the MDAMB231 cell line, which is KRAS transformed,(7) and another one using the MMTV-PyMT mouse model of breast cancer, which induces Ras, Src and PI3K activation.(37) Importantly, RAS has not been found to be frequently activated in breast cancer.(38)
Here, we found that different subtypes of breast cancer vary markedly in their dependence on autophagy, with TNBC cell lines displaying particular sensitivity to autophagy inhibition by ATG gene knockdown or chloroquine treatment when compared to luminal and non-transformed cell lines (Figures 1–3). These cells were dependent on autophagy for survival even in complete media, i.e. in the absence of stresses, which are known to stimulate autophagy to maintain cellular homeostasis. Conversely in autophagy-independent MCF7 cells autophagy was needed for survival upon starvation but not in unstressed conditions. Autophagy dependence was also independent of RAS pathway status since the only TNBC cell line that we used that is known to be RAS mutated is the MDAMB231 cell line.(39) Instead, we found that STAT3 constitutive activity is regulated by autophagy only in some cells and that this is required and sufficient for autophagy-dependence for survival (Figures 5–6). This differential requirement for autophagy has effects on tumor treatment because mice with chloroquine-treated MDAMB231 xenografts had a better event free survival (when compared to their matched controls) than those with MCF7 tumors (Figure 7A) and autophagy inhibition with chloroquine treatment synergized with chemotherapy in cell lines that were autophagy dependent and not in the ones that were autophagy independent (Figure 7B).
The effect of autophagy inhibition during cancer treatment remains a controversial issue.(1, 4, 40) Although numerous studies have shown a significant decrease in cell number with autophagy inhibition in combination with radiation or chemotherapy in breast cancer cell lines (41, 42) and in tumor xenografts;(43–45) other studies show a limited or null effect.(46) While it makes sense that autophagy inhibition may be more beneficial when used with particular cancer therapies (e.g. those that themselves directly activate the autophagy pathway), our data suggests that in addition, only some cancer cells will really benefit from its suppression during treatment. Particularly, we propose that triple-negative breast cancers (TNBC) with high constitutive levels of STAT3 phosphorylation will be autophagy dependent and will therefore respond the most to autophagy inhibition alone or in combination with chemotherapy. Therefore, synergy with chemotherapy and autophagy inhibition will work best in such tumor cells. The mechanism by which autophagy promotes STAT3 phosphorylation remains to be determined.
Recent studies have suggested a correlation between the autophagic marker LC3B and patient outcome in breast cancer, where breast cancers with the highest LC3B expression were found to show increased markers for cell proliferation, aggressiveness and had the worst outcome. (47, 48) Regarding breast cancer subtypes, Lazova et al found that only luminal A tumors correlated between LC3B expression and poor outcome, while Chen et al found this correlation only in TNBC. While both studies suggest that increased LC3B total staining could be used as a marker of increased autophagy, this is not necessarily true since increased total LC3 or autophagosome number could also reflect a block in autophagic flux. (49) Our results suggest that regardless of the levels of autophagy in breast cancer cells, molecular markers like activated STAT3, could be used to determine which breast cancers would benefit the most from autophagy inhibition.
This work has potential therapeutic implications since TNBC has the worst prognosis among all breast cancers. There are currently no specific therapies for patients with TNBC and, although they can be treated with chemotherapy, they tend to relapse and metastasize early.(11) Our results suggest that this subtype of the disease may be most responsive to autophagy inhibition and that screening for high levels of constitutive STAT3 activation in tumors may serve as a selection strategy for treatment with autophagy inhibitors.
Supplementary Material
Acknowledgments
The authors appreciate the contribution to this research made by the following University of Colorado Cancer Center Research Cores: Protein Production/Mab/Tissue Culture, Flow Cytometry, Functional Genomics and Research Histology Core (E. Erin Smith, April Otero, Jenna Van Der Volgen).
Financial Support: Supported by NIH grant CA150925 and the following Shared Resources supported by P30 CA046934 Protein Production/Mab/Tissue Culture, Flow Cytometry, Functional Genomics, Histology. Supported also (in part) by a research grant from the Cancer League of Colorado, Inc. (PM); Elope, Inc. St. Baldrick’s Foundation Scholar Award (CMG and JML) and NIH/NICHD Child Health Research Career Development Award (K12 HD068372) (JML).
Footnotes
The authors have no conflicts of interest to disclose.
References
- 1.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012 doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debnath J. The multifaceted roles of autophagy in tumors-implications for breast cancer. J Mammary Gland Biol Neoplasia. 2011;16:173–87. doi: 10.1007/s10911-011-9223-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 4.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–30. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–78. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeSantis C, Siegel R, Bandi P, Jemal A. Breast cancer statistics, 2011. CA Cancer J Clin. 2011;61:409–18. doi: 10.3322/caac.20134. [DOI] [PubMed] [Google Scholar]
- 9.Prat A, Parker JS, Fan C, Cheang MC, Miller LD, Bergh J, et al. Concordance among gene expression-based predictors for ER-positive breast cancer treated with adjuvant tamoxifen. Ann Oncol. 2012 doi: 10.1093/annonc/mds080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7:683–92. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- 12.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(−) stem cell-like breast cancer cells in human tumors. J Clin Invest. 2011;121:2723–35. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 14.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–88. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 15.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 16.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hillion J, Dhara S, Sumter TF, Mukherjee M, Di Cello F, Belton A, et al. The high-mobility group A1a/signal transducer and activator of transcription-3 axis: an achilles heel for hematopoietic malignancies? Cancer Res. 2008;68:10121–7. doi: 10.1158/0008-5472.CAN-08-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–72. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 19.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porter CC, Kim J, Fosmire S, Gearheart CM, van Linden A, Baturin D, et al. Integrated genomic analyses identify WEE1 as a critical mediator of cell fate and a novel therapeutic target in acute myeloid leukemia. Leukemia. 2012;26:1266–76. doi: 10.1038/leu.2011.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J, Tan AC. BiNGS!SL-seq: a bioinformatics pipeline for the analysis and interpretation of deep sequencing genome-wide synthetic lethal screen. Methods Mol Biol. 2012;802:389–98. doi: 10.1007/978-1-61779-400-1_26. [DOI] [PubMed] [Google Scholar]
- 22.Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolde R. pheatmap: Pretty Heatmaps. R package version 0.7.4 ed2012.
- 24.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 25.Teicher B. In Vivo Tumor Response End Points. In: Teicher B, editor. Tumor Models in Cancer Research. Humana Press; 2002. pp. 593–616. [Google Scholar]
- 26.Lee IH, Kawai Y, Fergusson MM, Rovira, Bishop AJ, Motoyama N, et al. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science. 2012;336:225–8. doi: 10.1126/science.1218395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147:223–34. doi: 10.1016/j.cell.2011.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang YP, Tsai CC, Huang WC, Wang CY, Chen CL, Lin YS, et al. Autophagy facilitates IFN-gamma-induced Jak2-STAT1 activation and cellular inflammation. J Biol Chem. 2010;285:28715–22. doi: 10.1074/jbc.M110.133355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noman MZ, Janji B, Kaminska B, Van Moer K, Pierson S, Przanowski P, et al. Blocking hypoxia-induced autophagy in tumors restores cytotoxic T-cell activity and promotes regression. Cancer Res. 2011;71:5976–86. doi: 10.1158/0008-5472.CAN-11-1094. [DOI] [PubMed] [Google Scholar]
- 31.Kang R, Loux T, Tang D, Schapiro NE, Vernon P, Livesey KM, et al. The expression of the receptor for advanced glycation endproducts (RAGE) is permissive for early pancreatic neoplasia. Proc Natl Acad Sci U S A. 2012;109:7031–6. doi: 10.1073/pnas.1113865109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia R, Bowman TL, Niu G, Yu H, Minton S, Muro-Cacho CA, et al. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001;20:2499–513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- 33.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006;13:1235–42. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 35.Maes H, Rubio N, Garg AD, Agostinis P. Autophagy: shaping the tumor microenvironment and therapeutic response. Trends Mol Med. 2013 doi: 10.1016/j.molmed.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, Mackay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013 doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 37.Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 2011;25:1510–27. doi: 10.1101/gad.2051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maycotte P, Thorburn A. Autophagy and cancer therapy. Cancer Biol Ther. 2011;11:127–37. doi: 10.4161/cbt.11.2.14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bristol ML, Di X, Beckman MJ, Wilson EN, Henderson SC, Maiti A, et al. Dual functions of autophagy in the response of breast tumor cells to radiation: cytoprotective autophagy with radiation alone and cytotoxic autophagy in radiosensitization by vitamin D 3. Autophagy. 2012;8:739–53. doi: 10.4161/auto.19313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milani M, Rzymski T, Mellor HR, Pike L, Bottini A, Generali D, et al. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 2009;69:4415–23. doi: 10.1158/0008-5472.CAN-08-2839. [DOI] [PubMed] [Google Scholar]
- 43.Rao R, Balusu R, Fiskus W, Mudunuru U, Venkannagari S, Chauhan L, et al. Combination of pan-histone deacetylase inhibitor and autophagy inhibitor exerts superior efficacy against triple-negative human breast cancer cells. Mol Cancer Ther. 2012;11:973–83. doi: 10.1158/1535-7163.MCT-11-0979. [DOI] [PubMed] [Google Scholar]
- 44.Cufi S, Vazquez-Martin A, Oliveras-Ferraros C, Corominas-Faja B, Cuyas E, Lopez-Bonet E, et al. The anti-malarial chloroquine overcomes Primary resistance and restores sensitivity to Trastuzumab in HER2-positive breast cancer. Sci Rep. 2013;3:2469. doi: 10.1038/srep02469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee KH, Hsu EC, Guh JH, Yang HC, Wang D, Kulp SK, et al. Targeting energy metabolic and oncogenic signaling pathways in triple-negative breast cancer by a novel adenosine monophosphate-activated protein kinase (AMPK) activator. J Biol Chem. 2011;286:39247–58. doi: 10.1074/jbc.M111.264598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bristol ML, Emery SM, Maycotte P, Thorburn A, Chakradeo S, Gewirtz DA. Autophagy inhibition for chemosensitization and radiosensitization in cancer: do the preclinical data support this therapeutic strategy? J Pharmacol Exp Ther. 2013;344:544–52. doi: 10.1124/jpet.112.199802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lazova R, Camp RL, Klump V, Siddiqui SF, Amaravadi RK, Pawelek JM. Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin Cancer Res. 2012;18:370–9. doi: 10.1158/1078-0432.CCR-11-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen S, Jiang YZ, Huang L, Zhou RJ, Yu KD, Liu Y, et al. The residual tumor autophagy marker LC3B serves as a prognostic marker in local advanced breast cancer after neoadjuvant chemotherapy. Clin Cancer Res. 2013;19:6853–62. doi: 10.1158/1078-0432.CCR-13-1617. [DOI] [PubMed] [Google Scholar]
- 49.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.