Abstract
DmpFG is a bifunctional enzyme comprised of an aldolase subunit, DmpG, and a dehydrogenase subunit, DmpF. The aldehyde intermediate produced by the aldolase is channeled directly through a buried molecular channel in the protein structure from the aldolase to the dehydrogenase active site. In this study, we have investigated the binding of a series of progressively larger substrates to the aldolase, DmpG, using molecular dynamics. All substrates investigated are easily accommodated within the active site, binding with free energy values comparable to the physiological substrate 4-hydroxy-2-ketovalerate. Subsequently, umbrella sampling was utilized to obtain free energy surfaces for the aldehyde intermediates (which would be generated from the aldolase reaction on each of these substrates) to move through the channel to the dehydrogenase DmpF. Small substrates were channeled with limited barriers in an energetically feasible process. We show that the barriers preventing bulky intermediates such as benzaldehyde from moving through the wild-type protein can be removed by selective mutation of channel-lining residues, demonstrating the potential for tailoring this enzyme to allow its use for the synthesis of specific chemical products. Furthermore, positions of transient escape routes in this flexible channel were determined.
Introduction
The channeling of substrates between spatially distinct active sites within multienzyme complexes is a topic of considerable interest. This process is known to occur in at least two distinct ways—using either an electrostatic highway on the surface of the protein, or a molecular channel buried within the protein structure (1,2). Substrate channeling via a buried molecular channel is advantageous, not only in the context of increased reaction rates and efficiency, but also for the isolation of both volatile and toxic intermediates from the bulk-media of the cell (3,4). One enzyme complex for which substrate channeling has been proposed is DmpFG (4-hydroxy-2-ketovalerate aldolase-aldehyde dehydrogenase (acylation)) (5). DmpFG is a microbial enzyme comprised of two subunits DmpG and DmpF (5) as shown in Fig. 1 A. DmpFG catalyzes the final two steps of the meta-cleavage pathway of catechol and its methylated substituents. This pathway breaks down toxic waste products such as naphthalenes, salicylates, and benzoates to harmless metabolites (6,7).
Figure 1.
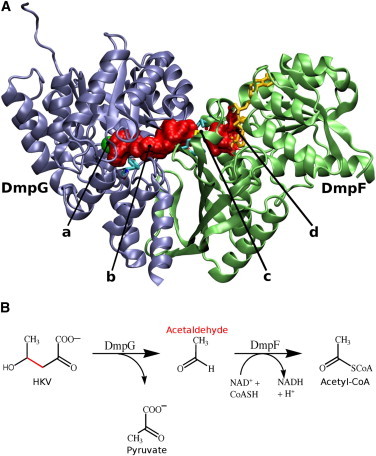
(A) The DmpFG hetero-dimer with the DmpG (blue) and DmpF (green) subunits. (a) The aldolase active site in DmpG with the central Mn2+ ion (green). (b) The solvent-accessible surface of a water-filled intramolecular channel reaching from the aldolase to the dehydrogenase active site (red). (c) The three residues of the hydrophobic triad at the channel’s exit and (d) the dehydrogenase active site in DmpF. (B) The overall reaction catalyzed by DmpFG. The toxic intermediate acetaldehyde formed in the aldolase active site is transported down an intramolecular channel to the dehydrogenase active site in DmpF (5). To see this figure in color, go online.
DmpG, the aldolase, catalyses the cleavage of the substrate HKV (4-hydroxy-2-ketovalerate) to acetaldehyde with the release of pyruvate (Fig. 1 B). Acetaldehyde must then reach the dehydrogenase active site in DmpF; however, it is both labile and toxic, and therefore release into the bulk solvent would not be advantageous to the organism. An alternative route to the second active site has been proposed from examination of the crystal structure of DmpFG, which revealed a 29 Å long water-filled channel linking the aldolase and dehydrogenase active sites (5). It has previously been shown by Smith et al. (8) using molecular dynamics simulations that it is energetically feasible for acetaldehyde to move from one active site to the other within DmpFG when NAD+, a coenzyme, is bound to DmpF. A hydrophobic triad (HT), observed in the crystal structure of DmpFG, is of interest in the context of channeling (5). The HT is composed of three bulky residues: Ile172, Ile196, and Met198, which together appeared to form a gate between the channel and the second active site. The crystal structure of DmpFG showed differences in this region of the structure in the presence and absence of the NAD+ cofactor, which were interpreted to facilitate the passage of the intermediate from the channel and into the dehydrogenase active site (5).
In recent years, DmpFG and orthologous aldolase-dehydrogenases have begun to feature more extensively in the literature (8–16). While structures of MhpEF (15) and HsaF-HsaG (16) have recently been determined, DmpFG has the best-characterized structure so far (5). As such, it provides a unique tool for investigating the dynamics of the channeling event that can be coupled with the data obtained from the extensive kinetic, biochemical, and site-directed mutagenesis studies carried out on orthologs such as BphI-BphJ (9–13), where BphI and BphJ have 57 and 58% sequence identity with DmpG and DmpF, respectively. BphI-BphJ are two enzymes in the polychlorinated biphenyls degradation pathway of Burkholderia xenovorans LB400 that catalyze the conversion of 4-hydroxy-2-ketoacids to a coenzyme A derivative and pyruvate. Again, a toxic aldehyde intermediate needs to move between the two spatially distinct active sites. Studies have shown that the aldehyde intermediate is channeled from the BphI active site to that of BphJ within the enzyme complex (9,11). More recently, it has also been shown that channeling occurs within DmpFG (14) and two other homologous aldolase-dehydrogenase complexes: TTHB246-TTHB247 (17), from Thermus thermophilus HB8, where TTHB246 and THB247 have 51 and 56% sequence identity with DmpG and DmpF, respectively; and HsaF-HsaG (16), from Mycobacterium tuberculosis, where HsaF and HsaG have 49 and 56% sequence identity with DmpG and DmpF, respectively.
Subsequent studies on BphI-BphJ have investigated whether aldehydes larger than acetaldehyde can move through the channel from one active site to the other (11). It was found that aliphatic aldehydes up to six carbons in length were efficiently channeled. Specifically, it was found that both acetaldehyde and propionaldehyde exhibited channeling efficiencies of 95%. Similarly, the much bulkier isobutyraldehyde channeled with 92% efficiency as opposed to 83% efficiency for the longer, but less bulky, butyraldehyde. This implies that even the most constricted regions within the channel are flexible enough to allow the passage of larger intermediates. In contrast, studies on TTHB246-TTHB247 showed that acetaldehyde was channeled with a much higher efficiency than propionaldehyde, possibly implying a more constricted channel (17).
These studies have provided much insight into the process of substrate channeling within this class of enzymes. The ability of BphI-BphJ to channel aldehydes much larger than those that would be produced by the polychlorinated biphenyls degradation pathway raises questions about how the channel can accommodate these large molecules and what the size limitations are for this process. It also opens up potential pathways for these enzymes to be used for the tailored synthesis of enantiomerically pure long-chain substrates. Carere et al. (11) has effectively used BphI-BphJ channel mutants to prevent the passage of larger aldehydes through the channel. More recently, Baker et al. (17) enhanced the channeling efficiency of propionaldehyde through TTHB246-TTHB247 by a point mutation.
Given the channeling studies for BphI-BphJ and TTHB246-TTHB247, we investigated whether the channel within DmpFG could accommodate aldehydes larger than acetaldehyde and whether it could be enhanced to channel specific aldehydes of different sizes and shapes. Furthermore, we studied whether changes to the enzyme could be made without affecting the behavior of this enzyme complex. In our study of DmpFG, we used molecular dynamics to investigate the positioning and energetic feasibility for long-chain substrates to bind within the DmpG active site. Molecular dynamics was also used to investigate the movement of the corresponding aldehydes through the channel within DmpFG, characterizing the energy barriers to this process. Finally, a channel mutant was designed to change the channeling capabilities of DmpFG.
Methods
DmpFG-substrate systems
The DmpFG-substrate system with NAD+ bound to DmpF was utilized as described by Smith et al. (8) The initial holo-enzyme coordinates were obtained from the Protein Data Bank (PDB:1NVM) and HKV was positioned in the active site of DmpG. Before collecting data, a series of equilibration steps were performed. The computed positions for all hydrogen atoms were energy-minimized for 5000 steps whereas nonhydrogen atoms remained fixed. Water and ions were then minimized for 5000 steps and subsequently equilibrated with 50 ps of molecular dynamics (MD) simulation whereas the protein and substrates were kept fixed. This was followed by 20,000 steps of minimization. Harmonic restraints were applied to the protein atoms and gradually decreased over 250 ps from 20.0, 10.0, 5.0, and 2.5 to 0.5 kcal/mol/Å2. This was followed by 250 ps of equilibration with no harmonic restraints on the protein.
All alternative substrates (Fig. 2), including HK6 (4-hydroxy-2-oxohexanoate, HK7 (4-hydroxy-2-oxoheptanoate), HK17 (4-hydroxy-2-oxo-isoheptanoate), and HKB (4-hydroxy-2-oxo-4-benzyl-butanoate) were generated using the initial HKV structure and energy minimized for 10,000 steps in vacuum before being positioned in the active site of DmpG. Each protein-substrate system was energy-minimized for 10,000 steps before 6 ns of equilibration. HKB, as the bulkiest substrate, was minimized more rigorously as follows: the system was energy-minimized for 20,000 steps in the active site of DmpG, and the system was equilibrated for 250 ps of MD simulation with harmonic restraints of 10 kcal/mol/Å2 on each atom of HKB, excluding the atoms of the benzyl ring, which were free to move. The system was equilibrated for a further 250 ps of MD simulation with the harmonic restraints reduced to 1 kcal/mol/Å2. After this, 6 ns of MD simulation were obtained with no harmonic restraints, as done for the other substrates.
Figure 2.
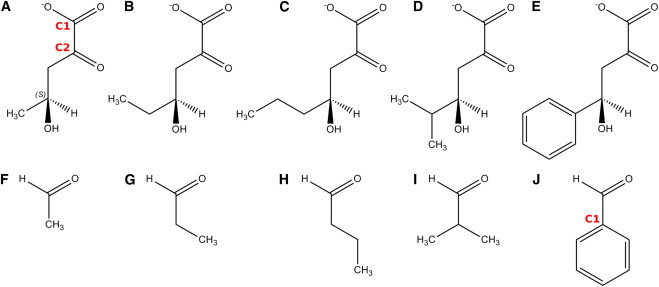
Alternative substrates of DmpG and the corresponding aldehyde products. (A) The S isomer of 4-hydroxy-2-ketovalerate (HKV). (B) 4-hydroxy-2-oxohexanoate (HK6). (C) 4-hydroxy-2-oxoheptanoate (HK7). (D) 4-hydroxy-2-iso-oxoheptanoate (HKI7). (E) 4-hydroxy-2-oxo-4-benzylbutanoate (HKB). (F) Acetaldehyde. (G) Propionaldehyde. (H) Butyraldehyde. (I) Isobutyraldehyde. (J) Benzaldehyde. To see this figure in color, go online.
The final modeled position of HKV was used to position both enolate (the precursor of pyruvate) and acetaldehyde in the DmpG active site. Acetaldehyde was fixed in position and the system was equilibrated for another 10 ps. The same protocol was used to position each aldehyde within the DmpG active site. Benzaldehyde was equilibrated more rigorously due to its bulky nature. Harmonic restraints of 1 kcal/mol/Å2 were applied to the aldehyde moeity of benzaldehyde while both C1 of the benzaldehyde ring (Fig. 2 J) and the methyl carbon of the enolate were restrained with a force of 0.5 kcal/mol/Å2. The system was then energy-minimized for 50,000 steps before being equilibrated for 250 ps. All simulations were performed using NAMD (18) and the CHARMM27 force field (19) using 2-fs timesteps with bonds to hydrogen kept rigid with the RATTLE algorithm. Additional parameters were required for the central Mn2+, enolate, and HKV and these were determined as described previously in Smith et al. (8). Constant temperature (310 K) and pressure (1 atm) were maintained using Langevin dynamics and a Langevin piston. The particle-mesh Ewald method was used to compute the complete electrostatics of the system (20). All molecular graphics were generated using the software VMD (21).
Free energy perturbation
Alchemical free energy perturbation (22–27) was utilized to determine the relative free energy of binding for each substrate equilibrated in the DmpG active site. Free energy perturbation has been applied extensively to determine the binding energy of ligands in proteins and many of the results obtained have been in close agreement with experimental values (27–33). In this study, the PSFGEN plug-in was utilized with the script ALCHEMIFY (performed in the software NAMD; Beckman Institute, Urbana, IL) (34) to generate dual-topology hybrid molecules of HKV with each of the alternate substrate side chains branching from carbon C2 (see Fig. S1 in the Supporting Material). This allows the enolate moiety to remain bound to the central Mn2+ ion while the side chain is morphed from the physiological substrate to each of the alternate substrates.
Two substrate-protein starting structures were used for each of these simulations: one equilibrated with HKV and one equilibrated with the alternative substrate. In each case, the equilibrated substrate was morphed to the other substrate, and then the process was reversed. This resulted in four free energy values being obtained for each substrate, which were averaged to obtain a mean value and a standard error for each protein-substrate system. In these simulations, λ-windows varied in size from 0.025 to 0.1 and were run for 2–4 ns after at least 100 ps of equilibration. To ascertain that the simulations were of an appropriate length, the convergence of the free energy value obtained for each λ-window was determined for both R-HKV and HKB by sampling three time points: 0.5, 1.5, and 2 ns (see Fig. S2).
Relative free energy values for each substrate were subsequently obtained in a water box with dimensions of 30 × 30 × 30 Å with 150 mM NaCl ions. Forty evenly spaced λ-windows were utilized and each was run for 1 ns. Three repeats in both the forward and reverse direction were obtained for each hybrid substrate, allowing both a mean and a standard error to be calculated.
Umbrella sampling
Umbrella sampling (35,36) was subsequently applied to investigate the channeling of aldehyde intermediates through the channel buried within DmpFG, including acetaldehyde, propionaldehyde, butyraldehyde, isobutyraldehyde, and benzaldehyde. One-dimensional free energy profiles were previously determined by Smith et al. (8) for acetaldehyde in the presence and absence of NAD+ using the method of metadynamics (37,38). It was noted in our previous study that acetaldehyde can escape the channel before fully sampling the free energy space of interest. Although repeats and mean free energy profiles were obtained, the ability of the intermediate to escape the channel makes gaining well-converged free-energy profiles difficult. With umbrella sampling and ever-increasing computing power, we can force the intermediate to remain at fixed locations along the channel until the free energy surface is completely converged. In this study, metadynamics (37,38) was initially applied to obtain positions for each aldehyde along the length of the channel through DmpFG, providing starting points for each umbrella sampling window.
Two collective variables were utilized to define the aldehyde’s position in the channel. The first one was the dynamic projection (distanceZ) of the center of mass of each respective aldehyde onto the axis defined by Mn2+, in the DmpG active site, and the α-carbon atom of Cys132 in the DmpF active site. Thus, the collective variable defines the position of each aldehyde along the channel with the origin at the midpoint of the channel. The second collective variable (distanceXY) defines the distance of the intermediate from the channel axis defined above. Umbrella sampling windows were positioned at 0.5 or 1 Å intervals along the length of the channel (with >40 windows per profile), with a force constant of either 5 or 10 kcal/mol/Å applied to the distanceZ collective variable and checked for a high level of overlap between adjacent windows. No force was applied to the distanceXY collective variable. 50 ns of simulation time were obtained for each umbrella sampling window.
The data was subsequently analyzed using the weighted histogram analysis method (39) with the softwares WHAM and WHAM-2d (University of Rochester Medical Center, Rochester, NY) (40) to obtain one- and two-dimensional free energy plots for each aldehyde with a tolerance of 0.0001. Free energy profiles through the channel were subsequently obtained by integrating in the R direction from 0 to 10 Å, effectively excluding the regions of the free energy surface that encompass escape routes. The convergence of these free energy profiles was determined by calculating the free energy profile using progressively larger intervals of 10 ns (see Fig. S3 and Fig. S4). Furthermore, to determine the statistical error in the final free energy profiles, each data set was shuffled, split into five, and then five more separate free energy profiles were obtained allowing the standard error to be determined (see Fig. S5 and Fig. S6). All free energy surfaces and profiles were aligned by calculating the average value from −7 to 0 Å and shifting the plots so they share the same average in this region.
Mutant systems
A mutant DmpFG system was designed to tailor the channeling capabilities of DmpFG specifically for the passage of benzaldehyde. In this mutant, Ile159 from the DmpF binding pocket was mutated to alanine (I159A) using the plug-in PSFGEN. Ile159 was selected because it is the bulkiest residue between the HT and Cys132, and as such, replacing it with a smaller residue is expected to lower the energy barrier faced by benzaldehyde as it enters this region. This in turn should increase the likelihood of benzaldehyde successfully traversing the channel. A two-dimensional free energy surface was then determined using umbrella sampling, as described above.
Sequence alignments
Sequence identities were obtained for DmpG and DmpF with their respective orthologs: BphI, TTHB246; HsaF and BphJ, TTHB247; and HsaG, using the protein-protein software Basic Local Alignment Search Tool (BLAST; National Center for Biotechnology Information (NCBI), National Institutes of Health, Bethesda, MD) (41,42). The multiple alignments were obtained using the Constraint-based Multiple Alignment Tool (COBALT; NCBI, National Institutes of Health) and the data was displayed using the protein sequence editor ALINE (43).
Results and Discussion
Substrate binding in DmpG
The R and the S forms of HKV were both accommodated within the DmpG active site as shown in Fig. 3, A and B, and as discussed by Smith et al. (8). However, whereas the hydroxyl group of S-HKV is oriented toward His21 in an appropriate position for catalysis, the hydroxyl of R-HKV is oriented in the opposite direction and coordinates with one of its own carboxylate oxygens. This implies that if R-HKV is able to bind, it would be unable to successfully complete a reaction, because steric hindrance from Tyr291 prevents this group from reorienting itself within the active site. This raises the question of whether R-HKV is actually likely to bind to the DmpG active site in the first place. The binding free energy of R-HKV was determined relative to S-HKV using free energy perturbation, yielding a value of 0.0 ± 0.8 kcal/mol and indicating that it is energetically feasible for R-HKV to bind in the active site of DmpG. Furthermore, because the relative binding energy values are so close in magnitude, it is possible that, assuming R-HKV can gain access to the active site, R-HKV could be a competitive inhibitor of the DmpG aldolase reaction, because once it binds to DmpG it is unable to undergo a reaction and it may not be easily displaced by S-HKV.
Figure 3.
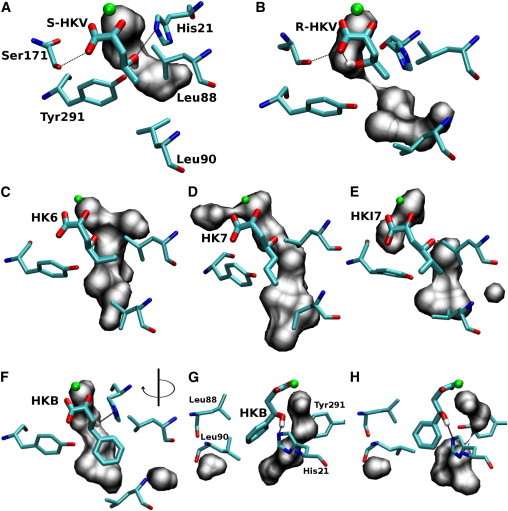
Alternative substrates in the DmpG active site. Snapshots from MD simulations showing the Mn2+ ion (green), a water-filled cavity (gray surface), and (A) S-HKV with hydroxyl group oriented toward His21 in a position favorable for catalysis. (B) R-HKV with hydroxyl group oriented toward its carboxylate oxygen and away from His21 in a position unfavorable for reaction. (C) HK6 in active site (note positions of Leu88 and Leu90). (D) HK7, which extends toward the channel but is still some distance from Leu90. (E) HKI7, the bulkiest aliphatic substrate that still has sufficient room in the active site. (F) HKB with benzyl group at an angle of −50°. (G) HKB at the same timepoint as shown in panel F but rotated such that the orientation of the benzyl ring in the active site can be seen more clearly. (H) HKB at ∼5 ns when the benzyl ring is at ∼−150°. To see this figure in color, go online.
Simulations for 6 ns of equilibration, with a range of alternative substrates, showed that each one can be accommodated within the DmpG active site, as shown in Fig. 3. The bulky side chains within the active site, such as Tyr291 and Leu88, are free to rotate away from the substrate, leaving sufficient space for both the substrate and water molecules (gray surface). These water molecules indicate a route for the aldehyde product of this reaction to enter the channel. It should be noted that HK7, the longest aliphatic substrate used in this trial, orients in such a way that its carbon chain extends toward the channel entrance (Fig. 3). Even bulky HKI7 can be accommodated within this space although it is in much closer proximity to Leu88 than the other aliphatic substrates.
Due to the presence of its benzyl group, HKB is much bulkier than all of the aliphatic substrates; and, as such, it was expected that its motion would be constricted due to steric hindrance from the active site residues. However, over the 6 ns of equilibrium MD simulation, it was noted that the benzyl ring was free to rotate. Two of these positions are shown in Fig. 3, G and H. The fact that HKB can be accommodated so easily within the active site, and that the benzyl ring can actually rotate, indicates that the residues surrounding the DmpG active site are highly flexible.
It was found that all of the substrates, including branched HKI7 and bulky HKB, could bind favorably to the DmpG active site, as shown in Table 1. The relative binding energies did not change greatly as the size and length of the molecule increased. Considering the uncertainty of these values, the most we can accurately draw from these results is that each of these substrates can bind to DmpG with a free energy close in magnitude to the physiological substrate HKV.
Table 1.
Relative free energy of transfer
| Substrate | ΔGprot (kcal/mol) | ΔGsolv (kcal/mol) | ΔΔG (kcal/mol) |
|---|---|---|---|
| HK6 | 43.4 ± 1.0 | 43.4 ± 0.1 | 0.0 ± 1.0 |
| HK7 | 31.9 ± 0.9 | 33.2 ± 0.2 | 1.3 ± 0.9 |
| HKI7 | 75.9 ± 1.3 | 74.2 ± 0.2 | −1.7 ± 1.3 |
| HKB | 88.3 ± 0.9 | 87.9 ± 0.2 | −0.4 ± 0.9 |
| R-HKV | 0.5 ± 0.8 | 0.5 ± 0.2 | 0.0 ± 0.8 |
Transfer from vacuum into the DmpG binding site, ΔGprot, the free energy of transfer from vacuum into bulk water, ΔGsolv, and the overall difference in binding free energy, ΔΔG, are shown for each substrate in comparison to S-HKV. Standard error values from four to six independent calculations are also included.
Channeling of aldehydes in DmpFG
Free energy surfaces were obtained for the channeling of a series of aliphatic aldehydes through DmpFG in an attempt to understand the limitations of this process in wild-type (WT) DmpFG (Figs. 4 and 5). The transport of acetaldehyde was the most energetically favorable, with an overall change in free energy of −3.3 kcal/mol to move from the first to the second active site. All of the barriers encountered by acetaldehyde were low, and as such were not expected to impede the movement of acetaldehyde from one active site to the other. Propionaldehyde, as a slightly larger aldehyde, faced more pronounced barriers than acetaldehyde, and was channeled with a total change in free energy of −2.3 kcal/mol. The largest energy barrier encountered by each of these aldehydes is located just before entering the dehydrogenase active site (5.6–8.5 Å), with values of 2.2 and 3.6 kcal/mol for acetaldehyde and propionaldehyde, respectively.
Figure 4.
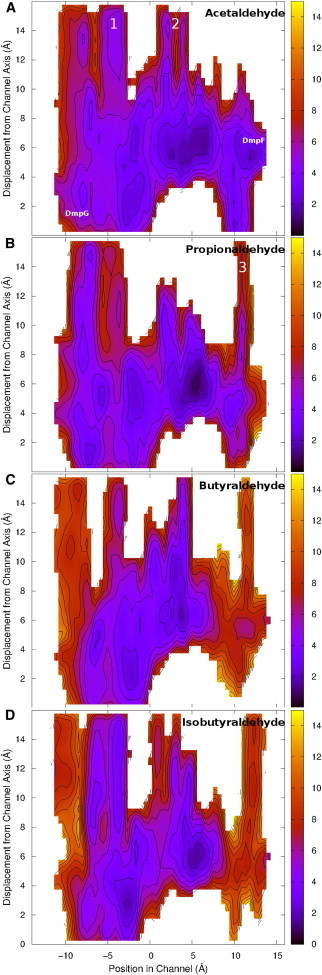
Free energy surfaces obtained for the passage of aldehydes through the channel. The x axis denotes the position along the channel from the aldolase active site in DmpG, to the dehydrogenase active site in DmpF (where zero on the x axis is the midpoint of the channel defined relative to the position of Mn2+ in DmpG and the Cys132 Cα carbon in DmpF). The y axis refers to the distance away from the channel axis and the z value is the free energy at each position with 1 kcal/mol contours. (A) Acetaldehyde. This plot also indicates the starting position of the aldehyde within the DmpG active site and the position of the DmpF active site: (B) propionaldehyde; (C) butyraldehyde; (D) isobutyraldehyde. Escape routes are annotated as 1 and 2 (A) and 3 (B). To see this figure in color, go online.
Figure 5.
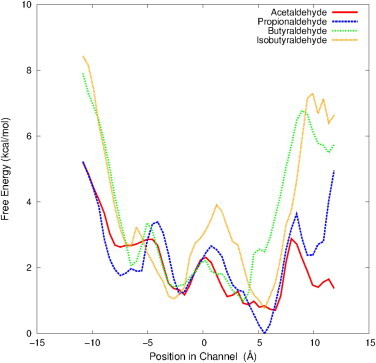
Free energy profiles obtained for the passage of acetaldehyde (red), propionaldehyde (blue), butyraldehyde (green), and isobutyraldehyde (orange) through the channel in DmpFG from the aldolase active site in DmpG to the dehydrogenase active site in DmpF. The plots were obtained by integrating across each free energy surface from 0 to 10 Å in the R direction. The statistical error of these profiles are not shown because the values are extremely small on these graphs (see Fig. S5 in the Supporting Material). To see this figure in color, go online.
Butyraldehyde, the longest aliphatic aldehyde in this trial, faced the highest energy barrier at this point in the channel with a value of 5.7 kcal/mol. The substantial increase in barrier size noted here for butyraldehyde can be attributed to the mobility of its long carbon chain. In its extended conformation, the width of this molecule would be essentially the same as acetaldehyde and propionaldehyde; however, when the chain is not fully extended, the molecule is broader—increasing the entropic cost of the channeling process if it must then readjust into the extended conformation to pass through the constricted regions of the channel. Although it is expected that this barrier would slow down the channeling process, the overall energy change for channeling butyraldehyde from one active site to the other was −1.8 kcal/mol—indicating that this event is still energetically feasible.
The free energy surface obtained for isobutyraldehyde, a bulky branched molecule, had energy barriers that were significantly higher than those observed for the aliphatic aldehydes. The barrier before the dehydrogenase active site had a free energy value of 6.5 kcal/mol. Furthermore, the overall energy change of this process was −1.6 kcal/mol, indicating that isobutyraldehyde is less likely than all of the aliphatic aldehydes investigated to move through the channel. Of interest, Carere et al. (11) has shown that isobutyraldehyde is channeled through BphI-BphJ with an efficiency of 92%. In fact, the efficiency value they obtained is significantly higher than that observed for butyraldehyde in their experiments. However, in the case of DmpFG, these energy surfaces indicate that the transport of isobutyraldehyde from one active site to the other is less likely than that of butyraldehyde.
Although we expected that DmpFG would have the same channeling efficiencies as BphI-BphJ due to the high sequence identity, it has been noted by Baker et al. (17) for another ortholog, TTHB246-TTHB247, that whereas it can channel acetaldehyde with an efficiency of 94%, the efficiency of channeling rapidly drops off to 57% for propionaldehyde. In this case, the low channeling efficiency was assigned to an alanine residue in TTHB246 that corresponds to Gly322 in BphI and Gly323 in DmpG, as shown in Fig. S7. A specific difference between DmpF and its orthologs BphJ, TTHB247, and HsaG is that whereas DmpF has a methionine (Met198) at the entrance to the dehydrogenase active site, the other orthologs all have a leucine at the equivalent position (see Fig. S8).
When the structure of HsaF-HsaG was obtained it was noted, on comparison with the structure of DmpFG (5), that although methionine and leucine both occupied equivalent spatial positions in each ortholog, methionine extended further into the region of the dehdyrogenase active site (16). This could explain why it is less energetically favorable for the bulky isobutyraldehyde to be channeled than butyraldehyde in DmpFG, despite the channeling efficiencies previously determined for BphI-BphJ (11). Further to this, DmpFG, BphI-BphJ, and TTHB246-TTHB247 were assigned to different clades based on sequence alignments with 24 orthologous aldolases and dehydrogenases (17), so some differences in channeling efficiency and in each enzyme’s behavior would not be unexpected.
Escape routes from a leaky channel
A question of great importance in the context of channeling enzymes is whether or not the channeling process is efficient—that is, how much of the intermediate successfully traverses the channel. In the context of DmpFG (14), BphI-BphJ (9–11,13) and TTHB246-TTHB247 (17), it has been shown that at least 5% of the substrate escapes the channel before completing the dehydrogenase reaction. In contrast, HsaF-HsaG, the orthologous aldolase dehydrogenase from Mycobacterium tuberculosis, was found to channel acetaldehyde and propionaldehyde with efficiencies of 99 and 98% (16). This study on DmpFG has allowed us to identify both the position at which escape occurs as well as the energy barriers that must be crossed to leave the channel in this manner.
In the case of DmpFG, the free energy surfaces indicate that there are three major regions of escape, all of which involve the intermediate leaving the side of the channel rather than the ends. These escape routes show up as low energy pathways extending to the top of the free energy surfaces. The first of these (Fig. 6 A), Escape Route 1, is located adjacent to Leu90 at −4.5 Å. Escape Route 2, the second large region, is located at ∼3 Å just before the HT. The final common route of escape (Escape Route 3) is located in the dehydrogenase active site at ∼12 Å. This is indicated clearly in Fig. 6 D. Although this final escape route was present for both propionaldehyde and butyraldehyde, it is considered very unlikely that the aldehyde will leave the channel in this manner, because the barriers are high, and upon reaching the active site, the aldehyde is likely to undergo a reaction.
Figure 6.
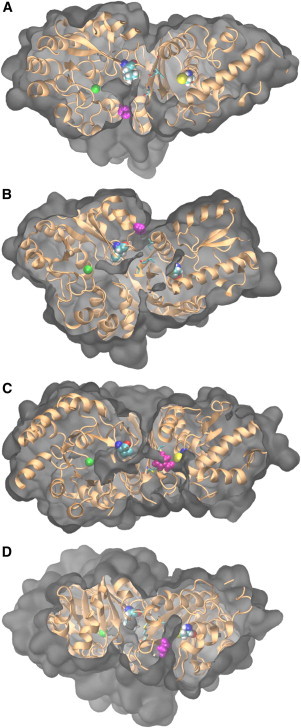
MD snapshots of the protein-intermediate systems obtained with angles and clipping planes selected to highlight the escape routes from the channel indicated in Fig. 4, A and B. (Gray) Protein surface; (orange) protein backbone. Each aldehyde intermediate (magenta), and the central Mn2+ ion (green). Leu90 and Cys132 are shown in VDW representation whereas Ile172, Ile196, and Met198 (the residues of the HT), are shown in licorice representation. (A) Propionaldehyde leaving from Escape Route 1, located adjacent to Leu90. (B and C) Escape Route 2, located between Leu90 and the HT for acetaldehyde in WT DmpFG and for benzaldehyde in I159A mutant DmpFG, respectively. (D) Propionaldehyde leaving from Escape Route 3, located at the dehydrogenase active site. To see this figure in color, go online.
A general trend observed from these free energy surfaces is that as the molecule gets larger, the region through which it can escape becomes more constricted. However, given that the barriers to transport through the channel also increase, escape is still an energetically viable process. For example, Carere et al. (11) found that butyraldehyde was channeled through BphI-BphJ with an efficiency of 83%, indicating that it can traverse that channel but that a significant amount of the intermediate escapes before completing the enzymatic reaction. Given the results we have obtained for DmpFG, this is not surprising, because the potential routes of escape from the channel at −4 and 4 Å present lower energy barriers of 4 and 3 kcal/mol, respectively, relative to the barrier of 5 kcal/mol, which must be crossed to proceed into the second active site (Fig. 4 D).
Tailoring the enzyme
Carere et al. (11) found that aliphatic aldehydes up to six carbons in length could be efficiently channeled by BphI-BphJ but no aromatic aldehydes have ever been investigated. In the context of determining the limits of channeling within DmpFG, we investigated the potential of this enzyme to channel benzaldehyde. We figured that benzaldehyde may not be channeled due to its bulky nature and would provide a good case study for the development of channel mutants that would widen the channel and allow benzaldehyde to cross. This is in contrast to Carere et al. (11), who used a double mutant to reduce or abolish channeling within the enzyme BphI-BphJ by constricting the channel aperture.
Benzaldehyde faces a large barrier to traverse the channel as shown in Fig. 7 A. The first barrier begins at −1.5 Å and peaks at 3 Å near the HT. The second barrier, starting from 5.5 Å, continues into the dehydrogenase active site region with a total change in energy of 7.4 kcal/mol, indicating that benzaldehyde is unlikely to enter the dehydrogenase active site. This barrier can partially be attributed to Ile159, a bulky residue at the entrance to the dehydrogenase active site that sterically blocks benzaldehyde. The overall free energy change of this process, +4.2 kcal/mol, coupled with the large energy barrier, indicates that benzaldehyde is unlikely to be channeled through WT DmpFG.
Figure 7.
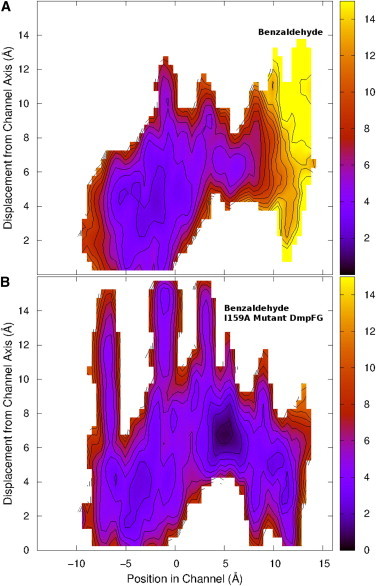
Free energy surfaces obtained for the passage of benzaldehyde through the channel within DmpFG from the aldolase active site in DmpG, to the dehydrogenase active site in DmpF through (A) WT and (B) I159A mutant DmpFG. To see this figure in color, go online.
In an attempt to make the channeling of benzaldehyde energetically feasible and to demonstrate the possibilities for tailoring channeling enzymes such as DmpFG for specific purposes, a mutant system was made. Ile159 from the DmpF binding pocket was mutated to alanine (I159A). Figs. 7 B and 8 clearly show that the barrier into the dehydrogenase active site region has been virtually eliminated such that acetaldehyde is now transported from one active site to the other in a downhill process with an overall energy change of −3.5 kcal/mol. The fact that it is now energetically feasible for this large aromatic aldehyde to be successfully channeled within mutant DmpFG, and that the free energy value is comparable to that obtained for the physiological intermediate, indicates the great potential for the manipulation of this class of enzymes for tailored synthesis. This result, combined with practical site-directed mutation studies performed on both BphI-BphJ and TTHB246-TTHB247 (17), demonstrates the great potential for tailoring these enzymes for the transport of specific aldehyde intermediates. Site-directed mutagenesis studies of BphI have been able to both alter the stereospecificity of the aldolase reaction from S to R (44) and remove all stereospecific control (12) by utilizing different point mutations.
Figure 8.
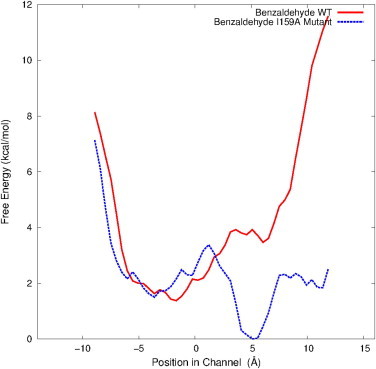
Free energy profiles obtained for the passage of benzaldehyde through the channel within DmpFG from the aldolase active site in DmpG, to the dehydrogenase active site in DmpF for WT (red) and I159A mutant (blue) DmpFG. The statistical errors of these profiles are not shown because the values are extremely small on these graphs (see Fig. S6 in the Supporting Material). To see this figure in color, go online.
Furthermore, the specificity of the aldol addition reaction has also been altered in favor of longer chain aldehydes, such as butyraldehyde and pentaldehyde, in the aldol addition reaction (12). These results are a direct demonstration of how site-directed mutagenesis supports the use of these enzymes for the tailored channeling and synthesis of specific chemical products.
Concluding remarks
In this study, we have demonstrated that it is energetically feasible for a series of substrates, R and S-HKV, HK6, HK7, HKI7, and HKB to bind within the active site of DmpG. These binding energies were very similar in magnitude, irrespective of the size and length of the side chain, indicating that the largest component of the binding energy comes from the interaction of the enolate moiety to the bound Mn2+ ion. We subsequently investigated the energetic feasibility of the respective aldehydes being channeled through DmpFG.
There appear to be two competing factors that influence channeling efficiency. The first is that if the energy barriers are all low, and the overall process is downhill, the transport will be fast and the aldehyde will have a greater chance of reaching the active site in DmpF before escape. The second factor is the size of the aldehyde in question; although a larger substrate faces larger energy barriers and will be transported more slowly, it is also more difficult for the molecule to escape the confines of the channel, increasing its chances of moving from one active site to the other. This is well demonstrated by the free energy profiles obtained for acetaldehyde and propionaldehyde. Both small aldehydes faced low barriers throughout the channeling process and the overall free energy change was negative, indicating that they could both be channeled through DmpFG in a spontaneous event.
In contrast, although butyraldehyde and isobutyraldehyde could be channeled with a negative change in free energy, both of these aldehydes faced large barriers—indicating that this event is now energetically more challenging, and as such the likelihood of it occurring is lower than the smaller aldehydes. However, due to their bulky nature they are also less likely to escape the channel than both acetaldehyde and propionaldehyde, meaning that this event is likely to occur but with a lower rate than that observed for the smaller aldehydes and so both classes of aldehyde may show a high channeling efficiency, but would show different channeling rates.
A notable feature of the channel in DmpFG is its flexible and transient nature. Although a continuous pathway is evident in the crystal structure, during our simulations side chains lining the pore are able to move, transiently blocking and opening the channel. As can be seen in Fig. 6, the channel and escape routes appear and disappear in different trajectory snapshots. However, due to the inherent flexibility of the channel, this does not appear to impede the motion of aldehyde intermediates between the DmpF and DmpG active sites.
Although HKB was able to bind favorably in the active site of WT DmpG, it was found that the subsequent channeling of its intermediate, benzaldehyde, was extremely unlikely because the overall energy change of this process was prohibitive. The mutation of Ile159 to alanine removed the largest barrier faced by benzaldehyde, allowing this aldehyde to traverse the channel in an energetically feasible process. These free energy profiles effectively show how the channeling process can be enhanced in this interesting class of enzymes for the transport of specific aldehyde products.
Acknowledgments
The authors gratefully acknowledge an award under the Merit Allocation Scheme on the National Computational Infrastructure facility at the Australian National University and computer time from iVEC (Interactive Virtual Environments Centre), a government-funded supercomputing resource for Western Australia.
Supporting Material
References
- 1.Milani M., Pesces A., Ascenzi P. Substrate channeling. Biochem. Mol. Biol. Educ. 2003;31:228–233. [Google Scholar]
- 2.Miles E.W., Rhee S., Davies D.R. The molecular basis of substrate channeling. J. Biol. Chem. 1999;274:12193–12196. doi: 10.1074/jbc.274.18.12193. [DOI] [PubMed] [Google Scholar]
- 3.Huang X., Holden H.M., Raushel F.M. Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu. Rev. Biochem. 2001;70:149–180. doi: 10.1146/annurev.biochem.70.1.149. [DOI] [PubMed] [Google Scholar]
- 4.Weeks A., Lund L., Raushel F.M. Tunneling of intermediates in enzyme-catalyzed reactions. Curr. Opin. Chem. Biol. 2006;10:465–472. doi: 10.1016/j.cbpa.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Manjasetty B.A., Powlowski J., Vrielink A. Crystal structure of a bifunctional aldolase-dehydrogenase: sequestering a reactive and volatile intermediate. Proc. Natl. Acad. Sci. USA. 2003;100:6992–6997. doi: 10.1073/pnas.1236794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shingler V., Powlowski J., Marklund U. Nucleotide sequence and functional analysis of the complete phenol/3,4-dimethylphenol catabolic pathway of Pseudomonas sp. strain CF600. J. Bacteriol. 1992;174:711–724. doi: 10.1128/jb.174.3.711-724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agarry S., Durojaiye A. Microbial degradation of phenols: a review. Int. J. Environ. Pollut. 2008;32:12–28. [Google Scholar]
- 8.Smith N.E., Vrielink A., Corry B. Biological channeling of a reactive intermediate in the bifunctional enzyme DmpFG. Biophys. J. 2012;102:868–877. doi: 10.1016/j.bpj.2012.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baker P., Pan D., Seah S.Y.K. Characterization of an aldolase-dehydrogenase complex that exhibits substrate channeling in the polychlorinated biphenyls degradation pathway. Biochemistry. 2009;48:6551–6558. doi: 10.1021/bi9006644. [DOI] [PubMed] [Google Scholar]
- 10.Wang W., Baker P., Seah S.Y.K. Comparison of two metal-dependent pyruvate aldolases related by convergent evolution: substrate specificity, kinetic mechanism and substrate channeling. Biochemistry. 2010;49:3774–3782. doi: 10.1021/bi100251u. [DOI] [PubMed] [Google Scholar]
- 11.Carere J., Baker P., Seah S.Y.K. Investigating the molecular determinants for substrate channeling in BphI-BphJ, an aldolase-dehydrogenase complex from the polychlorinated biphenyls degradation pathway. Biochemistry. 2011;50:8407–8416. doi: 10.1021/bi200960j. [DOI] [PubMed] [Google Scholar]
- 12.Baker P., Carere J., Seah S.Y.K. Probing the molecular basis of substrate specificity, stereospecificity, and catalysis in the class II pyruvate aldolase, BphI. Biochemistry. 2011;50:3559–3569. doi: 10.1021/bi101947g. [DOI] [PubMed] [Google Scholar]
- 13.Baker P., Carere J., Seah S.Y.K. Substrate specificity, substrate channeling, and allostery in BphJ: an acylating aldehyde dehydrogenase associated with the pyruvate aldolase BphI. Biochemistry. 2012;51:4558–4567. doi: 10.1021/bi300407y. [DOI] [PubMed] [Google Scholar]
- 14.Smith N.E., Tie W.J., Vrielink A. Mechanism of the dehydrogenase reaction of DmpFG and analysis of inter-subunit channeling efficiency and thermodynamic parameters in the overall reaction. Int. J. Biochem. Cell Biol. 2013;45:1878–1885. doi: 10.1016/j.biocel.2013.05.028. [DOI] [PubMed] [Google Scholar]
- 15.Fischer B., Boutserin S., Talfournier F. Catalytic properties of a bacterial acylating acetaldehyde dehydrogenase: evidence for several active oligomeric states and coenzyme A activation upon binding. Chem. Biol. Interact. 2013;202:70–77. doi: 10.1016/j.cbi.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Carere J., McKenna S.E., Seah S.Y.K. Characterization of an aldolase-dehydrogenase complex from the cholesterol degradation pathway of Mycobacterium tuberculosis. Biochemistry. 2013;52:3502–3511. doi: 10.1021/bi400351h. [DOI] [PubMed] [Google Scholar]
- 17.Baker P., Hillis C., Seah S.Y.K. Protein-protein interactions and substrate channeling in orthologous and chimeric aldolase-dehydrogenase complexes. Biochemistry. 2012;51:1942–1952. doi: 10.1021/bi201832a. [DOI] [PubMed] [Google Scholar]
- 18.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacKerell A.D., Jr., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 20.Essmann U., Perera L., Berkowitz M.L. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 21.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 22.Chipot C., Pearlman D.A. Free energy calculations. the long and winding gilded road. Mol. Simul. 2002;28:1–12. [Google Scholar]
- 23.Chipot C., Pohorille A. Theory and Applications in Chemistry and Biology. Springer; Berlin, Germany: 2007. Calculating free energy differences using perturbation theory in free energy calculations. [Google Scholar]
- 24.Zwanzig R.W. High-temperature equation of state by a perturbation method. I. Nonpolar gases. J. Chem. Phys. 1954;22:1420–1426. [Google Scholar]
- 25.Pearlman D.A. A comparison of alternative approaches to free energy calculations. J. Phys. Chem. 1994;98:1487–1493. [Google Scholar]
- 26.Simonson T., Archontis G., Karplus M. Free energy simulations come of age: protein-ligand recognition. Acc. Chem. Res. 2002;35:430–437. doi: 10.1021/ar010030m. [DOI] [PubMed] [Google Scholar]
- 27.Kollman P.A. Free energy calculations: applications to chemical and biochemical phenomena. Chem. Rev. 1993;93:2395–2417. [Google Scholar]
- 28.Merz K.M., Kollman P.A. Free energy perturbation simulations of the inhibition of thermolysin: prediction of the free energy of binding of a new inhibitor. J. Am. Chem. Soc. 1989;111:5649–5658. [Google Scholar]
- 29.Bash P.A., Singh U.C., Kollman P.A. Free energy calculations by computer simulation. Science. 1987;236:564–568. doi: 10.1126/science.3576184. [DOI] [PubMed] [Google Scholar]
- 30.Morgan B.P., Scoltz J.M., Bartlett P.A. Differential binding energy: a detailed evaluation of the influence of hydrogen-bonding and hydrophobic groups of the inhibition of thermolysin by phosphorous-containing inhibitors. J. Am. Chem. Soc. 1991;113:297–307. [Google Scholar]
- 31.Tropsha A., Hermans J. Application of free energy simulations to the binding of a transition-state-analogue inhibitor to HIV protease. Protein Eng. 1992;5:29–33. doi: 10.1093/protein/5.1.29. [DOI] [PubMed] [Google Scholar]
- 32.Cieplak P., Kollman P.A. Peptide mimetics as enzyme inhibitors: use of free energy perturbation calculations to evaluate isosteric replacement for amide bonds in a potent HIV protease inhibitor. J. Comput. Aided Mol. Des. 1993;7:291–304. doi: 10.1007/BF00125504. [DOI] [PubMed] [Google Scholar]
- 33.Reddy M.R., Bacquet R.J., Freer S. Calculation of solvation and binding free energy differences for folate-based inhibitors of the enzyme thymidylate synthase. J. Am. Chem. Soc. 1992;114:10117–10122. [Google Scholar]
- 34.Henin, J. ALCHEMIFY, Ver. 1.4. http://www.edam.uhp-nancy.fr/Alchemify. Accessed June 24, 2012.
- 35.Torrie G.M., Valleau J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: umbrella sampling. J. Comput. Phys. 1977;23:187–199. [Google Scholar]
- 36.Valleau J.P., Torrie G.M. Statistical Mechanics. Plenum Press; New York: 1977. A guide for Monte Carlo for statistical mechanics. [Google Scholar]
- 37.Laio A., Gervasio F.L. Metadynamics: a method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Rep. Prog. Phys. 2008;71:126601–126622. [Google Scholar]
- 38.Laio A., Parrinello M. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S., Rosenberg J.M., Kollman R.A. Multidimensional free-energy calculations using the weighted histogram analysis method. J. Comput. Chem. 1995;16:1339–1350. [Google Scholar]
- 40.Grossfield, A. WHAM: an implementation of the weighted histogram analysis method, Ver. 2.0.7. University of Rochester Medical Center, Rochester, NY. http://membrane.urmc.rochester.edu/content/wham/. Accessed April 19, 2013.
- 41.Altschul S.F., Madden T.L., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul S.F., Gish W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 43.Bond C.S., Schüttelkopf A.W. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. D Biol. Crystallogr. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- 44.Baker P., Seah S.Y.K. Rational design of stereoselectivity in the class II pyruvate aldolase BphI. J. Am. Chem. Soc. 2012;134:507–513. doi: 10.1021/ja208754r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


