Abstract
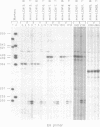
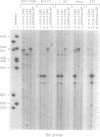
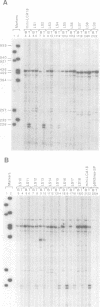
Tissue-specific alternative splicing is an important mechanism for controlling gene expression. Exons 4, 5 and 6 of the human leukocyte common antigen (LCA) gene are included in B cell mRNA but excluded from thymocyte mRNA by differential splicing. In order to study this tissue-specific alternative splicing, we constructed mini-genes that contain only a few of the LCA exons and the SV40 promoter. Mouse B cells and thymocytes were transfected with these mini-gene constructs and the structures of mRNAs were determined by primer extension analysis. The results show that the same primary transcript is spliced alternatively in B cells and thymocytes. This finding suggests that there is a tissue-specific trans-acting factor that regulates the alternative splicing of LCA pre-mRNA. By making various deletion mutants, cis-elements necessary for tissue-specific splicing were confined within the alternatively spliced exons and their immediate flanking intron sequences. Furthermore, linker scanning analysis shows that there are at least three distinct cis-elements within the LCA exon 4 sequence that are required for tissue-specific alternative splicing. Possible mechanisms of LCA alternative splicing are discussed.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akbar A. N., Terry L., Timms A., Beverley P. C., Janossy G. Loss of CD45R and gain of UCHL1 reactivity is a feature of primed T cells. J Immunol. 1988 Apr 1;140(7):2171–2178. [PubMed] [Google Scholar]
- Alt F. W., Bothwell A. L., Knapp M., Siden E., Mather E., Koshland M., Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3' ends. Cell. 1980 Jun;20(2):293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- Alt F. W., Yancopoulos G. D., Blackwell T. K., Wood C., Thomas E., Boss M., Coffman R., Rosenberg N., Tonegawa S., Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984 Jun;3(6):1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay A. N., Jackson D. I., Willis A. C., Williams A. F. Lymphocyte specific heterogeneity in the rat leucocyte common antigen (T200) is due to differences in polypeptide sequences near the NH2-terminus. EMBO J. 1987 May;6(5):1259–1264. doi: 10.1002/j.1460-2075.1987.tb02362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Beyer A. L., Christensen M. E., Walker B. W., LeStourgeon W. M. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1977 May;11(1):127–138. doi: 10.1016/0092-8674(77)90323-3. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart R. E., Andreadis A., Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Nadal-Ginard B. Developmentally induced, muscle-specific trans factors control the differential splicing of alternative and constitutive troponin T exons. Cell. 1987 Jun 19;49(6):793–803. doi: 10.1016/0092-8674(87)90617-9. [DOI] [PubMed] [Google Scholar]
- Charbonneau H., Tonks N. K., Walsh K. A., Fischer E. H. The leukocyte common antigen (CD45): a putative receptor-linked protein tyrosine phosphatase. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7182–7186. doi: 10.1073/pnas.85.19.7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. D., Grabowski P. J., Sharp P. A., Dreyfuss G. Heterogeneous nuclear ribonucleoproteins: role in RNA splicing. Science. 1986 Mar 28;231(4745):1534–1539. doi: 10.1126/science.3952495. [DOI] [PubMed] [Google Scholar]
- Chu G., Hayakawa H., Berg P. Electroporation for the efficient transfection of mammalian cells with DNA. Nucleic Acids Res. 1987 Feb 11;15(3):1311–1326. doi: 10.1093/nar/15.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement L. T., Yamashita N., Martin A. M. The functionally distinct subpopulations of human CD4+ helper/inducer T lymphocytes defined by anti-CD45R antibodies derive sequentially from a differentiation pathway that is regulated by activation-dependent post-thymic differentiation. J Immunol. 1988 Sep 1;141(5):1464–1470. [PubMed] [Google Scholar]
- Cooper T. A., Cardone M. H., Ordahl C. P. Cis requirements for alternative splicing of the cardiac troponin T pre-mRNA. Nucleic Acids Res. 1988 Sep 12;16(17):8443–8465. doi: 10.1093/nar/16.17.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Eperon L. P., Graham I. R., Griffiths A. D., Eperon I. C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988 Jul 29;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- Hall L. R., Streuli M., Schlossman S. F., Saito H. Complete exon-intron organization of the human leukocyte common antigen (CD45) gene. J Immunol. 1988 Oct 15;141(8):2781–2787. [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Kakizuka A., Kitamura N., Nakanishi S. Localization of DNA sequences governing alternative mRNA production of rat kininogen genes. J Biol Chem. 1988 Mar 15;263(8):3884–3892. [PubMed] [Google Scholar]
- Laski F. A., Rio D. C., Rubin G. M. Tissue specificity of Drosophila P element transposition is regulated at the level of mRNA splicing. Cell. 1986 Jan 17;44(1):7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Evans R. M., Rosenfeld M. G. Splice commitment dictates neuron-specific alternative RNA processing in calcitonin/CGRP gene expression. Cell. 1987 Feb 13;48(3):517–524. doi: 10.1016/0092-8674(87)90202-9. [DOI] [PubMed] [Google Scholar]
- Leff S. E., Rosenfeld M. G., Evans R. M. Complex transcriptional units: diversity in gene expression by alternative RNA processing. Annu Rev Biochem. 1986;55:1091–1117. doi: 10.1146/annurev.bi.55.070186.005303. [DOI] [PubMed] [Google Scholar]
- Lefrancois L., Thomas M. L., Bevan M. J., Trowbridge I. S. Different classes of T lymphocytes have different mRNAs for the leukocyte-common antigen, T200. J Exp Med. 1986 May 1;163(5):1337–1342. doi: 10.1084/jem.163.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayrand S. H., Pedersen N., Pederson T. Identification of proteins that bind tightly to pre-mRNA during in vitro splicing. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3718–3722. doi: 10.1073/pnas.83.11.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto C., Letvin N. L., Boyd A. W., Hagan M., Brown H. M., Kornacki M. M., Schlossman S. F. The isolation and characterization of the human helper inducer T cell subset. J Immunol. 1985 Jun;134(6):3762–3769. [PubMed] [Google Scholar]
- Morimoto C., Letvin N. L., Distaso J. A., Aldrich W. R., Schlossman S. F. The isolation and characterization of the human suppressor inducer T cell subset. J Immunol. 1985 Mar;134(3):1508–1515. [PubMed] [Google Scholar]
- Nabeshima Y., Fujii-Kuriyama Y., Muramatsu M., Ogata K. Alternative transcription and two modes of splicing results in two myosin light chains from one gene. Nature. 1984 Mar 22;308(5957):333–338. doi: 10.1038/308333a0. [DOI] [PubMed] [Google Scholar]
- Nagoshi R. N., McKeown M., Burtis K. C., Belote J. M., Baker B. S. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988 Apr 22;53(2):229–236. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- Omary M. B., Trowbridge I. S., Battifora H. A. Human homologue of murine T200 glycoprotein. J Exp Med. 1980 Oct 1;152(4):842–852. doi: 10.1084/jem.152.4.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S., Choi Y. D., Matunis M. J., Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988 Feb;2(2):215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- Ralph S. J., Thomas M. L., Morton C. C., Trowbridge I. S. Structural variants of human T200 glycoprotein (leukocyte-common antigen). EMBO J. 1987 May;6(5):1251–1257. doi: 10.1002/j.1460-2075.1987.tb02361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Rudd C. E., Morimoto C., Wong L. L., Schlossman S. F. The subdivision of the T4 (CD4) subset on the basis of the differential expression of L-C/T200 antigens. J Exp Med. 1987 Dec 1;166(6):1758–1773. doi: 10.1084/jem.166.6.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y., Tung J. S., Shen F. W., Boyse E. A. Alternative use of 5' exons in the specification of Ly-5 isoforms distinguishing hematopoietic cell lineages. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5364–5368. doi: 10.1073/pnas.84.15.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt P., Gattoni R., Keohavong P., Stévenin J. Alternative splicing of E1A transcripts of adenovirus requires appropriate ionic conditions in vitro. Cell. 1987 Jul 3;50(1):31–39. doi: 10.1016/0092-8674(87)90659-3. [DOI] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Solnick D., Lee S. I. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol. 1987 Sep;7(9):3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somasekhar M. B., Mertz J. E. Exon mutations that affect the choice of splice sites used in processing the SV40 late transcripts. Nucleic Acids Res. 1985 Aug 12;13(15):5591–5609. doi: 10.1093/nar/13.15.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Streuli M., Hall L. R., Saga Y., Schlossman S. F., Saito H. Differential usage of three exons generates at least five different mRNAs encoding human leukocyte common antigens. J Exp Med. 1987 Nov 1;166(5):1548–1566. doi: 10.1084/jem.166.5.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli M., Krueger N. X., Hall L. R., Schlossman S. F., Saito H. A new member of the immunoglobulin superfamily that has a cytoplasmic region homologous to the leukocyte common antigen. J Exp Med. 1988 Nov 1;168(5):1523–1530. doi: 10.1084/jem.168.5.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli M., Matsuyama T., Morimoto C., Schlossman S. F., Saito H. Identification of the sequence required for expression of the 2H4 epitope on the human leukocyte common antigens. J Exp Med. 1987 Nov 1;166(5):1567–1572. doi: 10.1084/jem.166.5.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli M., Morimoto C., Schrieber M., Schlossman S. F., Saito H. Characterization of CD45 and CD45R monoclonal antibodies using transfected mouse cell lines that express individual human leukocyte common antigens. J Immunol. 1988 Dec 1;141(11):3910–3914. [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T., Rudd C. E., Schlossman S. F., Morimoto C. Induction of suppression following autologous mixed lymphocyte reaction; role of a novel 2H4 antigen. Eur J Immunol. 1987 Jan;17(1):97–103. doi: 10.1002/eji.1830170117. [DOI] [PubMed] [Google Scholar]
- Terry L. A., Brown M. H., Beverley P. C. The monoclonal antibody, UCHL1, recognizes a 180,000 MW component of the human leucocyte-common antigen, CD45. Immunology. 1988 Jun;64(2):331–336. [PMC free article] [PubMed] [Google Scholar]
- Woollett G. R., Barclay A. N., Puklavec M., Williams A. F. Molecular and antigenic heterogeneity of the rat leukocyte-common antigen from thymocytes and T and B lymphocytes. Eur J Immunol. 1985 Feb;15(2):168–173. doi: 10.1002/eji.1830150211. [DOI] [PubMed] [Google Scholar]
- Ziff E. B. Transcription and RNA processing by the DNA tumour viruses. Nature. 1980 Oct 9;287(5782):491–499. doi: 10.1038/287491a0. [DOI] [PubMed] [Google Scholar]