Abstract
X-ray free-electron laser (XFEL) sources enable the use of crystallography to solve three-dimensional macromolecular structures under native conditions and free from radiation damage. Results to date, however, have been limited by the challenge of deriving accurate Bragg intensities from a heterogeneous population of microcrystals, while at the same time modeling the X-ray spectrum and detector geometry. Here we present a computational approach designed to extract statistically significant high-resolution signals from fewer diffraction measurements.
The ~40 femtosecond-duration XFEL pulse can deliver diffraction information on time scales that outrun radiation damage, allowing macromolecular reaction dynamics to be studied under functional physiological conditions1–3, while the small beam focus size permits the investigation of extremely small and weakly diffracting microcrystals4–6. Unlike single-crystal X-ray diffraction (XRD) experiments performed at conventional synchrotron radiation (SR) sources, XFEL studies destroy the sample with a single pulse, requiring the full data set to be assembled from a series of still diffraction shots of individual microcrystals, a technique known as serial femtosecond crystallography (SFX).
As with conventional crystallography, the objective of SFX is to obtain a complete set of structure factor amplitudes through the measurement of Bragg spot intensities (coherent scattering of X-rays described by Bragg’s law) to as high a diffraction angle as possible. The high-resolution signal is ultimately limited by noise, and the background (e.g. from solvent) often dominates the diffraction pattern for all but the most intense low-resolution (low angle) Bragg spots7. At SR sources, accurate sampling of the diffraction at the limit of detectability is accomplished by optimally modeling the diffraction experiment, including the relationship between real space (the crystal) and reciprocal space (the diffracted X-ray collected on the detector). The most intense Bragg spots are used to deduce the best-fitting lattice model (indexing), which is then used to predict exactly which pixels on each image to examine for Bragg spot integration, even though a signal may not be visually discernable from background. The same fundamental approach is applicable to the analysis of XFEL data. Here, we describe such a data processing approach for XFEL data, which enables weak signals to be measured from many fewer crystal specimens than possible with the previously available program CrystFEL8. The method has been added to our open source software suite, cctbx.xfel9. A primer and tutorial may be found at http://cci.lbl.gov/xfel, with code archived at http://cctbx.sf.net.
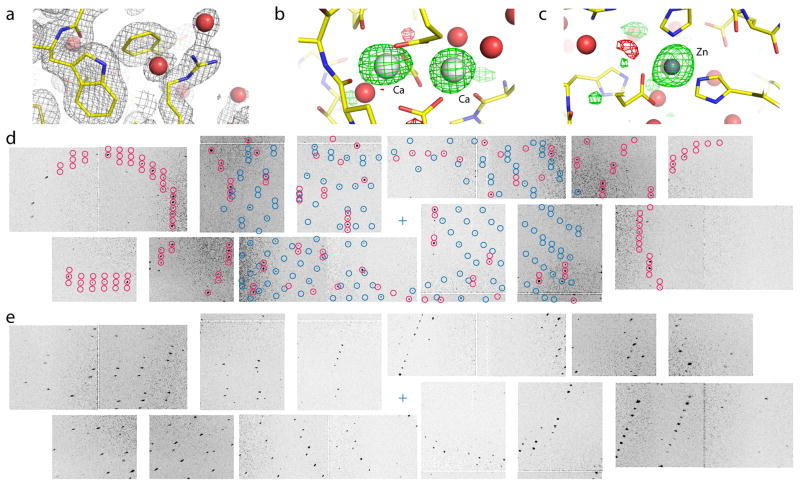
We tested our method for processing SFX diffraction patterns against data collected at the Coherent X-ray Imaging (CXI) instrument of the Linac Coherent Light Source (LCLS), using the Cornell-SLAC pixel array detector (CSPAD). We derived a structural model for the metalloprotein thermolysin (Fig. 1a, Supplementary Tables 1 and 2) that was comparable in quality to structures determined by conventional SR XRD at a similar resolution of 2.1 Å. The electron density of the native calcium and zinc ions (omitted from the phasing model) in the difference map (Figs. 1b, c) indicates that the metal positions are determined by the processed data and are not the result of bias from the phasing model. We also reprocessed 1.9 Å-resolution lysozyme data10 (Supplementary Tables 1 and 3) previously processed with the software suite CrystFEL8 to compare the two programs.
Figure 1. Thermolysin structure determination at 2.1 Å resolution.
(a) 2mFo−DFc electron density contoured at 1 σ (gray mesh) with water molecules shown as red spheres. (b) mFo−DFc difference density map contoured at +3 σ (green mesh) and −3 σ (red mesh) showing binding sites for two of the four Ca ions and (c) the single Zn ion. (d) Detail of two crystal lattices found on the same diffraction image. Modeled spot positions assigned to the different lattices are shown in red and blue, respectively. The sample-detector distance of 135 mm corresponds to a resolution of 2.15 Å at the edges. (e) Detail from a different diffraction image. Increasing radial spot elongation is observed with distance from the beam center (blue cross).
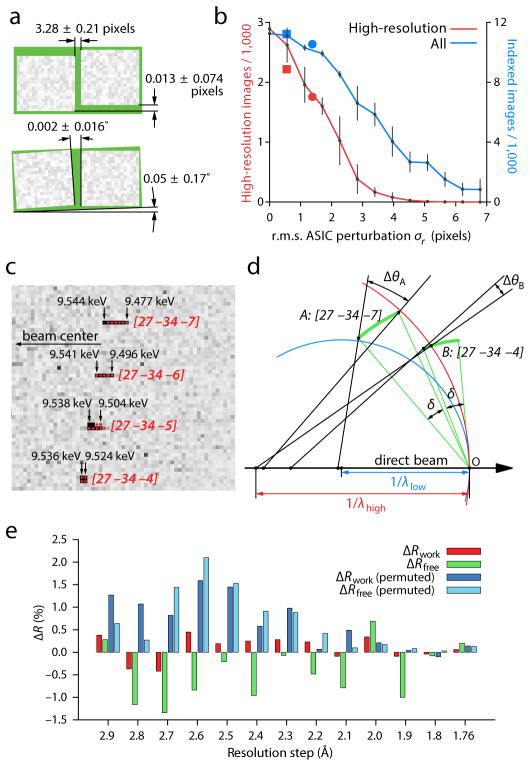
We found that cctbx.xfel was able to process about twice as many diffraction lattices from individual crystals as previously reported for CrystFEL10 (Supplementary Table 1). The indexing algorithm11, which identifies unit cell dimensions and crystal orientations, searches for directional vectors that describe the observed rows of Bragg spots, from which three are chosen to form the unit cell. Several factors make this a difficult problem. Firstly, the CSPAD detector consists of 64 pixel array readouts (Figs. 1d, e) that are periodically disassembled. Thus the metrology (the relative positions and orientations) of the readouts must be redetermined with sufficient accuracy (Fig. 2a), as even small subpixel offsets can diminish the number of images from which lattices can be indexed (Fig. 2b). Secondly, the destruction of each crystal after one XFEL shot removes the ability to view the diffracted lattice from various directions, hindering the selection of unit cell basis vectors. To compensate, we supply additional information to the indexing algorithm in the form of a target unit cell, from either an isomorphous crystal form or a preliminary round of indexing. This target unit cell permits us to choose a group of three vectors that best fits the known cell’s lengths and angles, thus increasing the number of successfully indexed images. A final factor is the high density of crystals delivered to the X-ray beam, which often produces diffraction patterns containing more than one lattice (Fig. 1d). While software exists for modeling multiple lattices in SR diffraction12, 13, previous XFEL approaches14 effectively filter these data away, by requiring that 80% of observed spots be covered by a single model. However, we find it straightforward to treat XFEL data with two lattices. The full set of bright candidate Bragg spots is used to derive the first lattice. Candidate spots falling on this lattice are then removed, and the remaining subset is used to find the second lattice, as previously described for SR data12. Spot overlaps among multiple lattices were rare, so the minimal inaccuracies in the integrated signal due to overlap were ignored.
Figure 2. Calibration and validation.
(a) Aggregate relative positions (top) and rotations (bottom) of 32 pairs of application-specific integrated circuits (ASICs), each pair bump-bonded to a pixel array sensor of the CSPAD detector. The two ASICs on each sensor are manufactured to be aligned along the long axis, separated by a 3.0-pixel gap. These calibration results bear out this expectation within the tolerances shown. (b) Impact of positional accuracy on the indexing and integration success rate. Perturbing the ASICs away from their true positions reduces both the total number of indexed images (blue) and the number of images that contain successfully integrated reflections at high (1.8–2.2 Å) resolution (red). Error bars are the standard deviation from five different sets of perturbations drawn from a twodimensional normal distribution with a standard deviation σr. Separate perturbations were drawn for each ASIC. Squares: failure to apply final subpixel corrections from iterative least squares refinement. Circles: failure to apply nearest-whole pixel corrections. (c) Detail of four Bragg reflections on a thermolysin diffraction pattern, showing pronounced (seven pixel) radial elongation for the [27 –34 –7] reflection and lesser elongation for those nearby. Solution of Bragg’s law for each pixel (black arrows) identifies the spread of photon energies that contribute to each reflection. Red disks delineate integration masks from a three-parameter model with wavelength limits λhigh = 1.297 (9.556 keV) and λlow = 1.313 (9.443 keV), and full-width mosaic spread δ = 0.174°. (d) Reciprocal space diagram indicating how different-shaped reflections arise. Reciprocal lattice points (arcs) all have a constant angular extent δ due to their mosaic spread. Points are in reflecting condition if they are within the zone between the high-energy (red) and low-energy (blue) Ewald spheres. Therefore, a greater fraction of the [27 –34 –7] mosaic distribution is within the reflecting condition, leading to a reflection that subtends a greater radial angle Δθ. (e) Paired refinements of the thermolysin structure. Red and green bars indicate the change in Rwork and Rfree, respectively, as higher-resolution data are added to the refinement. Dark and light blue bars show changes to the R-factors when the newly added high-resolution structure factors are randomly permuted. The data are interpreted as containing statistically significant signal for the resolution shells where ΔRfree is continuously negative, i.e. out to 2.1 Å.
The outcome of data integration depends critically on the ability to exactly target the pixels that actually contain signal. A too-inclusive model will capture adjacent pixels that contain only background noise, thus diluting the statistical significance of the measurement. Conversely, overly discriminating models fail to include all of the signal. A crucial first step for data processing, therefore, is to tailor the model to the data at hand. An explanation of why there is a need for new data-modeling algorithms, beyond what is implemented by CrystFEL, is presented in the Supplementary Note. In short, microscopic “mosaic” domains in the crystal produce Bragg spots shaped like concentric arcs, while the spread of energies in the self-amplified spontaneous emission (SASE) pulse streaks spots radially.
For cctbx.xfel we tested two approaches to model the Bragg spots. Although spots vary in size and shape across the lattice (Fig. 1e), they tend to be locally similar. This suggests that an empirical approach can be used whereby integration masks are chosen based on the shapes of nearby bright spots. We chose this method—which captures spot shapes of all extremes, both concentric arcs and radial streaks—as the default treatment for data analysis (Supplementary Tables 1 – 3). A deeper inspection of the data (Fig. 2c) revealed cases where Bragg reflections adjacent to each other nonetheless have very distinct radial widths. These differing widths are explained by the fact that for the full spread of SASE energies to be recorded in the diffraction pattern, Bragg’s law demands that the crystal contains microscopic (mosaic) domains with a distribution of either orientations or unit cell dimensions. Wide spots are produced for reflections that satisfy Bragg’s law for the full distribution of mosaic domains (given the crystal orientation and range of incident energies), while narrow spots are seen for those reflections that only satisfy the reflecting condition for a subset of domains (Fig. 2d). Modeling three parameters (high and low bandpass limits, mosaicity) predicts approximately which pixels to target for signal integration (red dots, Fig. 2d). The key benefit of this second, parametric approach is that it roughly accounts for the size and shape differences of adjacent Bragg spots, thus helping the integration mask conform to the actual signal. While the three-parameter model does not give an exact match to the spot shape (Fig. 2d), refinement of additional parameters could improve the approach.
We next tested how best to determine the resolution limits of the data set. An important consequence of shot-to- shot variability is that each lattice diffracts to a different limiting angle. Before merging the data into a single set of structure factors, we constructed Wilson plots (integrated Bragg spot intensity vs. diffraction angle bin) in order to determine a separate cutoff angle for each lattice. Once the data had been merged we employed an iterative paired-refinement technique15 to determine the overall highest resolution shell with a measurable information content (Fig. 2e). Remarkably, we found that at the highest resolution proven to contain statistically significant signal (2.1 Å), only 1700 lattices contributed to the thermolysin diffraction data, with an average multiplicity of observation of only 4.5 per structure factor (Supplementary Table 2). The size of this selected subset is much smaller than for previous high-resolution XFEL crystallography experiments; past experiments using CrystFEL have required >104 crystals to obtain reliable structure factors6, 10, 16. In cases where only 102–103 diffracting crystals were available, data merging has only been partially successful5, 17. Thus our results with cctbx.xfel are encouraging as XFEL progress has been limited by both the difficulty of preparing enough crystal specimens, and the limited data acquisition time at the light source.
In summary, our new developments implemented in cctbx.xfel include optimal indexing and retention of data from multiple lattices, separate determination of the resolution cutoff for individual lattices, better descriptions of the Bragg spot shape, and accurate detector geometry to permit well-conforming spot shape models. By carefully discriminating between image pixels known to contain diffraction signal, and the surrounding pixels containing only background noise, we were able to derive accurate structure factors with substantially fewer crystal specimen exposures.
We plan future software developments to further improve the final merged set of structure factors. As illustrated in Fig. 2d, a present limitation is that XFEL Bragg diffraction gives only a partial measurement of the structure factor, as the crystal is not fully rotated through the reflecting condition. We intend to implement postrefinement models18, 19 to allow the correction of intensity measurements to their full-spot equivalent. Such a correction requires a detailed knowledge of the incident spectrum. In Fig. 2e the range of X-rays is presented as a top-hat function, but in fact the SASE spectrum is stochastic and finely textured20. While the X-ray spectra were not available for the data shown here, single shot measurement of the spectrum is possible20 and will be incorporated into our method in the future. Taken all together, our method will make it easier for XFEL-based experiments to measure small structure factor differences, such as those from anomalous scattering that will enable the de novo determination of macromolecular structures. While SFX is presently a challenging technique, its potential payoff in terms of enabling specialized structural and dynamical studies of macromolecules is enormous.
Online methods
Sample preparation
Lyophilized thermolysin from Bacillus stearothermophilus (Hampton Research) was resuspended in 0.05 M NaOH at a concentration of 25 mg/ml. 300 μl of the protein stock was mixed in a 1:1 ratio with 40% PEG 2000, 100 mM MES pH 6.5, 5 mM CaCl2. Crystallization occurred within minutes. The obtained crystals were transferred into 10% PEG 2000, 100 mM MES pH 6.5, 5 mM CaCl2 (buffer A) and then stepwise into buffer A containing 10, 15, 20 and 30% (w/v) glycerol, respectively. Thermolysin concentration was determined spectrophotometrically using an absorbance value A=1.83 (1 mg/ml) at 277 nm21, and a molecular mass of 34.6 kDa22. The final protein concentration of the crystal suspension was found to be 20–24 mg/ml. The average size of the obtained crystals was 2 × 3 × 1 μm3. As judged by microscope images of various batches the size distribution is very narrow. Assuming an average crystal volume of 6 μm3, 12 monomers per unit cell and a nominal unit cell volume of 1 × 106 Å3, 6 × 105 unit cells/crystal gives a concentration of about 3.4 × 1010 crystals/ml.
Thermolysin data collection
Diffraction experiments were carried out at the CXI instrument at LCLS23. We previously reported the use of a nanoflow liquid injector that markedly reduces the requirements on sample amount24, 25. The suspension of thermolysin crystals was injected into the interaction region by this electrospun liquid jet, using a 1 m long silica capillary of 50 μm inner diameter, 150 μm outer diameter, outer diameter tapered at both ends (New Objective) with one end in a pressurized cell outside the vacuum chamber of the CXI instrument, dipping into a vial with 100 μl of the crystal suspension. A potential of +2500V (relative to a counter electrode below the interaction region) was applied to the suspension by means of a bare Pt electrode inside the sample vial. The flow rate was on the order of 0.5 μl/min by applying a backing pressure of 124.1 kPa to the suspension.
The CXI instrument was operated at energies of 9.56 and 9.77 keV (Supplementary Table 1), the beam intensity was 6×1011 photons/pulse, with a mean pulse duration of 47 fs and a frequency of 120 Hz. The beam was focused to a size of 2.25 μm2 FWHM at the interaction point. Diffraction was measured utilizing the front CSPAD detector26 of the CXI instrument. The detector has a pixel size of 110 × 110 μm2 and a total of 1516 × 1516 pixels.
Resolution of this particular experiment was limited by geometric factors and not the intrinsic strength of the diffraction signal. Several combinations of sample-to-detector distance and incident wavelength were utilized for data collection, but with the most aggressive choice (detector distance = 135 mm, λ = 1.30 Å), geometric limits were 2.15 Å at the detector edge and 1.75 Å in the corner, thus accounting for the falloff in data completeness at high resolution in Supplementary Table 2.
Raw data streams have been deposited into the Coherent X-ray Imaging Data Bank27 (CXIDB; http://cxidb.org) under accession ID 23, along with an exact list of the images that were merged (Supplementary Tables 1 and 2) to form the structure factor intensities. A tutorial on accessing information from the raw data files is presented at http://cci.lbl.gov/xfel.
Lysozyme data
To afford a fair comparison between CrystFEL and cctbx.xfel our only tractable option was to reprocess raw data that had been previously analyzed by the CrystFEL software developers. We obtained data from the CXIDB, which archives the raw data streams from the Boutet et al. 1.9 Å-resolution structure determination of lysozyme10 under accession ID 17. To select data for the comparison, we chose only those run numbers (305–327) that yielded the 12,247 images used in the Boutet paper, as documented in a list maintained at the CXIDB Web site (Supplementary Table 1). For this particular experiment the CXI instrument was operated at 9.39 keV and the pulse duration was 40 fs. With a detector distance of 93 mm, the geometric limits were 1.74 Å at the detector edge and 1.46 Å in the corner, both well beyond the 1.9 Å resolution limit that we imposed in order to perform a direct comparison with the published results.
Data processing
Data were processed with our package cctbx.xfel9. After subtraction of a dark-run average image, bright candidate Bragg spots were chosen with the Spotfinder component of cctbx28, with settings being adjusted by trial and error specifically for these data; e.g., the minimum spot area was set at 2 square pixels, and the criteria for accepting spots was set to allow spot picking to an outer resolution limit of about 2.5 Å for thermolysin and 1.9 Å for lysozyme. Images were indexed (unit cell dimensions and crystal orientations determined) with the Rossmann DPS algorithm29, 30 as implemented in our program LABELIT11. Unit cells dimensions modeled by the indexing algorithm varied from crystal to crystal, with population means and standard deviations for thermolysin reported in Supplementary Table 1. A small number of thermolysin lattices (233, ~2%) did not conform to hexagonal Bravais symmetry using our standard criteria31; these were removed from further processing and are not included in the reported population. Similarly, 321 non-tetragonal lysozyme lattices were removed (~1%). For previous data analyses with photosystem II3, 32 we also removed lattices whose unit cell lengths were highly non-isomorphous (differing by >10%) compared to the mean, in order to avoid merging data from non-identical crystal structures33, 34. However, for the thermolysin and lysozyme data, none of the unit cell lengths were rejected as outliers.
Improving indexing by using a target unit cell
As stated in the main text, the destruction of each crystal after one XFEL shot makes indexing difficult. Accuracy is much greater at SR sources, where it is possible to mount the crystal on a goniometer and view the diffracted lattice from two different crystal orientations approximately 90° apart11. In contrast, the liquid jet method delivers samples in random, unknown, orientations. Furthermore, the XFEL diffraction images examined here varied extensively in quality (resolution and number of Bragg spots), with a less successful indexing outcome from poorer images. With degraded data, the DPS algorithm can fail by choosing three candidate unit cell axes that individually appear to describe periodicity in the diffraction pattern, but when combined do not adequately cover the lattice. To avoid this failure mode, we supplied additional information to the indexing algorithm in the form of a target unit cell taken from isomorphous crystal forms (PDB codes 2TLI for thermolysin and 4ET8 for lysozyme). Groups of three candidate axes from the DPS algorithm are evaluated to find the best fit to the known cell lengths and angles. By requiring this approximate similarity, we increased the number of successfully indexed images from about 8000 to about 11600 for thermolysin. A similar approach was used previously by others to identify the lattice within noisy data35, 36. We expect that this method will be generally applicable to XFEL data and not limited to cases where an isomorphous crystal form is known. Data can be treated in two passes, first to determine a consensus unit cell from the highest-quality diffraction images where indexing is readily achieved, and secondly to use this consensus cell as a target for indexing the entire data set. In support of this idea, we note that the population standard deviation of the thermolysin unit cell lengths (Supplementary Table 1) is quite narrow (0.3–0.4%), and even for previous low-resolution PS II data3 the standard deviations (0.9–1.9%) were reasonably low.
Relationship between indexing and hit rates
In a previous paper we described the use of cctbx.xfel to provide detailed feedback on the diffraction quality within minutes of data acquisition9. For this initial analysis, the Spotfinder component of cctbx28 is used to classify a diffraction pattern as a “hit” if it contains 16 or more candidate Bragg spots with dark-subtracted peak heights above 450 analog-digital units (on the CSPAD high-gain setting) out to a resolution limit of 4.0 Å. This peak height criterion is chosen by trial and error to best identify Bragg spots for the thermolysin dataset, and the level can easily be changed in a configuration file for other datasets. Supplementary Figure 1 shows the final outcome: 77% of the initial low-resolution “hits” are successfully integrated and merged into structure factors; with a slightly lower success rate (65%) for hits containing the lowest number of candidate spots. Reasons for the residual failure rate are still to be determined, and will likely vary from case to case in future experiments.
Empirical approach to modeling the spot shape
Bragg spots from both datasets (thermolysin is illustrated in Fig. 1e) were observed to vary in size and shape both within a single lattice and also from image to image. Therefore, the previously published CrystFEL model that treats spots as uniformly round and equally-sized in reciprocal space14 was judged to be a poor fit to this data. As described in the Supplementary Note, the underlying phenomenon treated by that model (large λ/a ratio, where λ is the wavelength of the incident light, and a is the crystal width) does not apply for high-resolution experiments. In fact, it is not possible to identify a single criterion to describe the spot shape throughout the data sets; some images exhibit concentric arcs consistent with mosaic spread37 (not shown), while other images contain elongation that is chiefly radial (Fig. 1e). We do note however that whatever the behavior, spots tend to be locally similar in size and shape within each lattice (with one exception, see below). This suggests an empirical approach to determining the spot model. First, easily identified high-intensity Bragg spots (using the program Spotfinder28) are used to index the lattice. Next, at each predicted lattice position on the image, a mask is constructed consisting of a union of the ten nearest spot shapes from the Spotfinder set, similar to the approach taken by some SR data reduction programs38. This mask determines the set of pixels to be used for signal summation (integration). Taking a union of all nearby spot masks helps to increase the number of pixels assigned to each Bragg spot, to avoid missing pixels that actually contain signal. This is necessary because the predicted spot positions are slightly inaccurate due to the use of a monochromatic model; in fact the incident light has a 0.5–1.0% bandpass39 (as described in the paragraph immediately below). This simple empirical approach was used to derive all the structure factor measurements in Supplementary Tables 1–3.
Parametric approach to modeling the spot shape
Given the theoretical framework of Bragg’s law, it is possible to interpret the shape and size of Bragg spots in terms of more fundamental experimental properties including the spectral dispersion, the crystal size, and the internal crystal disorder40–47. Thus, while the above empirical approach is adequate for the present, a deeper understanding of XFEL Bragg spot shapes may be possible. In images of both thermolysin (Figs. 1e, 2c) and lysozyme we observe radial spot elongation that is most pronounced at higher diffraction angles. This is consistent with the protein crystals acting as spectral analyzers, such that each Bragg reflection disperses the broad bandpass SASE pulse (typically 0.2–0.5% bandpass)39 over a radial line up to several pixels wide. Furthermore, we observe that reflections adjacent to each other (Fig. 2c) can nonetheless have very distinct radial widths. The explanation is rooted in the fact that for a spread of energies to be recorded in the diffraction pattern, Bragg’s law demands that the crystal contain microscopic (mosaic) domains with a distribution of either orientations or unit cell dimensions. Fig. 2d represents each Bragg spot as a spherical cap in reciprocal space (shown as an arc) representing a spread of orientations, as has been done previously48. In our experiment, wide spots are produced for reflections that satisfy Bragg’s law for the full distribution of mosaic domains in the crystal (given the crystal orientation and range of incident energies), while narrow spots are seen for those reflections that only satisfy the reflecting condition for a subset of microscopic domains (Fig. 2d). By modeling three parameters (high and low bandpass limits, plus mosaicity) we were able to predict approximately which pixels to target for signal integration for each Bragg reflection (red dots, Fig. 2d). The key benefit of this approach is that it roughly accounts for the size and shape differences of adjacent Bragg spots, reducing the inclusion of non-signal pixels in the integration mask, and thus helping to extract weak signals. While the three-parameter model in Fig. 2d does not give an exact match to the spot shape, we believe that further development will improve the approach. Important additional parameters that could be refined include the spectral shape and unit cell variation, while others such as crystal size and beam divergence are probably negligible for experiments performed at the CXI 1 μm focus.
Signal integration and error estimation
Signal intensity I for each Bragg spot was integrated over a set of pixels determined by empirical mask construction as described above. A surrounding set of pixels, twice the size of the signal set, and separated from it by a guard zone two pixels wide, was designated for measuring the local background. This background set was used to fit a least-squares plane for background subtraction as described49. The estimated variance σ2(I) of the signal measurement was based on counting statistics49, using a rough estimate for the CSPAD high-gain value of 7.5 analog-to-digital units per photon. Integrated intensities were then corrected for polarization50. It was realized that the data set contained numerous intensity measurements at large negative multiples of σ (I), from which we concluded that Poisson statistics did not adequately model the experimental error. Error estimates from each diffraction pattern were therefore inflated by assuming that negative values of I/σ (I) are actually decoy measurements (noise only) with a Gaussian distribution centered at zero and with a standard deviation of 1, thus providing a lower bound on modeling errors. This inflation factor is determined separately for each image, and acts to increase the initially determined errors from counting statistics. Negative I values were then removed from the data set, and data on each image were scaled to a reference data set derived from an isomorphous structure (section immediately below). When later merging multiple measurements of the same Miller index, the error was modeled simply by propagating the per-measurement σ (I) values in quadrature. Since the systematic error contributions for XFEL data are not fully understood, no other systematic correction or error normalization was attempted. The error model derived here is believed to be entirely different than that used in CrystFEL, therefore the respective I/σ (I) values for the two programs in Supplementary Tables 1–3 cannot be compared.
Scaling
Integrated intensities from separate images were scaled to intensities derived from an isomorphous reference structure (PDB codes 2TLI for thermolysin and 4ET8 for lysozyme); this scaling step helped to account for specimen-to-specimen variation in crystal size and pulse power. For projects where no isomorphous reference structure is available, we propose an iterative procedure wherein the data are merged once without scaling to gain an approximate set of merged intensities, which are then used as the reference for rejecting poorly correlated images in the next round.
Different resolution cutoffs for each lattice
An important consequence of shot-to-shot variability is that each lattice diffracts to a different limiting angle; this can be illustrated even within a single image (Fig. 1d) where one lattice (red) extends to higher resolution than a second one (blue). For data reduction, we choose a separate limit for integrating each lattice. Integration relies on having an accurate crystal orientation model, which in turn depends on the set of bright candidate Bragg spots found in our case by the program Spotfinder28. For example, if Spotfinder spots extend only to 4 Å on a particular image, the orientational model is not accurate enough to predict the positions of weak spots at 2.5 Å resolution. We have verified this general result through studies on simulated data (results not shown). A very conservative approach is therefore used for integration: for each image separately the radius of integration is extended slightly past the Spotfinder limit, and a Wilson plot is constructed (integrated Bragg spot intensity vs. diffraction angle bin), to identify a resolution limit at which average intensity falls below average noise (based on counting statistics). The radius is increased until such a crossover point is found, at which point it is concluded that either there is no more signal to be found, or the model has diverged from the data. When merging multiple measurements together, it would be counterproductive to include high-resolution integrated measurements from beyond this limit where there is no signal, as this would degrade the overall statistical significance. Allowing separate resolution cutoffs for each image leads to a final merged data set with high multiplicity of observation at low resolution and lower multiplicity at high resolution (Supplementary Table 2), yet there is confidence that the highest resolution shell contains real signal.
The quality of the reflections merged in this fashion was assessed by calculating the correlation coefficient of semi-datasets merged from odd- and even-numbered images (CC1/2)15. We note that our multiplicity statistics (Supplementary Tables 2 and 3) differ from previously published high-resolution XFEL analyses6 that report uniform multiplicity counts over all resolution bins, which is the result of applying a single global resolution limit.
Validation of the resolution cutoff
As the data quality gradually decreases at the highest resolution (Supplementary Table 2), it would be advantageous to derive a convenient statistical “rule of thumb” to determine the highest resolution that contains valid, merged structure factors. There must be some reasonable cutoff as the multiplicity of observation and the internal correlation coefficient CC1/2 decrease, but it needs to be established which cutoff values should be chosen. To provide an objective criterion, we employed the iterative paired-refinement technique suggested by Karplus & Diederichs15. Each iteration compares the result of two atomic structure refinements, the first using data only out to a conservative resolution limit, and the second including reflections in the next, higher-resolution shell. The two models are then evaluated against the smaller, low-resolution set of reflections, and the two reliability factors are computed (Rwork and Rfree 51). As long as Rfree decreases, the added data contribute useful information to the refinement. An increase in Rwork but unchanged Rfree indicates that the model has become less overfit. As a negative control, the model is refined a third time adding the same higher-resolution intensities, but with randomly permuted (incorrect) Miller indices in the shell. Analysis of the thermolysin data starting at 3.0 Å, and progressing in steps of 0.1 Å towards the highest-resolution limit (1.76 Å) shows that there is significant information (i.e., Rfree decreases) out to at least 2.1 Å (Fig. 2e), while randomly permuted Miller indices nearly always increase the R-factors, as expected. At the 2.1 Å cutoff, the average observational multiplicity of each structure factor is only 4.5, and the correlation coefficient between semi-datasets is 17.0%.
Relationship between resolution and accurate detector model
The empirical and parametric approaches to constructing Bragg spot profiles as outlined above place very stringent requirements on the geometrical modeling (metrology) of the detector. Many diffraction patterns (Fig. 1) exhibit Bragg spots that are only one or two square pixels in area, particularly at low resolution. For spot modeling to work as proposed, therefore, the position of each pixel in space must be known to substantially better accuracy than the pixel dimension, however this is a difficult goal for current XFEL detectors due to their unique construction as a mosaic of pixel array sensors26, 52. We took a bootstrapping approach starting with approximately known sensor positions, followed by the use of Bragg observations from the entire data set (either thermolysin or lysozyme), to derive more accurate sensor positions and orientations by iterative non-linear least squares positional refinement (section immediately below). This improved metrology allowed us to model the Bragg spots with an r.m.s. deviation (observed spot position vs. modeled position) of 0.65 and 1.00 pixels for thermolysin and lysozyme, respectively. Any well-diffracting set of protein crystals would have sufficed for this procedure; it was not necessary for the unit cell or structure to be known ahead of time.
To assess the general importance of accurate detector metrology we carried out an analysis in which the accurately refined sensor positions were intentionally perturbed (Fig. 2b). Indexing success depended weakly on metrology (half of the images could still be indexed with a positional perturbation of 3.5 pixels); but high-resolution integration is strongly dependent, with a 30% loss of high-resolution signal resulting from a perturbation of just a single pixel. This is exactly as expected; our empirically-determined integration masks conform very tightly to the spot shape, therefore for the method to work the positions of individual detector tiles need to be accurately known.
We arrive at the same conclusion, by a different route, if we simply reverse the refinement steps of our detector calibration. This outcome (for the thermolysin data) is also plotted in Fig. 2b. Reversing the final step of iterative non-linear least squares positional refinement leaves us with sensor positions 0.55 pixels away from their true positions, with consequent loss in both high-resolution and overall data. Reversing the penultimate step (where we determine the nearest whole-integer pixel positions without any sensor rotations) puts the sensors 1.38 pixels away from true, with a further degradation in the results.
Refinement of the detector geometry model (metrology)
The CSPAD detector utilized at the CXI instrument is laid out in a mosaic arrangement consisting of four groups (quadrants) of eight silicon pixel-array sensors26. As the quadrants can be translated on mechanical rails, a coarse determination of their relative positions must be made before any Bragg patterns can be analyzed. Pseudo powder patterns were synthesized for this purpose by summing a large number of thermolysin diffraction images, all recorded at the same sample-detector distance. A graphical application was written, permitting the manual adjustment of the quadrant locations to align the observed powder rings with overlaid circular fiducial rings. This program is also suitable for calibrating the detector quadrants with silver behenate53 powder patterns.
Prior to the experiment, the sensor positions and orientations (within each quadrant) were characterized optically at the LCLS to within tens of μm, but this calibration did not necessarily achieve the accuracy required for spot modeling, nor did it probe the actual readouts that are bump-bonded to the sensors. Each sensor is bonded to a pair of side-by-side 194 × 185 pixel application-specific integrated circuits (ASICs)26. Detailed positions and orientations of the 64 ASIC readouts were refined by non-linear least squares refinement of the target functional
where robs is the observed detector position of the Bragg spot centroid determined with the program Spotfinder28, rcalc is the modeled position after indexing, and the sum is over all Spotfinder spots (on all images and ASICs) that correspond to modeled spots. Variable parameters in the refinement included the positions and rotations of all ASICs, the position of the direct beam and crystal-to-detector distance for each crystal shot, and the orientation and unit cell dimensions for each crystal. Correct performance of this algorithm was monitored by considering the refined placement of pairs of ASICs bonded to the same silicon sensor, which are thought to be exactly aligned by a mechanical guide piece during the manufacture process. These internal controls derived from the thermolysin data (Fig. 2a) show that the ASIC pairs are mutually aligned to an r.m.s. rotation of 0.016° and an r.m.s. displacement perpendicular to the long sensor axis of 0.074 pixels; we interpreted these values as the accuracy limits of our refinement method. The tolerances were similar for the lysozyme data, 0.030° and 0.072 pixels respectively. In addition, we found that on the particular detector used for thermolysin, the 32 sensors had an r.m.s. tilt of 0.17° in the plane of the detector, and that the separation between same-sensor ASIC pairs varied with an r.m.s. deviation of 0.21 pixels (Fig. 2a).
Refinement of the detector distance
We calibrated the absolute distance between crystal sample and imaging detector to an accuracy of about 1 mm. Fortunately the indexing algorithm and indeed the entire data processing pipeline is robust to this level of uncertainty, with small errors in the distance being absorbed by other modeled parameters (unit cell dimensions, wavelength). We determined the distance by grid search around an initial estimate: an entire run collected at a fixed distance was reprocessed several times with calibration offsets differing by 0.5 mm, which were then scored by counting the number of images successfully indexed (Supplementary Figure 2). Offsets of ±8 mm from the best value reduced the indexing rate by roughly a factor of 2.
An alternate distance calibration is possible by observing circular powder patterns from silver behenate as noted above, and the cctbx.xfel software can faciliate this analysis. Such a calibration might offer improved accuracy as it uses a recognized standard, however, as a practical matter given the time constraints of collecting data at LCLS, it was more efficient to simply use the thermolysin or lysozyme data itself to estimate the distance as shown in Supplementary Figure 2.
Structure solution
Merged structure factors were phased by molecular replacement using Phaser54 within the Phenix55 system. For thermolysin, the search model consisted of thermolysin (PDB code 2TLI56) from which all non-protein atoms were removed; for lysozyme the model was taken from PDB code 4ET810. New models were built into the resulting maps using phenix.autobuild57, and further refined using phenix.refine58. Refinement statistics are shown in Supplementary Table 1. The molecular clashscore (number of bad all-atom overlaps per thousand atoms) and Ramachandran stereochemical statistics were calculated with MolProbity59.
Crystallographic R factors for the refined thermolysin model are comparable in quality to synchrotron structures that have been determined at a similar resolution (2.1 Å). To determine this we used the program phenix.r_factor_statistics60, 61 to print the R factor distribution from 2271 Protein Data Bank Structures at resolutions in the range 2.05–2.15 Å. Our thermolysin values of Rwork = 22.2% and Rfree = 26.5% are within one standard deviation of the mean (Rwork = 20.1 ± 2.4%; Rfree = 24.6 ± 2.6%). The R factor distribution was derived by taking coordinates, structure factors, and R-free flags from the Protein Data Bank, and using the Phenix toolbox to derive the R factors. As a result, the distributions can be directly compared with our refinements, which were also performed with Phenix.
Similarly, for the 1.9 Å lysozyme structure, we considered 3578 Protein Data Bank Structures at resolutions in the range 1.85–1.95 Å. Our Phenix-refined values of Rwork = 18.7% and Rfree = 22.9% for the cctbx.xfel structure factors, and Rwork = 17.7% and Rfree = 22.0% for the CrystFEL structure factors, are each within one standard deviation of the mean (Rwork = 19.3 ± 2.3%; Rfree = 23.2 ± 2.6%).
The structure factors and model for thermolysin have been deposited with the Protein Data Bank under accession code 4OW3.
Supplementary Material
Acknowledgments
This work was supported by US National Institutes of Health (NIH) grants GM095887 and GM102520 and Director, Office of Science, US Department of Energy (DOE) under contract DE-AC02-05CH11231 for data-processing methods (N.K.S.); Director, DOE Office of Science, Office of Basic Energy Sciences (OBES), Chemical Sciences, Geosciences and Biosciences Division (CSGB) under contract DE-AC02-05CH11231 (J.Y. and V.K.Y.); NIH grant GM055302 (V.K.Y.); NIH grant P41GM103393 (U.B. and T.-C.W.), and DOE Office of Biological and Environmental Research (M.L. and T.-C.W.). Sample injection was supported by LCLS (M.J.B., D.W.S.) and the AMOS program, CSGB Division, OBES, DOE (M.J.B.), and through the SLAC National Accelerator Laboratory (SLAC) Laboratory Directed Research and Development Program (M.J.B., H.L.). J.M. was supported by the Artificial Leaf Project Umeå (K&A Wallenberg Foundation), the Solar Fuels Strong Research Environment Umeå (Umeå University), Vetenskapsrådet and Swedish Energy Agency (Energimyndigheten). Experiments were carried out at the LCLS at SLAC, an Office of Science User Facility operated for the DOE by Stanford University. We thank A. Perazzo, M. Dubrovin, I. Ofte, and A. Salnikov (LCLS) for collaboration on data analysis, and C. Kenney (SLAC) for expertise related to the CSPAD detector.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
Accession codes
Protein Data Bank: 4OW3 (structure factors and model for thermolysin); Coherent X-ray Imaging Data Bank: ID23 (raw data streams for thermolysin).
Author Contributions
J. Hattne, J.K., J.Y., U.B., V.K.Y., P.D.A., N.K.S. conceived of the new data processing methods and analyzed the data;
J. Hattne, N.E., R.J.G., A.S.B., R.W.G.-K., P.H.Z., M.M., P.D.A., N.K.S. wrote the data processing software;
U.B., J.Y.,V.K.Y., J.K., R.A.-M., J.M., A.Z., N.K.S., G.J.W., S.B., A.R.F., A.M., D.M., D.W.S., W.E.W., M.J.B. designed the experiment;
R.T., C.G., J. Hellmich, D.D., A.L., G.H., J.K., A.Z. prepared samples;
S.B., J.E.K., M.M., M.M.S., G.J.W. operated the CXI instrument;
M.J.B., H.L., R.G.S., J.K., J.M., B.L.-K., S.G., R.T., C.G., J. Hellmich, J.S., D.W.S., A.M., G.J.W. developed, tested and ran sample delivery system;
R.A.-M., U.B., M.J.B., S.B., N.E., R.J.G., P.G., C.G., S.G., G.H., J. Hattne., J. Hellmich, J.K., J.E.K., H.L., A.L., B.L.-K., D.M., M.M., J.M.,N.K.S., M.M.S., J.S., R.G.S., D.S., R.T., T.-C.W., G.J.W., V.K.Y., J.Y., A.Z. performed the LCLS experiment;
J. Hattne, N.E., J.K., J.Y., U.B., V.K.Y., P.D.A., N.K.S. wrote the manuscript with input from all authors.
References
- 1.Neutze R, et al. Nature. 2000;406:752–757. doi: 10.1038/35021099. [DOI] [PubMed] [Google Scholar]
- 2.Alonso-Mori R, et al. Proc Natl Acad Sci USA. 2012;109:19103–19107. doi: 10.1073/pnas.1211384109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kern J, et al. Science. 2013;340:491–495. doi: 10.1126/science.1234273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman HN, et al. Nature. 2011;470:73–77. doi: 10.1038/nature09750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koopmann R, et al. Nat Methods. 2012;9:259–262. doi: 10.1038/nmeth.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Redecke L, et al. Science. 2013;339:227–230. doi: 10.1126/science.1229663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourenkov GP, Popov AN. Acta Crystallogr D Biol Crystallogr. 2006;62:58–64. doi: 10.1107/S0907444905033998. [DOI] [PubMed] [Google Scholar]
- 8.White TA, et al. J Appl Crystallogr. 2012;45:335–341. [Google Scholar]
- 9.Sauter NK, et al. Acta Crystallogr D Biol Crystallogr. 2013;69:1274–1282. doi: 10.1107/S0907444913000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boutet S, et al. Science. 2012;337:362–364. doi: 10.1126/science.1217737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauter NK, Grosse-Kunstleve RW, Adams PD. J Appl Crystallogr. 2004;37:399–409. doi: 10.1107/S0021889804005874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauter NK, Poon BK. J Appl Crystallogr. 2010;43:611–616. doi: 10.1107/S0021889810010782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powell HR, Johnson O, Leslie AG. Acta Crystallogr D Biol Crystallogr. 2013;69:1195–1203. doi: 10.1107/S0907444912048524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirian RA, et al. Acta Crystallogr A. 2011;67:131–140. doi: 10.1107/S0108767310050981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karplus PA, Diederichs K. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirian RA, et al. Opt Express. 2010;18:5713–5723. doi: 10.1364/OE.18.005713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson LC, et al. Nat Methods. 2012;9:263–265. doi: 10.1038/nmeth.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Winkler FK, Schutt CE, Harrison SC. Acta Crystallogr A. 1979;35:901–911. [Google Scholar]
- 19.Rossmann MG, et al. J Appl Crystallogr. 1979;12:570–581. [Google Scholar]
- 20.Zhu D, et al. Appl Phys Lett. 2012;101:034103. [Google Scholar]
- 21.Inouye K. J Biochem. 1992;112:335–340. doi: 10.1093/oxfordjournals.jbchem.a123901. [DOI] [PubMed] [Google Scholar]
- 22.Titani K, et al. Nature. 1972;238:35–37. doi: 10.1038/newbio238035a0. [DOI] [PubMed] [Google Scholar]
- 23.Boutet S, Williams GJ. New J Phys. 2010;12:035024. [Google Scholar]
- 24.Sierra RG, et al. Acta Crystallogr D Biol Crystallogr. 2012;68:1584–1587. doi: 10.1107/S0907444912038152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogan MJ. Anal Chem. 2013;85:3464–3471. doi: 10.1021/ac303716r. [DOI] [PubMed] [Google Scholar]
- 26.Hart P, et al. Proc of SPIE. 2012;8504:85040C. [Google Scholar]
- 27.Maia FRNC. Nat Methods. 2012;9:854–855. doi: 10.1038/nmeth.2110. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, et al. J Appl Crystallogr. 2006;39:112–119. [Google Scholar]
- 29.Steller I, Bolotovsky R, Rossmann MG. J Appl Crystallogr. 1997;30:1036–1040. [Google Scholar]
- 30.Rossmann MG, van Beek CG. Acta Crystallogr D Biol Crystallogr. 1999;55:1631–1640. doi: 10.1107/s0907444999008379. [DOI] [PubMed] [Google Scholar]
- 31.Sauter NK, Grosse-Kunstleve RW, Adams PD. J Appl Crystallogr. 2006;39:158–168. doi: 10.1107/S0021889804005874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kern J, et al. Proc Natl Acad Sci USA. 2012;109:9721–9726. doi: 10.1073/pnas.1204598109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giordano R, et al. Acta Crystallogr D Biol Crystallogr. 2012;68:649–658. doi: 10.1107/S0907444912006841. [DOI] [PubMed] [Google Scholar]
- 34.Diederichs K, Karplus PA. Acta Crystallogr D Biol Crystallogr. 2013;69:1215–1222. doi: 10.1107/S0907444913001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paithankar KS, et al. Acta Crystallogr D Biol Crystallogr. 2011;67:608–618. doi: 10.1107/S0907444911015617. [DOI] [PubMed] [Google Scholar]
- 36.White TA, et al. Acta Crystallogr D Biol Crystallogr. 2013;69:1231–1240. doi: 10.1107/S0907444913013620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nave C. Acta Crystallogr D Biol Crystallogr. 1998;54:848–853. doi: 10.1107/s0907444998001875. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 39.Emma P, et al. Nature Photon. 2010;4:641–647. [Google Scholar]
- 40.Greenhough TJ, Helliwell JR. J Appl Crystallogr. 1982;15:338–351. [Google Scholar]
- 41.Greenhough TJ, Helliwell JR. J Appl Crystallogr. 1982;15:493–508. [Google Scholar]
- 42.Greenhough TJ, Helliwell JR, Rule SA. J Appl Crystallogr. 1983;16:242–250. [Google Scholar]
- 43.Ren Z, Moffat K. J Appl Crystallogr. 1995;28:461–481. [Google Scholar]
- 44.Dauter Z. Acta Crystallogr D Biol Crystallogr. 1999;55:1703–1717. doi: 10.1107/s0907444999008367. [DOI] [PubMed] [Google Scholar]
- 45.Diederichs K. Acta Crystallogr D Biol Crystallogr. 2009;65:535–542. doi: 10.1107/S0907444909010282. [DOI] [PubMed] [Google Scholar]
- 46.Schreurs AMM, Xian X, Kroon-Batenburg LMJ. J Appl Crystallogr. 2009;43:70–82. [Google Scholar]
- 47.Porta J, et al. Acta Crystallogr D Biol Crystallogr. 2011;67:628–638. doi: 10.1107/S0907444911017884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolotovsky R, Coppens P. J Appl Crystallogr. 1997;30:65–70. [Google Scholar]
- 49.Leslie AGW. Acta Crystallogr D Biol Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 50.Kahn R, et al. J Appl Crystallogr. 1982;15:330–337. [Google Scholar]
- 51.Brünger AT. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 52.Strüder L, et al. Nucl Instrum Methods Phys Res A. 2010;614:483–496. [Google Scholar]
- 53.Huang TC, et al. J Appl Crystallogr. 1993;26:180–184. [Google Scholar]
- 54.McCoy AJ, et al. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams PD, et al. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.English AC, et al. Proteins: Structure, Function, and Genetics. 1999;37:628–640. [PubMed] [Google Scholar]
- 57.Terwilliger TC, et al. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Afonine PV, et al. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen VB, et al. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Urzhumtseva L, et al. Acta Crystallogr D Biol Crystallogr. 2009;65:297–300. doi: 10.1107/S0907444908044296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Afonine PV, et al. J Appl Crystallogr. 2010;43:669–676. doi: 10.1107/S0021889810015608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.